Abstract
Anthocyanins are water-soluble colored pigments found in terrestrial plants and are responsible for the red, blue, and purple coloration of many flowers and fruits. In addition to the plethora of health benefits associated with anthocyanins (cardioprotective, anti-inflammatory, antioxidant, and antiaging properties), these compounds have attracted widespread attention due to their promising potential as natural food colorants. Previously, we reported the biotransformation of anthocyanin, specifically cyanidin 3-O-glucoside (C3G), from the substrate (+)-catechin in Escherichia coli. In the present work, we set out to systematically improve C3G titers by enhancing substrate and precursor availability, balancing gene expression level, and optimizing cultivation and induction parameters. We first identified E. coli transporter proteins that are responsible for the uptake of catechin and secretion of C3G. We then improved the expression of the heterologous pathway enzymes anthocyanidin synthase (ANS) and 3-O-glycosyltransferase (3GT) using a bicistronic expression cassette. Next, we augmented the intracellular availability of the critical precursor UDP-glucose, which has been known as the rate-limiting precursor to produce glucoside compounds. Further optimization of culture and induction conditions led to a final titer of 350 mg/liter of C3G. We also developed a convenient colorimetric assay for easy screening of C3G overproducers. The work reported here constitutes a promising foundation to develop a cost-effective process for large-scale production of plant-derived anthocyanin from recombinant microorganisms.
INTRODUCTION
Plant natural products currently have extensive applications in the food and pharmaceutical industries (1, 2). Among other things, their unique bioactivities and structures allow them to serve as antimicrobial agents, therapeutic drugs, condiments, and natural dyes (3–5). However, most of these resources remain largely untapped due to the limited availability of these plants, which is dependent upon regional and seasonal factors (6). Other major obstacles to reaping the full benefit of natural products include low yields of the active compounds in crude plant extracts, difficulty of purification, high cost and turnaround time of chemical synthesis due to their complex chemical structures, and regioselectivity issues associated with chemical synthesis (7). The engineering of recombinant microorganisms capable of producing these heterologous compounds is a feasible solution to overcome these limitations. Extensively studied organisms, such as Escherichia coli and Saccharomyces cerevisiae, are especially attractive for this task due to the ease and wide availability of tools for genetic modification and process optimization (8, 9). Coupled with recent advances in the metabolic engineering and synthetic biology of these model organisms, systematic approaches, such as computational modeling, genomics, proteomics, and transcriptomics, can be used to predict their complex behavior (10, 11). This allows for the exploitation of microorganisms to function as “microfactories” to produce specific compounds that are of high market value in various industrial sectors (12). However, in order for a strain to be viable for industrial scale-up, the engineered strain must be able to achieve high yields, high production rates, and high final titers of the desired compounds (13).
In recent years, anthocyanins have drawn tremendous interest due to their potential as natural food colorants (14). In the food industry, it is common practice to use artificial food colorants (AFCs) to improve the appearance of products and make them more palatable to consumers. However, there is increasing concern over the usage of AFCs, as research has shown possible health risks associated with their consumption. Particularly, consuming AFCs can cause behavioral changes in children with and without attention deficit hyperactivity disorder (ADHD), with the latter group exhibiting greater sensitivity to AFCs (15, 16). In response to growing demand from health-conscious consumers for natural food colorants, anthocyanins can provide a possible alternative to artificial food dyes. Apart from being natural, these polyphenols also possess a huge range of health benefits, many of which are associated with their potent antioxidant properties (17, 18). In fact, anthocyanins' antioxidant properties are reported to be equipotent to those of quercetin or catechin gallates, both of which are major flavonoids found in green tea. The antioxidant properties of these compounds can be attributed to their common catechol ring backbones, which dramatically increase the molecule's radical-scavenging ability (19).
Anthocyanins are naturally synthesized from flavanones, starting with the conversion of flavanones to dihydroflavonols by flavanone 3β-hydroxylase (FHT); the dihydroflavonols are then converted by dihydroflavonol 4-reductase (DFR) to leucoanthocyanidins. Subsequently, catechins are formed by the action of leucoanthocyanidin reductase (LAR). Anthocyanidin synthase (ANS), a 2-oxoglutarate and ferrous iron-dependent oxygenase, then utilizes either catechins or leucoanthocyanidins as substrates to form the unstable cyanidin intermediates before structure stabilization, in which the intermediates are converted to anthocyanins by the activity of UDPG-flavonoid 3-O-glycosyltransferase (3GT) (Fig. 1A).
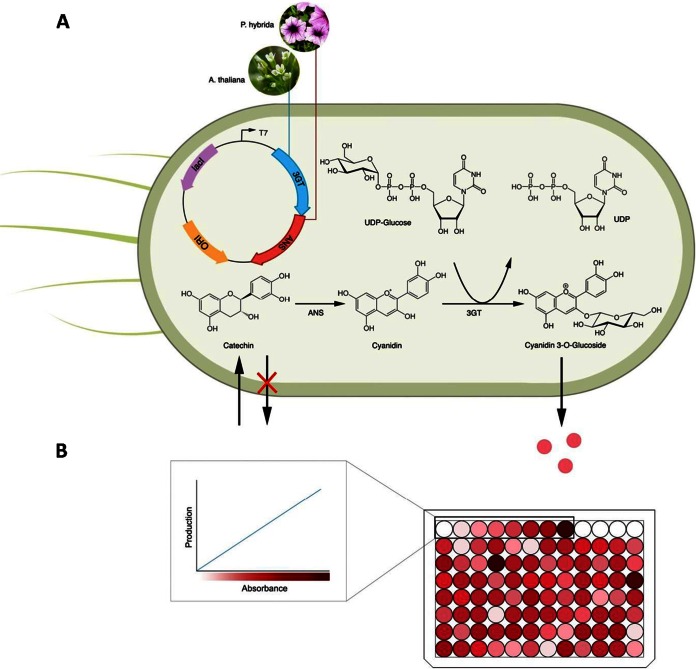
FIG 1.
Engineered pathway for the biosynthesis of colored anthocyanin, cyanidin 3-O-glucoside (C3G), in E. coli. (A) Mechanism catalyzing the formation of cyanidin from (+)-catechin by anthocyanidin synthase (ANS) and the transfer of a glucose group from UDP-glucose to cyanidin to yield C3G by 3-O-glycosyltransferase (3GT). (B) C3G-producing strains can be efficiently screened by using a colorimetric assay.
Ten years ago, we reported the biosynthesis of natural anthocyanins from a recombinant strain of E. coli for the first time (20). We achieved cyanidin-3-O-glucoside (C3G) production of up to 110 mg/liter with the optimization of medium pH, gene expression cassette, and supply of UDP-glucose (21, 22). In this paper, we present the various engineering strategies to further improve the titer of C3G. Toward the understanding of the detailed mechanism of anthocyanin biosynthesis in E. coli, we have identified transporter proteins facilitating the efficient transportation of catechin and C3G to increase substrate availability and remove product inhibition. We then balanced the expression of the two critical enzymes (ANS and 3GT) using a bicistronic expression cassette. Combined with a convenient colorimetric assay (Fig. 1B), we rapidly optimized the culture condition and induction parameters, leading to a C3G titer of 350 mg/liter, which is about a 3.3-fold improvement over our previous efforts and is the highest production level reported to date. This work, coupled with the recently reported high-yield, high-titer production of catechin in E. coli, can lead to a commercially viable process for the sustainable production of anthocyanins (23).
MATERIALS AND METHODS
Strains and media.
E. coli BL21*, used for recombinant protein production, and plasmid cloning materials were purchased from Invitrogen. DNA manipulations were performed according to standard recombinant DNA procedures (24). Restriction enzymes and T4 DNA ligase were purchased from New England BioLabs. All PCR amplification and cloning reactions were performed using Accuzyme DNA polymerase from Bioline. Plasmid DNA was prepared from stock strains using a Zyppy Plasmid MiniPrep kit, while fragment DNA was isolated by gel extraction using a Zymoclean Gel DNA Recovery kit (Zymo Research). Genomic DNA of E. coli was isolated using the PureLink Genomic DNA minikit. Plasmid-bearing cultures were grown in media containing the antibiotics ampicillin (70 μg/ml), kanamycin (40 μg/ml), chloramphenicol (20 μg/ml), and/or streptomycin (40 μg/ml) when needed. (+)-Catechin was purchased from TCI America.
Plasmid and strain constructions.
Plasmids pACYCDuet-1, pCDFDuet-1, pCOLADuet-1, and pETDuet-1 were purchased from Novagen. Plasmids pCDF-3A, pET-pgm-galU, and pCOLA-cmk-ndk were provided by MK laboratory stocks. The ycjU gene and the transporter proteins encoded by tnaB, marA, tolC, acrAB, and yadH were isolated from E. coli K-12 and separately cloned into the multiple-cloning sites of pACYC downstream of the T7 promoter. Positive transformants were verified by colony PCR and further verified by DNA sequencing.
Target gene knockouts were carried out using the λ-red recombinase/FLP system as previously described (25), with a few modifications. Briefly, E. coli harboring pKD46 plasmid was induced with 50 mM arabinose and grown at 30°C to an optical density at 600 nm (OD600) of 0.6 before being made electrocompetent by washing three times with ice-cold 10% (vol/vol) glycerol. Concentrated electrocompetent cells were then electroporated using a Bio-Rad Genepulser X-Cell system with 0.75 μg of PCR-amplified FRT-flanked kanamycin cassette with 50-bp homologous flanking regions. Next, cells were incubated at 37°C for 1 h and plated on kanamycin selection plates. All deletions were verified by colony PCR. Positive mutants were then transformed with pCP20 plasmid for the excision of antibiotic markers. A complete list of plasmids and strains can be found in Table 1.
TABLE 1.
Plasmids and strains used in the present study
| Plasmid or strain | Relevant properties or genetic marker | Source or reference |
|---|---|---|
| Plasmids | ||
| pACYCDuet | p15A ori (pACYC184), Cmr | Novagen |
| pCOLADuet | ColA ori, Kmr | Novagen |
| pCDFDuet | CloDF13 ori, Strr | Novagen |
| pETDuet | ColE1, Ampr | Novagen |
| pCDF-3A | pCDF carrying A. thaliana 3GT and P. hybrida ANS fusion protein | 21 |
| pCDF-3AO | pCDF carrying A. thaliana 3GT and P. hybrida ANS in an operon | This study |
| pET-pgm-galU | pETDuet carrying E. coli pgm and galU | 21 |
| pCOLA-cmk-ndk | pCOLADuet carrying E. coli cmk and ndk | 22 |
| pACYC-ycjU | pACYCDuet carrying E. coli ycjU | This study |
| pACYC-tnaB | pACYCDuet carrying E. coli tnaB | This study |
| pACYC-marA | pACYCDuet carrying E. coli marA | This study |
| pACYC-tolC | pACYCDuet carrying E. coli tolC | This study |
| pACYC-arcAB-tolC | pACYCDuet carrying E. coli arcAB and tolC | This study |
| pACYC-yadH | pACYCDuet carrying E. coli yadH | This study |
| Strains | ||
| E. coli BL21* | F− ompT hsdSB (rB− mB−) gal dcm rne131 (DE3) | Invitrogen |
| BL21* Δudg | E. coli BL21* Δudg | 22 |
| BL21* ΔtolC | E. coli BL21* ΔtolC | This study |
| BL21* 3AO | E. coli BL21* pCDF-3AO | This study |
| BL21* 3A | E. coli BL21* pCDF-3A | 21 |
C3G fermentations.
For E. coli strain BL21* harboring the genes that encode the anthocyanin biosynthetic pathway in the pCDF plasmid, preinocula were grown overnight in LB broth and then diluted to an OD600 of 0.1 in 50 ml of fresh LB for culturing in shake flasks. Cells were incubated at 37°C to an OD600 of 0.8 before they were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and grown for another 3 h at 30°C for protein overexpression. Next, cells were collected by centrifugation and resuspended in M9 medium (pH 5), which was one-fifth of the initial LB volume containing the necessary antibiotics, 1% glucose, 1 mM (+)-catechin, 1 mM IPTG, 0.1 mM 2-oxoglutarate, 2.5 mM sodium ascorbate, and 0.1 mM orotic acid. Incubation was continued at 30°C for 48 h prior to analysis of the heterologous products. For high-performance liquid chromatography (HPLC) analysis, 500-μl fermentation samples were centrifuged and 30 μl of the supernatant was injected into an HPLC to quantify C3G.
For high-level C3G production, an optimized fermentation approach was followed in which an overnight preinoculum culture with an OD600 of 0.1 was used to inoculate 100 ml of LB in a shake flask and incubated at 37°C until an OD600 of 3.2 was reached; thereafter, the culture was induced with 1 mM IPTG and grown for another 3 h at 30°C. Next, cells were concentrated to an OD600 of 15 in 10 ml of M9 medium (pH 5) containing 1% glucose, 2.5 mM (+)-catechin, 5 mM IPTG, 0.1 mM 2-oxoglutarate, 2.5 mM sodium ascorbate, and 0.1 mM orotic acid. Additional catechin (1 mM) and 1% glucose were used for supplementation after 24 h of fermentation.
HPLC analysis.
Cell-free culture medium from fermentations (30 μl) was analyzed by HPLC using an Agilent 1100 series instrument and a reverse-phase ZORBAX SB-C18 column (4.6 by 150 mm) maintained at 25°C. A 0.1% (vol/vol) concentration of formic acid in acetonitrile and 0.1% (vol/vol) formic acid in water were used as the mobile phases at a flow rate of 1.0 ml per min. The mobile-phase composition profile was as follows (percentile refers to volume-per-volume acetonitrile concentration): 10 to 50% for 10 min, 40 to 60% for 5 min, and 60 to 10% for 2 min. C3G was quantified by monitoring absorbance at 520 nm.
Colorimetric assay and C3G purification.
The red-colored C3G was measured with a microplate reader at an absorbance of 512 nm and quantified using a standard curve. For simple purification, C3G-containing culture medium was extracted with an equal volume of ethyl acetate, vortexed vigorously for 1 min, and centrifuged to separate the polar and nonpolar layers.
RESULTS AND DISCUSSION
Identifying C3G-associated E. coli transporter proteins.
In plants, anthocyanins are synthesized via the flavonoid biosynthetic pathway in the cytosol, which is a pH-neutral environment (26). Prevacuolar compartments containing anthocyanins are then mass transported into the central vacuole by merging with the central vacuole in the epidermal cells and subsequently ruptured to release the colored compound (27). For storage in the vacuole, anthocyanins must be stabilized under low-pH conditions for prolonged accumulation and prevention of degradation (28). Prokaryotic organisms such as E. coli lack many organelles found in plants, especially the vacuole for storage of anthocyanins. In a previous study, Yan et al. emulated the low-pH environment in the vacuole by lowering the pH of M9 minimal medium to 5 for stable C3G production in E. coli (21). Next, we resolved to identify native transport proteins responsible for transporting C3G across the cell membrane. The rationale for this study was based on, first, the claim that the intracellular accumulation of C3G accounted for more than 50% of the total yield and, second, the observation of cell culture medium in which centrifuged cell pellets were more intensely colored than the extracellular medium (Fig. 2A). We postulated that either there was a transport limitation for C3G or C3G was taken up by the cells following sequestration to the intracellular medium (Fig. 2B). The latter hypothesis was based on the comparison between C3G titers in the extracellular medium at 24 and 48 h, in which the titers consistently decreased over time while the centrifuged cell pellets turned more red (Fig. 3, control).
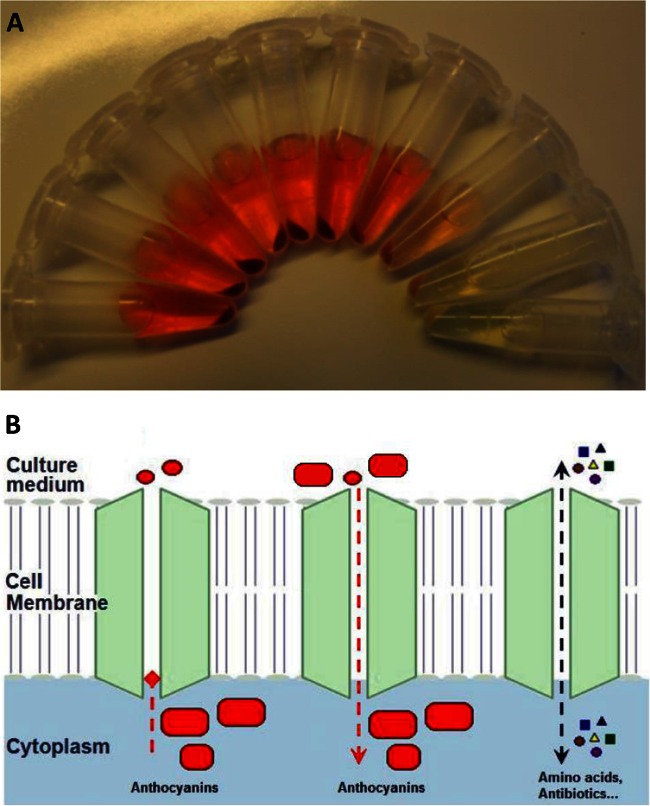
FIG 2.
Significant amount of cyanidin 3-O-glucoside (C3G) accumulated in the intracellular medium. (A) Cell pellets and the extracellular medium of different C3G-producing strains; (B) picture demonstration of two potential scenarios: C3G remains mostly in the cytoplasm (left), sequestered C3G is transported back into the cells (middle), and amino acids and antibiotics have specific transporter proteins for their uptake and efflux across the cell membrane (right).
FIG 3.
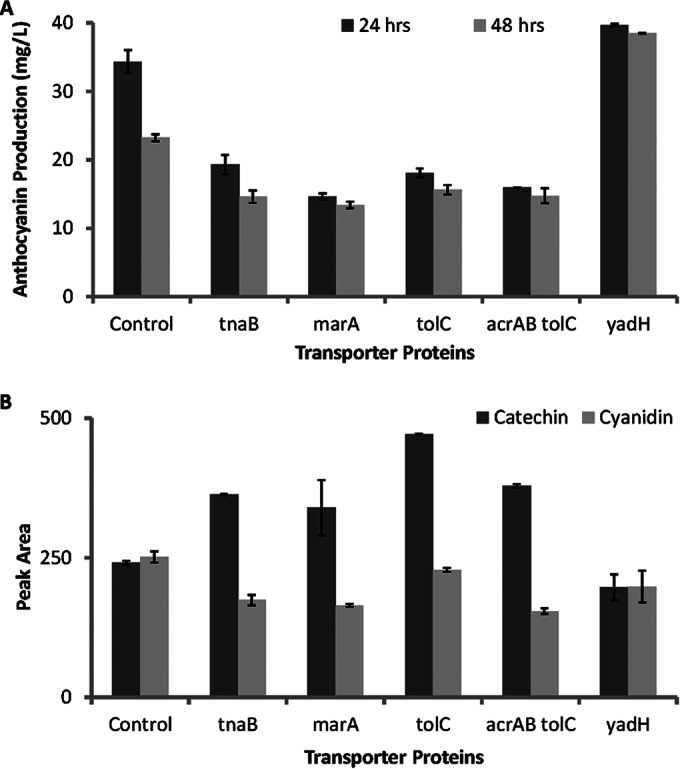
Extracellular cyanidin 3-O-glucoside (C3G), cyanidin, and (+)-catechin with the overexpression of different transporter proteins. (A) C3G accumulation after 24 and 48 h; (B) peak area of intermediate cyanidin and substrate catechin in the extracellular medium after 24 h of cultivation. Error bars represent standard deviations between biological and technical triplicates.
Transport proteins are generally classified into three categories: uptake pumps, efflux pumps, and regulators. In microorganisms, they are responsible for conferring multiple-drug resistance and protection against harmful substances such as detergents and disinfectants (29). In this case, we were interested in identifying transport proteins that are specific for the efflux of C3G and, to a lesser extent, the uptake of catechin for the Gram-negative E. coli strain BL21*. Five transporter proteins consisting of four efflux pumps, encoded by acrAB, tolC, aaeB, and yadH, and one uptake pump, encoded by tnaB, as well as one regulator, encoded by marA, were selected based on prior reports on secondary metabolite transporters in microorganisms (29, 30).
All the aforementioned transport proteins were overexpressed in the host strain harboring both the heterologous anthocyanin pathway and UDP-glucose pathway expression plasmids. Out of all the transporter proteins tested, only overexpression of the protein encoded by yadH improved the extracellular production of C3G by 15% (Fig. 3A), indicating that C3G might be a substrate for the YadH efflux pump. Further quantification of extracellular catechin and cyanidin revealed a significantly lower substrate uptake by all transport proteins except the YadH protein than with the control strain. Specifically, overexpressing the tolC product led to the lowest consumption of catechin, while the amount of cyanidin intermediate in the extracellular medium was comparable to that with the control strain (Fig. 3B). We postulated that the overexpression of tolC, an efflux pump for many common hydrophobic molecules, might facilitate the secretion of catechin. As expected, tolC knockout in BL21* expressing the heterologous anthocyanin biosynthetic pathway (pCDF-3A) was able to improve C3G production by 55%, to 66 mg/liter. Coupling of both tolC knockout and yadH overexpression further improved the extracellular C3G production, to 69 mg/liter (Table 2, fusion). The effect of combining a high intracellular concentration of substrate and effective secretion of product was visible in the culture broth (see Fig. S1 in the supplemental material, BL21*ΔtolC and BL21*ΔtolCyadH). These data reaffirmed the complexity of the E. coli efflux system: catechin and C3G have very similar molecular backbones, with minor differences in the C-ring and the addition of a glucoside functional group; however, they have very distinct routes of efflux. Also, maintaining a high intracellular pool of substrates by knocking out a substrate-specific efflux pump can significantly enhance biotransformation efficiency (31).
TABLE 2.
Effects of transporter gene modifications and expression systems on production of cyanidin 3-O-glucoside
| Strain | Transporter gene modification | Product titera (mg/liter) |
|
|---|---|---|---|
| Fusion, pCDF-3A | Operon, pCDF-3AO | ||
| BL21* | —b | 20.7c | 94.2 ± 2.4 |
| BL21* + pET-pgm-galU + pCOLA-cmk-ndk | —b | 42.2 ± 2.2 | 120 ± 12.1 |
| ΔtolC | 65.7 ± 4.4 | 120 ± 0.2 | |
| ΔtolC + pACYC-yadH | 69.0 ± 2.5 | 126 ± 2.4 | |
All values are averages from three trials, with standard deviations.
—, no plasmid transformed.
Reference 21.
Optimization of expression construct.
In the past, a 17% improvement in C3G titers was achieved by using a fusion protein of 3GT and ANS compared to the titers achieved with a dual-promoter system in a pCDFDuet plasmid (21). It was hypothesized that the translational fusion of both catalytic proteins facilitated the transport of the highly unstable intermediate cyanidin directly from ANS to 3GT and resulted in a higher level of C3G production. As an alternative, a new expression cassette was proposed in which ANS and 3GT were arranged in a bicistronic configuration driven by a single T7 promoter, with 3GT placed upstream of ANS. An operon-based gene configuration allows the coordinated expression of functionally related proteins, as both genes are transcribed and regulated together. For both genes, the native ribosome binding site (RBS) from the pCDF vector and the distance between the RBS and the start codon were retained to minimize the variation in translational efficiency. By comparing solely the extracellular production prior to any further genetic modifications, a titer of 94.2 mg/liter was achieved, compared to 17.7 mg/liter (2 promoters) and 20.7 mg/liter (fusion protein, BL21* 3A), representing at least a 4.5-fold improvement (Table 2). It has been proved that the translation initiation rate for a given mRNA is 6-fold greater during transcription than after its release, which could be explained by a cotranscriptional and cotranslational mechanism: a longer “transcription distance” allows more time for ribosomes to occupy the RBS region during transcription (32–34). Therefore, the relative expression of 3GT would be higher than that of ANS because of the relatively longer “transcription distance.” Presumably, a stronger expression of 3GT could more efficiently drive the carbon flux toward the end product C3G while preventing the accumulation of unstable cyanidin.
It was previously confirmed that UDP-glucose availability was the metabolic bottleneck for high-level C3G biosynthesis, and previous efforts on improving this rate-limiting precursor by simultaneous overexpression of the enzymes encoded by pgm, galU, cmk, and ndk in the host strain have yielded promising results (22). When these UDP-glucose pathway enzymes were overexpressed, BL21* 3AO yielded 120 mg/liter of C3G, compared to 42 mg/liter for BL21* 3A, representing a 2.9-fold improvement (Table 2).
Surprisingly, coupling the new expression system with the overexpression of yadH and deletion of tolC did not yield any significant improvement in extracellular C3G production (Table 2, operon). These data suggest that the transport of substrates and products may not have been a significant bottleneck in the C3G-overproducing strain when the pathway was expressed in the operon format and UDP-glucose limitation was alleviated.
Augmentation of intracellular UDP-glucose.
In addition to pgm, galU, cmk, and ndk, we sought to further improve UDP-glucose availability to support high-level C3G production. The phosphoglucomutases encoded by pgm and ycjU catalyze the same reversible reaction from glucose-6-phosphate to glucose-1-phosphate but act on different anomers, α and β, respectively (35). It was of interest to compare the activities of these enzymes in catalyzing the reversible reaction of glucose-6-phosphate to glucose-1-phosphate in vivo. After 17 h of cultivation, ycjU overexpression alone resulted in a 24% improvement over the wild type, to 104.5 mg/liter (Table 3). In combination with the enzymes encoded by cmk and ndk, the overexpression of the enzyme encoded by ycjU resulted in a C3G titer of 120.2 mg/liter, or 12% higher than with coexpression with the enzymes encoded by pgm and galU. This result suggested that the enzyme encoded by galU is not a bottleneck enzyme in the UDP-glucose pathway. Further overexpression of the YcjU enzyme on top of the Pgm, GalU, Cmk, and Ndk enzymes did not yield any significant titer improvement.
TABLE 3.
Cyanidin 3-O-glucoside production after 17 h with overexpression of UDP-glucose pathway genes
| Strain | Overexpressed gene(s) | Product titera (mg/liter) |
|---|---|---|
| BL21* 3AO | —b | 84.6 ± 1.2 |
| ycjU | 104.5 ± 8.1 | |
| pgm, galU, cmk, ndk | 107.2 ± 0.6 | |
| ycjU, cmk, ndk | 120.2 ± 1.3 | |
| ycjU, pgm, galU | 114.9 ± 2.3 | |
| ycjU, pgm, galU, cmk, ndk | 122.3 ± 0.3 |
All values are averages three trials, with standard deviations.
—, no plasmid transformed.
Colorimetric screen and assay for C3G production.
One major advantage that anthocyanins possess compared to other flavonoids is their associated color properties, especially at low pHs. Anthocyanins in red cabbage have been used as a pH indicator, where they turn from red (very acidic) to purple (neutral) to greenish yellow (basic). One other industrially important molecule with similar colorimetric property that has long been used in the metabolic engineering and synthetic biology fields is lycopene. This hydrophobic molecule, commonly found in tomatoes, can be monitored colorimetrically by measuring its absorbance at 470 nm (36). However, unlike for anthocyanins, the absolute lycopene quantification is tedious, as it requires the extraction of cell culture with organic solvents.
In order to demonstrate that the color intensity of C3G, under acidic conditions, is proportional to the production, C3G standards were measured at an absorbance of 512 nm. As expected, a positive linear trend was observed (see Fig. S2A and B in the supplemental material). To assess the accuracy of the calibration curve, a set of fermentations varying in culture conditions was constructed in a 24-well plate. After 2 days of fermentation, cell-free culture broths were measured by both spectrophotometry and HPLC. A good correlation was observed between the actual and predicted production (see Fig. S2C). We have demonstrated that this C3G colorimetric assay could be used as a medium-throughput method for identifying overproducers with the added benefit of product quantification. Unlike the lycopene screening assay, in which plate-based photometric screening is used, one disadvantage of the C3G colorimetric assay is the necessary cultivation of individual colonies, which drives up assay time while decreasing throughput. Upon plating of transformants on agar plates supplemented with all necessary substrates, inducers, and cofactors, no color accumulation was observed either intra- or extracellularly after 48 h of incubation (data not shown). Regardless, cultivating C3G-producing strains in liquid culture is useful for quick assays for studying and optimizing media and fermentation conditions. The current assay time of 48 h can be significantly reduced with more extensive engineering of the reporter strain as well as improving medium and culture conditions.
Utilizing this convenient colorimetric assay, we took a sequential approach to optimize substrate, inducer, and cofactor concentrations and culture pH (see Fig. S3 in the supplemental material). After incubation of the cultures for 48 h, the culture conditions of wells containing higher C3G titers were noted. Two notable modifications to fermentation conditions were derived from this colorimetric assay: an increase in initial IPTG concentration to 5 mM instead of 1 mM and an increase in biotransformation OD600 in M9 to 15 instead of 4.
To the best of our knowledge, this is the first report of the use of anthocyanins for microtiter plate-based photometric screening and optimization. Similar to lycopene, C3G in the colorimetric assay can be used as a screening molecule for desirable phenotypes in applications such as combinatorial engineering (37, 38). The calibration curve generated could be utilized for rapid C3G concentration quantification without the need for complex and time-consuming analytical tools.
Optimization of culture conditions.
Additional modifications in shake flask fermentation, specifically substrate concentration, carbon source concentration, and induction at OD600, were implemented to further elevate C3G titers. With the improved genetic construct utilizing an operon system, HPLC and glucose analysis showed that the initial catechin (1 mM) and glucose (1%, wt/vol) used to supplement M9 medium were rapidly exhausted after 24 h (data not shown). Subsequent shake flask fermentations optimizing substrate concentration suggested that a concentration of 2.5 mM catechin was necessary to maintain substrate-driving force. Glucose concentration, on the other hand, was maintained at 1%, as a decline in overall C3G production was observed with both lower (0.5%) and higher (up to 3%) initial glucose concentrations (data not shown). This effect can possibly be attributed to the inverse correlation between glucose concentration and IPTG-induced protein expression. In an IPTG-inducible T7 promoter system, optimal transcriptional initiation requires both IPTG binding to the lac repressor and high catabolite gene activator protein (CAP)/cyclic AMP (cAMP) complex binding upstream of the promoter. A high glucose concentration can result in low cAMP levels, subsequently disrupting the formation of the CAP/cAMP complex and hindering downstream transcription (39). In order to balance the need for good protein expression under glucose-limiting conditions while maintaining cell viability, fermentation with glucose pulsing was adopted instead. Additional 1% glucose and 1 mM catechin were used for supplementation by the end of first day to maintain both cell viability and substrate-driving force, while minimizing the detrimental effect of catabolic repression. By increasing the initial substrate concentration via substrate and glucose feeding, C3G production improved significantly in all three strains with overexpression of different UDP-glucose pathway enzymes (Fig. 4A). The most noticeable improvement was seen in strain BL21* 3AO overexpressing the enzymes encoded by ycjU, cmk, and ndk, which showed over a 70% improvement in titer, to 204.3 mg/liter.
FIG 4.
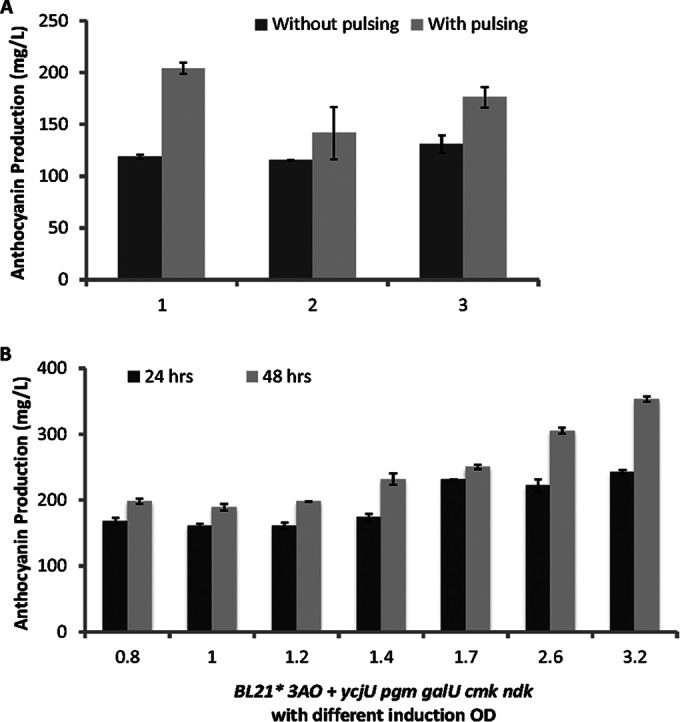
Cyanidin 3-O-glucoside (C3G) production as a result of optimization of culture conditions, including substrate concentration, carbon source concentration, and induction OD600 for strains harboring pCDF-3AO plasmids with the overexpression of different UDP-glucose pathway enzymes. (A) C3G production when cultures were pulsed with 1 mM catechin and 1% glucose at the end of the first day; (B) C3G production when induction OD600 was increased from an initial value of 0.8 to 3.2. Error bars represent standard deviations between biological and technical triplicates.
Next, induction OD600 was investigated, in which different induction ODs were tested as alternatives to the manufacturer-recommended mid-log-phase induction OD600 (0.8) for strain BL21*. Our results show that as induction OD600 was increased from 0.8 to 1.2, C3G production remained the same. However, production increased when induction occurred later in the growth phase (OD600 greater than 1.4) (Fig. 4B). The highest production was attained at the highest induction OD600, 3.2; the production was 350 mg/liter. Induction OD600 was not further increased as the strains had reached stationary phase, at 3.2 (data not shown). The beneficial effect of induction at a higher OD600 could possibly be attributed to better protein expression during a nutrient-depleted state. To date, the reported titer of 350 mg/liter is the highest reported anthocyanin production, a 3.5-fold increase in C3G production over the previously reported highest production, or a 7-fold improvement if compared solely to extracellular production. It should be noted that the actual amount of C3G produced could be higher than our reported value, as anthocyanins exist in equilibrium as a mixture of the red flavylium cation, blue quinoidal base, colorless hemiketals, and chalcone structures (18). Further optimization of medium conditions (pH, temperature, oxygen, light, metals for complex formation, and copigments) to favor the formation of the flavylium cation could potentially improve C3G titers using the engineered E. coli strain.
Simple extraction method for C3G purification.
Solvent extraction has been a common method for extraction of phenolic compounds. Since anthocyanins are polar molecules, a simple process for purification of C3G was demonstrated using the organic solvent ethyl acetate. By a single-step extraction, C3G (in the polar phase) can be separated from catechin and the intermediate, cyanidin (in the organic phase) (Fig. 5; see also Fig. S4 in the supplemental material). Remaining polar impurities, including UDP-glucose, organic acids, and proteins, in the cell culture medium can be subsequently purified using either solid-phase extraction or liquid-liquid extraction (40).
FIG 5.
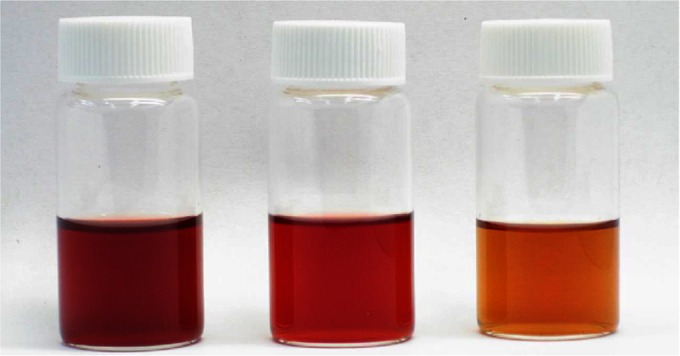
A titer of 350 mg/liter of cyanidin 3-O-glucoside (C3G) was achieved using engineered E. coli. (Left) Cell culture after 2 days of cultivation in clear M9 minimal medium; (middle) partially purified anthocyanin after extraction with ethyl acetate; (right) impurities extracted in organic phase with ethyl acetate.
Conclusions.
Unlike most natural compounds, anthocyanins offer an easy and direct colorimetric screen without the need of any coupled assay for product detection (41). We have shown for the first time the development of a microtiter plate assay capable of screening anthocyanin overproducers by optimizing fermentation conditions. By optimizing the media and strain, the 48-h culturing time can be significantly reduced to increase throughput. To further improve the biotransformation efficiency, a more combinatorial engineering approach can be utilized, e.g., direct evolution of ANS and 3GT enzymes to improve their biochemical properties and coupled with synthetic biology approaches that optimize the genotype of the recombinant strain. Apart from visual screening to identify improved variants, the titers can also be assessed in a more quantitative manner.
Overall, we set out to further improve the C3G production of the original heterologous two-enzyme pathway in E. coli BL21*. A more efficient system was identified utilizing an expression system in which both enzymes were expressed in a bicistronic operon. Metabolic engineering efforts aimed at improving the availability of known rate-limiting cofactors often led to higher product formation (42, 43). In this study, we focused on augmenting the intracellular pool of UDP-glucose by overexpressing another E. coli endogenous phosphoglucomutase, the enzyme encoded by ycjU instead of the initial pgm. The production with YcjU enzyme overexpression alone is better than that with coexpression of the Pgm and GalU enzymes. Lastly, by improving culture conditions through higher initial substrate concentration, substrate and glucose pulsing, and inducing at stationary phase, a C3G titer of 350 mg/liter was attained, representing a yield of 22.3 g of C3G per g of catechin. This optimized system, coupled with recent advances in producing high-yield, high-titer (+)-catechin, offers a process that can lead to a commercially viable production of the vibrant natural colorant C3G (23, 44). In addition, C3G can be semipurified after one-step solvent extraction, and its coloration is comparable to that of synthetic dye.
In conjunction with recent advances in understanding a plant's ability to stabilize and accumulate anthocyanin, novel methods to improve thermal and pH stability of anthocyanins have been developed. Some notable examples include the use of the copigment quercetagetin to stabilize anthocyanins derived from grape skin in a pH range of 3.0 to 5.0 (45) and the addition of yeast mannoproteins that form complexes with anthocyanins to improve their half-life and decrease their degradation constant at neutral pH (pH 7.0) while maintaining their oxidant capacity (46). These developments, in addition to classic metabolic engineering strategies, can guide our future design in engineering an industrial E. coli strain capable of overproducing anthocyanins to support the market demand for natural food colorants in a cost-effective way.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge financial support from Chromadex.
We appreciate C. Santos, A. Qualley, M. De Mey, and A. Parayil for discussions and comments on the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01448-15.
REFERENCES
- 1.Xu P, Bhan N, Koffas MA. 2013. Engineering plant metabolism into microbes: from systems biology to synthetic biology. Curr Opin Biotechnol 24:291–299. doi: 10.1016/j.copbio.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 2.Bhan N, Xu P, Koffas MAG. 2013. Pathway and protein engineering approaches to produce novel and commodity small molecules. Curr Opin Biotechnol 24:1137–1143. doi: 10.1016/j.copbio.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 3.Mora-Pale M, Sanchez-Rodriguez SP, Linhatrdt RJ, Dordick JS, Koffas MAG. 2013. Metabolic engineering and in vitro biosynthesis of phytochemicals and non-natural analogues. Plant Sci 210:10–24. doi: 10.1016/j.plantsci.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Mora-Pale M, Sanchez-Rodriguez SP, Linhardt RJ, Dordick JS, Koffas MAG. 2014. Biochemical strategies for enhancing the in vivo production of natural products with pharmaceutical potential. Curr Opin Biotechnol 25:86–94. doi: 10.1016/j.copbio.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Lin Y, Jain R, Yan Y. 2014. Microbial production of antioxidant food ingredients via metabolic engineering. Curr Opin Biotechnol 26:71–78. doi: 10.1016/j.copbio.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Lin Y, Yan Y. 2014. Biotechnological production of plant-specific hydroxylated phenylpropanoids. Biotechnol Bioeng 111:1895–1899. doi: 10.1002/bit.25237. [DOI] [PubMed] [Google Scholar]
- 7.Chemler JA, Koffas MA. 2008. Metabolic engineering for plant natural product biosynthesis in microbes. Curr Opin Biotechnol 19:597–605. doi: 10.1016/j.copbio.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen J, Jewett MC. 2008. Impact of systems biology on metabolic engineering of Saccharomyces cerevisiae. FEMS Yeast Res 8:122–131. doi: 10.1111/j.1567-1364.2007.00302.x. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen J, Larsson C, van Maris A, Pronk J. 2013. Metabolic engineering of yeast for production of fuels and chemicals. Curr Opin Biotechnol 24:398–404. doi: 10.1016/j.copbio.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 10.Förster J, Famili I, Fu P, Palsson B, Nielsen J. 2003. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res 13:244–253. doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bro C, Regenberg B, Förster J, Nielsen J. 2006. In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab Eng 8:102–111. doi: 10.1016/j.ymben.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Ververidis F, Trantas E, Douglas C, Vollmer G, Kretzschmar G, Panopoulos N. 2007. Biotechnology of flavonoids and other phenylpropanoid-derived natural products. Part II: reconstruction of multienzyme pathways in plants and microbes. Biotechnol J 2:1235–1249. [DOI] [PubMed] [Google Scholar]
- 13.Willrodt C, Karande R, Schmid A, Jusling M. 2015. Guiding efficient microbial synthesis of non-natural chemicals by physicochemical properties of reactants. Curr Opin Biotechnol 35:52–62. doi: 10.1016/j.copbio.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Mateus N, De Freitas V. 2008. Anthocyanins as food colorants, p 283–304. In Gould K, Davies KM, Winefield C (ed), Anthocyanins: biosynthesis, functions, and applications. Springer, New York, NY. [Google Scholar]
- 15.McCann D, Barrett A, Cooper A, Crumpler D, Dalen L, Grimshaw K, Kitchin E, Lok K, Porteous L, Prince E, Sonuga-Barke E, Warner JO, Stevenson J. 2007. Food additives and hyperactive behaviour in 3-year-old and 8/9-year-old children in the community: a randomised, double-blinded, placebo-controlled trial. Lancet 370:1560–1567. doi: 10.1016/S0140-6736(07)61306-3. [DOI] [PubMed] [Google Scholar]
- 16.Stevens LJ, Kuczek T, Burgess JR, Stochelski MA, Arnold LE, Galland L. 2013. Mechanisms of behavioral, atopic, and other reactions to artificial food colors in children. Nutr Rev 71:268–281. doi: 10.1111/nure.12023. [DOI] [PubMed] [Google Scholar]
- 17.Espín JC, García-Conesa MT, Tomás-Barberán FA. 2007. Nutraceuticals: facts and fiction. Phytochemistry 68:2986–3008. doi: 10.1016/j.phytochem.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 18.Santos-Buelga C, Mateus N, De Freitas V. 2014. Anthocyanins. Plant pigments and beyond. J Agric Food Chem 62:6879–6884. [DOI] [PubMed] [Google Scholar]
- 19.Pietta PG. 2000. Flavonoids as antioxidants. J Nat Prod 63:1035–1042. doi: 10.1021/np9904509. [DOI] [PubMed] [Google Scholar]
- 20.Yan Y, Chemler J, Huang L, Martens S, Koffas MA. 2005. Metabolic engineering of anthocyanin biosynthesis in Escherichia coli. Appl Environ Microbiol 71:3617–3623. doi: 10.1128/AEM.71.7.3617-3623.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan Y, Li Z, Koffas MA. 2008. High-yield anthocyanin biosynthesis in engineered Escherichia coli. Biotechnol Bioeng 100:126–140. doi: 10.1002/bit.21721. [DOI] [PubMed] [Google Scholar]
- 22.Leonard E, Yan Y, Fowler ZL, Li Z, Lim CG, Lim KH, Koffas MA. 2008. Strain improvement of recombinant Escherichia coli for efficient production of plant flavonoids. Mol Pharm 5:257–265. doi: 10.1021/mp7001472. [DOI] [PubMed] [Google Scholar]
- 23.Zhao S, Jones JA, Lachance D, Bhan N, Khalidi O, Venkataraman S, Wang Z, Koffas MAG. 2015. Improvement of catechin production in Escherichia coli through combinatorial metabolic engineering. Metab Eng 28:43–53. doi: 10.1016/j.ymben.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 25.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakajima J, Tanaka Y, Yamazaki M, Saito K. 2001. Reaction mechanism from leucoanthocyanidin to anthocyanidin 3-glucoside, a key reaction for coloring in anthocyanin biosynthesis. J Biol Chem 276:25797–25803. doi: 10.1074/jbc.M100744200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Wang L, Deroles S, Bennett R, Davies K. 2006. New insight into the structures and formation of anthocyanic vacuolar inclusions in flower petals. BMC Plant Biol 6:29–35. doi: 10.1186/1471-2229-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Springob K, Nakajima J, Yamazaki M, Saito K. 2003. Recent advances in the biosynthesis and accumulation of anthocyanins. Nat Prod Rep 20:288–303. doi: 10.1039/b109542k. [DOI] [PubMed] [Google Scholar]
- 29.Piddock LJV. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev 19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burse A, Weingart H, Ullrich MS. 2004. The phytoalexin-inducible multidrug efflux pump AcrAB contributes to virulence in the fire blight pathogen, Erwinia amylovora. Mol Plant Microbe Interact 17:43–54. doi: 10.1094/MPMI.2004.17.1.43. [DOI] [PubMed] [Google Scholar]
- 31.Fujii T, Fujii Y, Machida K, Ochiai A, Ito M. 2009. Efficient biotransformations using Escherichia coli with tolC acrAB mutations expressing cytochrome P450 genes. Biosci Biotechnol Biochem 73:805–810. doi: 10.1271/bbb.80627. [DOI] [PubMed] [Google Scholar]
- 32.Lim HN, Lee Y, Hussein R. 2011. Fundamental relationship between operon organization and gene expression. Proc Natl Acad Sci U S A 108:10626–10631. doi: 10.1073/pnas.1105692108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu P, Wang W, Li L, Bhan N, Zhang F, Koffas MAG. 2014. Design and kinetic analysis of a hybrid promoter-regulator system for malonyl-CoA sensing in Escherichia coli. ACS Chem Biol 9:451–458. doi: 10.1021/cb400623m. [DOI] [PubMed] [Google Scholar]
- 34.Xu P, Li L, Zhang F, Stephanopoulos G, Koffas M. 2014. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc Natl Acad Sci U S A 111:11299–11304. doi: 10.1073/pnas.1406401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuznetsova E, Proudfoot M, Gonzalez CF, Brown G, Omelchenko MV, Borozan I, Carmel L, Wolf YI, Mori H, Savchenko AV, Arrowsmith CH, Koonin EV, Edwards AM, Yakunin AF. 2006. Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J Biol Chem 281:36149–36161. doi: 10.1074/jbc.M605449200. [DOI] [PubMed] [Google Scholar]
- 36.Dietrich JA, McKee AE, Keasling JD. 2010. High-throughput metabolic engineering: advances in small-molecule screening and selection. Annu Rev Biochem 79:563–590. doi: 10.1146/annurev-biochem-062608-095938. [DOI] [PubMed] [Google Scholar]
- 37.Cress BF, Toparlak OD, Guleria S, Lebovich M, Stieglitz JT, Englaender JA, Jones JA, Linhardt RJ, Koffas MA. 30 March 2015. CRISPathBrick: modular combinatorial assembly of type II-A CRISPR arrays for dCas9-mediated multiplex transcriptional repression in E. coli. ACS Synth Biol doi: 10.1021/acssynbio.5b00012. [DOI] [PubMed] [Google Scholar]
- 38.Jones JA, Toparlak OD, Koffas MA. 2015. Metabolic pathway balancing and its role in the production of biofuels and chemicals. Curr Opin Biotechnol 33:52–59. [DOI] [PubMed] [Google Scholar]
- 39.Novy R, Morris B. 2001. Use of glucose to control basal expression in the pET system. Innovations 13:13–15. [Google Scholar]
- 40.Castañeda-Ovando A, de Lourdes Pacheco-Hernández M, Páez-Hernández ME, Rodríguez JA, Galán-Vidal CA. 2009. Chemical studies of anthocyanins: a review. Food Chem 113:859–871. doi: 10.1016/j.foodchem.2008.09.001. [DOI] [Google Scholar]
- 41.Santos CNS, Stephanopoulos G. 2008. Melanin-based high-throughput screen for l-tyrosine production in Escherichia coli. Appl Environ Microbiol 74:1190–1197. doi: 10.1128/AEM.02448-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim CG, Fowler ZL, Hueller T, Schaffer S, Koffas MA. 2011. High-yield resveratrol production in engineered Escherichia coli. Appl Environ Microbiol 77:3451–3460. doi: 10.1128/AEM.02186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu P, Ranganathan S, Fowler ZL, Maranas CD, Koffas MA. 2011. Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-CoA. Metab Eng 13:578–587. doi: 10.1016/j.ymben.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 44.Chemler JA, Fowler ZL, McHugh KP, Koffas MA. 2010. Improving NADPH availability for natural product biosynthesis in Escherichia coli by metabolic engineering. Metab Eng 12:96–104. doi: 10.1016/j.ymben.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 45.Xu H, Liu X, Yan Q, Yuan F, Gao Y. 2015. A novel copigment of quercetagetin for stabilization of grape skin anthocyanins. Food Chem 166:50–55. doi: 10.1016/j.foodchem.2014.05.125. [DOI] [PubMed] [Google Scholar]
- 46.Wu J, Guan Y, Zhong Q. 2015. Yeast mannoproteins improve thermal stability of anthocyanins at pH 7.0. Food Chem 172:121–128. doi: 10.1016/j.foodchem.2014.09.059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.