Abstract
Trichothecenes are fungal sesquiterpenoid compounds, the majority of which have phytotoxic activity. They contaminate food and feed stocks, resulting in potential harm to animals and human beings. Trichoderma brevicompactum and T. arundinaceum produce trichodermin and harzianum A (HA), respectively, two trichothecenes that show different bioactive properties. Both compounds have remarkable antibiotic and cytotoxic activities, but in addition, trichodermin is highly phytotoxic, while HA lacks this activity when analyzed in vivo. Analysis of Fusarium trichothecene intermediates led to the conclusion that most of them, with the exception of the hydrocarbon precursor trichodiene (TD), have a detectable phytotoxic activity which is not directly related to the structural complexity of the intermediate. In the present work, the HA intermediate 12,13-epoxytrichothec-9-ene (EPT) was produced by expression of the T. arundinaceum tri4 gene in a transgenic T. harzianum strain that already produces TD after transformation with the T. arundinaceum tri5 gene. Purified EPT did not show antifungal or phytotoxic activity, while purified HA showed both antifungal and phytotoxic activities. However, the use of the transgenic T. harzianum tri4 strain induced a downregulation of defense-related genes in tomato plants and also downregulated plant genes involved in fungal root colonization. The production of EPT by the transgenic tri4 strain raised levels of erg1 expression and reduced squalene accumulation while not affecting levels of ergosterol. Together, these results indicate the complex interactions among trichothecene intermediates, fungal antagonists, and host plants.
INTRODUCTION
A number of Trichoderma species are known for the ability to act as important biocontrol agents against phytopathogenic fungi (1, 2). Trichoderma species are able to act as biofertilizers, to induce plant defense responses, and to increase tolerance to abiotic stresses (3, 4). Trichoderma arundinaceum and T. brevicompactum also produce trichothecenes, sesquiterpenoid compounds which are harmful to plants and to the animals which consume contaminated food or feed stocks. Several studies on the structure-activity relationships of Fusarium trichothecene toxins in alga, plant, and animal models (5–7) indicate that certain Fusarium trichothecenes and intermediates have different toxic effects. Using an Arabidopsis leaf assay, Desjardins and coworkers found that toxicity varied >200-fold between different trichothecene compounds, while trichodiene (TD), a hydrocarbon precursor, was not phytotoxic (6).
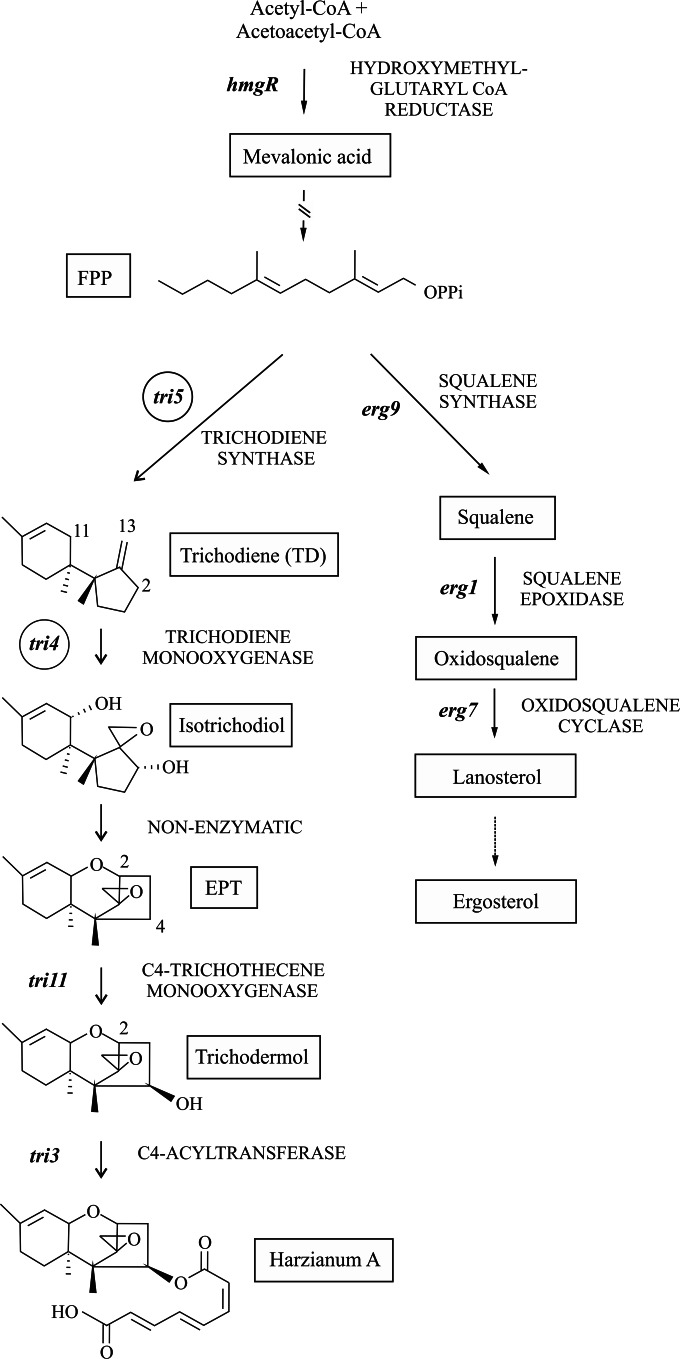
Biosynthesis of trichothecenes starts with the cyclization of farnesyl diphosphate (FPP), catalyzed by the terpene cyclase trichodiene synthase encoded by tri5 (8), to produce TD. In Trichoderma, TD is then oxygenated at C-2, C-11, and C-13 by a P450 monooxygenase encoded by tri4 (9, 10) to give the intermediate isotrichodiol, which is nonenzymatically converted to 12,13-epoxytrichothec-9-ene (EPT). The oxygenation of these three carbons is a characteristic that Trichoderma shares with Myrothecium, Stachybotrys, and Trichothecium. In Trichoderma, EPT is hydroxylated at C-4 by a cytochrome P450 monooxygenase encoded by tri11, giving rise to trichodermol, which serves as a substrate for acetylation or esterification by the Tri3 protein to produce trichodermin in T. brevicompactum or harzianum A (HA) in T. arundinaceum (Fig. 1).
FIG 1.
Schematic representation of the ergosterol and HA biosynthetic pathways. Chemical structures of HA and the intermediates have been included. The tri4 and tri5 genes, belonging to the trichothecene pathway, which were expressed in the T. harzianum strain to produce EPT, are labeled with circles. Genes and enzymes are indicated in italic and capital letters, respectively.
HA and trichodermin have been studied widely in the last few decades (11–16), and their biosynthetic pathways have been detailed almost completely (9). Research has expanded to examine the structure-function relationships of the various trichothecenes and their intermediates on plants and other fungi. Cytotoxicity of HA on human tumor cell lines (17) and the importance to cytotoxicity of the moiety of the C-4 side chain of trichothecenes (18) have been reported. Trichodermin, which carries an ester group in the C-4 side chain, is more toxic than trichodermol, which contains a hydroxyl group in that position (18). In recent years, the role of HA in the biocontrol activity of Trichoderma arundinaceum has been investigated. Disruption and/or silencing of the tri4 and tri5 genes in this fungus resulted in an elimination or reduction of HA production and in a reduction in biocontrol ability. These mutants also showed a decrease in the ability to induce the expression of tomato defense-related genes. The results of these studies led to the conclusion that HA plays an important function in the sensitization of Trichoderma-pretreated plants to the fungal pathogen Botrytis cinerea (10, 19).
To further the knowledge of the Trichoderma trichothecene biosynthetic pathway, the study of the effect of the trichothecene intermediates on the interaction with plants and fungal antagonists was recently undertaken. TD, representing the first committed step in the trichothecene biosynthetic pathway, induces in vivo expression of plant defense genes, mainly those related to jasmonate (JA)/ethylene (ET), and also induces B. cinerea virulence genes in confrontation experiments (20). In addition, TD reduces in vitro aboveground growth and root development in tomato plants, illustrating that more complex trichothecene structures may not be necessary to induce such responses. However, TD is a volatile organic compound (VOC) that is almost undetectable in the wild-type strain, since it is rapidly processed to the next step in the pathway of HA synthesis (20). EPT, with the typical trichothecene epoxy structure, is the first nonvolatile intermediate in Trichoderma trichothecene biosynthesis, and its effect on the interaction of fungal pathogens and/or plants has not been studied previously.
In the present work, we studied the role that EPT plays in gene expression in tomato plants, whether added as a pure compound or as produced by Trichoderma mutants. Wild-type T. harzianum does not produce trichothecenes and lacks the genes necessary for trichothecene production; therefore, it serves as an ideal recipient for transgenic expression of trichothecene genes. We used heterologous expression techniques to transform a transgenic strain of T. harzianum overexpressing tri5 to a strain overexpressing both tri5 and tri4. The double mutant was capable of transforming FPP into TD and then into EPT. The effect of EPT production on Trichoderma-tomato interactions was compared with the effects of exogenous trichodermol and HA, the next intermediate and the final product of the biosynthetic pathway, respectively. This allowed us to determine the relationship between the complexity of Trichoderma trichothecenes and their biological activities. We also determined the effects of tri4 activity and subsequent production of EPT on tomato-fungal pathogen interactions. Finally, we determined how the production of EPT affects the balance of other terpene intermediates in the producer fungus.
MATERIALS AND METHODS
Strains and culture conditions.
Trichoderma harzianum T34-tri5.27 (called T34-5.27 henceforth), constructed from T. harzianum CECT 2413 (T34) by expression of the T. arundinaceum tri5 gene (20), was used as a recipient for plasmid pb1Tatri4, designed to overexpress the T. arundinaceum IBT 40837 (Ta37) tri4 gene (see below), or for plasmid pJL43b1, the empty vector from which the plasmid pb1Tatri4 was constructed. Escherichia coli DH5α (Invitrogen, Carlsbad, CA) was used as the plasmid host. Selected transformants carrying tri4 were named T34-5.27-tri4.1, T34-5.27-tri4.2, and T34-5.27-tri4.3. The control transformant carrying plasmid pJL43b1 was named T34-5.27-b1.
Botrytis cinerea B05.10 (21) was used in the antifungal assays and in the in vivo tomato assays to determine the effect of tri4 gene overexpression on the size of the lesions caused by this pathogen in plants inoculated with the Trichoderma transformants. B05.10 was maintained on MEA (2% glucose, 2% malt extract, 1% peptone, 2% agar, pH 5.6) medium and grown at 21°C for 5 to 7 days, with a photoperiod of 16 h of light and 8 h of dark.
Fusarium sporotrichioides CECT 20166 was used in the antifungal assays and was maintained on MEA medium and grown at 28°C for 4 to 7 days.
Solanum lycopersicum var. Marmande (Semillas Rocalba S.A., Girona, Spain) was used for fungus-plant and metabolite-plant interaction studies.
Quantification of ergosterol-squalene.
A two-step growth procedure was followed. Spores were inoculated into CM medium (0.5% malt extract, 0.5% yeast extract, 0.5% glucose) at a concentration of 1 × 106 spores ml−1 and grown for 24 h (20). Ten milliliters of the CM culture-grown mycelia were transferred to 50 ml of minimal medium (MM) (22) and grown for 96 h at 28°C and 250 rpm. Mycelia were used for extraction and quantification of ergosterol-squalene as previously reported (23, 24). All measurements were made in triplicate.
Nucleic acid extraction and manipulation.
Total RNA was isolated from mycelia of Trichoderma cultures or tomato leaves/aerial parts/roots that were frozen with liquid nitrogen and ground in a mortar. RNA was extracted by the phenol-SDS method (25) and treated with a DNase and RNase protector (Fermentas, Vilnius, Lithuania).
cDNAs were synthesized by using 1 μg of total RNA and a reverse transcription system based on the use of an oligo(dT)15 primer (Promega, Madison, WI). cDNAs were quantified using a Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE).
Construction of pb1Tatri4, transformation of strain T34-5.27, and selection of transformants.
The T. arundinaceum tri4 open reading frame (ORF) was amplified by PCR with the oligonucleotides Tri437-5 (5′-ATG TTG GAC ATC AAC GCG-3′) and Tri437-3b (5′-TCA AAG CTT CTT GAT GAC CT-3′) by using Pfu polymerase (Fermentas, Vilnius, Lithuania), with T. arundinaceum IBT 40837 genomic DNA as the template. The resulting fragment was ligated to pTAcbh digested with NcoI, filled by use of the Klenow fragment, and dephosphorylated to obtain the pTAT4g plasmid (7,123 bp). Plasmid pTAcbh contains the T. harzianum tss1 gene promoter and the transcriptional terminator of the Trichoderma reesei cellobiohydrolase 2 gene. A 4,029-bp EcoRI/EheI fragment from pTAT4g, treated with the Klenow fragment, was ligated to pJL43b1 (26) which had been digested with HindIII, filled in by use of the Klenow fragment, and dephosphorylated. Plasmid pJL43b1 (4,488 bp) contained the phleomycin resistance gene of Streptoalloteichus hindustanus expressed under the control of the gpdA gene promoter of Aspergillus nidulans and the CYC1 transcriptional terminator of Saccharomyces cerevisiae. The resulting plasmid, pb1Tatri4, had a final size of 8,517 bp (see Fig. S1 in the supplemental material).
Transformation of T34-5.27 protoplasts was carried out as described previously (27), with the exception that 100 μg/ml of phleomycin and Czapek medium with 1 M sorbitol were used. The T34-5.27 strain was already resistant to hygromycin, as that marker was transferred with the Ta37 tri5 gene expression cassette into the host T34 strain.
Oligonucleotides T437CT (5′-ACA ATA CGG GCG TGG GTC AT-3′) and T437EbNt (5′-GGT ATT AAA TCC TCG GTG CTC GTT-3′) were used to analyze the transformants containing the pb1Tatri4 plasmid and also to amplify an 837-bp fragment internal to tri4 for use as a probe in the Southern experiments.
Detection and quantification of TD and EPT.
Conidia from the different Trichoderma strains were inoculated onto YEPD (1% Bacto yeast extract [Difco Laboratories, Detroit, MI], 1% Bacto peptone [Difco], and 5% glucose [Sigma-Aldrich Co., St. Louis, MO]) and grown at 28°C and 200 rpm for 4 days. Fungal cultures were extracted with ethyl acetate, and the extracts were analyzed by gas chromatography-mass spectrometry (GC-MS) (10). TD and EPT were quantified using standard curves. TD for the standard curve was purified from F. sporotrichioides Tri4 mutant strain F15 (28). EPT for the standard curve was purified from F. sporotrichioides transformant 4-4-18, a transgenic strain expressing Myrothecium roridum Tri4 (29).
Tomato plant assays.
Surface-sterilized tomato seeds were inoculated (10) with strain T34-5.27-b1 or T34-5.27-tri4.2 and sowed in commercial loamy field soil (Kekkilä 50/50; Projar S.A., Valencia, Spain) (refer to reference 10 for details of the soil composition). Pots were incubated in a greenhouse at 21 ± 2°C, with a photoperiod of 16 h of light and 8 h of dark, and watered as needed. After 4 weeks, the tomato leaves were inoculated with 15 μl of B05.10 conidial suspension (5 × 105 conidia ml−1 in germination buffer [20 mM glucose, 20 mM KH2PO4]). After 4 days of incubation, leaves with or without pathogen inoculation were collected to extract RNA in order to analyze the expression of tomato marker genes involved in defense responses or development (10).
Tomato-Trichoderma and tomato-Botrytis hydroponic cultures for analysis of the effect of EPT production on the ability to colonize tomato roots.
For the tomato hydroponic cultures (10, 30, 31), sterile tomato seeds were placed inside Phytatray II boxes (Sigma, St. Louis, MO) (30 seeds per box) on a sterile gauze sheet over a sterile stainless steel screen, which held them 1 cm above 100 ml of liquid MS medium, and maintained at 21°C in a plant growth chamber with controlled light and humidity conditions, as indicated above, for 2 weeks. Spores (106 ml−1) from the T34-5.27-b1 control strain, the T34-5.27-tri4.2 transformant, and, when indicated, strain B05.10, were used to inoculate 250-ml flasks containing 100 ml of CM medium. Each strain was grown at 28°C and 250 rpm in darkness for 24 h. Twenty-milliliter aliquots of Trichoderma and Botrytis mycelial suspensions were used to inoculate 100 ml of fresh potato-dextrose broth (PDB; Becton Dickinson, Heidelberg, Germany) and Czapek medium, respectively, and incubated for 48 h at 28°C and 250 rpm. Mycelia were harvested by filtration, washed with sterile water, and added to Phytatray II boxes that contained 2-week-old tomato plants (1.5 g of mycelium from each fungal strain was used). Tomato-Trichoderma (control or T34-5.27-tri4.2 transformant) and Tomato-Botrytis hydroponic cultures were maintained at 21°C and 80 rpm for 20 h. Finally, roots were recovered, cleaned with a direct cold Milli-Q sterile water jet, and used for RNA extraction and cDNA synthesis followed by quantitative PCR (qPCR) gene analysis.
In vitro analysis of the effects of EPT, trichodermol, and HA on tomato growth and on the expression of plant defense-related genes.
Sterilized tomato seeds (five per plate) (32) were placed on 150-mm-diameter petri dishes containing 60 ml of Murashige & Skoog (MS) medium (Sigma-Aldrich, St. Louis, MO) [0.49% MS basal medium, 1% sucrose, 0.05% 2-(N-morpholino)ethanesulfonic acid (MES), pH 5.7, 0.8% agar] and incubated in a greenhouse at 21 ± 2°C, with a photoperiod of 16 h of light and 8 h of dark, for 4 days to allow germination. Four germinated tomato seeds were placed on another 150-mm-diameter petri dish with 50 ml of solidified MS medium containing 0.1, 1, or 4 μg ml−1 EPT, trichodermol, or HA diluted in 250 μl of acetone (for EPT and trichodermol) or 250 μl of acetonitrile (for HA). Acetone (250 μl) or acetonitrile (250 μl) was used as a control for EPT/trichodermol or HA, respectively. Each petri dish was sealed with adhesive tape and Parafilm, placed in a slanted position, and incubated in a greenhouse at 21 ± 2°C for 6 days. Plants were then photographed to observe differences in the main and secondary root lengths and in aboveground growth. RNAs were extracted from the aerial parts of the plants, and cDNAs were analyzed by qPCR to determine the effect of EPT, trichodermol, or HA treatment on the expression of tomato genes involved in defense responses.
qPCR experiments.
In order to perform comparative gene expression studies by qPCR, oligonucleotides specific to the tomato plant defense-related genes were used (33, 34). These included PR1b1 and PR-P2 (salicylic acid [SA]-related genes) and PINI, PINII, and TomLoxA (jasmonic acid [JA]-related genes). Additionally, the expression of the following three genes related to plant development was also analyzed: SUCS, encoding a sucrose synthase; ACCS, encoding a 1-aminocyclopropane-1-carboxylic acid (ACC) synthase; and GAI, encoding a DELLA gibberellic acid-insensitive protein. Oligonucleotides used to analyze the expression of these three tomato development-related genes were designed for the present work (see Table S1 in the supplemental material). Expression of the T34 hmgR (encoding the hydroxymethyl glutaryl coenzyme A [CoA] reductase), erg9 (squalene synthase [SQS]), erg1 (squalene epoxidase [SE]), and erg7 (oxidosqualene cyclase) genes, which are involved in the biosynthesis of terpene and ergosterol (Fig. 1) (20), was also determined. For T34 gene expression studies, the α-actin gene and gpd were used as reference genes, according to GeNorm software (35) results. For tomato defense-related gene expression and colonization assays, the α-actin and GAPDH (glyceraldehyde 3-phosphate dehydrogenase) genes were used as the housekeeping genes, respectively. qPCRs were carried out by using a StepOnePlus system (Applied Biosystems, Foster City, CA). The reaction mixtures were set up in a total volume of 20 μl, with the following components per reaction mixture: 10 μl Power SYBR green PCR master mix (Applied Biosystems, Foster City, CA), 0.4 μl 10 μM forward primer, 0.4 μl 10 μM reverse primer, 5 μl cDNA, and H2O to 20 μl. Relative expression values and the significance of the differences between the gene expression levels were calculated using REST 2009 software (36). For each primer pair used in this work, we performed a standard curve assay, using 320, 160, 80, 40, 20, and 10 ng of cDNA for Trichoderma and Botrytis genes and 160, 80, 40, 20, 10, and 5 ng of cDNA for tomato genes, to determine the PCR amplification efficiency (E value). The E values for all primers used were between 0.9 and 1.1. Each measurement was made in triplicate.
Quantification of tri4 gene copy number in T34-5.27-tri4 transformants by qPCR.
Genomic DNAs from the three tri4 transformants were diluted 10−2 in sterile distilled water, and the DNA concentrations were quantified using a Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE). A standard curve assay was performed by using concentrations of 32, 16, 8, 4, 2, and 1 ng/μl for each genomic DNA, along with oligonucleotides for the α-actin and tri4 genes (see Fig. S2 in the supplemental material). For each genomic DNA sample, using the equations for these calibration curves, the amounts corresponding to the α-actin and tri4 genes (in nanograms per microliter) for a given genomic DNA concentration were calculated. By determining the ratio between the nanograms per microliter of tri4 and the nanograms per microliter of the α-actin gene, the tri4 gene copy number could be determined, assuming that there was one copy of the α-actin gene per genome.
Statistical analysis.
Analysis of variance (ANOVA) and Kruskal-Wallis tests were performed with IBM SSPS Statistics 19 software and were used to compare the data regarding production of TD-EPT and squalene-ergosterol between the strains analyzed in the present work.
Antifungal assays. (i) Antibiograms of EPT, trichodermol, and HA against B05.10.
Strain B05.10 was grown on MEA medium at 21°C for 7 days, with a photoperiod of 16 h of light and 8 h of dark. Spores were recovered using sterile water and a bent, sterile glass rod, centrifuged at 4,000 rpm for 10 min, and resuspended in water to a concentration of 1 × 108 spores ml−1. Spores were added at a concentration of 1 × 105 spores ml−1 to a bottle containing 100 ml of molten MEA medium containing 1% agar cooled to 50°C. Twenty milliliters of the medium containing the fungal spores was spread in a petri dish with a 9-cm diameter. Once the medium was solidified, 5 plugs of 7 mm in diameter were removed from each plate, and 60-μl samples of each EPT, trichodermol, and HA dilution were added to the holes. The central hole in each plate was filled with acetone or acetonitrile as a control. Plates were then kept at 4°C for 5 h, to allow for metabolite diffusion, and finally incubated at 21°C for 24 to 48 h to detect and measure the diameters of the areas of inhibition.
(ii) Growth assay on cellophane membranes.
Seven-millimeter-diameter potato-dextrose agar (PDA) plugs of T34-5.27-b1 (control) or tri4 transformants were individually placed at the center of petri dishes containing PDA or MEA medium on cellophane sheets (25- to 30-kDa cutoff). After 2 days of incubation at 28°C, the membranes were removed from the plates, and a single, 7-mm-diameter mycelial plug of the plant pathogen Fusarium sporotrichioides (from PDA plates) or B. cinerea (from MEA plates) was placed at the center of the plate. In parallel, the pathogens were grown on PDA or MEA (control plates). Each pathogen was tested on three plates. Growth diameters were calculated every 24 h for 96 h.
(iii) Antifungal assays on tomato leaves.
Leaves from plants grown from sterilized seeds in loam were used to carry out an antifungal assay against B05.10. Leaves from 4-week-old plants were detached and placed in petri dishes lined with sterile wet filter paper. Fifteen-microliter aliquots of B05.10 conidial suspensions (5 × 105 spores ml−1) in germination buffer were placed on the leaf surfaces. After approximately 4 h (when the B. cinerea conidial suspension had dried), 15 μl of Trichoderma filter-sterilized broth (or PDB medium, in control assays) was placed over each B05.10 conidial suspension spot, and the plates were incubated in a greenhouse at 21°C for 72 h. Two inoculations per leaf were made on six leaves per treatment, and there were two replicates for each experiment.
RESULTS
Selection of T. harzianum T34-5.27 transformants with the Ta37 tri4 gene.
In order to produce the trichothecene EPT in Trichoderma harzianum, plasmid pb1Tatri4 was used to transform the transgenic strain T34-5.27, which carries tri5 from T. arundinaceum. Transformants were selected by their resistance to phleomycin. After the selection rounds, 10 transformants were selected to be analyzed by PCR and Southern hybridization. Transformants T34-5.27-tri4.1, T34-5.27-tri4.2, and T34-5.27-tri4.3, which showed the expected PCR and Southern blot bands, were selected for further studies (see Fig. S1 in the supplemental material).
qPCR analysis to calculate the numbers of tri4 gene copies integrated into the genomes of the selected transformants showed 4 or 5 copies in T34-5.27-tri4.1, 9 or 10 copies in T34-5.27-tri4.2, and 8 or 9 copies in T34-5.27-tri4.3 (see Fig. S2 in the supplemental material).
Production of EPT by T34-5.27-tri4 transformants.
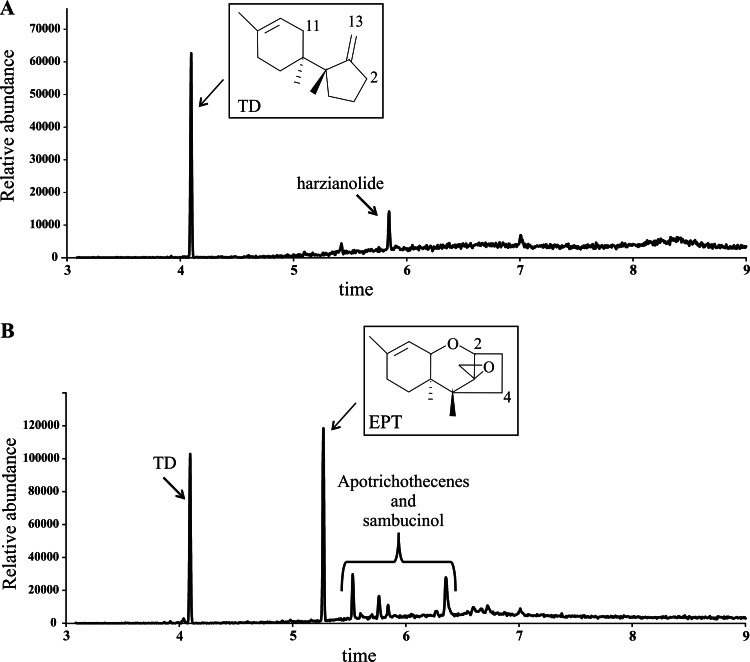
After 96 h of growth on YEPD medium, GC-MS analysis of culture extracts detected TD production (peak at 4.09 min) by the host strain T34-5.27-b1 and all the transformants. Only the tri4 transformants produced EPT (peak at 5.27 min) (Fig. 2). In addition, apotrichothecenes and sambucinol, which are shunt metabolites of trichodiene, were detected in the tri4 transformants (Fig. 2).
FIG 2.
GC-MS analyses of 96-h culture broths of T34-5.27-b1 (A) and T34-5.27-tri4.2 (B). The chemical structures of TD and EPT are shown. GC-MS analyses of the T34-5.27-tri4.1 and T34-5.27-tri4.3 transformants were very similar to that of T34-5.27-tri4.2. Note that additional compounds, i.e., harzianolide (arrow) and apotrichothecenes and sambucinol (bracket), were also detected.
The production of TD and EPT was quantified for 6- and 8-day-old cultures, and as might be expected, higher levels of these compounds were detected in 8-day-old cultures. In addition, a strong but not statistically significant reduction in the level of TD was observed in transformants T34-5.27-tri4.1 to T34-5.27-tri4.3 compared to T34-5.27-b1 (control) (Table 1). Interestingly, production of TD and EPT did not correlate with the tri4 gene copy number calculated for each transformant: higher levels of TD and EPT were observed in transformant 1, which showed a smaller number of tri4 copies integrated into its genome.
TABLE 1.
Production of TD and EPT in 6- and 8-day-old cultures of three tri4-overexpressing transformants and the control transgenic strain T34-5.27-b1
| Time of culture (days) | Strain | Compound concn (μg/ml of culture) (mean ± SD)a |
|
|---|---|---|---|
| TD | EPT | ||
| 6 | T34-5.27-b1 | 8.35 ± 0.66 | |
| T34-5.27-tri4.1 | 7.76 ± 1.05 | 11.95 ± 1.90 | |
| T34-5.27-tri4.2 | 6.31 ± 4.92 | 10.47 ± 1.64 | |
| T34-5.27-tri4.3 | 8.73 ± 2.44 | 8.29 ± 1.19 | |
| 8 | T34-5.27-b1 | 42.62 ± 17.54 | |
| T34-5.27-tri4.1 | 19.67 ± 6.65 | 36.43 ± 10.51 | |
| T34-5.27-tri4.2 | 14.88 ± 11.3 | 29.74 ± 3.79 | |
| T34-5.27-tri4.3 | 27.14 ± 7.48 | 28.31 ± 4.86 | |
Data were analyzed by ANOVA (n = 3). For each time point, the values are not significantly different (P < 0.05).
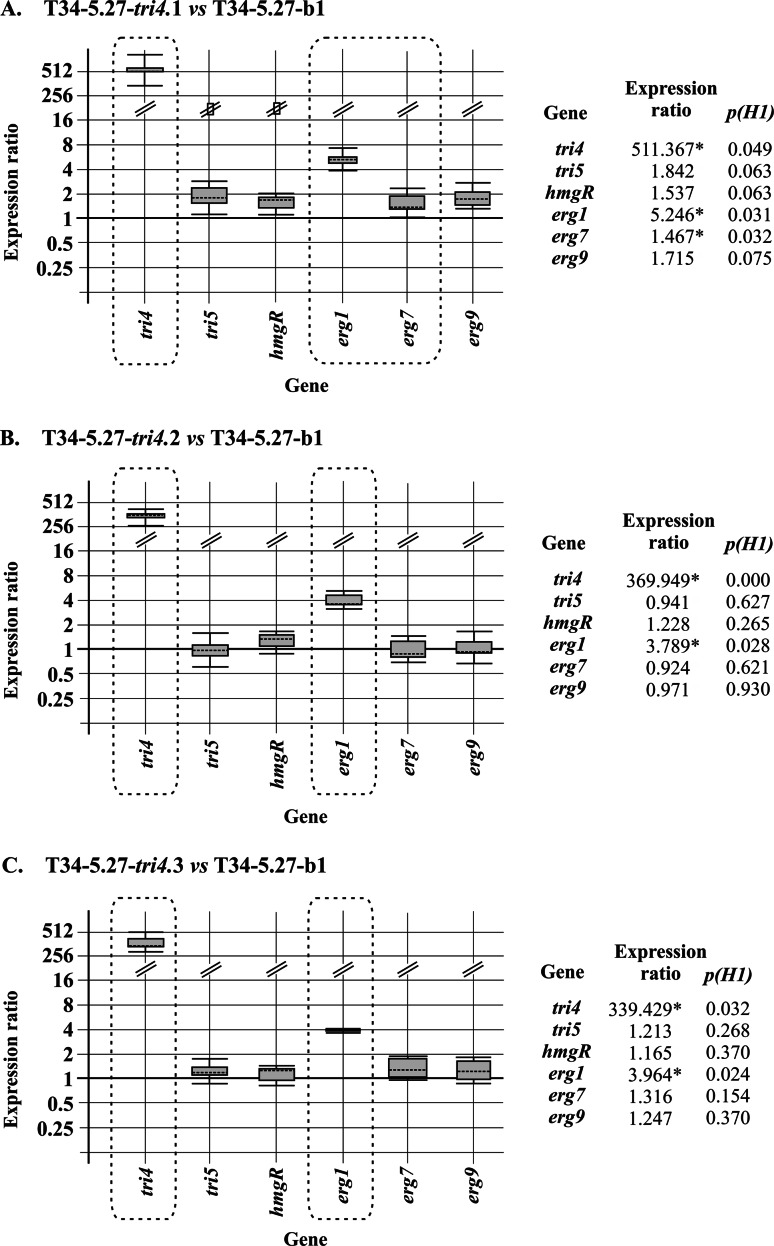
Expression of tri4 affects the expression of genes involved in the terpene biosynthetic pathway.
Analysis by qPCR of mycelia from the three selected transformants grown for 96 h in MM showed high levels of tri4 gene expression, indicating that the tss1 gene promoter used in the overexpression construct was properly recognized. The expression of other terpene biosynthetic genes was analyzed in tri4 transformants and compared with their expression in the recipient strain. Only the expression of erg1, encoding the squalene epoxidase (SE) enzyme, was significantly upregulated by tri4 overexpression, raising the values of expression 3.789 (P = 0.028)- and 5.246 (P = 0.031)-fold in transformants 2 and 1, respectively (Fig. 3).
FIG 3.
(A to C) qPCR analysis of expression levels of terpene biosynthetic genes in the three selected tri4-overexpressing transformants relative to the levels of expression of these genes in the control strain, T34-5.27-b1, in mycelia collected from 96-h cultures. Comparisons and graphic representations were carried out using REST software (36). Values indicated with asterisks (boxed in the graphic representation) correspond to genes significantly differentially expressed (P ≤ 0.05) in comparison with the reference condition. Strain Ta37 was used to compare the levels of expression of tri4 in the T34-5.27-tri4 transformants, since tri4 is not present in strain T34.
Expression of tri4 results in a drastic reduction in the level of squalene accumulation but does not affect the level of ergosterol.
Expression of tri4 and production of EPT resulted in reductions of the level of squalene, ranging from 38.4% in transformant 3 to 54.28% in transformant 2, when measured from mycelia grown for 96 h on MM. These reductions in the level of squalene correlated with the increases in the level of erg1 gene expression. However, the level of ergosterol was not significantly affected in the transformants in comparison with the recipient strain (Table 2).
TABLE 2.
Production of squalene and ergosterol by the control T34-5.27-b1 strain and transformants expressing the T. arundinaceum tri4 gene at 96 ha
| Strain | Dry wt (g)b | Squalene concn (mg/g dry wt of strain) | % variation | Ergosterol concn (mg/g dry wt of strain) |
|---|---|---|---|---|
| T34-5.27-b1 | 0.2309 ± 0.0015A | 0.2393 ± 0.0013A | 8.3057 ± 0.2989A | |
| T34-5.27-tri4.1 | 0.1780 ± 0.0127B | 0.1259 ± 0.0052B | −47.38 | 8.3048 ± 0.0972A |
| T34-5.27-tri4.2 | 0.1992 ± 0.0087B | 0.1094 ± 0.0099C | −54.28 | 7.9592 ± 0.1342A |
| T34-5.27-tri4.3 | 0.1758 ± 0.01B | 0.1474 ± 0.004D | −38.40 | 7.8440 ± 0.0776A |
Data are means ± standard deviations, unless indicate otherwise. Data were analyzed by ANOVA (n = 3). For each column, values followed by different superscript letters are significantly different (P < 0.05).
Total mycelial mass collected from a 50-ml culture.
In vitro effects of EPT, trichodermol, and HA on the growth of tomato plants and on the expression of tomato defense-related genes.
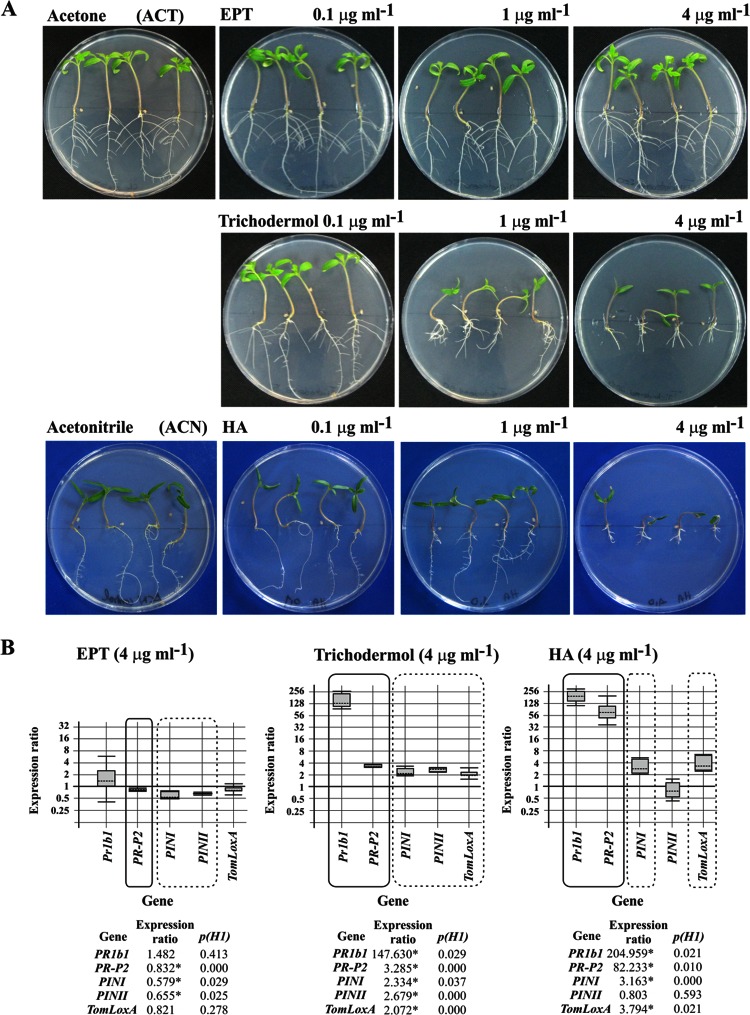
Exogenously added EPT did not significantly affect the in vitro growth of tomato plants at any of the concentrations assayed, which ranged from 0.1 to 4 μg ml−1 (Fig. 4). This result contrasts with that observed with trichodermol, the next intermediate in the HA biosynthetic pathway, which showed a strong phytotoxic effect at concentrations of 1 μg ml−1 and 4 μg ml−1, with a reduced development of the aerial parts and roots. Even plants grown with the lowest trichodermol concentration, 0.1 μg ml−1, had a reduction in lateral root development. The results observed in the presence of EPT also contrasted with those for HA, the final product of the pathway, which showed a significant phytotoxic effect on the aerial parts at 1 μg ml−1 and on both the aerial parts and the roots at 4 μg ml−1.
FIG 4.
(A) Photographs illustrating the effects of three concentrations of trichothecene (EPT), trichodermol, and HA on the growth and root architecture of tomato plants. (B) qPCR analysis of the levels of expression of five tomato defense-related genes in the aerial parts of tomato plants grown under the effects of EPT (4 μg ml−1), trichodermol (4 μg ml−1), and HA (4 μg ml−1) versus the levels of expression in control plants without any of these compounds. Comparative calculations and graphic representations were carried out as indicated in the legend to Fig. 3.
When the expression of tomato defense-related genes was analyzed, EPT did not show a remarkable effect on their level of expression; only PR-P2 (SA-related gene) and PINI and PINII (JA-related genes) were slightly but significantly downregulated, showing changes of 0.832 (P = 0.000)-, 0.579 (P = 0.029)-, and 0.655 (P = 0.025)-fold, respectively, in comparison with control plants. However, trichodermol significantly upregulated all of the genes analyzed, with those related to SA having more drastic increases in the level of expression, reaching levels of 147.630 (P = 0.029)- and 3.285 (P = 0.000)-fold for PR1b1 and PR-P2, respectively. Finally, HA showed an effect similar to that of trichodermol, with two major differences: a higher upregulation of SA-related genes, with relative expression ratios of 205 (P = 0.021)- and 82 (P = 0.010)-fold for PR1b1 and PR-P2, respectively, and no difference in expression of PINII in comparison with the level in control plants (Fig. 4B).
Analysis of antifungal activity of EPT and trichodermol.
In order to determine if EPT and trichodermol, two Ta37 trichothecene intermediates, have antifungal activity, antibiograms against B05.10 were determined by using concentrations of purified EPT of 0.1 to 5 mg ml−1 and of purified trichodermol of 0.1 to 1 mg ml−1. Purified HA at concentrations of 0.1 to 1 mg ml−1 was used as a control (Fig. 5). No inhibition area was observed for any of the concentrations of the trichothecene intermediates used, indicating that neither hydroxylation of TD at C-2, C-11, and C-13 nor hydroxylation of EPT at C-4 is sufficient to confer antifungal activity to these intermediates, and only the end product, HA in the case of T. arundinaceum, had detectable antifungal activity.
FIG 5.
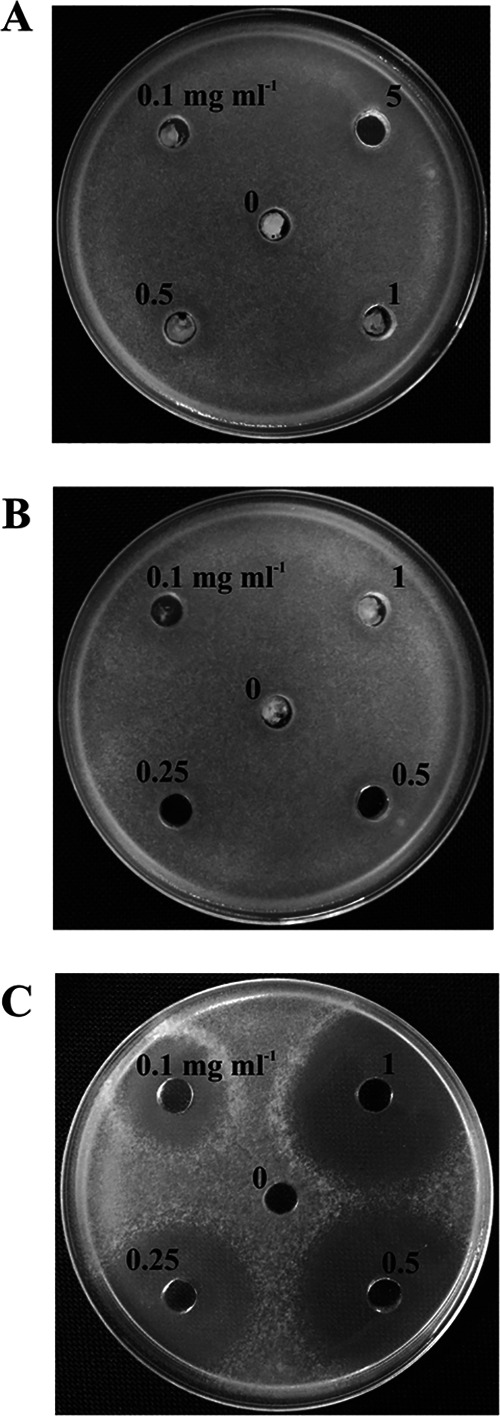
Antibiogram assay of EPT (12,13-epoxytrichothec-9-ene) (A), trichodermol (B), and HA (C) against B. cinerea B05.10. The concentrations used are indicated for each hole, in milligrams per milliliter. Dilutions were carried out in acetone, except for HA, in which case dilutions were made in acetonitrile. In the center of the plates (0), acetone or acetonitrile was loaded to show that the solvent did not affect B. cinerea growth.
Production of EPT does not affect the antifungal activity of Trichoderma.
T. harzianum T34-5.27-tri4 transformants were grown on cellophane membranes to allow the diffusion of extracellular compounds into the medium. After the removal of the membranes containing the mycelia, the effects of diffusible hydrolytic enzymes plus metabolites on the growth of two different plant-pathogenic fungi, F. sporotrichioides and B. cinerea, were determined. No additional effect on fungal growth was detected for the tri4-expressing transformants in comparison with the activity of the control strain, T34-5.27-b1 (data not shown). This result was confirmed by using an approach to determine the antifungal activity of the Trichoderma strains against B. cinerea on tomato leaves. We observed that 96-h culture broths of the T34-5.27-tri4.1 and T34-5.27-tri4.2 transformants inhibited the production of Botrytis lesions to levels similar to those in broths of the control strain T34-5.27-b1 (see Fig. S3 in the supplemental material).
Effect of in vivo EPT production on expression of tomato defense-related genes.
Tomato seeds inoculated with spores from T34-5.27-b1 and T34-5.27-tri4.2 were grown for 4 weeks, and several plant parameters were measured. The stem diameter and length were not significantly affected by inoculation with the transformant overexpressing tri4 in comparison with the control strain. The production of EPT also did not affect the size of lesions caused by B05.10 (data not shown).
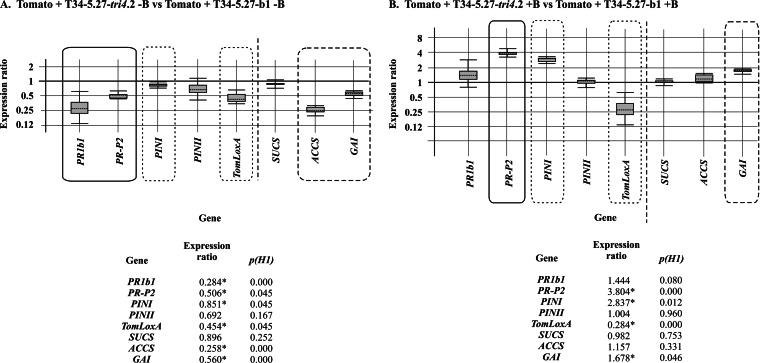
When the expression of tomato genes related to defense and development was analyzed by qPCR, we observed that most of the genes were downregulated or not affected by interaction with the T34-5.27-tri4.2 transformant in comparison with those in plants inoculated with the control strain. The genes involved in tomato defense were downregulated by tri4 expression, with values of 0.284 (P = 0.000)- and 0.851 (P = 0.045)-fold for PR1b1 and PINI, respectively. In parallel, the effect of EPT production on the expression of genes related to plant development was determined. The expression of SUCS, which encodes a sucrose synthase, was not affected, while ACCS, encoding a 1-aminocyclopropane-1-carboxylic acid (ACC) synthase, and GAI, which encodes a DELLA protein insensitive to gibberellic acid, were downregulated by factors of 0.258 (P = 0.000) and 0.560 (P = 0.000), respectively (Fig. 6). Interestingly, infection by B05.10 of plants inoculated with these Trichoderma strains resulted in upregulation of PR-P2 and PINI, by factors of 3.804 (P = 0.000) and 2.837 (P = 0.012), respectively, and in downregulation of TomLoxA, by a factor of 0.284 (P = 0.000), compared to the effects of just tri5. In the case of the development-related genes, only GAI was upregulated, by a factor of 1.678 (P = 0.046) (Fig. 6).
FIG 6.
qPCR analysis of the levels of expression of five tomato defense-related genes (PR1b1, PR-P2, PINI, PINII, and TomLoxA) and three development-related genes (SUCS, ACCS, and GA1) in leaves collected from tomato plants whose seeds were coated with conidia of T34-5.27-tri4.2 versus the expression levels of these genes in leaves from plants inoculated with the control T34-5.27-b1 strain (A) and in the same plants as in panel A, but infected with B05.10 (+B) after 4 weeks of growth (see Materials and Methods) (B). Calculations and graphic representations were performed as indicated in the legend to Fig. 3.
To further confirm these results, gene expression comparisons between T34-5.27-b1 and T34-5.27-tr4.2 were done in the presence or absence of B. cinerea. Trichoderma (with tri5 or with tri5-tri4) strongly upregulated PR1b1 and PR-P2, downregulated PINI, PINII, and TomLoxA, upregulated SUCS and ACCS, and slightly downregulated GAI (see Fig. S4 in the supplemental material). Comparing Fig. S4A with Fig. S4B suggests that tri4 overexpression may slightly upregulate PR1b1, PR-P2, and GAI compared to the levels observed with tri5 alone.
Effect of EPT production on the ability of T. harzianum to colonize tomato roots.
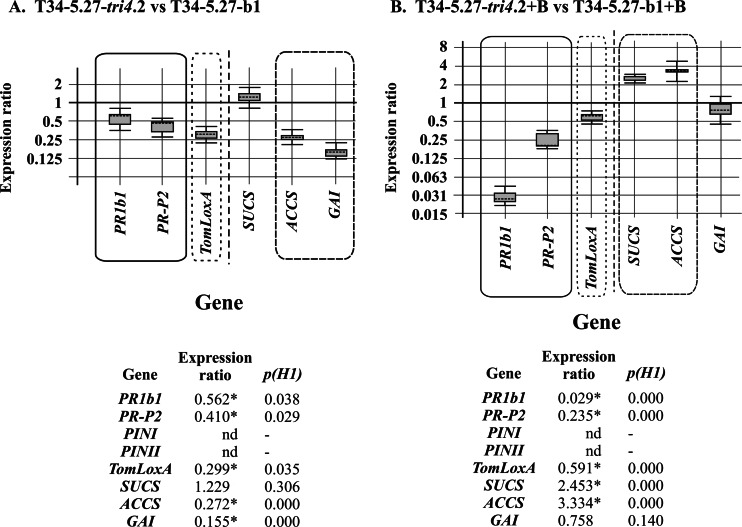
Tomato hydroponic cultures were inoculated with the transformants. Analysis by qPCR of the ratio of expression of the tomato housekeeping gene GAPDH (glyceraldehyde 3-phosphate dehydrogenase) to expression of the Trichoderma α-actin housekeeping gene resulted in a ratio of colonization of 0.82 for the T34-5.27-tri4.2 transformant, using a reference value of 1 for the colonization ability of T34-5.27-b1. This suggests that tri4 overexpression reduced the ability of Trichoderma to colonize tomato roots. These cDNAs were also used to determine the expression levels of plant defense- and development-related genes. We observed that PR1b1, PR-P2, and TomLoxA were downregulated in tomato roots colonized with T34-5.27-tri4.2 with respect to those colonized with T34-5.27-b1, by factors ranging from 0.299 (P = 0.035)- to 0.562 (P = 0.038)-fold, for TomLoxA and PR1b1, respectively. The levels of expression of the tomato development-related genes indicated that SUCS was not affected, while ACCS and GAI were downregulated, by factors of 0.272 (P = 0.000) and 0.155 (P = 0.000), respectively (Fig. 7A).
FIG 7.
qPCR analysis of the levels of expression of defense-related (PR1b1, PR-P2, and TomLoxA) and development-related (SUCS, ACCS, and GAI) genes in roots harvested from tomato hydroponic cultures inoculated with T34-5.27-tri4.2 relative to the levels of expression of these genes in roots inoculated with T34-5.27-b1 (A) and analysis of the levels of expression of these genes in roots harvested from tomato hydroponic cultures inoculated with T34-5.27-tri4.2 plus B05.10 (+B) versus their levels of expression in roots inoculated with T34-5.27-b1 plus B05.10 (+B) (B). The data in the lower panels include the relative numeric gene expression levels corresponding to the graphics illustrated above. Calculations and graphic representations were performed as indicated in the legend to Fig. 3. Note that expression of the PINI and PINII genes was not detected (nd) under the experimental conditions used in the present study.
In order to determine if B05.10 would affect the ability of Trichoderma to colonize roots, B05.10 mycelia, together with mycelia from the T34-5.27-tri4.2 strain, were added to the tomato hydroponic cultures. Using the same procedure as that indicated in the previous paragraph, a ratio of colonization of 0.69 was observed for the T34-5.27-tri4.2 strain. This indicated that the presence of B05.10 resulted in a lower colonization ratio by the T34-5.27-tri4.2 transformant. In these roots, the expression of tomato defense- and development-related genes showed a pattern significantly different from that observed for tomato roots grown without B. cinerea. PR1b1 and PR-P2 (SA-related genes) were strongly downregulated, by factors of 0.029 (P = 0.000) and 0.235 (P = 0.000), respectively, while TomLoxA was only slightly downregulated, with a relative expression value of 0.591 (P = 0.000) (Fig. 7B). In the case of the tomato development-related genes analyzed, the relative expression values followed an expression pattern that seemed to be almost opposite that observed without Botrytis: SUCS and ACCS were significantly upregulated, and GAI was not affected (Fig. 7B). This suggests that the presence of B. cinerea is important for induction of SUCS and ACCS.
These data indicate that expression of tri4 in roots reduced the ability of Trichoderma to colonize tomato roots and downregulated the expression of defense- and development-related genes, except for SUCS. The presence of Botrytis resulted in a still lower Trichoderma colonization ability and in a downregulation of all the plant defense-related genes analyzed. In contrast, most of the development-related genes analyzed, except for GAI, were upregulated.
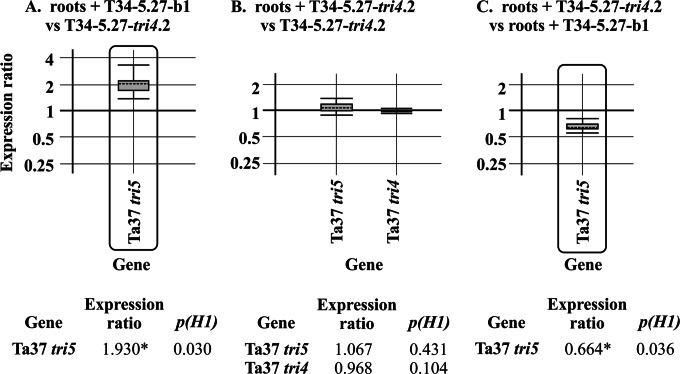
Expression of the tri4 and tri5 genes in tomato roots inoculated with the T34-5.27-tri4.2 transformant.
Roots from the hydroponic cultures used to determine the colonization ability of the T34-5.27-tri4.2 transformant, as well as those inoculated with the control T34-5.27-b1 strain, were also analyzed by qPCR to determine the levels of expression of the tri4 and tri5 genes. Expression of tri5 was detected in roots inoculated with both Trichoderma strains (Fig. 8A and B), reaching expression levels similar to those observed for the 96-h mycelia of the T34-5.27-tri4.2 strain, with only a slight upregulation of tri5 in roots inoculated with the control strain. As expected, tri4 expression was detected only in roots inoculated with the T34-5.27-tri4.2 transformant, reaching a level similar to that found in the mycelium of this transformant grown alone (Fig. 8B). Finally, expression of tri5 was slightly downregulated, by a factor of 0.664 (P = 0.036), in roots inoculated with the T34-5.27-tri4.2 transformant versus roots inoculated with the control strain (Fig. 8C).
FIG 8.
qPCR analysis of the relative expression levels of Trichoderma arundinaceum IBT 40837 (Ta37) tri5 in roots of tomato hydroponic cultures inoculated with T34-5.27-b1 (A) and of Ta37 tri5 and tri4 in roots of tomato hydroponic cultures inoculated with T34-5.27-tri4.2 (B), in both cases versus the level of expression in 96-h mycelia of the T34-5.27-tri4.2 strain. (C) Relative expression level of tri5 in roots of tomato hydroponic cultures inoculated with T34-5.27-tri4.2 versus the level detected in roots inoculated with the recipient strain T34-5.27-b1. Calculations and graphic representations were performed as indicated in the legend to Fig. 3. Note that the expression of tri4 was not detected in roots of tomato hydroponic cultures inoculated with strain T34-5.27-b1.
DISCUSSION
In order to measure the effects of intermediates of the HA biosynthetic pathway on plants and fungal antagonists, we chose to clone and insert individual trichothecene genes into T. harzianum, a species which does not produce trichothecenes. Insertion of T. arundinaceum tri5 into T. harzianum T34 resulted in production of trichodiene (20). Testing with the T34-5.27 transformants suggested that TD acts as a volatile signaling compound in the interactions of Trichoderma with plants and other microorganisms (20). In the present study, we inserted T. arundinaceum tri4 into the transgenic T34-5.27 strain in order to examine the effect of EPT on plant-fungus interactions. High levels of both TD and EPT were detected, indicating that the promoter region was recognized and the genes were transcribed and translated. It should be noted that the extraction procedure for EPT and TD might not recover TD that had volatilized in the culture flasks. It is unlikely that volatile TD is available for further processing to make EPT.
The levels of tri4 expression and EPT production did not match the predicted numbers of copies of the tri4 gene that were integrated into the tri4 transformant genomes. Transformant T34-5.27-tri4.2, with the largest number of tri4 copies (9 or 10 copies), showed a significantly lower level of tri4 expression as well as a lower level of TD or EPT production than that in transformant T34-5.27-tri4.1 (4 or 5 copies). This may indicate that only some of the tri4 copies integrated into the genome of each transformant are functional. The ability of T. harzianum to integrate multiple copies of transforming DNA into different loci of the fungal genome has been reported previously (27). The recombination processes needed for the integration of the transformation cassette into the fungal genome could be responsible for inactivation of several of the tri4 copies integrated, giving rise to truncated or aberrant copies of this gene.
In the presence of Tri4 activity, one might expect the level of TD to decrease, with a concomitant increase in EPT. However, our results show that the presence of Tri4 activity did not significantly reduce the amount of TD present. There was also no decrease in the amount of ergosterol present, although there was a decrease in the amount of the metabolic intermediate squalene, in the tri4 transformants. These results are supported by previous reports showing that a reduction in squalene synthase activity in T. reesei as a result of erg20 overexpression did not affect the level of ergosterol produced (37). These data may indicate that the production of EPT in the tri4-overexpressing transformants was the result of an increase in the channeling of FPP toward EPT rather than from a reduction in the level of accumulated TD. However, these data slightly contrast with those obtained when tri5 was expressed in a T34 strain. In that case, the production of TD resulted in a reduction in the level of ergosterol produced and an increase in the amount of squalene accumulated (20). In addition, production of TD did not result in a remarkable upregulation of the genes involved in the biosynthesis of ergosterol from FPP. However, in the present work, a significant increase in the expression of erg1 was detected in all tri4-overexpressing transformants analyzed, indicating the existence of a specific mechanism of erg1 upregulation by this trichothecene intermediate or by some other compound whose production would be induced by tri4. The role of squalene synthase (SQS), encoded by erg9, as a key enzyme in the regulation of ergosterol biosynthesis has been reported for S. cerevisiae (38). However, the results presented here, together with previous data (20, 39), led us to focus on the important role of the SE protein, encoded by erg1/ERG1, in the ergosterol biosynthetic pathway. This role would be even more important than that carried out by other key enzymes (e.g., SQS), as a flux-controlling step from FPP to sterol production, at least in Trichoderma. The increase in erg1 expression and the maintenance of the level of ergosterol production can be understood as a way to ensure the minimal level of FPP channeling toward ergosterol in a background strain producing TD and EPT. Interestingly, the erg9 expression level was not significantly affected by tri4 overexpression. A reduced upregulation of erg1 was also observed in the transformant of a T34 strain overexpressing tri5 compared with the T34 wild-type strain (20), indicating that production of TD plus EPT would require additional FPP and/or squalene.
Purified EPT did not have antifungal activity against the fungal phytopathogen B05.10 or phytotoxic activity against tomatoes. Trichodermol also did not have antifungal activity, but interestingly, it was strongly phytotoxic at concentrations as low as 1 μg ml−1 (3.99 μM). Unexpectedly, purified HA also showed a strong phytotoxic activity at concentrations above 1 μg ml−1. This result contrasts with previous data describing a lack of in vivo phytotoxicity of this compound (10, 17, 20) and indicates that the concentration of HA produced by T. arundinaceum probably does not reach a high enough level in culture to produce a toxic effect during in vivo experiments.
Expression of tri4 slightly affected the expression of plant defense-related genes, which is in agreement with the results obtained for tomato plants grown in the presence of different concentrations of EPT. However, trichodermol, which showed a strong phytotoxic effect in this study, resulted in the upregulation of all the tomato defense-related genes tested, especially the SA-related genes, particularly PR1b1, which showed an upregulation of 147-fold. Similarly, HA upregulated the SA-related genes analyzed, with relative expression ratios of up to 205- and 82-fold for PR1b1 and PR-P2, respectively. These responses have certain similarities to those observed in tomato plants infected only with B05.10, which showed a strong induction of SA-related genes (31). The production of specific phytotoxic compounds and direct attack by a pathogen are signals which plants detect and respond to, resulting in the upregulation of expression of defense-related genes, particularly SA-related genes. SA is involved in the induction of callose deposition in plant cell walls, resulting in resistance to fungal colonization (40). In this way, production of trichodermol might allow the plant to identify a particular producer fungus as a potential pathogen-like organism.
Interestingly, when plants inoculated with the Trichoderma strains analyzed in this work were further infected with B05.10, the expression of the defense-related genes changed. Prior to pathogen inoculation, PR1b1, PR-P2, and PINI were slightly downregulated in plants treated with the tri4-expressing transformant with respect to the levels in plants inoculated with the control strain T34-5.27-b1. After pathogen inoculation, two of these three marker genes were upregulated. These data indicate that infection with B05.10 affects the plant's response to EPT, probably by making plants more sensitive to this compound, resulting in an increased induction of plant defense-related genes. TomLoxA was downregulated under both conditions, before and after B05.10 infection. This gene has been related to the control of the spread of beneficial fungi in roots (41, 42). A reduction in the dissemination of T. harzianum might result from tri4 overexpression, which would indicate that a plant responds to these toxins and pathogen attacks by showing a certain degree of incompatibility to both invaders and then trying to resist colonization by the one that is penetrating through the roots.
In addition to the defense-related genes analyzed above, the SUCS, ACCS, and GAI genes, which are involved in plant development, were also studied. Sucrose synthase is capable of catalyzing both sucrose synthesis and degradation. Its concentration is higher in tissues that synthesize sucrose and also in sucrose-utilizing tissues. Thus, it is an enzyme primarily involved in plant growth. ACCS encodes 1-aminocyclopropane-1-carboxylic acid (ACC) synthase, which catalyzes the rate-limiting step in the ethylene (ET) biosynthetic pathway. An increase in ET production is invariably accompanied by increased ACC production, because of induction or activation of ACC synthase. ET is also involved in regulating plant responses to both biotic and abiotic stresses (43). GAI encodes a DELLA protein that is insensitive to gibberellic acid. DELLA proteins are negative regulators of the plant hormone gibberellin and thus repress plant growth. In parallel, these proteins would induce JA-dependent defense responses (44). As a result of our studies, we found that overexpression of tri4 in the T34-5.27 strain did not significantly affect the expression of SUCS, while ACCS and GAI were downregulated (Fig. 6A). The lack of differences in the SUCS expression level matched the lack of differences in growth between plants inoculated with the tri4 transformant and those inoculated with the control strain. Taking into account the Trichoderma-plant interaction model proposed by Hermosa et al. (44), downregulation of ACCS and GAI would result in a stimulation of plant growth and a reduction of JA-dependent defenses, which in turn should produce an increase of the SA defense response. However, the expression of the JA- and SA-related genes analyzed in the present study was generally downregulated, indicating that other mechanisms which are activated by the production of EPT may affect this response.
In summary, the production of EPT affected the balance of terpene intermediates involved in the biosynthesis of ergosterol by reducing the level of squalene. While erg1 was strongly upregulated by EPT, the final amount of ergosterol produced in tri4 transformants was not significantly affected. Exogenously added EPT did not show antifungal or phytotoxic activity and did not significantly affect the expression of tomato defense-related genes. However, the next intermediate in the Trichoderma trichothecene biosynthetic pathway, trichodermol, was highly phytotoxic and upregulated the expression of all the tomato defense-related genes analyzed, but it did not have antifungal activity. In vivo production of EPT during interaction with tomato plants inoculated with the tri4-expressing transformant and infected with B05.10 resulted in a downregulation of TomLoxA, which indicates an induction of resistance of the plant to both fungal (B. cinerea) attack and trichothecenes. The knowledge of the biological activities of the trichothecene intermediates will allow us to identify the biosynthetic steps in which these compounds acquire their toxicity and help us to understand the mechanism used by plants to respond to each of the trichothecene intermediates. Further studies will serve to assess the potential of trichothecene intermediates in multitrophic interactions and integrated pest management strategies as compounds able to modulate plant defenses against both pests and pathogens.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by grants from MICINN and MINECO (grants AGL2009-13431-C02 and AGL2012-40041-C02-02) and from the Junta de Castilla y León (grant LE125A12-2). M. G. Malmierca was granted an FPU fellowship by the Spanish Ministry of Science and Innovation (grant AP2007-02835).
We thank J. Álvarez from the University of León and J. Teresi and K. MacDonald from the Bacterial Foodborne Pathogens and Mycology Unit, USDA/ARS, for their excellent technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01626-15.
REFERENCES
- 1. Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. 2004. Trichoderma species—opportunistic, avirulent plant symbionts. Nat Rev Microbiol 2:43–56. doi: 10.1038/nrmicro797. [DOI] [PubMed] [Google Scholar]
- 2.Lorito M, Woo SL, Harman GE, Monte E. 2010. Translational research on Trichoderma: from 'omics to the field. Annu Rev Phytopathol 48:395–417. doi: 10.1146/annurev-phyto-073009-114314. [DOI] [PubMed] [Google Scholar]
- 3.Shoresh M, Harman GE, Mastouri F. 2010. Induced systemic resistance and plant responses to fungal biocontrol agents. Annu Rev Phytopathol 48:21–43. doi: 10.1146/annurev-phyto-073009-114450. [DOI] [PubMed] [Google Scholar]
- 4.Hermosa R, Viterbo A, Chet I, Monte E. 2012. Plant-beneficial effects of Trichoderma and of its genes. Microbiology 158:17–25. doi: 10.1099/mic.0.052274-0. [DOI] [PubMed] [Google Scholar]
- 5.Alexander NJ, McCormick SP, Zeigenhorn SL. 1999. Phytotoxicity of selected trichothecenes using Chlamydomonas reinhardtii as a model system. Nat Toxins 7:265–269. doi:. [DOI] [PubMed] [Google Scholar]
- 6.Desjardins AE, McCormick SP, Appell M. 2007. Structure-activity relationships of trichothecene toxins in an Arabidopsis thaliana leaf assay. J Agric Food Chem 55:6487–6492. doi: 10.1021/jf0709193. [DOI] [PubMed] [Google Scholar]
- 7.Bondy GS, McCormick SP, Beremand MN, Pestka JJ. 1991. Murine lymphocyte proliferation impaired by substituted neosolaniols and calonectrins—Fusarium metabolites associated with trichothecene biosynthesis. Toxicon 29:1107–1113. doi: 10.1016/0041-0101(91)90208-9. [DOI] [PubMed] [Google Scholar]
- 8.Hohn TM, Beremand PD. 1989. Isolation and nucleotide sequence of a sesquiterpene cyclase gene from the trichothecene-producing fungus Fusarium sporotrichioides. Gene 79:131–138. doi: 10.1016/0378-1119(89)90098-X. [DOI] [PubMed] [Google Scholar]
- 9.Cardoza RE, Malmierca MG, Hermosa MR, Alexander NJ, McCormick SP, Proctor RH, Tijerino AM, Rumbero A, Monte E, Gutiérrez S. 2011. Identification of loci and functional characterization of trichothecene biosynthetic genes in the filamentous fungus Trichoderma. Appl Environ Microbiol 77:4867–4877. doi: 10.1128/AEM.00595-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malmierca MG, Cardoza RE, Alexander NJ, McCormick SP, Hermosa R, Monte E, Gutiérrez S. 2012. Involvement of Trichoderma trichothecenes in the biocontrol activity and in the induction of plant defense-related genes. Appl Environ Microbiol 78:4856–4868. doi: 10.1128/AEM.00385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godtfredsen WO, Vangedal S. 1965. Trichodermin, a new sesquiterpene antibiotic. Acta Chem Scand 19:1088–1102. doi: 10.3891/acta.chem.scand.19-1088. [DOI] [PubMed] [Google Scholar]
- 12.Corley DG, Miller-Wideman M, Durley RC. 1994. Isolation and structure of harzianum A: a new trichothecene from Trichoderma harzianum. J Nat Prod 57:422–425. doi: 10.1021/np50105a019. [DOI] [PubMed] [Google Scholar]
- 13.Nielsen KF, Gräfenhan T, Zafari D, Thrane U. 2005. Trichothecene production by Trichoderma brevicompactum. J Agric Food Chem 53:8190–8196. doi: 10.1021/jf051279b. [DOI] [PubMed] [Google Scholar]
- 14.Rocha O, Ansari K, Doohan FM. 2005. Effects of trichothecene mycotoxins on eukaryotic cells: a review. Food Addit Contam 22:369–378. doi: 10.1080/02652030500058403. [DOI] [PubMed] [Google Scholar]
- 15.Jin HZ, Lee JH, Zhang WD, Lee HB, Hong YS, Kim YH, Lee JJ. 2007. Harzianums A and B produced by a fungal strain, Hypocrea sp F000527, and their cytotoxicity against tumor cell lines. J Asian Nat Prod Res 9:203–207. doi: 10.1080/10286020500531977. [DOI] [PubMed] [Google Scholar]
- 16.Degenkolb T, Dieckmann R, Nielsen KF, Gräfenhan T, Theis C, Zafari D, Chaverri P, Isamaiel A, Brückner H, von Döhren H, Thrane U, Petrine O, Samuels GJ. 2008. The Trichoderma brevicompactum clade: a separate lineage with new species, new peptabiotics, and mycotoxins. Mycol Prog 7:177–219. doi: 10.1007/s11557-008-0563-3. [DOI] [Google Scholar]
- 17.Lee HB, Kim Y, Jin HZ, Lee JJ, Kin C-J, Park JY, Jung HS. 2005. A new Hypocrea strain producing harzianum A cytotoxic to tumour cell lines. Lett Appl Microbiol 40:497–503. doi: 10.1111/j.1472-765X.2005.01719.x. [DOI] [PubMed] [Google Scholar]
- 18.Wu Q, Dohnal V, Kuca K, Yuan Z. 2013. Trichothecenes: structure-toxic activity relationships. Curr Drug Metab 14:641–660. doi: 10.2174/1389200211314060002. [DOI] [PubMed] [Google Scholar]
- 19.Malmierca MG, Cardoza RE, Alexander NJ, McCormick SP, Collado IG, Hermosa MR, Monte E, Gutiérrez S. 2013. Relevance of trichothecenes in fungal physiology: disruption of tri5 in Trichoderma arundinaceum. Fungal Genet Biol 53:22–33. doi: 10.1016/j.fgb.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Malmierca MG, McCormick SP, Cardoza RE, Alexander NJ, Monte E, Gutiérrez S. 12 May 2014. Production of trichodiene by Trichoderma harzianum alters the perception of this biocontrol strain by plants and antagonized fungi. Environ Microbiol doi: 10.1111/1462-2920.12506. [DOI] [PubMed] [Google Scholar]
- 21.Quidde T, Osbourn AE, Tudzynski P. 1998. Detoxification of α-tomatine by Botrytis cinerea. Physiol Mol Plant Pathol 52:151–165. doi: 10.1006/pmpp.1998.0142. [DOI] [Google Scholar]
- 22.Penttilä M, Nevalainen H, Ratto M, Salminen E, Knowles J. 1987. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61:155–164. doi: 10.1016/0378-1119(87)90110-7. [DOI] [PubMed] [Google Scholar]
- 23.Ghimire GP, Lee HC, Sohng JK. 2009. Improved squalene production via modulation of the methylerythritol 4-phosphate pathway and heterologous expression of genes from Streptomyces peucetius ATCC 27952 in Escherichia coli. Appl Environ Microbiol 75:7291–7293. doi: 10.1128/AEM.01402-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardoza RE, Hermosa MR, Vizcaíno JA, González FJ, Llobell A, Monte E, Gutiérrez S. 2007. Partial silencing of a hydroxy-methylglutaryl-CoA reductase encoding gene in Trichoderma harzianum CECT 2413 results in a lower level of resistance to lovastatin and a lower antifungal activity. Fungal Genet Biol 44:269–283. doi: 10.1016/j.fgb.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 25.Ausubel FM, Brent R, Kingston RE, Moore DJ, Smith A, Seidman JG, Struhl K. 1987. Current protocols in molecular biology. Wiley, New York, NY. [Google Scholar]
- 26.Gutiérrez S, Velasco J, Marcos AT, Fernández FJ, Fierro F, Barredo JL, Díez B, Martín JF. 1997. Expression of the cefG gene is limiting for cephalosporin biosynthesis in Acremonium chrysogenum as shown by promoter replacement studies. Appl Microbiol Biotechnol 48:606–614. doi: 10.1007/s002530051103. [DOI] [PubMed] [Google Scholar]
- 27.Cardoza RE, Vizcaíno JA, Hermosa R, Monte E, Gutiérrez S. 2006. A comparison of the phenotypic and genetic stability of recombinant Trichoderma spp. generated by protoplast- and Agrobacterium-mediated transformation. J Microbiol 44:383–395. [PubMed] [Google Scholar]
- 28.Hohn TM, Desjardings AE, McCormick SP. 1995. The tri4 of Fusarium sporotrichioides encodes a cytochrome P450 monooxygenase involved in trichothecene biosynthesis. Mol Gen Genet 248:95–102. doi: 10.1007/BF02456618. [DOI] [PubMed] [Google Scholar]
- 29.Trapp SC, Hohn TM, McCormick SP, Jarvis BB. 1998. Characterization of the gene cluster for biosynthesis of macrocyclic trichothecenes in Myrothecium roridum. Mol Gen Genet 257:421–432. doi: 10.1007/s004380050666. [DOI] [PubMed] [Google Scholar]
- 30.Rubio MB, Domínguez S, Monte E, Hermosa R. 2012. Comparative study of Trichoderma gene expression in interactions with tomato plants using high-density oligonucleotide microarrays. Microbiology 158:119–128. doi: 10.1099/mic.0.052118-0. [DOI] [PubMed] [Google Scholar]
- 31.Cardoza RE, Malmierca MG, Gutiérrez S. 2014. Overexpression of erg1 gene in Trichoderma harzianum CECT 2413: effect on the induction of tomato defence-related genes. J Appl Microbiol 117:812–823. doi: 10.1111/jam.12574. [DOI] [PubMed] [Google Scholar]
- 32.Malmierca MG, Barua J, McCormick SP, Izquierdo-Bueno I, Cardoza RE, Alexander NJ, Hermosa R, Collado IG, Monte E, Gutiérrez S. 2015. Novel aspinolide production by Trichoderma arundinaceum with a potential role in Botrytis cinerea antagonistic activity and plant defence priming. Environ Microbiol 17:1103–1118. doi: 10.1111/1462-2920.12514. [DOI] [PubMed] [Google Scholar]
- 33.Pinedo C, Wang CM, Pradier JM, Dalmais B, Choquer M, Le Pecheur P, Morgant G, Collado IG, Cane DE, Viaud M. 2008. Sesquiterpene synthase from the botrydial biosynthetic gene cluster of the phytopathogen Botrytis cinerea. ACS Chem Biol 3:791–801. doi: 10.1021/cb800225v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tucci M, Ruocco M, De Masi L, De Palma M, Lorito M. 2011. The beneficial effect of Trichoderma spp. on tomato is modulated by the plant genotype. Mol Plant Pathol 12:341–354. doi: 10.1111/j.1364-3703.2010.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pilsyk S, Perlinska-Lenart U, Gorka-Niec W, Graczyk S, Antosiewicz B, Zembek P, Palamarczyk G, Kruszewska JS. 2014. Overexpression of erg20 gene encoding farnesyl pyrophosphate synthase has contrasting effects on activity of enzymes of the dolichyl and sterol branches of mevalonate pathway in Trichoderma reesei. Gene 544:114–122. doi: 10.1016/j.gene.2014.04.073. [DOI] [PubMed] [Google Scholar]
- 38.Paradise EM, Kirby J, Chan R, Keasling JD. 2008. Redirection of flux through the FPP branch-point in Saccharomyces cerevisiae by down-regulating squalene synthase. Biotechnol Bioeng 100:371–378. doi: 10.1002/bit.21766. [DOI] [PubMed] [Google Scholar]
- 39.Veen M, Stahl U, Lang C. 2003. Combined overexpression of genes of the ergosterol biosynthetic pathway leads to accumulation of sterols in Saccharomyces cerevisiae. FEMS Yeast Res 4:87–95. doi: 10.1016/S1567-1356(03)00126-0. [DOI] [PubMed] [Google Scholar]
- 40.Alonso-Ramírez A, Poveda J, Martin I, Hermosa R, Monte E, Nicolás C. 2014. Salicylic acid prevents Trichoderma harzianum from entering the vascular system of roots. Mol Plant Pathol 15:823–831. doi: 10.1111/mpp.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.León-Morcillo RJ, Martín-Rodríguez M-R, Vierheilig H, Ocampo JA, García-Garrido JM. 2012. Late activation of the 9-oxylipin pathway during arbuscular mycorrhiza formation in tomato and its regulation by jasmonate signalling. J Exp Bot 63:3545–3558. doi: 10.1093/jxb/ers010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harel Y, Mehari ZH, Rav-David D, Elad Y. 2014. Systemic resistance to gray mold induced in tomato by benzothiadiazole and Trichoderma harzianum T39. Phytopathology 104:150–157. doi: 10.1094/PHYTO-02-13-0043-R. [DOI] [PubMed] [Google Scholar]
- 43.Iqbal N, Teivellini A, Masood A, Ferrante A, Khan NA. 2013. Current understanding on ethylene signaling in plants: the influence of nutrient availability. Plant Physiol Biochem 73:128–138. doi: 10.1016/j.plaphy.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 44.Hermosa R, Rubio MB, Cardoza RE, Nicolás C, Monte E, Gutiérrez S. 2013. The contribution of Trichoderma to balancing the costs of plant growth and defence. Int Microbiol 16:69–80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.