Abstract
Ixodes scapularis is the principal vector of Lyme disease on the East Coast and in the upper Midwest regions of the United States, yet the tick is also present in the Southeast, where Lyme disease is absent or rare. A closely related species, I. affinis, also carries the pathogen in the South but does not seem to transmit it to humans. In order to better understand the geographic diversity of the tick, we analyzed the microbiota of 104 adult I. scapularis and 13 adult I. affinis ticks captured in 19 locations in South Carolina, North Carolina, Virginia, Connecticut, and New York. Initially, ticks from 4 sites were analyzed by 454 pyrosequencing. Subsequently, ticks from these sites plus 15 others were analyzed by sequencing with an Illumina MiSeq machine. By both analyses, the microbiomes of female ticks were significantly less diverse than those of male ticks. The dissimilarity between tick microbiomes increased with distance between sites, and the state in which a tick was collected could be inferred from its microbiota. The genus Rickettsia was prominent in all locations. Borrelia was also present in most locations and was present at especially high levels in one site in western Virginia. In contrast, members of the family Enterobacteriaceae were very common in North Carolina I. scapularis ticks but uncommon in I. scapularis ticks from other sites and in North Carolina I. affinis ticks. These data suggest substantial variations in the Ixodes microbiota in association with geography, species, and sex.
INTRODUCTION
There are an estimated 300,000 cases of Lyme disease in the United States, with the majority of cases occurring in the Northeast and upper Midwest (1). Lyme disease is caused by the spirochete Borrelia burgdorferi sensu stricto, for which the black-legged tick, Ixodes scapularis, is the principal vector in the Midwest and on the East Coast. It also transmits other pathogens causing human diseases, including the pathogens causing anaplasmosis and babesiosis (2, 3).
Ixodes scapularis and a closely related species, Ixodes affinis, also occur in the Southeast, where Lyme disease is rare or absent. B. burgdorferi has been detected in 35 to 50% of I. scapularis ticks in the northeastern states (4) but is rarely found in ticks in the southern United States (5–9). In contrast, 63% of I. affinis ticks were found to have Borrelia DNA by PCR (10). I. affinis ticks feed on a wide range of hosts (8).
While there have been many studies of tick microbiomes (11–17), relatively little is known about the microbiota of Ixodes ticks. Efforts to identify microbial commensals and symbionts have been made using culture-based methods (18), cloning, and sequencing of gel-purified PCR products (4). Narasimhan et al. (19) recently used 454 pyrosequencing of the 16S rRNA gene to describe the microbiome of I. scapularis larvae and nymphs from Connecticut. Here, we assess how the microbiomes of adult Ixodes ticks vary with regard to geography, species, and sex.
MATERIALS AND METHODS
Tick sampling, collection sites, and processing.
Adult I. scapularis or I. affinis ticks were collected by flagging vegetation. In a few cases, ticks were collected from participants in a longitudinal study of North Carolina park and forestry rangers (20). Immediately after collection, the ticks were preserved in 85% ethanol and transported to laboratory facilities, where they were stored at −20°C until they were processed as described below. The sites are described in Table 1.
TABLE 1.
Ticks collected for this study
| Species | State | Site | No. of ticks sequenced by: |
|
|---|---|---|---|---|
| 454 | Illumina | |||
| Ixodes scapularis | Connecticut | Lake Gaillard | 8 | 4 |
| Ixodes scapularis | North Carolina | Bladen County | 0 | 2 |
| Ixodes affinis | North Carolina | Halifax County | 0 | 4 |
| Ixodes affinis | North Carolina | Jones County | 0 | 2 |
| Ixodes scapularis | North Carolina | Morrow Mountain, Stanly County | 0 | 1 |
| Ixodes scapularis | North Carolina | Pine Island, Currituck County | 8 | 3 |
| Ixodes scapularis | North Carolina | Tumble Creek, Bladen County | 0 | 1 |
| Ixodes scapularis | North Carolina | View Nicholson Road, Martin County | 8 | 7 |
| Ixodes affinis | North Carolina | Washington County | 0 | 2 |
| Ixodes scapularis | New York | Armonk | 10 | 2 |
| Ixodes scapularis | South Carolina | Savannah River | 0 | 11 |
| Ixodes scapularis | Virginia | Pearisburg, Giles County | 0 | 12 |
| Ixodes scapularis | Virginia | Pocahontas State Park | 0 | 10 |
| Ixodes scapularis | Virginia | York River State Park | 0 | 10 |
Ticks were shipped to our laboratory facilities for processing. For each collection site, black-legged ticks were sorted by sex, placed individually into 1.5-ml microcentrifuge tubes containing 95% ethanol (USA Scientific, Ocala, FL), and then stored at −80°C for subsequent extraction of genomic DNA.
Extraction of DNA from tick samples.
Genomic DNA was extracted from individual ticks using methods described previously (17, 21). The extracted DNA samples were further purified with a Wizard DNA cleanup system (Promega, Madison, WI, USA) and quantified with a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA). The purified DNA extracts were stored at −80°C for future use.
16S rRNA gene amplification for 454 analysis.
Bacterial gene amplification of the PCR products was performed as described previously by Ponnusamy et al. (17). 16S rRNA genes were amplified using primers 27F/534R targeting the V1 to V3 hypervariable regions and covering a sequence distance of ∼500 bp, multiplexed with 10-mer nucleotide bar codes, and sequenced using the 454 technology. PCR products of the expected size were excised from the agarose gel and gel purified using a QIAquick gel extraction kit (Qiagen, Inc., Valencia CA, USA). Gel-purified amplicon DNAs were quantified using a Quant-iT PicoGreen kit (Invitrogen, Carlsbad, CA, USA) and pooled for pyrosequencing. All samples with their respective bar codes were pooled (from each 1/4 of a plate) in equimolar amounts for 454 pyrosequencing on a Roche GS-FLX system at the Microbiome Facility, University of North Carolina School of Medicine, Chapel Hill, NC, USA. To evaluate the reproducibility of the amplification and sequencing process, PCR was conducted in three separate amplifications, and the sequence compositions were compared. Sequences generated from independent PCR runs were pooled, after it was determined that their compositions were nearly identical (data not shown). After sequence quality control, 644,031 sequences were present, of which 630,864 were assigned to 5,208 operational taxonomic units (OTUs). OTUs were picked via an open reference procedure (described below).
Illumina MiSeq analyses.
One hundred seventeen samples were processed and sequenced in a single direction using an Illumina MiSeq machine located at the University of California, San Diego. 16S rRNA genes were targeted using primers 515F/806R, available on the Earth Microbiome Project website (www.earthmicrobiome.org), and a total of 4,424,307 sequences were detected. An open reference OTU picking approach was used (22) to cluster sequences, utilizing the program uclust (23). First, sequences were prefiltered to remove reads with less than 60% homology to at least one 16S rRNA gene in the Greengenes collection (24, 25). Sequences which passed the prefilter were clustered against the sequences in the Greengenes August 2013 release. Sequences with at least a 97% identity match to a Greengenes OTU were counted as occurrences of that OTU. Of the remaining sequences which failed to cluster against a known OTU, 1% of them were selected as new seed sequences to cluster the failed reads against. Sequences that matched a de novo OTU seed at 97% identity or more were counted as occurrences of that OTU. A final PyNAST filtering step was used to eliminate de novo OTUs that were likely contaminants or a non-16S rRNA gene (26). OTUs with only a single occurrence were discarded from the data set, leaving 3,569,383 sequences clustered into 16,355 OTUs. A reference tree was constructed using reference sequences for known OTUs and representative sequences for the de novo OTUs and the FastTree program (v2.1.3) (27). Taxonomy was assigned to de novo OTUs using the uclust program (23).
Bioinformatics analysis.
Data were analyzed using QIIME software (v1.8.0) (28). Illumina and 454 data could not be combined because sequencing technology effects would overwhelm any ecological signals in the data, and thus, each data set was analyzed independently. The techniques used on both data sets included rarefaction, principal coordinates analysis (PCoA), supervised learning, alpha diversity analysis and comparison, and differential abundance testing. Data reported in Results are from the Illumina analyses, unless otherwise stated. The unrarefied Illumina OTU table is provided in Table S1 in the supplemental material, the rarefied (10,000 sequences per sample) Illumina table used for the analyses is Table S2 in the supplemental material, the 454 unrarefied table is provided in Table S3 in the supplemental material, and the rarefied (3,800 sequences per sample) 454 table is provided in Table S4 in the supplemental material. Table S5 in the supplemental material contains the mapping file, which contains the sample identifier and associated metadata (sex, collection latitude, etc.) for the Illumina data. Table S6 in the supplemental material has this information for the 454 data.
(i) Rarefaction.
Rarefaction was used prior to all analytical techniques. Rarefaction is a conservative approach that subsamples from libraries with high coverage to equalize the number of sequences present in each sample. Recent work has suggested alternatives to rarefaction for differential abundance testing but has not provided alternatives for beta diversity calculations (29). Since the majority of our analyses are based on beta diversity and because rarefaction is conservative but does not induce false-positive results (46), we opted for this strategy. The 454 OTU table was rarefied at 3,800 sequences per sample, although tables with 500 and 1,000 sequences per sample and a table with unrarefied sequences were also analyzed. The patterns did not change (data not shown). The Illumina OTU table was rarefied to 10,000 sequences per sample. Tables with 1,000 and 2,000 sequences per sample and a table with unrarefied samples were also analyzed. The patterns did not change (data not shown).
(ii) Beta diversity and PCoA.
Beta diversity was calculated using weighted and unweighted UniFrac methods (30). Principal coordinates analysis was conducted using QIIME, and the results were visualized using the EMPeror tool (31).
(iii) Alpha diversity.
Alpha diversity was calculated for all samples using the phylogenetic distance (32), Shannon entropy (in bits), and observed species. Phylogenetic distance is a measure of the total branch length that a sample (or a group of samples) has, Shannon entropy is a measure of the number and relative proportion of OTUs in a sample (or group of samples), and the number of observed species is just the number of OTUs that were found in a given community.
(iv) Supervised learning analysis.
Supervised learning is a subset of machine learning that builds predictive models of sample class membership on the basis of the features that those samples contain. In our study, the features are OTUs or taxonomically summarized OTUs, and the sample classes are parameters, such as tick origin (Connecticut, New York, North Carolina, etc.). We utilized a random forest (RF) approach that is implemented in R and QIIME (v1.8) and employed 5- or 10-fold cross validation. In brief, cross validation reduces overfitting by splitting the data from the study into 10 (or 5) units with sample class memberships that are as identical as possible (e.g., each unit contains two samples from ticks from a given state). Each forest learns on 9/10 units and predicts sample class membership on the 10th unit. The classification accuracy of the RF approach is evaluated by use of a statistic called the “error ratio.” The error ratio is simply computed as the number of mislabeled samples using random guesses/the number of mislabeled samples using the models of the decision trees. Thus, higher error ratios are better, and error ratios of ≥2 indicate that significant differences exist between the sample classes and that particular features are significantly predictive of the class of a sample (33).
(v) Semivariograms.
Semivariograms were developed to test spatial autocorrelation. They show the level of autocorrelation in samples from different locations. Here, they measure correlation in UniFrac distances over different spatial scales. In essence, we are comparing the spatial and UniFrac distance matrices generated and seeing if increasing distance in one measure correlates with increasing distance in the other. Various models, including linear, log, normal, and nugget models, are fit to the distance data to derive a qualitative understanding of the covariation. All semivariogram calculations were performed by use of the QIIME script plot_semivariogram.py.
Characterization of Enterobacteriaceae.
The rpoB gene of members of the family Enterobacteriaceae was amplified using a Kapa HiFi HotStart PCR kit. Four microliters of 1:10-diluted template DNA was mixed with 300 nM deoxynucleoside triphosphates and 300 nM (each) primers rpoB F (5′-CAGTTYATGGAYCAGAACAACCCG) and rpoB R (5′-ACGTTGCATGTTCGCACCCATCA), designed for this study. Cycling conditions were as follows: a 95°C initial denaturation for 3 min, followed by 35 cycles of denaturation at 98°C for 20 s, annealing at 59°C for 15 s, and extension at 72°C for 30 s, ending with a final extension at 72°C for 2 min. Samples were individually bar coded, prepared for sequencing using an Illumina Nextera XT sample preparation kit (Illumina, San Diego, CA), and sequenced at the University of North Carolina High-Throughput Sequencing Facility on an Illumina MiSeq platform using 2 × 300-bp chemistry. Paired-end reads were merged using the PEAR (v0.9.0) program (34), trimmed to remove read regions with a phred quality score of 25 or less, and filtered to remove all merged reads with a length of 400 bp or less. Merged-read haplotypes were processed using a custom clustering algorithm, which has previously been used to cluster deep sequencing amplicon data (35) (N. Hathaway and J. Bailey, submitted for publication). For each sample, reads with 1- and 2-base indels, as well as low-quality base call mismatches, were collapsed. A base was considered to be of low quality if its phred-scaled quality score was <20 or if the mean quality score for the 11 bases centered on the base in question was <15. Haplotype clusters were used for analysis if they comprised >0.5% of all reads for a given sample.
Characterization of Borrelia.
In order to better characterize samples identified to be Borrelia burgdorferi by the MiSeq system, we amplified and bidirectionally sequenced a portion of the plasmid-borne outer surface protein C (ospC) gene (36) for a subset of the samples that yielded Borrelia 16S rRNA gene sequences. ospC sequences were assembled using the Sequencher program (v5.3; Gene Codes Corp., Ann Arbor, MI) and aligned with reference sequences from GenBank using the ClustalX program, implemented in MEGA software (v6.0) (37). Consensus ospC sequences were compared with the sequences in the NCBI GenBank database by BLAST analysis on 23 April 2015.
Generation of geographic information system maps.
The 19 sampling sites were geocoded using the ArcGIS program (v10.2) (38) for use in spatial analysis. The prevalence of each of the 8 overall most common taxa at each sampling site was plotted.
Nucleotide sequence accession numbers.
The sequences have been uploaded to the Qiita database (http://qiita.ucsd.edu/) under the title “Variation in the Microbiota of Ixodes Ticks with Regard to Geography, Species, and Sex” and to the European Bioinformatics Institute (EBI) database (Illumina data, study accession number ERP011260 and submission accession number ERA462672; 454 data, study accession number ERP011240 and submission accession number ERA462591).
RESULTS
Alpha diversity.
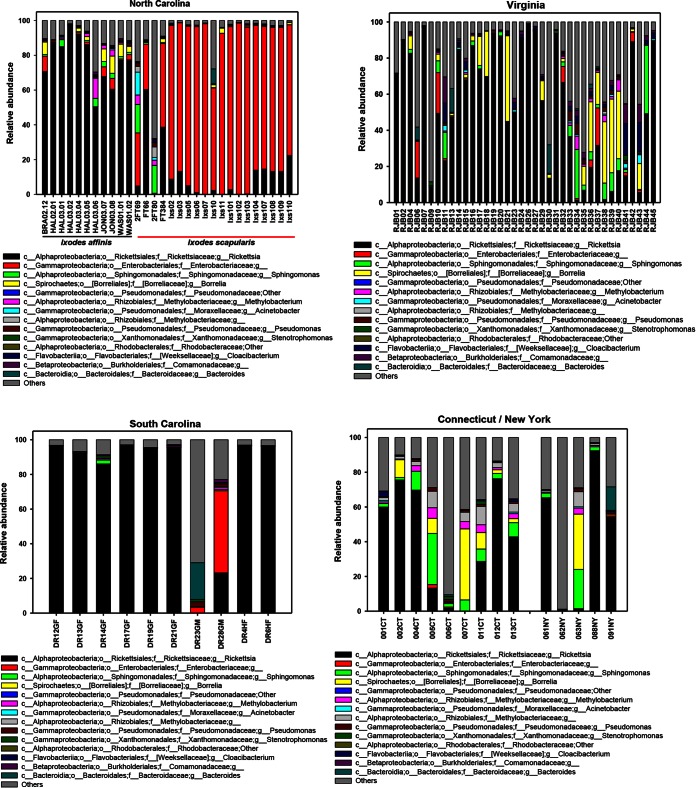
Tick microbiomes were dominated by Proteobacteria (81.7% of all reads), with individual ticks having between 49.33% and 96.69% of their reads mapping to a member of the Proteobacteria. Four other phyla, specifically, Actinobacteria, Bacteroidetes, Firmicutes, and Spirochaetes, were represented at frequencies ranging from 2 to 5%. Genera that were present at a ≥0.5% frequency are shown in Fig. 1.
FIG 1.
Genus-level abundance profiles for individual ticks (identified on the x axis) collected in North Carolina, Virginia, South Carolina, and Connecticut/New York (see Tables S5 and S6 in the supplemental material). Columns represent the percentage of MiSeq 16S rRNA gene sequences assigned to each genus. c, class; o, order; f, family; g, genus. Phyla, classes, orders, and families within brackets are names proposed by the curators of Greengenes.
Both I. scapularis and I. affinis ticks were collected from North Carolina. Their microbial compositions were significantly different, with the microbiota of I. affinis ticks being dominated by Rickettsia (77.28%) and the microbiota of I. scapularis ticks being dominated by an unknown genus in the family Enterobacteriaceae (73.45%) (Fig. 1). Notably, the high abundance of Enterobacteriaceae found in the North Carolina I. scapularis ticks was also found in the 454 data.
The alpha diversity of male ticks was significantly higher than that of female ticks (P < 0.01; Table 2). A similar result was found by 454 sequencing (not shown). Differences in Shannon entropy were more pronounced than those in phylogenetic diversity (PD), suggesting that male tick communities were both more diverse and more even. Part of this difference in alpha diversity is explained by phylum-level representation, where female ticks of both species had many more species of Proteobacteria (for I. scapularis ticks, 88.99% for female ticks and 72.79% for male ticks; for I. affinis ticks, 94.47% for female ticks and 84.29% for male ticks) and substantially fewer species of several other phyla. Results from the 454 data were similar, with Proteobacteria being detected in 98.55% of female ticks and 91.76% of male ticks and with significant alpha diversity comparisons being found.
TABLE 2.
Differences in alpha diversity between male and female ticksa
| Tick or parameter | Value (mean ± SD) |
|
|---|---|---|
| PD | Shannon entropy | |
| Male ticks | 22.25 ± 11.18 | 3.66 ± 2.11 |
| Female ticks | 17.26 ± 5.39 | 1.78 ± 1.18 |
| t statistic | 2.765 | 5.297 |
| P value | 0.009 | 0.001 |
Illumina data for I. scapularis and I. affinis ticks were pooled. See the text [e.g., “(iii) Alpha diversity”] for a more detailed description of PD and Shannon entropy.
Comparing I. scapularis alpha diversity by state of collection revealed a significantly higher Shannon entropy in Connecticut than North Carolina (P < 0.05; see Table S7 in the supplemental material). Rarefying at 2,000 sequences per sample allowed inclusion of more New York and Virginia samples, and at this level, both states had significantly higher Shannon scores than North Carolina. While there were large differences in PD between states, variance was also high, and a pairwise comparison revealed no significant differences (see Table S7 in the supplemental material). Interestingly, although the Shannon entropies of North Carolina and Connecticut ticks were different, their PDs were almost identical, suggesting that the OTUs in Connecticut ticks have a much more even representation.
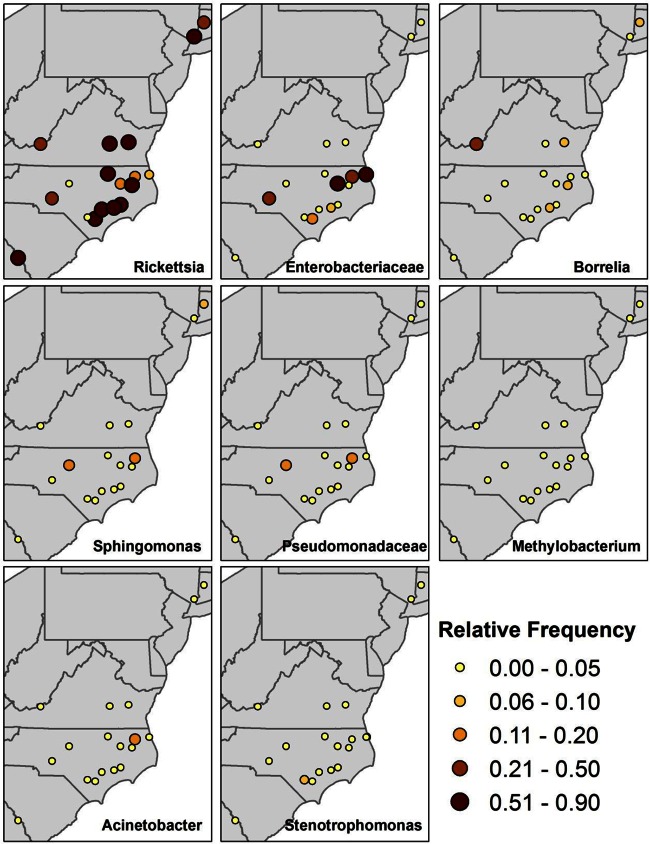
The overwhelming representation of 3 genera was visualized by overlaying the distribution of the most abundant genera on a map of sampling locations (Fig. 2). Rickettsia spp. dominated most sites, with Enterobacteriaceae dominating North Carolina sites with I. scapularis ticks. A single site (Pearisburg, Virginia) showed high levels of Borrelia, while 4 other sites in North Carolina, Virginia, and Connecticut showed a >5% relative abundance of Borrelia.
FIG 2.
Relative frequency of the 8 most common genera according to the site of tick collection. The size and color of each dot indicates the percentage of total 16S rRNA gene sequences assigned to a specific genus being mapped obtained at that site.
Beta diversity.
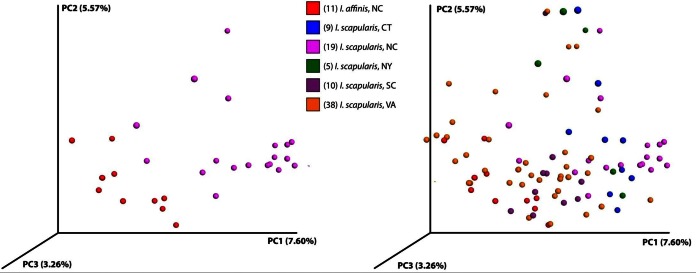
Using weighted and unweighted UniFrac methods, as well as other metrics like the Hellinger distance, we clustered ticks using principal coordinates analysis (PCoA). Illumina data did not show a clear separation of ticks by sex, state, or site of capture. The 454 data showed an extremely clear clustering, but that was likely an artifact of having so few sampling sites (see Fig. S1 in the supplemental material). In the Illumina data, a clear separation was seen between I. affinis and I. scapularis ticks captured in North Carolina (Fig. 3, left). Surprisingly, the North Carolina I. affinis ticks appeared to cluster more closely with I. scapularis ticks from other states (Fig. 3, right).
FIG 3.
Principal component analysis of unweighted UniFrac distances between ticks. (Left) I. scapularis (purple) and I. affinis (red) ticks captured in North Carolina cluster separately; (right) data for all ticks show that the North Carolina I. affinis ticks are closer to I. scapularis ticks from the other sites.
To test isolation by distance, we created semivariograms of the data, comparing the physical distance between sample locations and the weighted UniFrac distance (Fig. 4). The linear trend suggests a correlation between physical distance and community distance; the farther away that two ticks are, the more distinct their microbiota are. We also fit several other models to the data, including Gaussian and exponential models. The exponential and Gaussian models showed very similar behaviors. The sill (the range on the x axis where the fit line has not yet flattened) is very small, from approximately 0 to 30 km. The range (the range on the x axis where the fit line is flat) occurs at approximately 30 km. These alternate models suggest that differences in tick microbiota saturate with distance; ticks that are geographically close share microbes, but beyond a short distance there is not increasing dissimilarity with distance.
FIG 4.
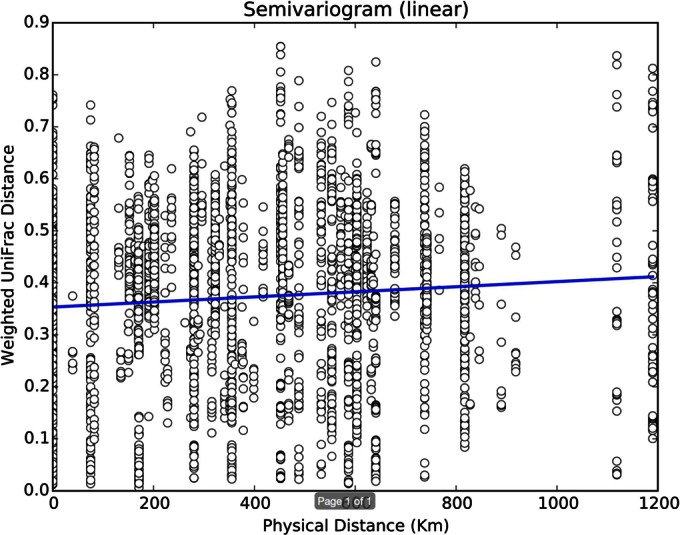
Semivariogram showing the linear relationship between the weighted UniFrac distance and the geographic distance for all pairs of ticks.
Comparing the abundance of I. scapularis OTUs by sex, we found 5 distinct (but unannotated) OTUs in the Rickettsia which were significantly higher in females. One of these OTUs (Greengenes OTU ID no. 112194) had a mean count of 6,021 per female tick, constituting over 60% of female tick sequences, whereas the mean count per male tick was 2,299. In a comparison by state of collection, we found 43 OTUs which had significantly different abundances between classes after Bonferroni multiple-hypotheses correction (Table 2). High-abundance OTUs that were different between classes were found in the Alpha-, Beta-, and Gammaproteobacteria. These data also confirm significant geographic diversity in the Ixodes microbiome.
Supervised learning.
Supervised learning was used to determine whether tick state of origin or sex could be determined from the microbial consortia. We used a random forest approach to sample classification for I. scapularis ticks based on sex and state of collection. The classifier had significant success for both, with error ratios of 2.98 and 2.44 for state and sex, respectively, being determined. Supervised learning results for the 454 data produced even better classification ratios, but these were likely the product of the small sample size. When I. affinis ticks were included, the error ratios dropped to 2.88 and 1.99, respectively. The most important features—the features that the classifier uses most successfully—for prediction of sample sex were in the Rickettsia.
While beta diversity did not reveal clustering patterns, the supervised learning results suggested that that there are state- and sex-specific microbial consortia. Not all state sources of the ticks were equally well identified by the classifier. All 5 ticks from New York State were attributed to another state. Interestingly, when the intrastate versus interstate distances were compared, New York ticks were the only ones with an intrastate mean distance greater than the mean interstate distance for all ticks (see Fig. S2 in the supplemental material). The RF classifier likely has trouble with New York ticks because the average distance between any two New York ticks was greater than the distance from a New York tick to a tick in another state. In summary, this supervised learning analysis confirms that there are microbiome features characteristic of ticks from different geographic locations and ticks of different sexes.
Ixodes affinis ticks from North Carolina have microbial compositions that are more similar to those of I. scapularis ticks from Virginia than to those of I. scapularis ticks from North Carolina. Supervised learning analysis, conducted to differentiate both species and state simultaneously, correctly classified 17/19 North Carolina I. scapularis ticks, with the 2 misclassified ticks being assigned to North Carolina I. affinis or Connecticut I. scapularis. Conversely, of the 11 I. affinis ticks from North Carolina, 3 were classified as North Carolina I. affinis ticks and 8 were classified as Virginia I. scapularis ticks (see Table S8 in the supplemental material). This finding comports with the descriptive differences evident in Fig. 1.
Characterization of Enterobacteriaceae.
At the Pine Island and View Nicholson Road sites in North Carolina, the majority of reads were from organisms in the Enterobacteriaceae family. In the Illumina data, 87.4% and 83.5% of the reads for Pine Island and View Nicholson Road were for members of the family Enterobacteriaceae, respectively, and in the 454 data, these values were 84.0% and 69.7%, respectively. One OTU within this family (Greengenes OTU ID no. 281015) comprised 95.31% of all the reads consisting of the Enterobacteriaceae in the Illumina data. In the 454 data, one OTU (Greengenes OTU ID no. 691423) comprised 76.91% of all reads consisting of the Enterobacteriaceae. We attempted to further identify the organism represented by Greengenes OTU ID no. 691423.
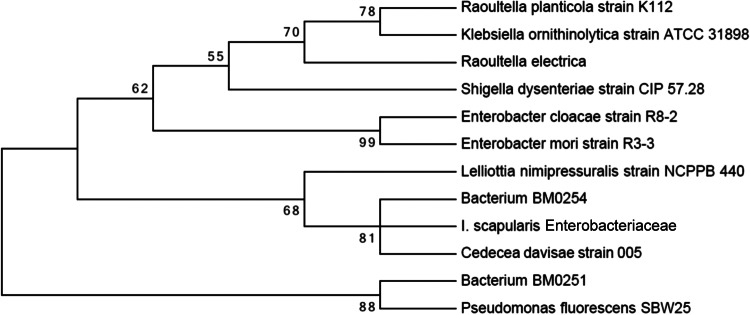
In a BLAST search, the 221-bp reference sequence for Greengenes OTU ID no. 691423 that was created during 454 OTU picking was found to be 100% homologous to 98 previously reported Enterobacteriaceae sequences belonging to a variety of genera (BLAST analysis was performed on 3 August 2014). In order to better classify this taxon, we amplified a segment of rpoB from 14 ticks (6 collected in North Carolina and 8 collected in Connecticut). Amplicon sequencing produced 25,791,347 paired-end reads. After read pairs were merged to create a single haplotype, 740,217 reads met stringent length (≥400 bases) and quality (read-averaged phred score, ≥25) criteria. Of these, the sequences of 160,628 reads matched the rpoB sequence and were included in further analyses. Among the 14 tick isolates sequenced, we achieved a minimum rpoB coverage depth of 3,931 reads, a median depth of 9,040 reads, and a maximum depth of 29,091 reads. After the reads were cleaned, they were clustered, resulting in over 99% of reads from each isolate clustering to a single rpoB haplotype. The sequence of the rpoB haplotype was 98% identical to the sequence of the bacterium strain BM0254 rpoB gene (GenBank accession no. JX471764.1) and 94% identical to the sequence of the Cedecea davisae rpoB gene (GenBank accession no. EU010119.1) and clustered with them phylogenetically (Fig. 5). BM0254 has previously been found in mosquitoes, while C. davisae has previously been found in tsetse flies and cockroaches.
FIG 5.
Maximum likelihood phylogenetic tree showing the relationship between the Enterobacteriaceae isolates from I. scapularis ticks found in this study and other related Enterobacteriaceae. The tree was constructed using the MEGA program (v6) (http://www.megasoftware.net/mega.php) with default settings and 1,000 bootstraps.
Characterization of Borrelia.
From the Illumina data, Borrelia reads represented >5% of reads in ticks from 5 of the 18 sites (Fig. 2) and 3.2% of the reads overall. Surprisingly, the highest prevalence of Borrelia (18.5%) was found in Pearisburg, VA. In the 454 data, Borrelia spp. were found in ticks at a frequency of less than 5% at all 4 sites and Borrelia sequences made up 1.4% of the reads overall. The ticks at the 4 sites where Borrelia spp. were found had similar levels of Borrelia spp. from the Illumina data and the 454 data: 4.2% and 3%, respectively, at Armonk, NY; 0.6% and 0.3%, respectively, at Pine Island, NC; 0.3% and 0.0%, respectively, at View Nicholson Road, NC; and 8.2% and 2.4%, respectively, at Lake Galliard, CT.
For the 454 data, a single OTU (Greengenes OTU ID no. 4319519) comprised 83% of the Borrelia reads. All top 100 BLAST hits in GenBank (accessed 7 August 2014) for the 237-bp reference sequence created during 454 OTU picking for this sequence (99% identity and above) were members of the B. burgdorferi sensu lato group.
The same OTU comprised 97.49% of all Borrelia reads in the Illumina data. ospC sequence data from 10 PCR-positive ticks sampled near Pearisburg, VA, yielded four sequence types which were highly similar or identical to published genotypes for Borrelia burgdorferi sensu stricto, and these had GenBank accession numbers AF065143 (n = 2, 99% similarity), CP002268 (n = 1, 100% similarity), DQ437470 (n = 1, 100% similarity), and AE000792 (n = 6, 99 to 100% similarity). One tick from this site that yielded Borrelia sequences from Illumina analysis (relative abundance, 0.018) failed to produce amplicons during nested PCR for ospC and the 16S rRNA-23S rRNA intergenic spacer region, suggesting that an extremely low Borrelia abundance in ticks may not be detectable by conventional PCR.
Characterization of Rickettsiales.
In the Illumina data, one OTU (Greengenes OTU ID no. 112194) comprised 97.26% of all reads in the genus Rickettsia. In the 454 data, one OTU (Greengenes OTU ID no. 316724) comprised 97.28% of all reads in the genus Rickettsia. A BLAST search of the 221-bp reference sequence created during 454 OTU picking against the rickettsial endosymbiont of Ixodes scapularis (REIS; gi|239946612|ref|NZ; GenBank accession no. CM000770.1|) showed 99% identity.
Anaplasma spp. were not highly prevalent, comprising a maximum of 3.6% of reads at one site (York River State Park, VA). In both the Illumina and the 454 data, the majority of the Anaplasma reads (76.70% and 78.89%, respectively) were represented by a single OTU (Greengenes OTU ID no. 152987). The 218-bp reference sequence created during 454 OTU picking was 100% identical to 93 Anaplasma phagocytophilum sequences present in GenBank (accessed 7 August 2014).
DISCUSSION
Here we show that the microbiomes of Ixodes ticks from the eastern United States vary by geographic location as well as by species and sex. There were two previous studies of I. scapularis larval and nymph microbiota that, like the current study, showed a high prevalence of Rickettsia (4, 14). Neither study reported similarities with REIS, probably because the REIS sequences are not yet available in GenBank and can be searched for only via their unassembled reads. The most common taxa found in our study partially overlapped those found in other studies with Ixodes ticks. For example, in an analysis of I. scapularis larval and nymphal microbiomes by Narasimhan et al. (19), Sphingomonas, Pseudomonas, Methylobacterium, Borrelia, and Anaplasma were common, as in this study. In a study of I. persulcatus ticks from China (39), Rickettsia, Pseudomonadeae, Sphingomonas, and Borrelia were among the most common bacteria, similar to the findings presented here. Neither of these 2 studies (19, 39), however, reported the presence of Enterobacteriaceae. In contrast, both Rickettsia and Enterobacteriaceae were present at high levels in all stages of I. ricinus ticks (12). All these differences could be the result of geographic variation, as was observed here.
For this study, we initially analyzed the microbiomes of ticks from 4 sites using the 454 platform. Subsequently, we analyzed the microbiomes of ticks from 15 additional sites (as well as the ticks from the other 4 sites) using the Illumina platform. For the analyses with both data sets that were possible, the concordance of the results was high. Ticks had similar levels of the abundant phyla and a similar representation of key genera. The alpha diversity differences between male and female ticks were perfectly recapitulated, and supervised learning with the two data sets showed similar agreement of the results.
The composition of the microbiota was affected by both geography and sex. The microbiota from females was less diverse, possibly because they had higher relative burdens of Rickettsia, especially 5 OTUs. The increased burden of rickettsial populations is similar to that found by us (17) and by Clay and Fuqua (14) in Amblyomma americanum ticks. The increased burden of Rickettsia in females could be an adaptation to transovarial transmission (40).
Of particular interest is our observation of an apparently clonal infection with Enterobacteriaceae in Ixodes ticks. Internal Gram-negative rods were first observed in I. scapularis ticks in 1998 (18). In our study, a single OTU, the OTU with Greengenes ID no. 691423, represented 95.31% of the 16S rRNA gene reads assigned to the Enterobacteriaceae. This bacterium was the most common microbe found in ticks from North Carolina and was >10% of the microbial population in both I. scapularis and I. affinis ticks from multiple sites (Fig. 1).
The sequence of Greengenes OTU ID no. 691423 had 100% homology to the sequences of multiple enterobacterial genera within GenBank. This finding underscores the need for sequence data from additional genes in order to better classify taxa of interest. For this purpose, we used the rpoB gene. Consistent with our findings with the 16S rRNA gene, all of our rpoB deep sequencing data clustered to a single sequence. Interestingly, this sequence showed the greatest homology to two taxa identified in insects: BM0254, which was identified in Anopheles gambiae mosquitoes (41), and Cedecea davisae, which was identified in cockroaches (42, 43). Thus, it is likely that the bacterium identified here and its close relatives may be important commensals of arthropods. It is possible that the tick-borne Enterobacteriaceae might have deleterious effects on humans or animals. C. davisae is a well-documented human and animal pathogen (44), but this possibility needs to be studied further.
The map of the major constituents of the microbiota (Fig. 1) is notable for two reasons. First, ticks from Pearisburg, VA, had the highest burden of Borrelia, and the burden was higher than that of ticks from the Northeast, all of which were confirmed to harbor B. burgdorferi sensu stricto. Lyme disease is endemic in the Northeast but has only recently emerged in the part of Virginia where Pearisburg is located (45). Second, ticks from only 5 sites had light Rickettsia burdens (<20%), and ticks from 3 of these sites were heavily infected with members of the family Enterobacteriaceae. This suggests a possible competitive interaction between taxa.
Supervised learning was used to corroborate our standard statistical approach. Supervised learning results support the finding that there are characteristic microbiota associated with tick species, sex, and location.
One weakness of this approach was our inability to assess the microbial burdens within each tick, so while we could assess the diversity of microbial communities, we could not assess their relative sizes. However, the concordance between different sequencing and analytical approaches to the assessment of diversity suggests that these findings were not biased by community size.
In summary, Ixodes gut microbiota populations are quite heterogeneous. The geographic differences may offer insight into the differences in the capacity of ticks to transmit pathogens. Future studies on the causes and consequences of these differences are needed.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nick Matinyan for help in the lab. We thank Natasha Butz, Andrea Azcarate-Peril, and the University of North Carolina Microbiome Core Facility for performing 454 sequencing. We thank John Kokas and Vanessa Vinci for assistance with tick collections.
The project was supported in part by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through grant award number UL1TR000083, by the North Carolina State University Center for Comparative Molecular Medicine and Translational Research through award number 2011-2611, and by the Howard Hughes Medical Institute.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01562-15.
REFERENCES
- 1.Hinckley AF, Connally NP, Meek JI, Johnson BJ, Kemperman MM, Feldman KA, White JL, Mead PS. 2014. Lyme disease testing by large commercial laboratories in the United States. Clin Infect Dis 59:676–681. doi: 10.1093/cid/ciu397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Layfield D, Guilfoile P. 2002. The prevalence of Borrelia burgdorferi (Spirochaetales: Spirochaetaceae) and the agent of human granulocytic ehrlichiosis (Rickettsiaceae: Ehrlichieae) in Ixodes scapularis (Acari: Ixodidae) collected during 1998 and 1999 from Minnesota. J Med Entomol 39:218–220. doi: 10.1603/0022-2585-39.1.218. [DOI] [PubMed] [Google Scholar]
- 3.Prusinski M, Kokas J, Hukey K, Kogut S, Lee J, Backenson P. 2014. Prevalence of Borrelia burgdorferi (Spirochaetales: Spirochaetaceae), Anaplasma phagocytophilum (Rickettsiales: Anaplasmataceae), and Babesia microti (Piroplasmida: Babesiidae) in Ixodes scapularis (Acari: Ixodidae) collected from recreational lands in the Hudson Valley Region, New York State. J Med Entomol 51:226–236. doi: 10.1603/ME13101. [DOI] [PubMed] [Google Scholar]
- 4.Moreno CX, Moy F, Daniels TJ, Godfrey HP, Cabello FC. 2006. Molecular analysis of microbial communities identified in different developmental stages of Ixodes scapularis ticks from Westchester and Dutchess Counties, New York. Environ Microbiol 8:761–772. doi: 10.1111/j.1462-2920.2005.00955.x. [DOI] [PubMed] [Google Scholar]
- 5.Adelson ME, Rao RV, Tilton RC, Cabets K, Eskow E, Fein L, Occi JL, Mordechai E. 2004. Prevalence of Borrelia burgdorferi, Bartonella spp., Babesia microti, and Anaplasma phagocytophila in Ixodes scapularis ticks collected in northern New Jersey. J Clin Microbiol 42:2799–2801. doi: 10.1128/JCM.42.6.2799-2801.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisen L, Eisen RJ, Mun J, Salkeld DJ, Lane RS. 2009. Transmission cycles of Borrelia burgdorferi and B. bissettii in relation to habitat type in northwestern California. J Vector Ecol 34:81–91. doi: 10.1111/j.1948-7134.2009.00010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CDC. 2010. Lyme disease incidence rates by states, 2005–2009. CDC, Atlanta, GA. [Google Scholar]
- 8.Maggi RG, Reichelt S, Toliver M, Engber B. 2010. Borrelia species in Ixodes affinis and Ixodes scapularis ticks collected from the coastal plain of North Carolina. Ticks Tick-Borne Dis 1:168–171. doi: 10.1016/j.ttbdis.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Clark KL, Oliver JH Jr, James AM, Durden LA, Banks CW. 2002. Prevalence of Borrelia burgdorferi sensu lato infection among rodents and host-seeking ticks in South Carolina. J Med Entomol 39:198–206. doi: 10.1603/0022-2585-39.1.198. [DOI] [PubMed] [Google Scholar]
- 10.Harrison BA, Rayburn WH Jr, Toliver M, Powell EE, Engber BR, Durden LA, Robbins RG, Prendergast BF, Whitt PB. 2010. Recent discovery of widespread Ixodes affinis (Acari: Ixodidae) distribution in North Carolina with implications for Lyme disease studies. J Vector Ecol 35:174–179. doi: 10.1111/j.1948-7134.2010.00074.x. [DOI] [PubMed] [Google Scholar]
- 11.Clay K, Klyachko O, Grindle N, Civitello D, Oleske D, Fuqua C. 2008. Microbial communities and interactions in the lone star tick, Amblyomma americanum. Mol Ecol 17:4371–4381. doi: 10.1111/j.1365-294X.2008.03914.x. [DOI] [PubMed] [Google Scholar]
- 12.Carpi G, Cagnacci F, Wittekindt NE, Zhao F, Qi J, Tomsho LP, Drautz DI, Rizzoli A, Schuster SC. 2011. Metagenomic profile of the bacterial communities associated with Ixodes ricinus ticks. PLoS One 6:e25604. doi: 10.1371/journal.pone.0025604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Overbeek L, Gassner F, van der Plas CL, Kastelein P, Nunes-da Rocha U, Takken W. 2008. Diversity of Ixodes ricinus tick-associated bacterial communities from different forests. FEMS Microbiol Ecol 66:72–84. doi: 10.1111/j.1574-6941.2008.00468.x. [DOI] [PubMed] [Google Scholar]
- 14.Clay K, Fuqua C. 2010. The tick microbiome: diversity, distribution and influence of the internal microbial community for a blood-feeding disease vector. InCritical needs and gaps in understanding prevention, amelioration, and resolution of Lyme and other tick-borne diseases: the short-term and long-term outcomes. Workshop report National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 15.Nakao R, Abe T, Nijhof AM, Yamamoto S, Jongejan F, Ikemura T, Sugimoto C. 2013. A novel approach, based on BLSOMs (batch learning self-organizing maps), to the microbiome analysis of ticks. ISME J 7:1003–1015. doi: 10.1038/ismej.2012.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Budachetri K, Browning RE, Adamson SW, Dowd SE, Chao C-C, Ching W-M, Karim S. 2014. An insight into the microbiome of the Amblyomma maculatum (Acari: Ixodidae). J Med Entomol 51:119–129. doi: 10.1603/ME12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponnusamy L, Gonzalez A, Van Treuren W, Weiss S, Parobek CM, Juliano JJ, Knight R, Roe RM, Apperson CS, Meshnick SR. 2014. Diversity of Rickettsiales in the microbiome of the lone star tick, Amblyomma americanum. Appl Environ Microbiol 80:354–359. doi: 10.1128/AEM.02987-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin PA, Schmidtmann ET. 1998. Isolation of aerobic microbes from Ixodes scapularis (Acari: Ixodidae), the vector of Lyme disease in the eastern United States. J Econ Entomol 91:864–868. doi: 10.1093/jee/91.4.864. [DOI] [PubMed] [Google Scholar]
- 19.Narasimhan S, Rajeevan N, Liu L, Zhao YO, Heisig J, Pan J, Eppler-Epstein R, Deponte K, Fish D, Fikrig E. 2014. Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the Lyme disease spirochete. Cell Host Microbe 15:58–71. doi: 10.1016/j.chom.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaughn MF, Funkhouser SW, Lin FC, Fine J, Juliano JJ, Apperson CS, Meshnick SR. 2014. Long-lasting permethrin impregnated uniforms: a randomized-controlled trial for tick bite prevention. Am J Prev Med 46:473–480. doi: 10.1016/j.amepre.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 21.Smith MP, Ponnusamy L, Jiang J, Ayyash LA, Richards AL, Apperson CS. 2010. Bacterial pathogens in Ixodid ticks from a Piedmont county in North Carolina: prevalence of rickettsial organisms. Vector Borne Zoonotic Dis 10:939–952. doi: 10.1089/vbz.2009.0178. [DOI] [PubMed] [Google Scholar]
- 22.Navas-Molina JA, Peralta-Sanchez JM, Gonzalez A, McMurdie PJ, Vazquez-Baeza Y, Xu Z, Ursell LK, Lauber C, Zhou H, Song SJ, Huntley J, Ackermann GL, Berg-Lyons D, Holmes S, Caporaso JG, Knight R. 2013. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol 531:371–444. doi: 10.1016/B978-0-12-407863-5.00019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 24.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMurdie PJ, Holmes S. 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vazquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. 2013. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2:16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biol Conserv 61:1–10. doi: 10.1016/0006-3207(92)91201-3. [DOI] [Google Scholar]
- 33.Knights D, Costello EK, Knight R. 2011. Supervised classification of human microbiota. FEMS Microbiol Rev 35:343–359. doi: 10.1111/j.1574-6976.2010.00251.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parobek CM, Bailey JA, Hathaway NJ, Socheat D, Rogers WO, Juliano JJ. 2014. Differing patterns of selection and geospatial genetic diversity within two leading Plasmodium vivax candidate vaccine antigens. PLoS Negl Trop Dis 8:e2796. doi: 10.1371/journal.pntd.0002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bunikis J, Garpmo U, Tsao J, Berglund J, Fish D, Barbour AG. 2004. Sequence typing reveals extensive strain diversity of the Lyme borreliosis agents Borrelia burgdorferi in North America and Borrelia afzelii in Europe. Microbiology 150:1741–1755. doi: 10.1099/mic.0.26944-0. [DOI] [PubMed] [Google Scholar]
- 37.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Environmental Systems Resource Institute. 2013. ArcMap 10.2. Environmental Systems Resource Institute, Redlands, CA. [Google Scholar]
- 39.Zhang XC, Yang ZN, Lu B, Ma XF, Zhang CX, Xu HJ. 2014. The composition and transmission of microbiome in hard tick, Ixodes persulcatus, during blood meal. Ticks Tick-Borne Dis 5:864–870. doi: 10.1016/j.ttbdis.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 40.Telford SR., III 2009. Status of the “East Side hypothesis” (transovarial interference) 25 years later. Ann N Y Acad Sci 1166:144–150. doi: 10.1111/j.1749-6632.2009.04522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez-Ceron L, Santillan F, Rodriguez MH, Mendez D, Hernandez-Avila JE. 2003. Bacteria in midguts of field-collected Anopheles albimanus block Plasmodium vivax sporogonic development. J Med Entomol 40:371–374. doi: 10.1603/0022-2585-40.3.371. [DOI] [PubMed] [Google Scholar]
- 42.Kaaya GP, Okech MA. 1990. Microorganisms associated with tsetse in nature: preliminary results on isolation, identification and pathogenicity. Int J Trop Insect Sci 11:443–448. doi: 10.1017/S1742758400012868. [DOI] [Google Scholar]
- 43.Pellegrini G, Levre E, Valentini P, Cadoni M. 1992. Cockroach infestation and possible contribution to the spread of some enterobacteria. Igiene Moderna 97:19–30. [Google Scholar]
- 44.Akinosoglou K, Perperis A, Siagris D, Goutou P, Spiliopoulou I, Gogos CA, Marangos M. 2012. Bacteraemia due to Cedecea davisae in a patient with sigmoid colon cancer: a case report and brief review of the literature. Diagn Microbiol Infect Dis 74:303–306. doi: 10.1016/j.diagmicrobio.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 45.Brinkerhoff RJ, Gilliam WF, Gaines D. 2014. Lyme disease, Virginia, USA, 2000–2011. Emerg Infect Dis 20:1661–1668. doi: 10.3201/eid2010.130782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss SJ, Xu Z, Amir A, Peddada S, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vazquez-Baeza Y, Birmingham A, Knight R. 2015. Effects of library size variance, sparsity, and compositionality on the analysis of microbiome data. PeerJ PrePrints 3: e1408. doi: 10.7287/peerj.preprints.1157v1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.