ABSTRACT
HIV-1 Env glycoprotein-mediated fusion is initiated upon sequential binding of Env to CD4 and the coreceptor CXCR4 or CCR5. Whereas these interactions are thought to be necessary and sufficient to promote HIV-1 fusion, other host factors can modulate this process. Previous studies reported potent inhibition of HIV-1 fusion by selective P2X1 receptor antagonists, including NF279, and suggested that these receptors play a role in HIV-1 entry. Here we investigated the mechanism of antiviral activity of NF279 and found that this compound does not inhibit HIV-1 fusion by preventing the activation of P2X1 channels but effectively blocks the binding of the virus to CXCR4 or CCR5. The notion of an off-target effect of NF279 on HIV-1 fusion is supported by the lack of detectable expression of P2X1 receptors in cells used in fusion experiments and by the fact that the addition of ATP or the enzymatic depletion of ATP in culture medium does not modulate viral fusion. Importantly, NF279 fails to inhibit HIV-1 fusion with cell lines and primary macrophages when added at an intermediate stage downstream of Env-CD4-coreceptor engagement. Conversely, in the presence of NF279, HIV-1 fusion is arrested downstream of CD4 binding but prior to coreceptor engagement. NF279 also antagonizes the signaling function of CCR5, CXCR4, and another chemokine receptor, as evidenced by the suppression of calcium responses elicited by specific ligands and by recombinant gp120. Collectively, our results demonstrate that NF279 is a dual HIV-1 coreceptor inhibitor that interferes with the functional engagement of CCR5 and CXCR4 by Env.
IMPORTANCE Inhibition of P2X receptor activity suppresses HIV-1 fusion and replication, suggesting that P2X signaling is involved in HIV-1 entry. However, mechanistic experiments conducted in this study imply that P2X1 receptor is not expressed in target cells or involved in viral fusion. Instead, we found that inhibition of HIV-1 fusion by a specific P2X1 receptor antagonist, NF279, is due to the blocking of virus interactions with both the CXCR4 and CCR5 coreceptors. The ability of NF279 to abrogate cellular calcium signaling induced by the respective chemokines showed that this compound acts as a dual-coreceptor antagonist. P2X1 receptor antagonists could thus represent a new class of dual-coreceptor inhibitors with a structure and a mechanism of action that are distinct from those of known HIV-1 coreceptor antagonists.
INTRODUCTION
HIV-1 currently remains a major public health concern worldwide. In the face of virus's ability to develop resistance to antiviral agents, new therapeutic approaches focusing on host factors required for HIV-1 replication hold promise. HIV-1 enters target cells via a multistep process that is initiated upon binding of the gp120 subunit of the Env glycoprotein to its receptor and coreceptor, CD4 and CXCR4/CCR5, respectively (reviewed in reference 1). Coreceptor engagement promotes critical conformational changes in gp41 leading to fusion of the viral and cellular membranes (reviewed in reference 2). Most HIV-1 isolates utilize either of the two coreceptors, while dual-tropic strains can use both CXCR4 and CCR5 for entry. CD4 and coreceptors are thought to be necessary and sufficient to support HIV-1 fusion; however, additional host factors have been reported to modulate this process (3–12). In recent years, purinergic receptors of the P2 family have been implicated in HIV-1 entry/infection (13–15). This notion is, to a large extent, based on the inhibitory effect of P2 antagonists. P2 receptors are widely expressed membrane proteins (16) that are activated by extracellular ATP and ATP/ADP (for the P2X and P2Y subfamilies, respectively). Mammals express seven P2X receptors (P2XRs) (P2XR1 to -7) and eight P2Y receptors (P2Y1, P2Y2, P2Y4, P2Y6, P2Y11, P2Y12, P2Y13, and P2Y14) (16–18). P2YRs are metabotropic G-protein-coupled receptors (GPCRs) (19, 20), whereas P2XRs are ionotropic receptors (21) that form homotrimers as well as heterotrimers with other members of the same family. P2 receptors are involved in numerous biological functions, such as inflammation, cell activation, and chemotaxis (16, 19, 22).
The involvement of P2X1 receptors in HIV-1 infection of macrophages was recently demonstrated (13). This inhibitor-based study implicated P2X1R, P2X7R, and P2Y1R in infection and replication of HIV-1, but only P2X1R appeared to be involved in HIV-1 entry/fusion. It was thus proposed that P2X1R facilitates HIV-1 entry via the downstream effects of elevated intracellular calcium concentrations. However, conflicting results were obtained by Schachter and colleagues, who reported that inhibition of ecto-ATPase activity resulting in increased concentrations of extracellular ATP attenuates HIV-1 infection in macrophages (23). The role of purinergic receptors in HIV-1 infection of CD4+/CXCR4+ target cells and CD4+ T cells has also been documented (14). The authors of that study concluded that ATP released by the pannexin-1 hemichannel activates the P2Y2 receptor and its downstream effector kinase Pyk2; the resulting depolarization of the plasma membrane was hypothesized to promote HIV-1 fusion. A more recent study by Swartz and colleagues implicated purinergic receptors in cell-free infection of CD4+ T lymphocytes and in cell-to-cell HIV-1 transmission (15). Their results showed that P2X1R but not P2Y antagonists block HIV-1 infection through both routes without interfering with virological synapse formation or CXCR4 recruitment by internalized virions.
We previously conducted a pilot screen for HIV-1 fusion inhibitors and found that the P2X1R antagonists NF449 and NF279 inhibited the fusion of both R5- and X4-tropic HIV-1 strains without affecting the expression levels of CD4 and coreceptors (24). This effect was HIV specific, since fusion of viruses pseudotyped with the vesicular stomatitis virus G glycoprotein (VSV-G) was insensitive to these inhibitors. In contrast, P2Y or P2X7 receptor antagonists did not suppress HIV-1 fusion (24). These findings further implicate P2X1R in HIV-1 fusion, but the mechanism by which these receptors regulate viral fusion remains unclear. Here we investigated the mechanism of HIV-1 fusion inhibition by NF279 and found no evidence of P2X1R involvement in virus entry. P2X1R expression could not be detected in target cells in which NF279 blocked HIV-1 fusion. Functional dissection of the HIV-1 fusion reaction revealed that NF279 targets the Env-coreceptor interaction step in both R5- and X4-tropic viruses. Furthermore, the ability of NF279 to suppress the CCR5 and CXCR4 agonist-mediated elevation of the cytosolic calcium concentration implies that this compound acts as a dual-coreceptor antagonist. Our results thus demonstrate that at micromolar concentrations required to inhibit HIV-1 fusion, NF279 (and maybe other P2X1R antagonists) prevents Env-coreceptor binding without affecting the upstream Env-CD4 binding step.
MATERIALS AND METHODS
Cells and culture media.
The following cell lines were obtained from the NIH AIDS Research and Reference Reagent Program (ARRRP): TZM-bl cells (donated by J. Kappes and X. Wu) (25), CEM-A cells (26), and CEM-NKR-CCR5-Luc cells (donated by J. Moore and C. Spenlehauer) (27). CEM-A and CEM-NKR-CCR5-Luc cells were cultured in RPMI 1640 medium (Cellgro; Mediatech Inc., Manassas, VA) supplemented with 10% fetal bovine serum (FBS; HyClone Laboratories, Logan, UT) and 100 U/ml penicillin-streptomycin (Gemini Bio-Products, Sacramento, CA). The CEM-A cell culture was supplemented with 100 μM hypoxanthine and 16 μM thymidine. CEM-NKR-CCR5 medium contained 0.8 mg/ml G418 (Mediatech). HEK 293T/17 cells (ATCC, Manassas, VA) and JGR.H11 cells (a gift from D. Kabat) were grown in Dulbecco modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% FBS and 100 U/ml penicillin-streptomycin. Growth medium for HEK 293T/17 cells was supplemented with 0.5 mg/ml G418. Jurkat-CCR5 cells (from L. Chernomordik, NICHD) were grown in complete RPMI 1640 medium supplemented with 0.5 mg/ml G418, 0.5 μg/ml hygromycin (Invitrogen, San Diego, CA), and 50 μg/ml gentamicin (Atlanta Biologicals, Lawrenceville, GA). NP2 cells stably expressing the dual-split protein 2 (DSP2) fragment of dual-split Renilla luciferase-green fluorescent protein (GFP) (designated N4X4-DSP2 cells) and HEK 293T cells stably expressing DSP1 (293T-DSP1 cells) (28) were gifts from H. Hoshino, N. Hosoya, and A. Iwamoto (University of Tokyo, Tokyo, Japan). NP2 cells were grown in minimal essential medium (MEM) (Cellgro) supplemented with 10% FBS, 100 U/ml penicillin-streptomycin, and 4 μg/ml blasticidin (Bioworld, Atlanta, GA).
Primary macrophage culture.
Peripheral blood was obtained from healthy volunteer donors according to a protocol approved by the Emory University Institutional Review Board (IRB). Peripheral blood mononuclear cells (PBMCs) were obtained from the buffy coat after Ficoll density centrifugation and were negatively selected by using magnetically activated cell sorting (MACS) monocyte isolation kit II (Miltenyi Biotec, Bergish Gladbach, Germany). Monocytes were differentiated into macrophages upon culturing for 7 days in RPMI 1640 medium supplemented with 10% FBS, 100 U/ml penicillin-streptomycin, 2 mM glutamine, 1% sodium pyruvate, 1% nonessential amino acids, and 5 ng/ml granulocyte-macrophage colony-stimulating factor (GM-CSF; Cell Sciences, Canton, MA, USA).
Plasmids.
The pCAGGS-HXB2-Env vector for X4-tropic envelope expression was provided by J. Binley (Torrey Pines Institute, CA), the pBal26-Env vector for R5-tropic envelope expression was a gift from P. Clapham (University of Massachusetts, Worchester, MA), and pHPG-R3A-Env was a gift from J. Hoxie (University of Pennsylvania). The pMDG-VSVG plasmid expressing VSV-G was a gift from J. Young (Salk Ins, La Jolla, CA). The HIV-1 proviral clone pR8ΔEnv lacking the env gene was obtained from D. Trono (University of Geneva, Geneva, Switzerland), and pcRev and PMM310-BlaM-Vpr expressing Rev and β-lactamase (BlaM)–Vpr, respectively, were obtained from the ARRRP (29). The pcDNA3.1-P2X1R-WT and pcDNA3.1-P2X1R-delL (L351-deleted) plasmids encoding wild-type human P2X1R and its dominant-negative mutant, respectively, were kindly provided by M. Hoylaerts (University of Leuven, Leuven, Belgium) (30).
Reagents and inhibitors.
The P2X1R inhibitors NF279 and NF449, the P2Y11R inhibitor NF340, the P2X7R inhibitor A740003, and thapsigargin were obtained from Tocris Bioscience (R&D Systems, Minneapolis, MN). ATP, oxidized ATP (oATP), α,β-methylene ATP (α,β-meATP), apyrase from potato, the CXCR4 antagonist AMD3100, the calcium ionophore A23187, and poly-l-lysine were obtained from Sigma-Aldrich (St. Louis, MO). The calcium ionophore ionomycin was obtained from Calbiochem (EMD Millipore, Billerica, MA). The cell-permeable calcium indicator Fluo4-AM was obtained from Invitrogen. The BMS-806 compound was synthesized by ChemPacific Corp. (Baltimore, MD). The C52L recombinant peptide was a kind gift from Min Lu (University of New Jersey). The human chemokine stromal cell-derived factor 1 alpha (SDF-1α) was from obtained Cell Sciences (Canton, MA), the synthesized CCR5 receptor agonist RANTES (regulated on activation, normal T cell expressed and secreted) was a kind gift of W. Lu (University of Maryland), and chemokine interferon-inducible T cell alpha chemoattractant (I-TAC)/CXCL11 was obtained from Novus Biologicals (Littleton, CO). Monomeric recombinant HIV-1 IIIB gp120 (CHO) and the CCR5 antagonist TAK-779 (31) were obtained from the ARRRP. The concentration of ATP was quantified by a luminescence assay, using the commercial ATPlite kit (Perkin-Elmer) according to the manufacturer's recommendations. Luminescence was measured by using a TopCount NXT plate reader, and ATP concentrations of samples were determined by comparison with ATP standards.
Virus production.
Pseudoviruses containing BlaM-Vpr were produced as described previously (10). Briefly, a 10-cm dish of HEK 293T/17 cells was transfected with JetPrime reagent (Polyplus Transfection, Illkirch-Graffenstaden, France) by using the following plasmids: the viral envelope glycoprotein vector (3 μg), pR8ΔEnv (2 μg), pMM130-BlaM-Vpr (2 μg), and pcRev (1 μg). Twelve hours after transfection, the medium was replaced with phenol red-free DMEM growth medium, and pseudoviruses were harvested 48 h after transfection. The supernatant was centrifuged for 5 min at 350 × g, filtered through a 0.45-μm filter membrane, and concentrated by using the LentiX reagent (Clontech Laboratories, Madison, WI) according to the manufacturer's recommendations. Pseudoviruses were then aliquoted and stored at −80°C. Infectious titers were determined by a β-galactosidase assay in TZM-bl cells (32).
Virus-cell fusion and viability assays.
TZM-bl, CEM-A, Jurkat, or CEM-NKR-CCR5-Luc cells (5 × 104 cells/well) were seeded into a Costar black clear-bottom 96-well plate (Corning, New York, NY) in complete phenol red-free medium the day before experimentation. Primary human macrophages were seeded 6 days before the experiment at a density of 4.5 × 104 cells per well. For Jurkat or CEM-NKR-CCR5-Luc cells, the plates were precoated with a poly-l-lysine solution (0.1 mg/ml). Viruses were added to cells at a multiplicity of infection (MOI) of ∼1 and centrifuged at 2,095 × g for 30 min at 4°C. Cells were washed and incubated for 90 min at 37°C in the absence or presence of the indicated inhibitors. A similar protocol was used for virus fusion with macrophages, except that BaL26 pseudoparticles were used at an MOI of ∼5 and cells were incubated for 2 h at 37°C. To create a temperature-arrested stage (TAS) of HIV-1 fusion, cells were incubated with viral pseudoparticles (pp) for 3 h at 20.5°C (TZM-bl cells with HXB2pp), 2 h at 18°C (TZM-bl cells with BaL26pp), or 2.5 h at 18°C (primary human macrophages with BaL26pp) and washed (or not) before incubation at 37°C. The fusion reaction was stopped by briefly chilling the plates on ice, and cells were loaded with the CCF4 acetoxymethyl ester (CCF4-AM) substrate (GeneBLAzer in vivo detection kit; Invitrogen), as described previously (10, 33). Cells were incubated overnight at 12°C, and the resulting changes in substrate fluorescence were measured by using a SpectraMax i3 plate reader (Molecular Devices, Sunnyvale, CA). The fusion activity was derived from the ratio of the blue and green emissions. Cell viability was measured by using a colorimetric CellTiter 96 AQueous assay (Promega) according to the manufacturer's recommendations.
Calcium flux measurements.
Cells were cultured on collagen-coated glass-bottom dishes (MatTek Corp., Ashland, MA) for 36 h in phenol red-free complete medium and loaded with 20 μM Fluo4-AM (Invitrogen) for 30 min at 37°C in complete medium buffered with 20 mM HEPES. Cells were washed two times, preincubated for 10 min in complete medium-HEPES in the presence of the indicated compounds, and imaged on a DeltaVision personal microscope (GE Healthcare). Images were acquired every 2 s for 4 to 8 min. After recording the baseline signal for ∼1 min, complete medium containing receptor agonists, as a control, or containing an agonist and an inhibitor/antagonist was added to the cells. At the end of the experiment, 2.5 μM A23187 was added to cells to determine the maximal Fluo4-AM fluorescence increase (as a loading control). Time-lapse images were analyzed by using ImageJ software (http://www.nih.gov/). The mean fluorescence from all cells in the field was determined after thresholding to remove background fluorescence and normalized to the signal elicited by A23187. For measurement of the calcium influx induced by chemokine-dependent coreceptor activation, the average normalized mean fluorescence from several (three to five) independent experiments was plotted after aligning the fluorescence traces to correct for differences in the time of agonist/antagonist addition. Due to a small fraction of cells exhibiting asynchronous calcium responses in the presence of gp120 or ATP in T cells, the mean fluorescence traces for each cell imaged in 3 experiments were normalized to the A23187 signal and plotted on the same graph.
Cell-cell fusion.
HEK 293T cells stably expressing DSP1 were transfected 24 h before the experiment with 3 μg of pCAGGS-HXB2-Env and 1 μg of pcRev using the JetPrime reagent. In parallel, cells expressing DSP2 were seeded onto a black-wall clear-bottom 96-well plate precoated with collagen. The next day, 293T-DSP1 cells were loaded with 40 μM Enduren (Promega) in Hanks balanced salt solution (HBSS) for 2 h at 37°C, detached by using nonenzymatic CellStripper solution (Cellgro), and overlaid onto cells expressing DSP2 in the absence or in the presence of the indicated inhibitors. Cell fusion was initiated by raising the temperature to 37°C for 2 h, and luciferase activity was measured by using a TopCount NXT plate reader (Perkin-Elmer).
P2XR expression.
Cells were lysed with radioimmunoprecipitation assay (RIPA) buffer (Sigma) supplemented with a protease inhibitor cocktail (Roche) and subjected to 10% SDS-PAGE. Proteins were transferred onto a nitrocellulose membrane at 4°C and incubated overnight at 4°C under agitation with primary rabbit antibodies to P2X1R (Abcam, San Francisco, CA) or mouse anti-tubulin antibodies (Sigma-Aldrich) diluted to 1:200 and 1:4,000, respectively, in phosphate-buffered saline (PBS) containing 0.1% Tween 20 and 2.5% milk. Membranes were washed three times and incubated with horseradish peroxidase (HRP)-conjugated mouse anti-rabbit (Cell Signaling Technology, Danvers, MA) or goat anti-mouse (Bio-Rad, Hercules, CA) antibodies and subjected to Western Lightning ECL analysis (Perkin-Elmer).
Statistical analysis.
Statistical significance was determined by using a two-tailed t test.
RESULTS
P2X1 receptor antagonists, but not P2X7 or P2Y11 receptor antagonists, inhibit HIV-1 fusion.
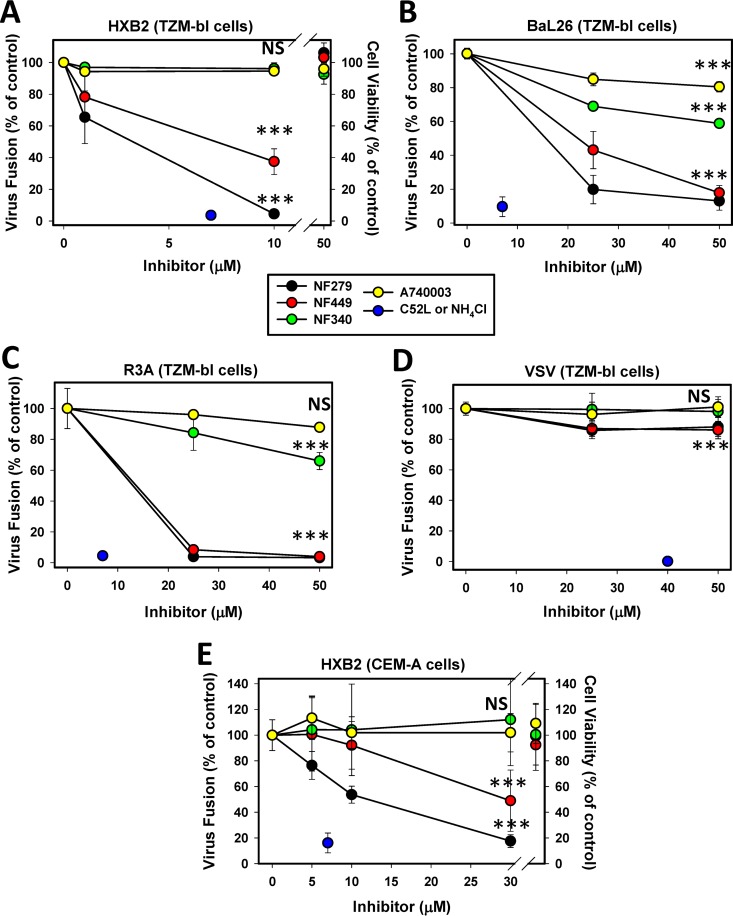
We first confirmed and expanded previously reported findings that P2X1R inhibitors selectively block HIV-1 fusion (13–15, 24). The effects of antagonists of P2X1 (NF279 and NF449), P2Y11 (NF340), and P2X7 (A740003) receptors on HIV-1 fusion with HeLa-derived and CD4+ T cells were tested. In agreement with our previously reported findings (24), the P2X1R antagonists NF279 and NF449 suppressed fusion between particles pseudotyped with CXCR4-tropic (HXB2), CCR5-tropic (BaL26), and dual-tropic (R3A) HIV-1 Env glycoproteins and TZM-bl cells in a dose-dependent manner (Fig. 1A to C). Both compounds were nearly equally potent and were more effective against fusion mediated by laboratory-adapted HXB2 Env than against fusion mediated by BaL26 or R3A Env. We have previously shown that this inhibition was not due to downregulation of CD4 or coreceptor expression (24). Both NF279 and NF449 inhibited HXB2 pseudovirus (HXB2pp) fusion with lymphoid CEM-A cells (Fig. 1E). However, high concentrations of NF279 were required to block fusion with CEM-A cells compared to TZM-bl cells, and NF449 was considerably less potent than NF279. NF279 also inhibited HXB2pp and BaL26pp fusion with CEM-A and Jurkat cells engineered to express CCR5 (data not shown).
FIG 1.
P2X1 receptor antagonists specifically inhibit HIV-1 fusion irrespective of coreceptor tropism. HIV-1 fusion with TZM-bl cells (A to D) and CEM-A cells (E) was measured by a BlaM assay using HXB2 or BaL26 pseudoviruses containing BlaM-Vpr. Pseudoviruses bearing X4-tropic HXB2 (A and E), R5-tropic Bal26 (B), or dual-tropic R3A (C) HIV-1 Env or VSV-G (D) were prebound to cells in the cold and allowed to fuse for 90 min at 37°C. The fusion reaction was carried out in either the absence or the presence of the P2 receptor antagonists NF279 or NF449 (for P2X1R), NF340 (for P2Y11R), and A740003 (for P2X7R). The resulting BlaM signals were normalized to the signal for the untreated control. Data are means and standard deviations of results from three independent experiments carried out in triplicate. TZM-bl (A) or CEM-A (E) cell viability following virus-cell coincubation in the absence or in the presence of the highest tested concentrations of P2 receptor antagonists is shown (right of the axis break). Data are mean normalized 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay readouts and standard deviations from two independent experiments carried out in triplicate. ***, P < 0.001; NS, not significant.
In contrast to P2X1R antagonists and consistent with our previously reported results (24), the P2X7 and P2Y11 receptor antagonists A740003 and NF340 exhibited marginal to no inhibitory activity against HIV-1 fusion with TZM-bl or CEM-A cells (Fig. 1). Importantly, none of the tested P2X1R, P2X7R, or P2Y11R antagonists considerably affected VSVpp fusion (Fig. 1D) or the viability of TZM-bl and CEM-A cells (Fig. 1A and E). These results appear to further support a specific role for P2X1R in HIV-1 entry/fusion.
Extracellular ATP or elevated cytosolic calcium levels are not required for HIV-1 fusion.
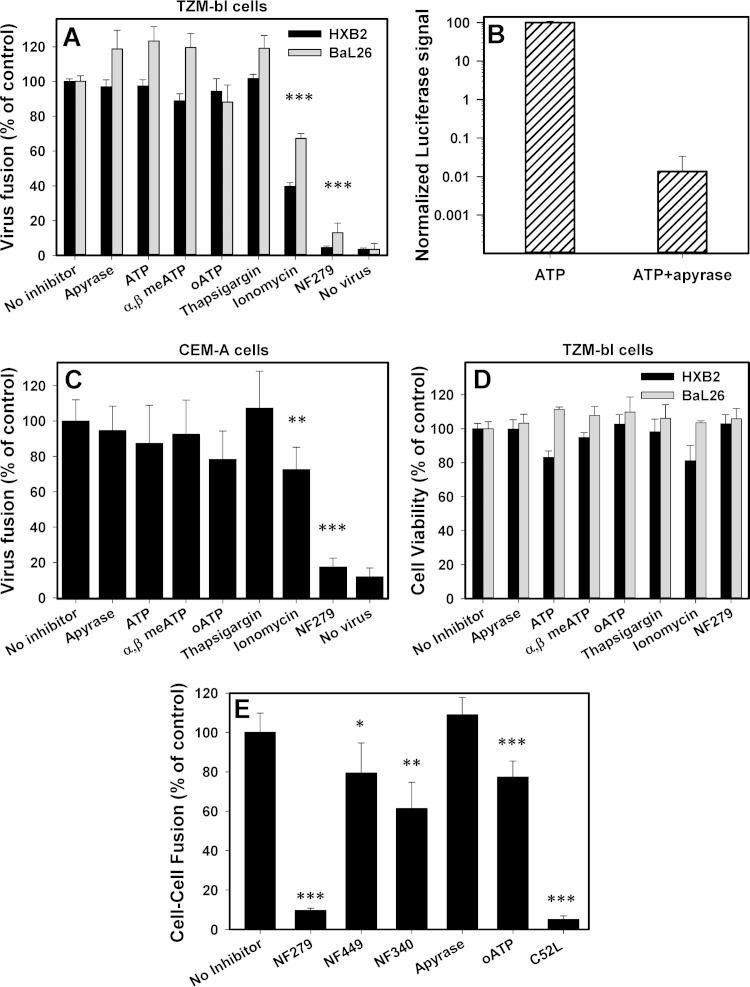
P2X receptors are activated upon the release of cellular ATP (e.g., see reference 34). We therefore asked whether depletion of extracellular ATP, presumably released by cells upon HIV binding (13, 14), or addition of exogenous ATP could modulate HIV-1 fusion. To address this question, virus-cell fusion experiments were carried out in the presence of a high dose of the ATP-degrading enzyme apyrase. Apyrase did not considerably affect HXB2pp or BaL26pp fusion with TZM-bl cells (Fig. 2A) at a concentration that effectively degraded 1 mM ATP within 30 min at 37°C (Fig. 2A and B). The addition of 0.5 μM ATP, a concentration that is known to activate P2X1R (35), did not augment virus-cell fusion. A relatively low multiplicity of infection was used in these experiments in order to avoid saturation of the BlaM signal. Even 100 μM ATP did not considerably facilitate HIV-1 fusion (data not shown). Likewise, neither apyrase nor exogenous ATP modulated HXB2pp fusion with CEM-A cells (Fig. 2C). The stable ATP analogs α,β-meATP, which is thought to selectively activate P2X1R (35), and oATP, a general inhibitor of purinergic receptors, did not noticeably affect fusion with either cell type. Whereas BaL26pp fusion was modestly promoted by ATP and α,β-meATP, this effect was also observed in samples treated with apyrase (Fig. 2A), suggesting that the modest enhancement of fusion in the presence of these reagents was not related to P2X1R activation.
FIG 2.
Extracellular ATP or an elevated cytosolic calcium concentration is not required for HIV-1 fusion. (A) Fusion of HXB2pp or BaL26pp with TZM-bl cells was allowed to proceed in the absence or in the presence of 10 U/ml of apyrase, 0.5 μM ATP or 100 μM α,β-meATP or oATP, 2 μM thapsigargin, 10 μM ionomycin, and 30 μM NF279. (B) Control for apyrase activity. ATP (1 mM) was incubated with 10 U/ml apyrase for 30 min at 37°C. The concentrations of ATP in mock-treated and apyrase-treated samples were quantified by using an ATPlite kit. (C) HXB2pp fusion with CEM-A cells under the conditions described above for panel A. Data are mean normalized BlaM activities (± standard deviations) from three independent experiments carried out in triplicate. (D) Cell viability measured in the same samples by an MTS assay. (E) HIV-1 Env-mediated cell-cell fusion assay. 293T-DSP1 cells transfected with HXB2 Env were coincubated with target N4X4-DSP2 cells in the absence or presence of 30 μM NF449 or NF340, 0.5 μM ATP, 100 μM oATP, 10 U/ml apyrase, or 7 μM C52L for 2 h at 37°C. Data points are the normalized mean luminescence values and standard deviations from three independent experiments carried out in triplicate. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Since much lower concentrations of ATP are required to activate P2X1R than P2X4R and P2X7R (35), the above-described results do not rule out the possibility that very low doses of ATP released locally through pannexin-1 hemichannels (36, 37) activate P2X1R. We therefore asked if downstream effects of P2X1R activation, specifically the elevation of the cytosolic calcium concentration, can modulate HIV-1 fusion. The calcium concentration in the cytoplasm during HIV-1 entry/fusion was increased by adding the calcium ionophore ionomycin or the sarco/endoplasmic reticulum calcium ATPase (SERCA) inhibitor thapsigargin. Thapsigargin did not significantly augment HXB2pp or BaL26pp fusion with TZM-bl or CEM-A cells, while ionomycin actually tended to inhibit fusion (Fig. 2A and C). Neither of the above-described conditions, including the presence of ionomycin or thapsigargin, affected cell viability (Fig. 2D). These results imply that elevation of the cytosolic calcium concentration alone does not promote HIV-cell fusion.
We also tested the effects of the above-described agents/inhibitors on HIV-1 Env-mediated cell-cell fusion, using a dual-split-protein assay (38, 39). In this assay, fusion is detected based upon de novo assembly of functional GFP and Renilla luciferase proteins from their respective complementary domains (designated DSP1 and DSP2) expressed separately in fusing partners. A robust luciferase signal was detected after coculture of HEK 293T cells stably expressing DSP1 that were transfected with HXB2 Env and target N4X4-DSP2 cells expressing DSP2 and CD4/CXCR4. NF279 effectively inhibited cell-cell fusion, while another P2X1R antagonist (NF449) and another P2Y11 antagonist (NF340) were less potent (Fig. 2E). Similar to virus-cell fusion, apyrase did not considerably affect the extent of cell-cell fusion, while oATP exhibited a mild deleterious effect. Our results thus show that P2X1R antagonists selectively inhibit HIV-cell and Env-mediated cell-cell fusion. However, NF279 is universally potent in both fusion assays involving different cell types, whereas NF449 exhibits variable potency. The different potencies of these two selective P2X1R antagonists against HIV-1 fusion raise concerns regarding a possible off-target effect of NF279.
Functional P2X receptors are present in epithelial but not lymphoid cell lines.
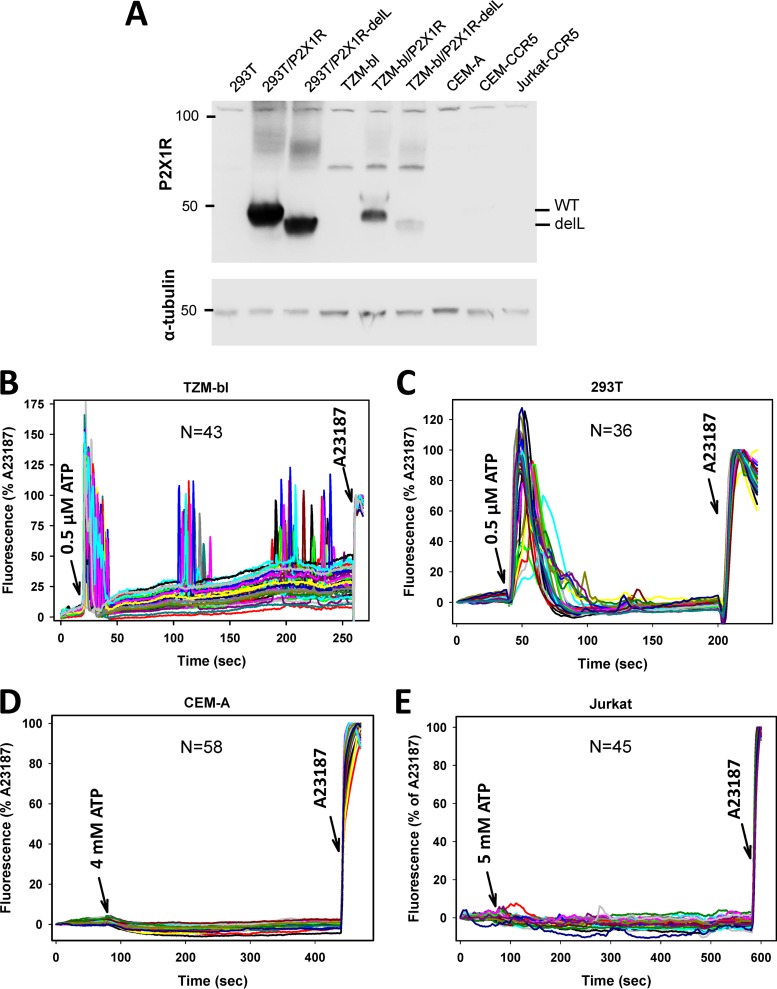
The insensitivity of HIV-1 fusion to exogenous ATP (Fig. 2) prompted us to examine the expression of the P2X1 receptor in TZM-bl, CEM-A, or Jurkat cells by Western blotting. None of these cell lines exhibited a detectable band corresponding to this receptor (Fig. 3A), but a robust P2X1R signal was present after ectopic expression of this receptor or its truncated dominant negative mutant (30) in HEK 293T or TZM-bl cells. The lack of P2X1R expression in epithelial cells is in agreement with data from previous reports (16, 34, 40). The undetectable levels of this receptor in lymphoid cells are somewhat surprising, since recent studies reported P2X1R expression in Jurkat and primary CD4+ T cells (14, 41). It is possible that the subclones of Jurkat and CEM-A cells used in our fusion experiments expressed exceptionally low levels of this receptor.
FIG 3.
Functional P2X receptors are present in epithelial but not lymphoid cell lines used in fusion experiments. (A) Western blotting does not detect P2X1 receptor expression in target cells of epithelial or lymphoid origin. (Top) P2X1R expression was examined in lysates of HEK 293T/17 (lane 1), TZM-bl (lane 4), and lymphoid (lanes 7 to 9) cells. As a positive control for P2X1R expression, lysates of HEK 293T/17 and TZM-bl cells transiently transfected with wild-type (WT) P2X1R (lanes 2 and 5) or the P2X1RdelL mutant (lanes 3 and 6) are shown. (Bottom) Loading control using α-tubulin antibodies. (B to E) Elevated intracellular calcium concentrations induced by ATP stimulation were measured in TZM-bl (B), HEK 293T/17 (C), CEM-A (D), and Jurkat-CCR5 (E) cells. Cells were loaded with the calcium indicator Fluo4, and fluorescence changes in complete growth medium at 37°C were visualized by time-lapse microscopy. Cells were imaged for ∼1 min to acquire the background signal, after which ATP was added at the indicated concentrations (left arrows). After 5 min of image acquisition, the calcium ionophore A23187 was added (right arrows) as a Fluo4 loading control. The curves show the normalized fluorescence intensities of individual cells from a representative experiment performed in triplicate. N is the number of traces shown in each panel.
We next asked if the cells used in the above-described fusion experiments express any functional P2X receptors by testing the ability of ATP to induce calcium signaling. Changes in intracellular calcium concentrations were monitored by imaging of cells loaded with the calcium indicator Fluo4-AM, as described in Materials and Methods. Exogenously added ATP elicited strong calcium signaling in TZM-bl and HEK 293T cells (Fig. 3B and C). Surprisingly, even very high (millimolar) concentrations of ATP did not elicit detectable responses in CEM-A or Jurkat cells (Fig. 3D and E). The lack of calcium signaling indicates that functional P2X1 receptors are not expressed on the lymphoid cell lines used in our experiments and thus argues against the involvement of these channels in HIV-1 fusion. T cells have been reported to respond to very high concentrations of exogenous ATP by increasing the cytosolic calcium levels (41, 42), but these responses likely involve P2X7 channels.
Collectively, the undetectable levels of P2X1R in epithelial and lymphoid cells and the lack of ATP-mediated signaling in lymphoid cells indicate that functional P2X1R is not required for HIV-1 fusion. These findings suggest that NF279 affects a different cellular target(s) at concentrations used to inhibit fusion.
P2X1R antagonists prevent functional engagement of CXCR4 and CCR5 coreceptors by HIV-1 Env.
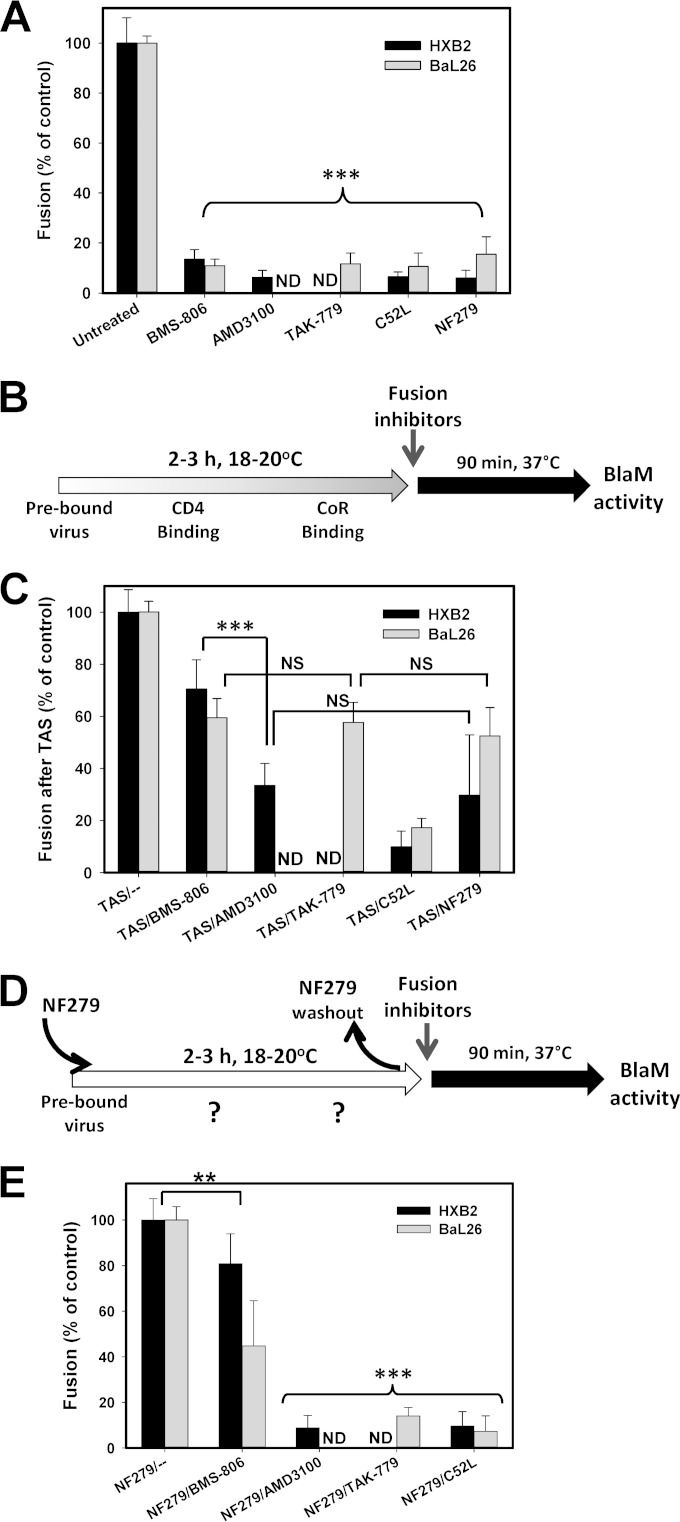
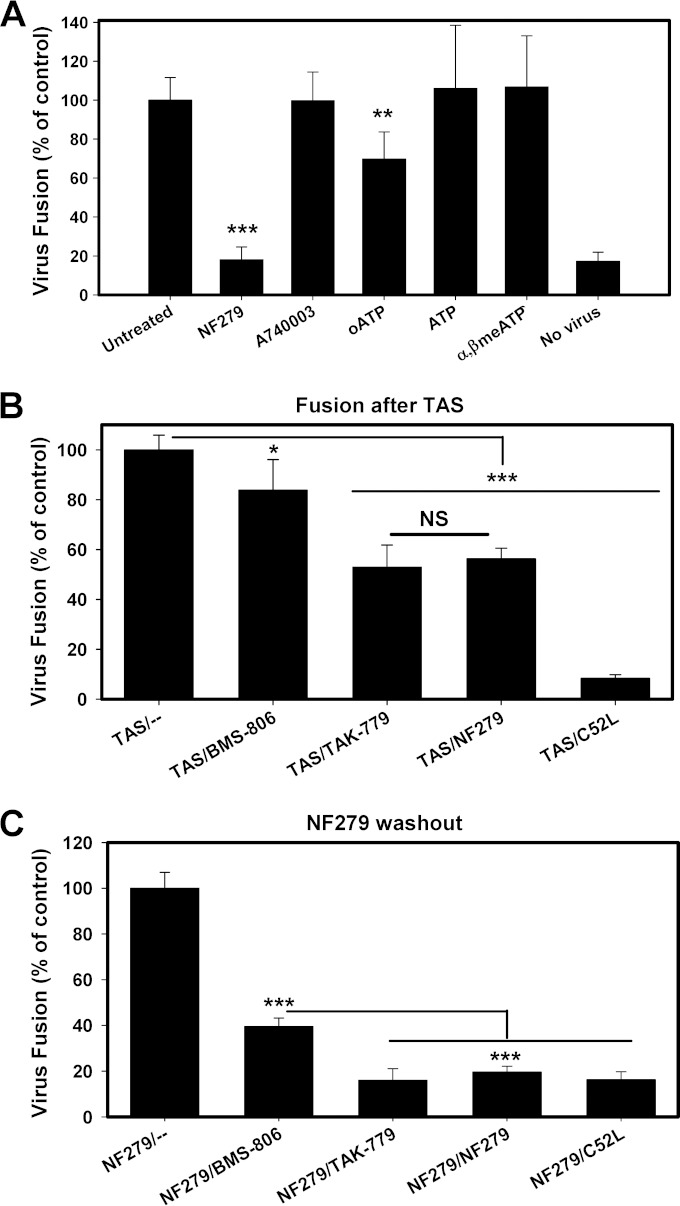
To define the mechanism by which NF279 blocks HIV-1 fusion, we reversibly arrested this process at an intermediate step referred to as a temperature-arrested stage (TAS) (10, 33, 43). This is achieved through a prolonged incubation of viruses with target cells at a temperature that is not permissive to fusion, typically ∼20°C; a subsequent temperature increase leads to fast and efficient fusion. At the TAS, a fraction of viruses functionally engages a requisite number of CD4 and coreceptor molecules, as evidenced by the acquired partial resistance of fusion to very high concentrations of inhibitors of receptor and coreceptor binding (10, 33, 43). We therefore created a TAS between HXB2pp or BaL26pp and TZM-bl cells and asked whether NF279 was still capable of blocking fusion after the formation of Env-CD4-coreceptor complexes.
The optimal conditions to create a TAS depend on the virus strain and on the CD4/coreceptor expression levels in target cells and therefore must be empirically optimized. In agreement with our previously reported results (10, 33), optimal TAS conditions for HXB2pp and BaL26pp were 2 to 3 h of preincubation at 20°C and 18°C, respectively (Fig. 4B). The TAS was probed by using concentrations of fusion inhibitors that effectively blocked uninterrupted HXB2pp and BaL26pp fusion at 37°C without the preincubation step (Fig. 4A). We found that, as expected, HIV-1 fusion at the TAS was somewhat more resistant to the CD4 inhibitor BMS-806 than to the CXCR4 or CCR5 antagonists AMD3100 and TAK-779 (Fig. 4C). Consistent with the lack of critical gp41 refolding at the TAS (44, 45), a 6-helix bundle inhibitor, the C52L peptide, abrogated HIV-cell fusion (Fig. 4C). HIV-1 fusion at the TAS remained partially sensitive to NF279, and the extent of inhibition was close to those achieved by AMD3100 and TAK-779 (Fig. 4C). The similar extents of fusion inhibition by specific coreceptor antagonists and NF279 indicate that the latter compound targets the coreceptor binding step of fusion, downstream of CD4 binding.
FIG 4.
P2X1R antagonists prevent functional engagement of CXCR4 and CCR5 by HIV-1 Env. HXB2pp or BaL26pp were prebound to TZM-bl cells in the cold, and fusion was initiated by shifting the temperature to 37°C for 90 min and measured by a BlaM assay. (A) Abrogation of HIV-1 fusion with TZM-bl cells with high concentrations of fusion inhibitors and NF279. The CD4 binding-site-targeting compound BMS-806 (10 μM); the CXCR4 and CCR5 binding inhibitors AMD3100 and TAK-779, respectively (both at 7 μM); the gp41 6-helix bundle inhibitor C52L (7 μM); and NF279 (10 μM for HXB2pp and 50 μM for BaL26pp) were added immediately prior to virus-cell coincubation at 37°C. (B) Schematic protocol for creating a temperature-arrested stage (TAS) of HIV-1 fusion prior to the addition of fusion inhibitors or NF279. CoR, coreceptor. (C) A TAS of fusion between HXB2pp or BaL26pp and TZM-bl cells was created as depicted in panel B, and fully inhibitory concentrations of fusion inhibitors or NF279 (at the concentrations described above for panel A) were added before raising the temperature to induce fusion. (D) Illustration of a TAS protocol similar to the one depicted in panel B, but incubation at a subthreshold temperature was carried out in the presence of a fully inhibitory concentration of NF279, after which time the antagonist was washed out and viruses/cells were incubated at 37°C in the absence or in the presence of high concentrations of fusion inhibitors. (E) Inhibition of HXB2pp and BaL26pp fusion with TZM-bl cells after preincubation at a subthreshold temperature in the presence of NF279, as depicted in panel D. Fusion inhibitors were added at the concentrations indicated above for panel A, immediately prior to shifting the temperature to 37°C. Data in panels A, C, and E are the normalized mean BlaM activities and standard deviations from three experiments carried out in triplicate. **, P < 0.01; ***, P < 0.001; NS, not significant; ND, not determined.
To further ascertain the exact step of HIV-1 fusion targeted by NF279, we employed an inhibitor substitution strategy, which we have previously implemented to define the mechanism of action of human α-defensin (46). In these experiments, HXB2pp or BaL26pp and target cells were preincubated at their respective subthreshold temperatures in the presence of a fully inhibitory concentration of NF279. At the end of incubation, NF279 was washed away, and a fully inhibitory concentration of CD4 or the coreceptor inhibitor was added prior to induction of fusion by shifting cells to 37°C (Fig. 4D). In this protocol, if NF279 blocks CD4 binding, fusion would remain sensitive to both BMS-806 and coreceptor inhibitors. If, on the other hand, coreceptor binding is blocked by NF279, then fusion is expected to remain sensitive to coreceptor antagonists but not to CD4 inhibitors. As shown in Fig. 4E, HXB2pp and BaL26pp fusion remained partially resistant to BMS-806, similarly to the conventional TAS protocol (Fig. 4C), showing that NF279 did not interfere with the Env-CD4 binding step. However, substitution of NF279 with AMD3100 and TAK-779 inhibited HXB2pp and BaL26pp fusion, respectively, to the levels observed with the 6-helix bundle inhibitor C52L (Fig. 4E). These findings clearly demonstrate that NF279 blocks HIV-1 Env interactions with both CXCR4 and CCR5.
NF279 inhibits HIV-1–coreceptor engagement in primary human macrophages.
To extend the above-described observations to more physiologically relevant cells, we examined the effects of P2 receptor inhibitors and ATP derivatives on BaL26pp fusion with monocyte-derived human macrophages (MDMs). These experiments were carried out by using methods similar to those described above for TZM-bl cells. Briefly, pseudoviruses were prebound to MDMs in the cold and incubated at 37°C for 2 h. This protocol resulted in a robust BlaM signal. Similarly to TZM-bl cells, NF279, but not ATP, α,β-meATP, or the P2X7R inhibitor A740003, suppressed HIV-1 fusion with MDMs (Fig. 5A). The only difference was that oATP modestly reduced the efficiency of fusion with MDMs. The inhibitory effects of oATP (however modest) and NF279 on HIV-1 fusion with MDMs are consistent with data from a previous study (13).
FIG 5.
NF279 inhibits HIV-1–coreceptor engagement in primary human macrophages. (A) Fusion of BaL26pp with MDMs was allowed to proceed in the absence or in the presence of 50 μM NF279 or 100 μM each A740003, ATP, α,β-meATP, and oATP. (B) A TAS of fusion between BaL26pp and MDMs was created, essentially as described in the legends of Fig. 4B and C. BaL26 pseudoviruses were prebound to MDMs and incubated for 2.5 h at a low temperature (18°C), after which time a fully inhibitory concentration of BMS-806, TAK-779, C52L, or NF279 (50 μM) was added. Fusion was then induced by raising the temperature to 37°C. (C) NF279 washout experiments were performed by creating a TAS of fusion between BaL26pp and MDMs in the presence of a fully inhibitory concentration of NF279, as described in the legends of Fig. 4D and E. Before the temperature was raised to allow fusion, NF279 was replaced with fully inhibitory concentrations of HIV-1 fusion inhibitors. Data in panels A to C are the normalized mean BlaM signals and standard deviations from two independent experiments carried out in triplicate. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant.
To address whether NF279 inhibits HIV-1 fusion with macrophages through interfering with Env-coreceptor binding, we examined the efficiency of NF279 inhibition after BaL26 Env forms ternary complexes with CD4 and CCR5 on macrophages. Using a protocol similar to that used to capture a TAS with TZM-bl cells, we observed that BaL26 successfully engaged CD4 and CCR5 on MDM cells at a reduced temperature. The formation of ternary complexes was apparent from the partial resistance of fusion to fully inhibitory doses of compounds targeting the CD4 and CCR5 binding steps of HIV-1 fusion (Fig. 5B). The identical degrees of fusion inhibition by saturating concentrations of NF279 and TAK-779 added at the TAS (Fig. 5B) support the notion that NF279 blocks the CCR5 binding step.
To verify this conclusion, NF279 washout experiments were performed, whereby MDMs were incubated for an extended period of time at a subthreshold temperature for fusion in the presence of NF279 (for the experimental protocol, see the legend of Fig. 4D). At this point, the NF279-imposed block of fusion could be readily reversed by simply washing off the inhibitor prior to raising the temperature (Fig. 5C). When NF279 was replaced with TAK-779, fusion with macrophages was abrogated, further confirming that NF279 inhibits the CCR5 engagement step. Interestingly, substitution of NF279 with BMS-806 partially inhibited BaL26pp fusion with TZM-bl cells and MDMs (Fig. 4E and 5C). Since we have previously shown that NF279 does not downregulate CD4 expression (24), it is possible that NF279 can also interfere with Env-CD4 binding, at least in the case of BaL26. Collectively, the above-described results with MDMs confirm and expand our conclusion that NF279 antagonizes Env-coreceptor interactions.
The P2X1R inhibitor NF279 is a dual CXCR4/CCR5 antagonist.
Functional dissection of the HIV-1 fusion steps targeted by NF279 (Fig. 4) shows that this compound blocks Env-coreceptor interactions but does not show whether this effect is due to binding to coreceptors or to the coreceptor binding site on gp120. We therefore examined the ability of NF279 to interact with CCR5 and CXCR4. Since small-molecule coreceptor antagonists are known to inhibit chemokine-mediated calcium signaling (e.g., see references 31 and 47), we asked if NF279 can also modulate this activity in TZM-bl and CEM-A cells. Elevation of the intracellular calcium concentration was visualized by loading the cells with Fluo4-AM and measuring the increases in cell fluorescence in response to antagonists/agonists. To control for the Fluo4-AM loading efficiency, 2.5 μM A23187 was always added at the end of each experiment to evoke maximal increases in fluorescence, and the agonist-mediated signals were normalized to the signal in the presence of the ionophore.
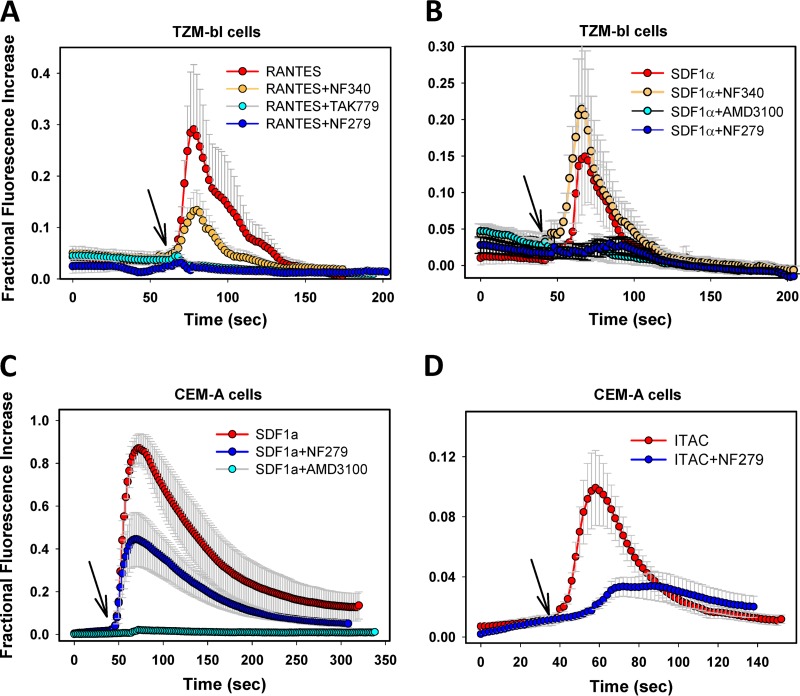
Both RANTES (CCR5 agonist) and SDF-1α (CXCR4 agonist) elicited robust transient increases of cytosolic calcium concentrations in TZM-bl cells (Fig. 6A and B; see also Movie S1 in the supplemental material). As expected, these responses were abrogated when RANTES or SDF-1α was applied together with the respective antagonists of CCR5 and CXCR4 coreceptors, TAK-779 and AMD3100. Importantly, NF279 abrogated Ca2+ influx mediated by either SDF-1α or RANTES, whereas the P2Y11 antagonist NF340 exhibited minor to no effects on cytosolic calcium levels (Fig. 6A and B; see also Movie S2 in the supplemental material). In CEM-A cells, SDF-1α application elicited extremely strong calcium responses at levels similar to those elicited by A23187 (Fig. 6C). AMD3100 fully blocked SDF-1α-mediated elevations of calcium concentrations in these cells, whereas high concentrations of NF279 partially inhibited this response. The antagonistic effect of NF279 was more apparent at lower doses of SDF-1α, whereas elevated calcium levels in CEM-A cells induced by a high dose of the agonist were only marginally inhibited (data not shown). Together, our results show that NF279 acts as a dual CXCR4/CCR5 antagonist in TZM-bl cells. In comparison, the ability of NF279 to antagonize CXCR4 signaling in CEM-A cells is limited, so it appears to act as a partial antagonist. Of note, incomplete inhibition of CXCR4 signaling in CEM-A cells seems to correlate with the lower potencies of NF279 and especially of NF449 against HXB2pp fusion with these cells than with TZM-bl cells (Fig. 1).
FIG 6.
The P2X1R inhibitor NF279 acts as a dual CXCR4/CCR5 antagonist. Calcium influx was induced upon CXCR4 and CCR5 activation with 25 nM SDF-1α and 300 nM RANTES, respectively, in TZM-bl cells (A and B) and CEM-A cells (C and D). Cells were loaded with the calcium indicator Fluo4 and imaged in complete medium for ∼1 min prior to the addition of chemokines in the absence or in the presence of 30 μM the P2 receptor antagonist NF279 or NF340 (arrows). In control experiments, SDF-1α and RANTES were applied together with 7 μM the CXCR4 antagonist AMD3100 or the CCR5 antagonist TAK-779 (see also Movies S1 and S2 in the supplemental material). Alternatively, CXCR7 on CEM-A cells was activated with 60 nM I-TAC in the absence or in the presence of NF279 (D). The curves represent the mean fluorescence intensities of TZM-bl (n = 50) or CEM-A (n = 70) cells normalized to the fluorescence signal after the addition of A23187 (2.5 μM) from three experiments conducted in triplicate (± standard deviations).
Considering that NF279 antagonizes calcium signaling mediated by both CCR5 and CXCR4, we asked if this compound could also interfere with the activities of other chemokine receptors. NF279 blocked Ca2+ influx elicited by the CXCR3/CXCR7 agonist I-TAC (interferon-inducible T cell alpha chemoattractant) in CEM-A cells (Fig. 6D). Of note, the average I-TAC-mediated calcium signal was much weaker than that induced by SDF-1α due to a small fraction of cells responding to I-TAC. Collectively, the above-described results indicate that NF279 interacts with chemokine receptors and not with the gp120 coreceptor binding site. This compound most likely blocks chemokine receptor signaling by binding to these proteins. The alternative possibility that NF279 blocks calcium influx by targeting a downstream step of chemokine receptor signaling appears unlikely considering that this compound (i) blocks HIV-1–coreceptor interactions and (ii) is relatively polar and thus should not permeate the plasma membrane.
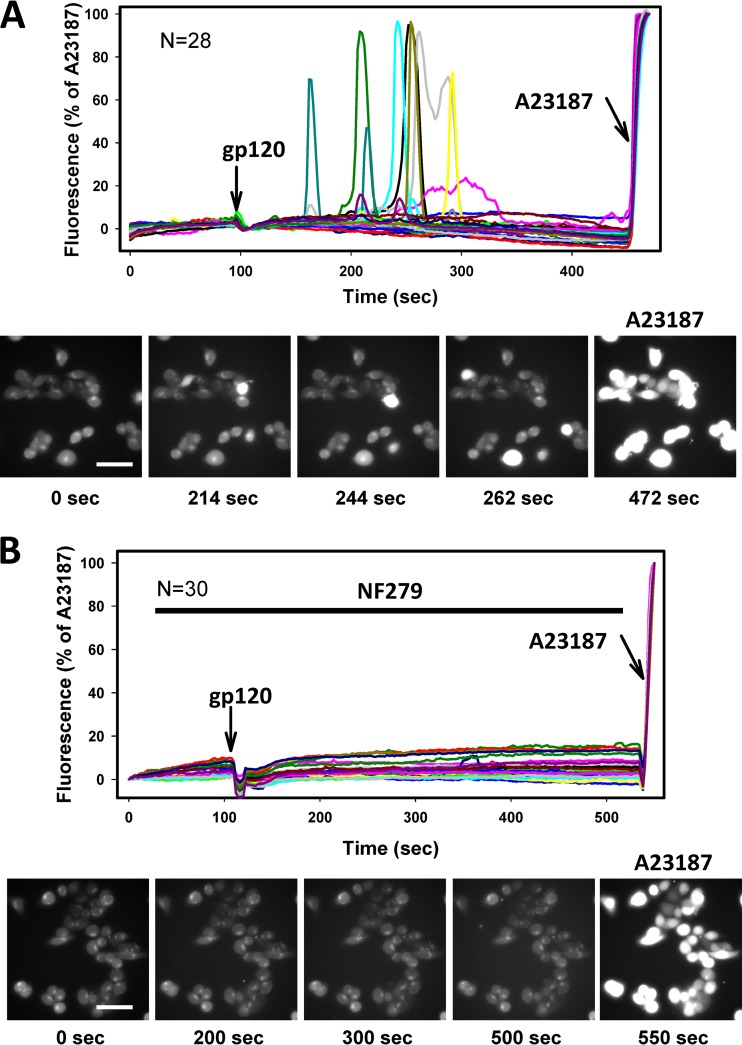
We next asked if NF279 could also interfere with gp120 signaling. The addition of recombinant monomeric gp120 derived from the X4-tropic IIIB strain to TZM-bl cells resulted in transient increases in cytosolic calcium levels. Although calcium responses were observed in only a fraction of cells, the cells responding to this treatment exhibited profound increases in fluorescence (Fig. 7A). In contrast, no calcium signaling was detected when gp120 was added together with NF279 (Fig. 7B), demonstrating that this compound antagonizes gp120-mediated activation of CXCR4. Our results thus show that NF279 is a promiscuous antagonist of several chemokine receptors that is capable of blocking functional interactions between gp120 and coreceptors.
FIG 7.
Calcium signaling induced by monomeric gp120 is inhibited by NF279. Changes in cytosolic calcium levels in TZM-bl cells were measured as described in Materials and Methods and the legend of Fig. 6. Recombinant gp120 (130 nM) (arrows) was applied to cells in the absence (A) or in the presence (B) of 30 μM NF279. Graphs in panels A and B show changes in the Fluo4 signals from individual cells (n = 28 and n = 30, respectively) after normalization to the signal elicited by 2.5 μM A23187. Fluorescence traces are from a representative experiment done in triplicate. Image panels below the graphs show snapshots of cells at the indicated time points (bar, 50 μm). The last image in each panel was acquired after the addition of A23187.
The determinants of NF279 and TAK-779 inhibitory activities are similar but distinct from those of other CCR5 antagonists.
Among a panel of CCR5 mutants examined by different laboratories, a G163R substitution at the interface between the second extracellular loop and the transmembrane domain 4 reduces the affinity of gp120 for CCR5 and thereby attenuates virus fusion/infection (48, 49). Nonetheless, JGR.H11 cells expressing high levels of the G163R CCR5 mutant support efficient HIV-1 fusion/infection (49). Since this mutation appears to selectively attenuate the antiviral activity of aplaviroc (48) but only modestly affects the activity of the dual CCR5 and CCR2b antagonist TAK-779 (50, 51), we asked whether the dual-coreceptor antagonist NF279 maintains the ability to block HIV-1 entry through the mutant coreceptor.
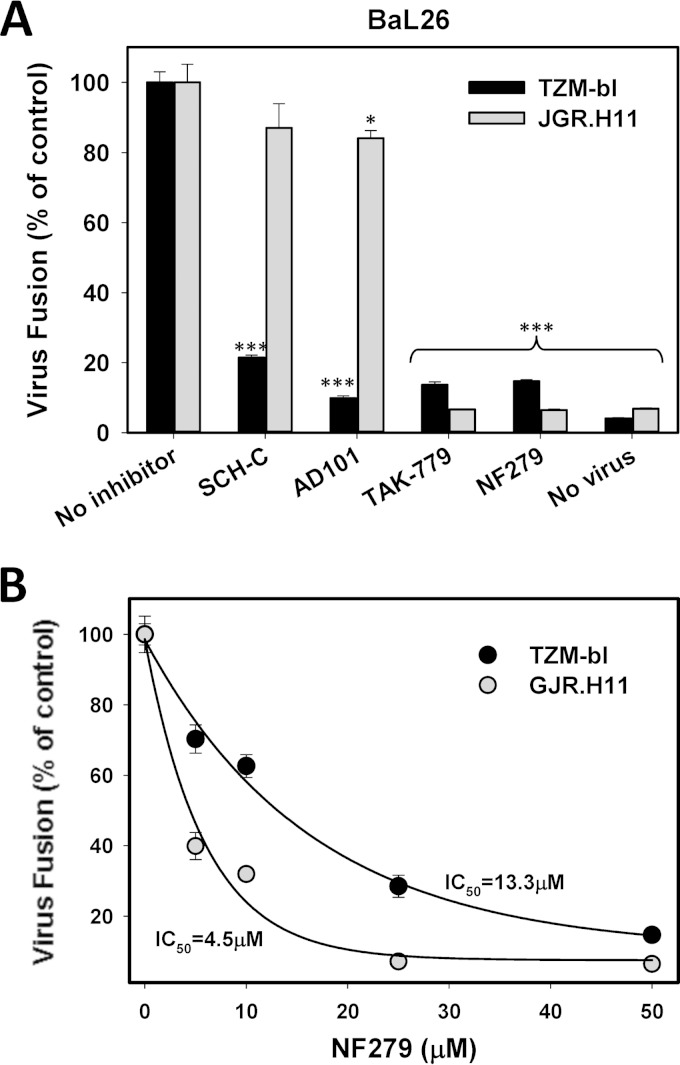
Fusion of BaL26pp with TZM-bl cells or with JGR.H11 cells expressing the G163R CCR5 mutant was allowed to proceed in the presence of different CCR5 antagonists. In agreement with data from a previous report (49), BaL26pp fusion with TZM-bl cells was twice as efficient as fusion to JGR.H11 cells (data not shown). Whereas high concentrations of the three CCR5 antagonists Schering C (Sch-C), AD101, and TAK-779 blocked fusion with TZM-bl cells, only TAK-779 antagonized HIV-1 fusion with cells expressing mutant CCR5 (Fig. 8A). Interestingly, similarly to TAK-779, NF279 blocked BaL26pp fusion with both cell lines. In fact, NF279 was ∼3-fold more potent against HIV-1 fusion in cells expressing the G163R CCR5 mutant than in those expressing wild-type CCR5 (Fig. 8B). These results reveal differences in the mechanisms of action of NF279 and specific CCR5 antagonists while highlighting similarities with the dual CCR5 and CCR2b antagonist TAK-779.
FIG 8.
NF279 and TAK-779, but not Sch-C or AD101, inhibit HIV-1 fusion mediated by the attenuated CCR5 mutant. Virus-cell fusion was initiated as described in Materials and Methods and measured by using a BlaM assay. (A) BaL26pp were allowed to fuse with TZM-bl or GJR.H11 cells in the absence or in the presence of the 5 μM the CCR5 antagonist Sch-C, AD101, or TAK-779 or in the presence of 50 μM NF279. *, P < 0.05; ***, P < 0.001. (B) Dose-response curves for NF279-mediated inhibition of BaL26pp fusion with TZM-bl or GJR.H11 cells. Data points are the mean normalized BlaM activities and standard deviations from two experiments conducted in triplicate. IC50, 50% inhibitory concentration.
DISCUSSION
To undergo fusion, HIV-1 strictly requires CD4 and cognate coreceptors. However, additional host factors and cell signaling may modulate this process (3–12). Specifically, several reports have implicated P2 receptors in HIV-1 entry/fusion (13–15, 24). It has been proposed that Env-CD4/coreceptor engagement provides a signal for ATP release from target cells, which in turn activates purinergic receptors and raises intracellular calcium levels. Exactly how these events aided HIV-1 entry remained unclear. Although plasma membrane depolarization resulting from calcium influx has been envisioned to promote HIV-1 fusion (14), we are unaware of direct experimental evidence linking HIV-1-mediated membrane depolarization to enhanced viral fusion.
Our results strongly argue against the involvement of P2 receptors in HIV-1 fusion. First, P2X1R expression on the cells used in our fusion assays was undetectable, and ATP failed to mediate calcium signaling in T cell lines supporting efficient HIV-1 entry. Second, data from mechanistic studies of P2X1R inhibitors imply that the observed inhibition of HIV-1 entry/fusion by NF279 is due to an off-target effect of high concentrations of this antagonist. Indeed, whereas 10 to 100 μM NF279 or NF449 is required to inhibit HIV-1 fusion with different target cells (13–15, 24), nanomolar concentrations of these compounds have been reported to block P2X1 channels (see reference 35 and references therein).
Several lines of evidence support the conclusion that NF279 (and possibly other P2X1R inhibitors) acts as a dual HIV-1 coreceptor antagonist. First, NF279 prevents functional gp120 binding to CXCR4 or CCR5 in fusion assays but loses potency once Env-CD4-coreceptor complexes are formed. This effect was observed in both HeLa-derived target cells and primary human macrophages. Second, NF279 potently inhibits specific CXCR4 and CCR5 ligand-mediated calcium responses in HeLa-derived cells and partially antagonizes SDF1-α-mediated CXCR4 signaling in CEM-A cells. The incomplete inhibition of CXCR4 signaling in CEM-A cells by NF279 may reflect a greater heterogeneity of this coreceptor due to posttranslational modifications that are different from those in TZM-bl cells. The existence of distinct pools of CCR5 and CXCR4 on the cell surface was previously documented (52–57). In addition, it appears that NF279 can promiscuously interfere with the function of other chemokine receptors, as exemplified by its ability to block calcium influx elicited by the CXCR3/CXCR7 agonist I-TAC in T cells. These broad-spectrum effects of NF279 on different chemokine receptors show that caution should be exercised when interpreting the results obtained with micromolar doses of this P2X1R antagonist.
Although pharmacological evidence supports the role of P2X1R in HIV-1 fusion (13–15, 24), small interfering RNA (siRNA) knockdown of P2X1R in target cells appears to actually promote HIV-1 Env-mediated cell-cell fusion (14). In contrast, depletion of pannexin-1, P2Y2 receptor, and Pyk2 kinase in target cells attenuated HIV-1 infection and Env-mediated cell-cell fusion (14). Unfortunately, the possibility that knockdown of these proteins can diminish CD4 and/or coreceptor expression in target cells was not addressed in that study. Additional evidence implicating P2Y2 came from experiments with nonspecific inhibitors of P2Y2 receptors and pannexin-1 and usually involved prolonged incubation (24 to 48 h) with high doses of inhibitors (14). Under our conditions, HIV-1 fusion was not inhibited by the P2Y11 inhibitor NF340 (Fig. 1) or the nonspecific P2Y2 inhibitor kaempferol, which has been employed previously (14) (data not shown). The lack of a considerable inhibitory effect of the P2Y receptor inhibitors on HIV-1 infection of CD4+ T cells was also observed previously (15). However, our results do not rule out the role of purinergic receptor signaling in HIV-1 infection. We surmise that long-term inactivation of P2 receptors or downstream signaling pathways can modulate postentry steps of HIV-1 infection, as documented previously (13, 14).
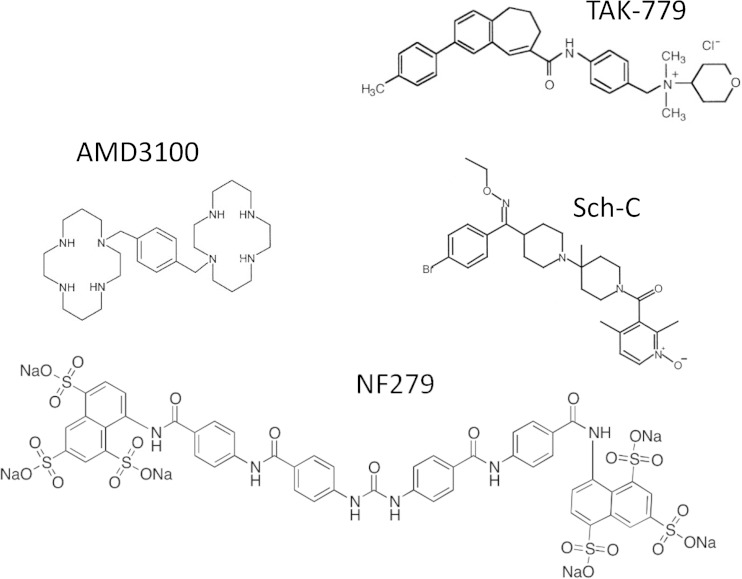
The mechanism by which conventional coreceptor antagonists block HIV-1 fusion has been extensively studied. It appears that these compounds bind to a conserved hydrophobic pocket and allosterically block the functional engagement of coreceptors by HIV-1 Env (e.g., see references 48 and 58–60). Our data imply that NF279 binds to CXCR4 and CCR5 and prevents the functional engagement of these coreceptors by HIV-1 via an unknown mechanism. Although the dual-coreceptor antagonist TAK-779 binds to the same hydrophobic pocket as that of highly specific CCR5 antagonists, the TAK-779-interacting residues do not seem to fully overlap residues interacting with vicriviroc or Sch-C (60). Several CCR5 residues, including G163, are involved in a hydrogen bond network that appears to be essential for maintaining proper protein conformation (48). Consequently, the G163R mutation markedly reduces the binding of antagonists to mutant CCR5 (48, 61). In fact, aplaviroc-CCR5 interactions have been mapped to several residues lining the conserved hydrophobic pocket and to G163 (48). The fact that both TAK-779 and NF279, but not other CCR5-specific antagonists, effectively inhibit HIV-1 fusion mediated by the G163R mutant (Fig. 8) highlights the similarities between TAK-779 and NF279. However, NF279 appears too large to fit into the hydrophobic pocket on coreceptors occupied by smaller antagonists (Fig. 9). This elongated symmetric molecule consists of two arms with three sulfates at each end, which is quite different from the structures and net charges of conventional coreceptor antagonists (Fig. 9). An intriguing possibility is that the NF279 sulfates can compete with the N-terminal tyrosine sulfates on CCR5 and CXCR4 for binding to gp120. Although we currently have no evidence for an NF279-gp120 interaction, the ability of NF279 to block CXCR4 and CCR5 signaling does not rule out its binding to gp120. Competition with coreceptors for gp120 binding is consistent with the ability of NF279 to inhibit HIV-1 fusion with cells expressing attenuated CCR5 carrying the G163R mutation near the second extracellular loop (Fig. 8). The mechanism by which NF279 and its analogs inhibit gp120-coreceptor interactions awaits further investigations.
FIG 9.
Structures of compounds NF279, TAK-779, Sch-C, and AMD3100.
Purinergic receptor inhibitors appear to lack significant toxicity in vivo and are currently being developed as drug candidates to treat rheumatoid arthritis and neuropathic pain (62, 63). However, while the dual-coreceptor-antagonistic activity of NF279 can appear desirable for blocking R5- and X4-tropic HIV-1 infection, the promiscuous inhibition of multiple chemokine receptors by this compound may not be a good trait for a drug candidate.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Hoylaert for the P2X1 expression vectors and David Kabat for the JGR.H11 cell line. We are grateful to David Kabat for stimulating discussions and Caleb Mason and other members of the Melikyan laboratory for critical readings of the manuscript.
This work was supported by NIH R01 grants GM108480 and GM054787 to G.B.M. and grant R01 AI058828 to P.S.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01178-15.
REFERENCES
- 1.Wilen CB, Tilton JC, Doms RW. 2012. Molecular mechanisms of HIV entry. Adv Exp Med Biol 726:223–242. doi: 10.1007/978-1-4614-0980-9_10. [DOI] [PubMed] [Google Scholar]
- 2.Melikyan GB. 2011. Membrane fusion mediated by human immunodeficiency virus envelope glycoprotein. Curr Top Membr 68:81–106. doi: 10.1016/B978-0-12-385891-7.00004-0. [DOI] [PubMed] [Google Scholar]
- 3.Jimenez-Baranda S, Gomez-Mouton C, Rojas A, Martinez-Prats L, Mira E, Ana Lacalle R, Valencia A, Dimitrov DS, Viola A, Delgado R, Martinez AC, Manes S. 2007. Filamin-A regulates actin-dependent clustering of HIV receptors. Nat Cell Biol 9:838–846. doi: 10.1038/ncb1610. [DOI] [PubMed] [Google Scholar]
- 4.Puri A, Rawat SS, Lin HM, Finnegan CM, Mikovits J, Ruscetti FW, Blumenthal R. 2004. An inhibitor of glycosphingolipid metabolism blocks HIV-1 infection of primary T-cells. AIDS 18:849–858. doi: 10.1097/00002030-200404090-00002. [DOI] [PubMed] [Google Scholar]
- 5.Barrero-Villar M, Barroso-Gonzalez J, Cabrero JR, Gordon-Alonso M, Alvarez-Losada S, Munoz-Fernandez MA, Sanchez-Madrid F, Valenzuela-Fernandez A. 2008. PI4P5-kinase Ialpha is required for efficient HIV-1 entry and infection of T cells. J Immunol 181:6882–6888. doi: 10.4049/jimmunol.181.10.6882. [DOI] [PubMed] [Google Scholar]
- 6.Barrero-Villar M, Cabrero JR, Gordon-Alonso M, Barroso-Gonzalez J, Alvarez-Losada S, Munoz-Fernandez MA, Sanchez-Madrid F, Valenzuela-Fernandez A. 2009. Moesin is required for HIV-1-induced CD4-CXCR4 interaction, F-actin redistribution, membrane fusion and viral infection in lymphocytes. J Cell Sci 122:103–113. doi: 10.1242/jcs.035873. [DOI] [PubMed] [Google Scholar]
- 7.Gordon-Alonso M, Rocha-Perugini V, Alvarez S, Moreno-Gonzalo O, Ursa A, Lopez-Martin S, Izquierdo-Useros N, Martinez-Picado J, Munoz-Fernandez MA, Yanez-Mo M, Sanchez-Madrid F. 2012. The PDZ-adaptor protein syntenin-1 regulates HIV-1 entry. Mol Biol Cell 23:2253–2263. doi: 10.1091/mbc.E11-12-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon-Alonso M, Yanez-Mo M, Barreiro O, Alvarez S, Munoz-Fernandez MA, Valenzuela-Fernandez A, Sanchez-Madrid F. 2006. Tetraspanins CD9 and CD81 modulate HIV-1-induced membrane fusion. J Immunol 177:5129–5137. doi: 10.4049/jimmunol.177.8.5129. [DOI] [PubMed] [Google Scholar]
- 9.Thali M. 2011. Tetraspanin functions during HIV-1 and influenza virus replication. Biochem Soc Trans 39:529–531. doi: 10.1042/BST0390529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. 2009. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 137:433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashi Y, Nemoto-Sasaki Y, Tanikawa T, Oka S, Tsuchiya K, Zama K, Mitsutake S, Sugiura T, Yamashita A. 2014. Sphingomyelin synthase 2, but not sphingomyelin synthase 1, is involved in HIV-1 envelope-mediated membrane fusion. J Biol Chem 289:30842–30856. doi: 10.1074/jbc.M114.574285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harouse JM, Bhat S, Spitalnik SL, Laughlin M, Stefano K, Silberberg DH, Gonzalez-Scarano F. 1991. Inhibition of entry of HIV-1 in neural cell lines by antibodies against galactosyl ceramide. Science 253:320–323. doi: 10.1126/science.1857969. [DOI] [PubMed] [Google Scholar]
- 13.Hazleton JE, Berman JW, Eugenin EA. 2012. Purinergic receptors are required for HIV-1 infection of primary human macrophages. J Immunol 188:4488–4495. doi: 10.4049/jimmunol.1102482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seror C, Melki MT, Subra F, Raza SQ, Bras M, Saidi H, Nardacci R, Voisin L, Paoletti A, Law F, Martins I, Amendola A, Abdul-Sater AA, Ciccosanti F, Delelis O, Niedergang F, Thierry S, Said-Sadier N, Lamaze C, Metivier D, Estaquier J, Fimia GM, Falasca L, Casetti R, Modjtahedi N, Kanellopoulos J, Mouscadet JF, Ojcius DM, Piacentini M, Gougeon ML, Kroemer G, Perfettini JL. 2011. Extracellular ATP acts on P2Y2 purinergic receptors to facilitate HIV-1 infection. J Exp Med 208:1823–1834. doi: 10.1084/jem.20101805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swartz TH, Esposito AM, Durham ND, Hartmann BM, Chen BK. 2014. P2X-selective purinergic antagonists are strong inhibitors of HIV-1 fusion during both cell-to-cell and cell-free infection. J Virol 88:11504–11515. doi: 10.1128/JVI.01158-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burnstock G, Knight GE. 2004. Cellular distribution and functions of P2 receptor subtypes in different systems. Int Rev Cytol 240:31–304. doi: 10.1016/S0074-7696(04)40002-3. [DOI] [PubMed] [Google Scholar]
- 17.Webb TE, Kaplan MG, Barnard EA. 1996. Identification of 6H1 as a P2Y purinoceptor: P2Y5. Biochem Biophys Res Commun 219:105–110. doi: 10.1006/bbrc.1996.0189. [DOI] [PubMed] [Google Scholar]
- 18.Janssens R, Boeynaems JM, Godart M, Communi D. 1997. Cloning of a human heptahelical receptor closely related to the P2Y5 receptor. Biochem Biophys Res Commun 236:106–112. doi: 10.1006/bbrc.1997.6895. [DOI] [PubMed] [Google Scholar]
- 19.Boarder MR, Weisman GA, Turner JT, Wilkinson GF. 1995. G protein-coupled P2 purinoceptors: from molecular biology to functional responses. Trends Pharmacol Sci 16:133–139. doi: 10.1016/S0165-6147(00)89001-X. [DOI] [PubMed] [Google Scholar]
- 20.Barnard EA, Webb TE, Simon J, Kunapuli SP. 1996. The diverse series of recombinant P2Y purinoceptors. Ciba Found Symp 198:166–180; discussion 180–168. [DOI] [PubMed] [Google Scholar]
- 21.Erb L, Liao Z, Seye CI, Weisman GA. 2006. P2 receptors: intracellular signaling. Pflugers Arch 452:552–562. doi: 10.1007/s00424-006-0069-2. [DOI] [PubMed] [Google Scholar]
- 22.Mei L, Du W, Gao W, Mei QB. 2010. Purinergic signaling: a novel mechanism in immune surveillance. Acta Pharmacol Sin 31:1149–1153. doi: 10.1038/aps.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schachter J, Delgado KV, Barreto-de-Souza V, Bou-Habib DC, Persechini PM, Meyer-Fernandes JR. 2015. Inhibition of ecto-ATPase activities impairs HIV-1 infection of macrophages. Immunobiology 220:589–596. doi: 10.1016/j.imbio.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Marin M, Du Y, Giroud C, Kim JH, Qui M, Fu H, Melikyan GB. 2015. High-throughput HIV-cell fusion assay for discovery of virus entry inhibitors. Assay Drug Dev Technol 13:155–166. doi: 10.1089/adt.2015.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tremblay M, Sullivan AK, Rooke R, Geleziunas R, Tsoukas C, Shematek G, Gilmore N, Wainberg MA. 1989. New CD4(+) cell line susceptible to infection by HIV-1. J Med Virol 28:243–249. doi: 10.1002/jmv.1890280408. [DOI] [PubMed] [Google Scholar]
- 27.Spenlehauer C, Gordon CA, Trkola A, Moore JP. 2001. A luciferase-reporter gene-expressing T-cell line facilitates neutralization and drug-sensitivity assays that use either R5 or X4 strains of human immunodeficiency virus type 1. Virology 280:292–300. doi: 10.1006/viro.2000.0780. [DOI] [PubMed] [Google Scholar]
- 28.Teeranaipong P, Hosoya N, Kawana-Tachikawa A, Fujii T, Koibuchi T, Nakamura H, Koga M, Kondo N, Gao GF, Hoshino H, Matsuda Z, Iwamoto A. 2013. Development of a rapid cell-fusion-based phenotypic HIV-1 tropism assay. J Int AIDS Soc 16:18723. doi: 10.7448/IAS.16.1.18723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tobiume M, Lineberger JE, Lundquist CA, Miller MD, Aiken C. 2003. Nef does not affect the efficiency of human immunodeficiency virus type 1 fusion with target cells. J Virol 77:10645–10650. doi: 10.1128/JVI.77.19.10645-10650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oury C, Toth-Zsamboki E, Van Geet C, Thys C, Wei L, Nilius B, Vermylen J, Hoylaerts MF. 2000. A natural dominant negative P2X1 receptor due to deletion of a single amino acid residue. J Biol Chem 275:22611–22614. doi: 10.1074/jbc.C000305200. [DOI] [PubMed] [Google Scholar]
- 31.Baba M, Nishimura O, Kanzaki N, Okamoto M, Sawada H, Iizawa Y, Shiraishi M, Aramaki Y, Okonogi K, Ogawa Y, Meguro K, Fujino M. 1999. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc Natl Acad Sci U S A 96:5698–5703. doi: 10.1073/pnas.96.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimpton J, Emerman M. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol 66:2232–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de la Vega M, Marin M, Kondo N, Miyauchi K, Kim Y, Epand RF, Epand RM, Melikyan GB. 2011. Inhibition of HIV-1 endocytosis allows lipid mixing at the plasma membrane, but not complete fusion. Retrovirology 8:99. doi: 10.1186/1742-4690-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burnstock G, Kennedy C. 2011. P2X receptors in health and disease. Adv Pharmacol 61:333–372. doi: 10.1016/B978-0-12-385526-8.00011-4. [DOI] [PubMed] [Google Scholar]
- 35.Coddou C, Yan Z, Obsil T, Huidobro-Toro JP, Stojilkovic SS. 2011. Activation and regulation of purinergic P2X receptor channels. Pharmacol Rev 63:641–683. doi: 10.1124/pr.110.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orellana JA, Velasquez S, Williams DW, Saez JC, Berman JW, Eugenin EA. 2013. Pannexin1 hemichannels are critical for HIV infection of human primary CD4+ T lymphocytes. J Leukoc Biol 94:399–407. doi: 10.1189/jlb.0512249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pelegrin P, Surprenant A. 2006. Pannexin-1 mediates large pore formation and interleukin-1beta release by the ATP-gated P2X7 receptor. EMBO J 25:5071–5082. doi: 10.1038/sj.emboj.7601378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kondo N, Marin M, Kim JH, Desai TM, Melikyan GB. 2015. Distinct requirements for HIV-cell fusion and HIV-mediated cell-cell fusion. J Biol Chem 290:6558–6573. doi: 10.1074/jbc.M114.623181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kondo N, Miyauchi K, Matsuda Z. 2011. Monitoring viral-mediated membrane fusion using fluorescent reporter methods. Curr Protoc Cell Biol Chapter 26:Unit 26.9. doi: 10.1002/0471143030.cb2609s50. [DOI] [PubMed] [Google Scholar]
- 40.Surprenant A, North RA. 2009. Signaling at purinergic P2X receptors. Annu Rev Physiol 71:333–359. doi: 10.1146/annurev.physiol.70.113006.100630. [DOI] [PubMed] [Google Scholar]
- 41.Woehrle T, Yip L, Elkhal A, Sumi Y, Chen Y, Yao Y, Insel PA, Junger WG. 2010. Pannexin-1 hemichannel-mediated ATP release together with P2X1 and P2X4 receptors regulate T-cell activation at the immune synapse. Blood 116:3475–3484. doi: 10.1182/blood-2010-04-277707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yip L, Woehrle T, Corriden R, Hirsh M, Chen Y, Inoue Y, Ferrari V, Insel PA, Junger WG. 2009. Autocrine regulation of T-cell activation by ATP release and P2X7 receptors. FASEB J 23:1685–1693. doi: 10.1096/fj.08-126458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melikyan GB, Markosyan RM, Hemmati H, Delmedico MK, Lambert DM, Cohen FS. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J Cell Biol 151:413–424. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melikyan GB, Egelhofer M, von Laer D. 2006. Membrane-anchored inhibitory peptides capture human immunodeficiency virus type 1 gp41 conformations that engage the target membrane prior to fusion. J Virol 80:3249–3258. doi: 10.1128/JVI.80.7.3249-3258.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raviv Y, Viard M, Bess J Jr, Blumenthal R. 2002. Quantitative measurement of fusion of HIV-1 and SIV with cultured cells using photosensitized labeling. Virology 293:243–251. doi: 10.1006/viro.2001.1237. [DOI] [PubMed] [Google Scholar]
- 46.Demirkhanyan LH, Marin M, Padilla-Parra S, Zhan C, Miyauchi K, Jean-Baptiste M, Novitskiy G, Lu W, Melikyan GB. 2012. Multifaceted mechanisms of HIV-1 entry inhibition by human alpha-defensin. J Biol Chem 287:28821–28838. doi: 10.1074/jbc.M112.375949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hatse S, Princen K, De Clercq E, Rosenkilde MM, Schwartz TW, Hernandez-Abad PE, Skerlj RT, Bridger GJ, Schols D. 2005. AMD3465, a monomacrocyclic CXCR4 antagonist and potent HIV entry inhibitor. Biochem Pharmacol 70:752–761. doi: 10.1016/j.bcp.2005.05.035. [DOI] [PubMed] [Google Scholar]
- 48.Maeda K, Das D, Ogata-Aoki H, Nakata H, Miyakawa T, Tojo Y, Norman R, Takaoka Y, Ding J, Arnold GF, Arnold E, Mitsuya H. 2006. Structural and molecular interactions of CCR5 inhibitors with CCR5. J Biol Chem 281:12688–12698. doi: 10.1074/jbc.M512688200. [DOI] [PubMed] [Google Scholar]
- 49.Kuhmann SE, Platt EJ, Kozak SL, Kabat D. 2000. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J Virol 74:7005–7015. doi: 10.1128/JVI.74.15.7005-7015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shiraishi M, Aramaki Y, Seto M, Imoto H, Nishikawa Y, Kanzaki N, Okamoto M, Sawada H, Nishimura O, Baba M, Fujino M. 2000. Discovery of novel, potent, and selective small-molecule CCR5 antagonists as anti-HIV-1 agents: synthesis and biological evaluation of anilide derivatives with a quaternary ammonium moiety. J Med Chem 43:2049–2063. doi: 10.1021/jm9906264. [DOI] [PubMed] [Google Scholar]
- 51.Lau G, Labrecque J, Metz M, Vaz R, Fricker SP. 2015. Specificity for a CCR5 inhibitor is conferred by a single amino acid residue: role of Ile198. J Biol Chem 290:11041–11051. doi: 10.1074/jbc.M115.640169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blanpain C, Vanderwinden JM, Cihak J, Wittamer V, Le Poul E, Issafras H, Stangassinger M, Vassart G, Marullo S, Schlndorff D, Parmentier M, Mack M. 2002. Multiple active states and oligomerization of CCR5 revealed by functional properties of monoclonal antibodies. Mol Biol Cell 13:723–737. doi: 10.1091/mbc.01-03-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berro R, Klasse PJ, Lascano D, Flegler A, Nagashima KA, Sanders RW, Sakmar TP, Hope TJ, Moore JP. 2011. Multiple CCR5 conformations on the cell surface are used differentially by human immunodeficiency viruses resistant or sensitive to CCR5 inhibitors. J Virol 85:8227–8240. doi: 10.1128/JVI.00767-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Colin P, Benureau Y, Staropoli I, Wang Y, Gonzalez N, Alcami J, Hartley O, Brelot A, Arenzana-Seisdedos F, Lagane B. 2013. HIV-1 exploits CCR5 conformational heterogeneity to escape inhibition by chemokines. Proc Natl Acad Sci U S A 110:9475–9480. doi: 10.1073/pnas.1222205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zaitseva M, Peden K, Golding H. 2003. HIV coreceptors: role of structure, posttranslational modifications, and internalization in viral-cell fusion and as targets for entry inhibitors. Biochim Biophys Acta 1614:51–61. doi: 10.1016/S0005-2736(03)00162-7. [DOI] [PubMed] [Google Scholar]
- 56.Baribaud F, Edwards TG, Sharron M, Brelot A, Heveker N, Price K, Mortari F, Alizon M, Tsang M, Doms RW. 2001. Antigenically distinct conformations of CXCR4. J Virol 75:8957–8967. doi: 10.1128/JVI.75.19.8957-8967.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee B, Sharron M, Blanpain C, Doranz BJ, Vakili J, Setoh P, Berg E, Liu G, Guy HR, Durell SR, Parmentier M, Chang CN, Price K, Tsang M, Doms RW. 1999. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J Biol Chem 274:9617–9626. doi: 10.1074/jbc.274.14.9617. [DOI] [PubMed] [Google Scholar]
- 58.Dragic T, Trkola A, Thompson DA, Cormier EG, Kajumo FA, Maxwell E, Lin SW, Ying W, Smith SO, Sakmar TP, Moore JP. 2000. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc Natl Acad Sci U S A 97:5639–5644. doi: 10.1073/pnas.090576697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tilton JC, Doms RW. 2010. Entry inhibitors in the treatment of HIV-1 infection. Antiviral Res 85:91–100. doi: 10.1016/j.antiviral.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 60.Kondru R, Zhang J, Ji C, Mirzadegan T, Rotstein D, Sankuratri S, Dioszegi M. 2008. Molecular interactions of CCR5 with major classes of small-molecule anti-HIV CCR5 antagonists. Mol Pharmacol 73:789–800. [DOI] [PubMed] [Google Scholar]
- 61.Maeda K, Das D, Yin PD, Tsuchiya K, Ogata-Aoki H, Nakata H, Norman RB, Hackney LA, Takaoka Y, Mitsuya H. 2008. Involvement of the second extracellular loop and transmembrane residues of CCR5 in inhibitor binding and HIV-1 fusion: insights into the mechanism of allosteric inhibition. J Mol Biol 381:956–974. doi: 10.1016/j.jmb.2008.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gunosewoyo H, Kassiou M. 2010. P2X purinergic receptor ligands: recently patented compounds. Expert Opin Ther Pat 20:625–646. doi: 10.1517/13543771003702424. [DOI] [PubMed] [Google Scholar]
- 63.Gum RJ, Wakefield B, Jarvis MF. 2012. P2X receptor antagonists for pain management: examination of binding and physicochemical properties. Purinergic Signal 8:41–56. doi: 10.1007/s11302-011-9272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.