A multistep differentiation protocol for inducing human induced pluripotent stem cells (hiPSCs) into OSR1+SIX2+ renal progenitors capable of reconstituting three-dimensional proximal renal tubule-like structures in vitro and in vivo was established. Renal subcapsular transplantation of hiPSC-derived renal progenitors ameliorated the acute kidney injury in mice induced by ischemia/reperfusion injury, significantly suppressing the elevation of blood urea nitrogen and serum creatinine levels and attenuating histopathological changes.
Keywords: Nephrons, Kidney, Cell- and tissue-based therapy, Induced pluripotent stem cells, Acute kidney injury, SIX2 protein, Renal progenitors
Abstract
Acute kidney injury (AKI) is defined as a rapid loss of renal function resulting from various etiologies, with a mortality rate exceeding 60% among intensive care patients. Because conventional treatments have failed to alleviate this condition, the development of regenerative therapies using human induced pluripotent stem cells (hiPSCs) presents a promising new therapeutic option for AKI. We describe our methodology for generating renal progenitors from hiPSCs that show potential in ameliorating AKI. We established a multistep differentiation protocol for inducing hiPSCs into OSR1+SIX2+ renal progenitors capable of reconstituting three-dimensional proximal renal tubule-like structures in vitro and in vivo. Moreover, we found that renal subcapsular transplantation of hiPSC-derived renal progenitors ameliorated the AKI in mice induced by ischemia/reperfusion injury, significantly suppressing the elevation of blood urea nitrogen and serum creatinine levels and attenuating histopathological changes, such as tubular necrosis, tubule dilatation with casts, and interstitial fibrosis. To our knowledge, few reports demonstrating the therapeutic efficacy of cell therapy with renal lineage cells generated from hiPSCs have been published. Our results suggest that regenerative medicine strategies for kidney diseases could be developed using hiPSC-derived renal cells.
Significance
This report is the first to demonstrate that the transplantation of renal progenitor cells differentiated from human induced pluripotent stem (iPS) cells has therapeutic effectiveness in mouse models of acute kidney injury induced by ischemia/reperfusion injury. In addition, this report clearly demonstrates that the therapeutic benefits come from trophic effects by the renal progenitor cells, and it identifies the renoprotective factors secreted by the progenitors. The results of this study indicate the feasibility of developing regenerative medicine strategy using iPS cells against renal diseases.
Introduction
Embryonic nephron progenitors have the capacity to differentiate into nephron-forming glomerular and renal tubular epithelia [1, 2]. The previous lineage tracing analyses verified that nephron progenitors exist in the metanephric mesenchyme derived from the intermediate mesoderm (IM) [2, 3]. Odd-skipped related 1 (Osr1) is one of the earliest markers specific for the IM [3–5]. We previously generated OSR1-green fluorescent protein (GFP) knock-in human induced pluripotent stem cell (hiPSC) lines by homologous recombination using bacterial artificial chromosome (BAC)-based vectors, and established robust methods for inducing OSR1+ IM cells using growth factors and small molecules [6, 7].
Osr1 is continuously expressed from the IM through nephron progenitors, although the expression also extends into the lateral plate mesoderm in early-stage mouse, chick, and fish embryos [3–5]. Another lineage analysis revealed that a homeodomain transcriptional regulator, Six2, is required to maintain a nephron progenitor population, ensuring the development of a full complement constituting nephrons. However, Six2 is also expressed in other fetal organs, such as the skeletal muscle, limbs, heart, eyes, and middle ears [2, 8]. Osr1 and Six2 interact synergistically to maintain nephron progenitor cells during kidney organogenesis [9]. Therefore, the combination of Osr1 and Six2 can be used as a marker to more specifically define nephron progenitors.
AKI results in a high mortality rate, especially in intensive care patients, with a mortality rate of more than 60% [10]. In addition, AKI has been reported as a cause of chronic kidney disease and a risk factor for cardiovascular diseases [11]. Despite the urgent need, the treatments for AKI remain to be developed [12]. Recently, human fetal nephron progenitor cells have been shown to participate in the repair of renal tissue in experimental animal models of renal failure [13], suggesting that nephron progenitors generated from hiPSCs could be used for the development of regenerative medicine against renal diseases. However, few studies have demonstrated to date the therapeutic effects of hiPSC-derived renal lineage cells against kidney disease [14].
In the present study, we established a protocol for differentiating hiPSCs into OSR1+SIX2+ renal progenitors that have the developmental potential to differentiate and form three-dimensional proximal renal tubule-like structures. Furthermore, we established a method for transplanting hiPSC-derived renal progenitors into the renal subcapsule, which ameliorated AKI in mice.
Materials and Methods
Cell Culture
Cell cultures were performed as described previously [6, 7]. hiPSCs (585A1, 585B1, 604A1, 604B1, 648A1, 648B1, 692D2, 606A1, 606B1, 610B1, 201B6, 201B7, 253G1 and 253G4) [15–18] and human embryonic stem cells (hESCs) (khES1, khES3, and H9) [19, 20] were grown on feeder layers of mitomycin C-treated mouse embryonic fibroblasts derived from embryonic day (E) 12.5 ICR mouse embryos or SNL feeder cells in medium containing primate ES medium (ReproCELL, Yokohama, Japan, http://www.reprocell.com) supplemented with 500 U/ml penicillin/streptomycin (PS; Invitrogen, Carlsbad, CA, http://www.invitrogen.com) and 4 or 5 ng/ml recombinant human basic fibroblast growth factor (Wako Chemical, Osaka, Japan, http://www.wako-chem.co.jp/english). For routine passaging, the hiPSC/ESC colonies were dissociated by an enzymatic method with CTK dissociation solution consisting of 0.25% trypsin (Invitrogen), 0.1% collagenase IV (Invitrogen), 20% knockout serum replacement (KSR, Invitrogen), and 1 mM CaCl2 in phosphate-buffered saline (PBS) and split at a ratio of 1:3 to 1:6.
BAC Recombineering
BAC recombineering is described in the supplemental online data.
Genetic Modification of hiPSCs
Genetic modification of hiPSCs is described in the supplemental online data.
TaqMan Polymerase Chain Reaction Assay
TaqMan polymerase chain reaction (PCR) is described in the supplemental online data.
Removal of PGK-Neo Cassette by Transient Creatinine-Recombinase Expression
Removal of PGK-neo cassette by transient creatinine (Cre) recombinase expression is described in the supplemental online data.
Single-Nucleotide Polymorphism Array Analysis
The single-nucleotide polymorphism (SNP) array analysis is described in the supplemental online data.
Karyotyping
Karyotyping is described in the supplemental online data.
Real-Time PCR and Real-Time Quantitative Reverse Transcription-PCR
Real-time PCR and real-time quantitative reverse transcription (RT)-PCR is described in the supplemental online data.
Flow Cytometry and Cell Sorting
Flow cytometry and cell sorting are described in the supplemental online data.
Immunostaining and Lectin Staining
Immunostaining and lectin staining are described in the supplemental online data.
Embryoid Body-Based Differentiation Protocol
For embryoid body (EB) formation from hiPSCs/ESCs, a 10-cm plate containing hiPSCs/ESCs at 70%–80% confluence was rinsed with PBS and treated with CTK dissociation solution for 4 minutes at 37°C. The CTK dissociation solution was rinsed with PBS and replaced with EB medium containing Dulbecco’s modified Eagle’s medium (DMEM)/F12 plus GlutaMAX (Invitrogen) supplemented with 0.1 mM nonessential amino acids (Invitrogen), 1,000 U/ml PS (Invitrogen), 55 μM 2-mercaptoethanol (Invitrogen), and 20% KSR. The cells were then scraped off with a cell scraper and distributed onto 6-cm plates coated with 0.2% gelatin to remove the feeder cells. After 1 hour, the cells were washed with stage 1 medium containing DMEM/F12 plus GlutaMAX supplemented with 500 U/ml PS and 2% fetal bovine serum (HyClone, Logan, UT, http://www.hyclone.com) [6] and transferred into low attachment dishes (Sumitomo Bakelite Co., Ltd., Tokyo, Japan, http://www.sumibe.co.jp) containing stage 1 medium with 100 ng/ml recombinant human/mouse/rat activin A (R&D Systems, Minneapolis, MN, http://www.rndsystems.com) and 1 μM CHIR99021 (Axon Medchem BV, Groningen, The Netherlands, https://us.axonmedchem.com). To differentiate cells toward the IM, EBs on culture day 3 were transferred onto 24-well plates coated with Synthemax II (Corning, Corning, NY, http://www.corning.com). The cells were then cultured with stage 2 medium (DMEM/F12 plus GlutaMAX, 0.1 mM nonessential amino acids, 500 U/ml PS, 55 μM 2-mercaptoethanol, and 10% KSR) supplemented with 100 ng/ml recombinant human bone morphogenetic protein (BMP) 7 (R&D Systems) and 1 μM CHIR99021. After 3 days (on day 6), the medium was changed to the stage 2 medium supplemented with 1 μM TTNPB (Santa Cruz Biotechnology, Santa Cruz, CA, http://www.scbt.com) and 5 ng/ml transforming growth factor (TGF)-β1 (PeproTech, Rocky Hill, NJ, http://www.peprotech.com) for stage 3 treatment, after which the cultures were refreshed with new medium containing the same factors on day 8. On day 11, the medium was switched to the stage 2 medium supplemented with 5 ng/ml TGF-β1 and 0.5 μM dorsomorphin homolog (DMH) 1 (Tocris Bioscience, Bristol, United Kingdom, http://www.tocris.com) for stage 4 treatment. Subsequently, the medium was refreshed with new medium and the above factors every 3 days.
Media Conditioned by Ureteric Bud Cells
Media conditioned by ureteric bud cells (UBCs) are described in the supplemental online data.
Organ Culture Experiments With E11.5 Spinal Cord or Metanephros
The organ culture experiments are described in the supplemental online data.
Coculture of OSR1+SIX2+ Cells With NIH3T3 Fibroblasts Expressing Wnt4
Coculture of OSR1+SIX2+ cells with NIH3T3 fibroblasts expressing Wnt4 is described in the supplemental online data.
Graft Preparation and Implantation Into Epididymal Fat Pads of Immunodeficient Mice
Graft preparation and implantation are described in the supplemental online data.
Graft Preparation and Implantation Into Immunodeficient Mice With Renal Disease
The Kyoto University Animal Care Committee approved all animal experiments. For transplantation, OSR1+SIX2+ renal progenitor cells (iPSC-RPs), OSR1−SIX2− cells, OSR1+SIX2− cells, and OSR1−SIX2+ cells obtained on culture days 25–28 were isolated by flow cytometry sorting, split into spindle-shaped bottom low adhesion 96-well plates (Sumitomo Bakelite) at a density of 1.0 × 105 cells per well with UBC-conditioned media supplemented with 50 ng/ml BMP7 and 10 μM Y-27632 for 24 hours and incubated with UBC-conditioned media supplemented with 50 ng/ml BMP7, 0.5 μM BIO, and 10 μM Y-27632 for an additional 24 hours. Undifferentiated hiPSCs (4A6C3-10) were split into spindle-shaped bottom, low-adhesion, 96-well plates at a density of 1.0 × 105 cells per well with primate ES cell media supplemented with 10 μM Y-27632 for 48 hours. After the cells were washed with saline to remove the media, 15 cell aggregates (approximately 1.5 × 106 cells) of each type of iPSC-RPs, OSR1−SIX2− cells, OSR1+SIX2− cells, OSR1−SIX2+ cells, and undifferentiated iPSCs were transplanted into the kidney subcapsules of the immunodeficient mice (NOD.CB17-Prkdcscid/J) with AKI.
The mouse AKI models were generated in a blind manner as described previously [21–23]. In brief, 6-week-old male immunodeficient mice were anesthetized via isoflurane inhalation and maintained at 37°C. After right nephrectomy through a flank incision, an atraumatic microvascular clamp (Natsume Seisakusho, Tokyo, Japan, http://www.nazme.co.jp) was used to occlude the left renal artery for 40 minutes. After releasing the clamp, the five types of cellular aggregates were transplanted; the control mice received a saline injection.
For transplantation into the renal parenchyma of the AKI mice, we injected 15 cell aggregates (approximately 1.5 × 106 cells) of iPSC-RPs after the same pretreatment with UBC-conditioned medium containing BMP7, BIO, and Y-27632 into the cortex of the host kidney just after ischemia/reperfusion (I/R) injury through a slim micropipette tip. The wound was sealed with Spongel (Astellas Parma, Tokyo, Japan, http://www.astellas.com) immediately to prevent hemorrhage.
The conditioned medium of OSR1+SIX2+ cells was generated as previously reported [24]. In brief, 1 × 105 OSR1+SIX2+ cells were cultured with 100 μl of serum-free DMEM/F12 for 48 hours, and the supernatant was separated from the cells by filtering through a 0.22-μm filtration unit. We injected 200 µl of the conditioned medium i.p. once per day from day 0 just after generating the AKI to day 7.
The peripheral blood plasma was assayed for the blood urea nitrogen (BUN) and Cre levels using the DRI-CHEM 7000VZ instrument (Fujifilm, Tokyo, Japan, http://www.fujifilmcom).
Renal Histopathology
After removing the renal tissue from the host mice, the kidneys were fixed in 10% neutral buffered formalin and embedded in paraffin. Thereafter, paraffin sections were sliced (3 μm) and subsequently stained with hematoxylin and eosin, periodic acid-Schiff, and Masson’s trichrome. Histological examinations were performed by the CMIC Bioresearch Center Co., Ltd. (Tokyo, Japan), for objective assessments. At least 10 nonoverlapping fields in 1 entire section of each kidney sample were randomly examined using a ×20 objective. The degree of tissue damage in the kidney cortex and outer medulla was assessed according to the commonly used and well-established method for AKI [25–27]. In brief, we graded the extent of tubular necrosis, urinary casts, tubular dilatation, and loss of tubular borders on a scale of 0 to 2, as follows: 0, none; 0.5, minimal; 1, mild; 1.5, moderate; and 2, marked. Interstitial fibrosis was also graded using the same category criteria.
Microarray Analysis
The microarray analysis is described in the supplemental online data.
Multiplex Protein Detection
Multiplex protein detection is described in supplemental online data.
Statistical Analysis
The data are shown as the mean ± SEM or 95% confidence intervals. We applied the Kruskal-Wallis test for comparisons between the two groups. For longitudinal data, the mixed effects model for repeated measures was used for each dependent variable [28]. The model included the treatment group, time, and treatment-by-time interaction as factors and random intercept for each subject, and the model parameters were estimated by restricted maximum likelihood. The covariance matrix was modeled in compound symmetry. The comparisons of interactions were formulated by one degree of freedom contrasts between the mean values for the groups. Multiple comparison type I error rate adjustment was based on a Bonferroni-adjusted criterion with a type I error rate of 0.05. All statistical analyses were performed using SAS software, version 9.4. (SAS Institute Inc., Cary, NC, http://www.sas.com).
Results
Generation of OSR1-GFP/SIX2-tdTomato Double Knock-In hiPSC Lines
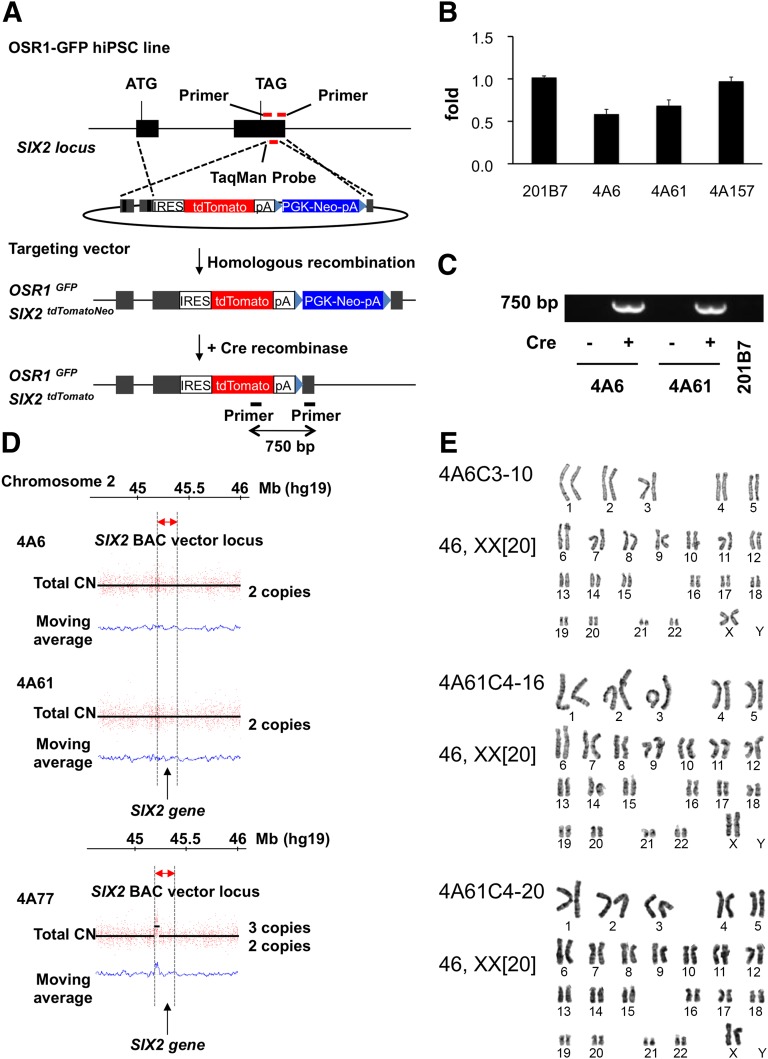
To monitor and quantitatively evaluate the differentiation of OSR1+SIX2+ renal progenitors from hiPSCs, we established OSR1-GFP/SIX2-tdTomato double knock-in hiPSC lines using our previously reported gene targeting strategy [6]. We constructed a BAC-based vector shown in Figure 1A. Because mutations in the SIX2 gene were identified as a possible cause of renal hypodysplasia [29], a tdTomato reporter gene fused with the internal ribosome entry site (IRES) was knocked-in after a stop codon to retain the intact SIX2 gene expression. The OSR1-GFP knock-in hiPSC line, 3D45, which we previously generated, was used as a parental line [6].Two clones, 4A6 and 4A61, were selected as candidate homologous recombinants from approximately 300 drug-resistant clones by TaqMan quantitative PCR analyses performed to detect the loss of a 3′UTR region of the SIX2 gene, which was replaced by an IRES-tdTomato-Neo cassette (Fig. 1B). The Neo cassette was then excised at the flanking loxP sites by the transient expression of Cre recombinase, which was confirmed by genomic PCR (Fig. 1C).
Figure 1.
The generation of OSR1-GFP/SIX2-tdTomato double knock-in hiPSC lines. (A): A schematic representation of the targeting strategy using BAC-based vectors to produce OSR1-GFP/SIX2-tdTomato double knock-in hiPSC lines. The black boxes represent two exons of the SIX2 gene. (B): The TaqMan quantitative polymerase chain reaction (PCR) analyses of genomic DNA from the drug-resistant lines and the parental line (201B7). Note that a value of 1 indicates 2 intact SIX2 loci, and 0.5 suggests 1 intact and 1 targeted locus. 4A157 is a drug-resistant clone without homologous recombination. The primer pair indicated in (A) (red bars) was used. (C): Genomic PCR was used to confirm the removal of the Neo cassette by Cre recombination. The primer pair indicated in (A) (black bars) was used. (D): The CN of SIX2 gene loci was analyzed by determining the number of single-nucleotide polymorphisms (SNPs) in the 3 hiPSC lines: 4A6, 4A61, and 4A77. 4A77 is a drug-resistant transgenic line without homologous recombination. The red dots and y-axis represent the signal intensity of each SNP probe in each panel, and the CNs (black lines) were detected using a hidden Markov model-based algorithm implemented in the CNAG/AsCNAR software program (Cancer Genomics Project, University of Tokyo, Tokyo, Japan). The blue lines represent the moving average of total CNs. (E): Normal karyotypes were found for the 3 OSR1-GFP/SIX2-tdTomato double knock-in hiPSC lines (4A6C3-10, 4A61C4-16, and 4A61C4-20) in which the Neo cassette was excised at the flanking loxP sites by transient expression of Cre recombinase. Abbreviations: bp, base pairs; BAC, bacterial artificial chromosome; CN, copy number; Cre, creatinine; GFP, green fluorescent protein; hiPSC, human inducible pluripotent stem cell; IRES, internal ribosome entry site.
We performed SNP array analyses to examine the copy numbers of the SIX2 gene [6] and found that the two clones had two loci, and noncandidate clones, such as 4A77, showed three copies (Fig. 1D). Thus, the analyses confirmed the generation of two knock-in lines with homologous recombination. We then used a G-banding analysis for the three clones after removing the Neo cassette (4A6C3-10, 4A61C4-16, and 4A61C4-20) and found that all three showed a normal karyotype (Fig. 1E).
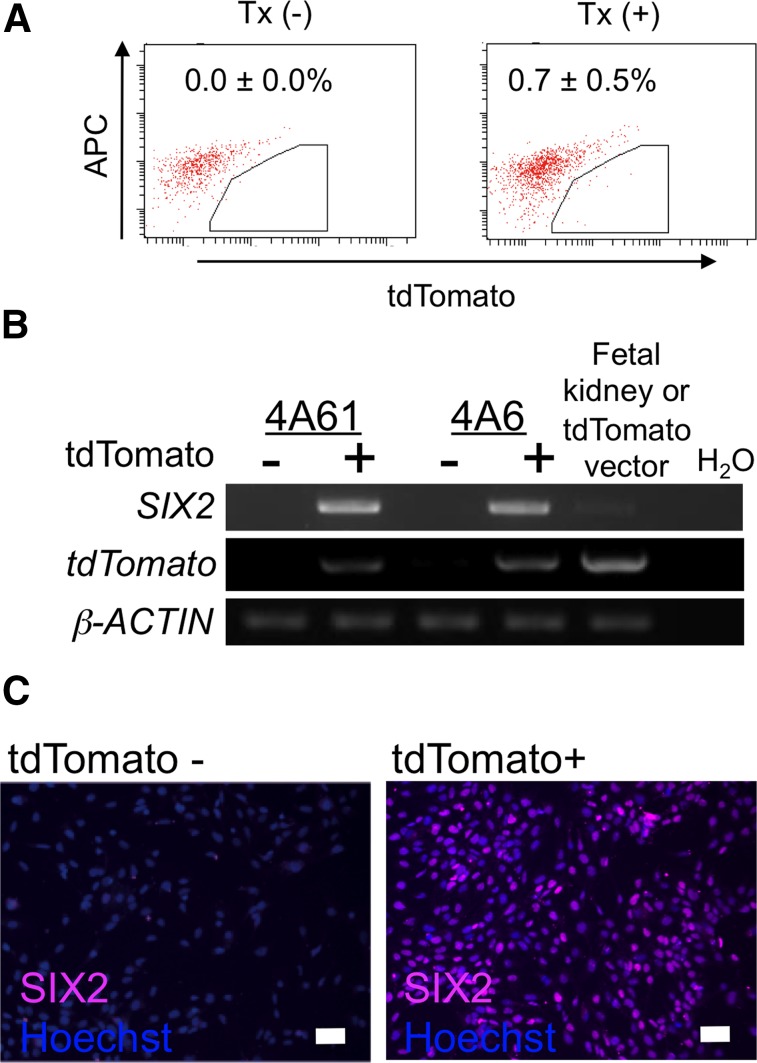
Next, tdTomato-positive cells were induced from 4A6C3-10 at low efficiency (0.7% ± 0.5%) by our EB-based IM differentiation method [6] (Fig. 2A) and were isolated by flow cytometry for RT-PCR analyses and immunostaining, which showed that only the tdTomato-positive cells expressed SIX2 and indicated a correlation between tdTomato and SIX2 expression (Fig. 2B, 2C). These results suggested that the reporter lines could be used to monitor both OSR1+ and SIX2+ cells differentiated from hiPSCs.
Figure 2.
Confirmation of the ability of OSR1-GFP/SIX2-tdTomato double knock-in human inducible pluripotent stem cell lines to monitor SIX2 expression. (A): The flow cytometric analyses of tdTomato-positive cells on culture day 19 induced with or without the factors used in our previously reported differentiation protocol for the intermediate mesoderm [6]. The data from three independent experiments are presented as the mean ± SEM (n = 3). (B): The results of the reverse transcription-polymerase chain reaction analyses of the tdTomato-positive and tdTomato-negative cell populations isolated on day 19 shown in (A). (C): Immunostaining using antibodies against SIX2 on the tdTomato-positive and tdTomato-negative cells isolated on day 19. Alexa Fluor 647 was used as the secondary antibody to eliminate any overlap with the GFP or tdTomato signals. Scale bars = 50 μm. Abbreviations: APC, allophycocyanin; GFP, green fluorescent protein; Tx, treatment with factors described in [6].
Establishment of the Protocol for Inducing hiPSCs Into OSR1+SIX2+ Cells
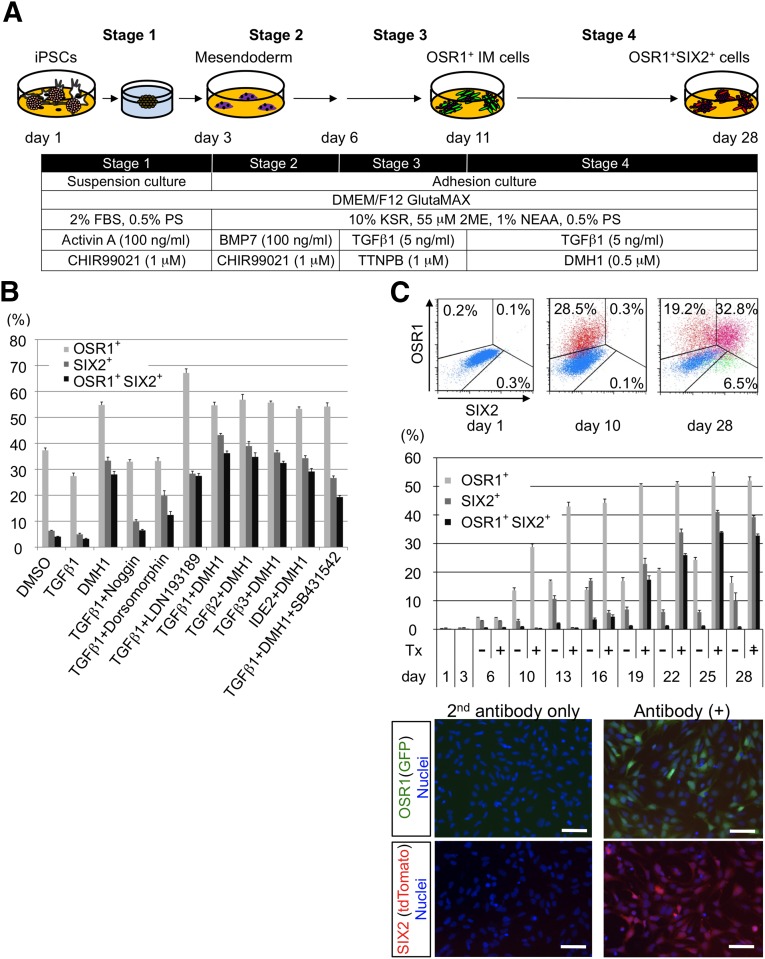
To establish protocols for the more efficient differentiation of OSR1+SIX2+ renal progenitors from hiPSCs through OSR1+ IM cells, we examined the effects of approximately 50 different growth factors and 8,000 small molecules on the OSR1+ IM cells induced by our EB-based IM differentiation method (data not shown). We previously reported that retinoic acid receptor (RAR) agonists efficiently differentiated hiPSCs into IM cells [7]. Thus, the α-RAR agonist, TTNPB, was added to the EB method, which more efficiently generated OSR1+ cells (data not shown). The screening of growth factors and chemicals revealed that the combination of BMP inhibitors and TGF-β isoforms was the most potent treatment leading to OSR1+SIX2+ cell induction (Fig. 3A, 3B). DMH1, a selective BMP antagonist [30], more potently induced OSR1+SIX2+ cells than the other BMP inhibitors we examined, including Noggin, dorsomorphin, and LDN-193189. Among the TGF-β isoforms, TGF-β1 was the most efficient inducer. A selective TGF-β receptor 1 inhibitor, SB431542, prevented the differentiation of OSR1+SIX2+ cells (Fig. 3B). We finally established an optimal protocol to induce the differentiation of OSR1+SIX2+ cells from hiPSCs-derived IM cells using a combined treatment with 1 μM TTNPB, 5 ng/ml TGF-β1, and 0.5 μM DMH1 (Fig. 3A).
Figure 3.
The establishment of methods for differentiating human iPSCs into OSR1+SIX2+ cells. (A): The differentiation method used to generate OSR1+SIX2+ cells with embryoid body-based three-dimensional cultures. (B): The differentiation efficiencies of OSR1+, SIX2+, and OSR1+SIX2+ cells were analyzed by flow cytometry on culture day 28. The treatments at stages 1, 2, and 3 were the same as those used in A. The factors and their concentrations used at stage 4 were as follows: 0.5 μM DMSO as a negative control, 5 ng/ml TGF-β1, 5 ng/ml TGF-β2, 5 ng/ml TGF-β3, 0.5 μM DMH1, 100 ng/ml Noggin, 0.5 μM DMH1, 0.5 μM LDN-193189, 50 μM IDE2, and 10 μM SB431542. Noggin, DMH1, LDN, and DMH1 are BMP inhibitors. IDE2 is a small molecule that activates TGF-β signaling. The data from five independent experiments are presented as the mean ± SEM (n = 5). (C): The periodic differentiation pattern of OSR1+SIX2+ cells was analyzed by flow cytometry. Upper: The two-dimensional distribution of the cell populations. Middle: The results of the time course analyses. The data from five independent experiments are presented as the mean ± SEM (n = 5). Day 1 indicates culture day 1 before treatment. Day 3 indicates day 3 after stage 1 treatment. Day 6 indicates day 6 after stage 2 treatment. On days 6–28, the cells were treated with stage 3 and 4 treatment or with DMSO at stages 3 and 4 (negative control). Representative anti-GFP and anti-DsRed (tdTomato) immunostaining images of OSR1+SIX2+ renal progenitors isolated on day 28 are shown in the lower panels. Scale bars = 50 μm. Abbreviations: BMP, bone morphogenetic protein; DMEM, Dulbecco’s modified Eagle’s medium; DMH1, dorsomorphin homolog 1; DMSO, dimethyl sulfoxide; FBS, fetal bovine serum; GFP, green fluorescent protein; KSR, knockout serum replacement; PS, penicillin/streptomycin; TGF, transforming growth factor; Tx+, treatment with stage 3 and 4 treatment; Tx−, treatment with negative control.
By periodically monitoring the OSR1+ and SIX2+ cells during the course of differentiation by flow cytometry and 4A6C3-10 cells, we found a gradual increase in the frequency of OSR1+SIX2+ cells up to more than 30% on culture day 28 (Fig. 3C; supplemental online Fig. 1), consistent with the results of the quantitative RT-PCR analyses (Fig. 4A). We found both GFP and tdTomato fluorescence signals in almost all the isolated OSR1+SIX2+ cells by augmenting the signals using anti-GFP and anti-DsRed immunostaining, which also confirmed the induction rate of the OSR1+SIX2+ cells (Fig. 3C).
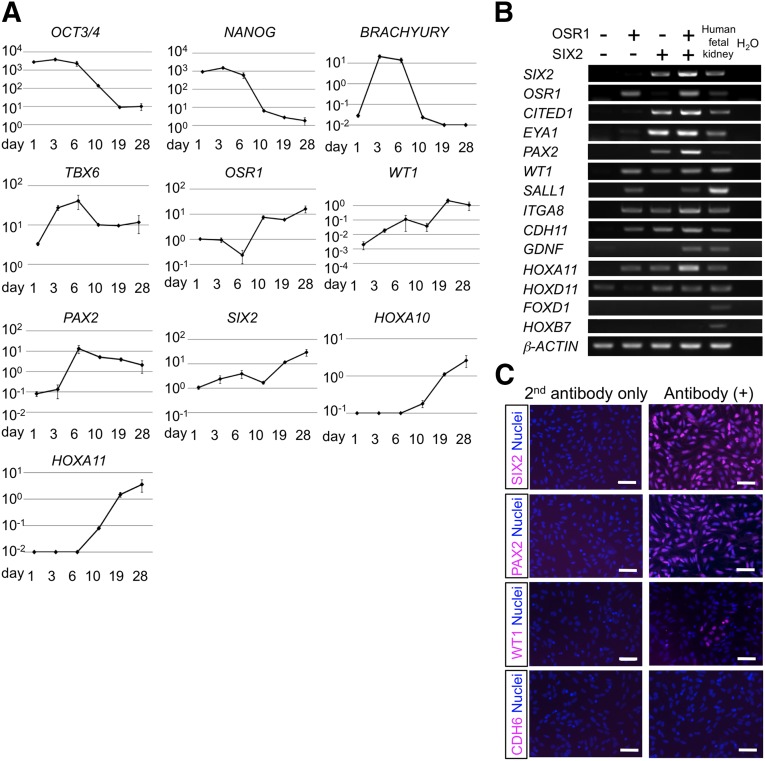
Figure 4.
The expression of renal lineage marker genes in the differentiation culture of human induced pluripotent stem cells (hiPSCs) into OSR1+SIX2+ renal progenitors. (A): The time course analyses of mRNA expression in the differentiation culture. The graphs indicate the expression of each transcript relative to β-ACTIN. OCT3/4 and NANOG are markers for undifferentiated hiPSCs, BRACHYURY and TBX6 are markers for posterior nascent mesoderm, OSR1 is a marker for intermediate mesoderm, WT1, PAX2, SIX2, HOXA10, and HOXA11 are markers for the metanephric mesenchyme. The data from three independent experiments are presented as the mean ± SEM (n = 3). (B): The expression of marker genes for nephron progenitors in OSR1+SIX2+ cells differentiated from an OSR1-GFP/SIX2-tdTomato double knock-in hiPSC line, 4A6C3-10, on culture day 28. (C): Immunostaining images of OSR1+SIX2+ cells isolated on day 28 against SIX2, PAX2, WT1, and CDH6. (B, C): Representative data obtained from three independent experiments shown. Scale bars = 50 μm.
Different hiPSC/ESC lines have been shown to vary in their differentiation potential [17, 31]. We examined our induction protocol for OSR1+SIX2+ cells with 18 different hiPSC/ESC lines [15–20] (supplemental online Fig. 2). Although the expression levels of OSR1 and SIX2 differed among the cell lines, our protocol more potently induced the expression of the two genes in multiple lines compared with 4A6C3-10.
We next examined the gene expression profiles of renal lineage markers in our hiPSC differentiation culture. As expected, the treated EBs expressed BRACHYURY and TBX6, markers for posterior nascent mesoderm, which has been reported to differentiate into IM [32], at stages 1 and 2 and OSR1 after stage 2. Thereafter, the expression of metanephric mesenchyme markers, WT1, PAX2, and SIX2, and the posterior HOX genes was observed (Fig. 4A). The OSR1+SIX2+ cells isolated by flow cytometry sorting on day 28 also expressed other nephron progenitor markers, including CITED1, EYA1, SALL1, ITGA8, CDH11, GDNF, and HOXD11, and expression of a stromal marker, FOXD1, and a nephric duct marker, HOXB7, was not detected (Fig. 4B). The OSR1+SIX2+ cells were also positive for PAX2 and WT1, but not for an early nephron marker CDH6, as analyzed using immunocytochemistry (Fig. 4C). Not all cells were WT1+, suggesting that the OSR1+SIX2+ cells generated by our EB-based differentiation protocol are heterogeneous. No marker gene expression for other nonrenal mesoderm, endoderm, or ectoderm tissues was detected in the OSR1+SIX2+ cells on day 28, indicating that these cells are renal lineage-committed cells (supplemental online Fig. 3).
hiPSC-Derived OSR1+SIX2+ Cells Have Developmental Potential as Tubulogenic Renal Progenitors
Because Six2+ nephron progenitors contribute to multiple epithelial cell types constituting nephrons [1, 2], we assessed the developmental potential of hiPSC-derived OSR1+SIX2+ cells isolated on day 28 using various methods. First, when the OSR1+SIX2+ cell aggregates without additional treatments were cocultured with mouse embryonic spinal cord and NIH3T3 fibroblasts expressing Wnt4, a well-known inducer of the metanephric mesenchyme for 7 days [1, 33], the aggregates did not develop tubule-like structures (data not shown).
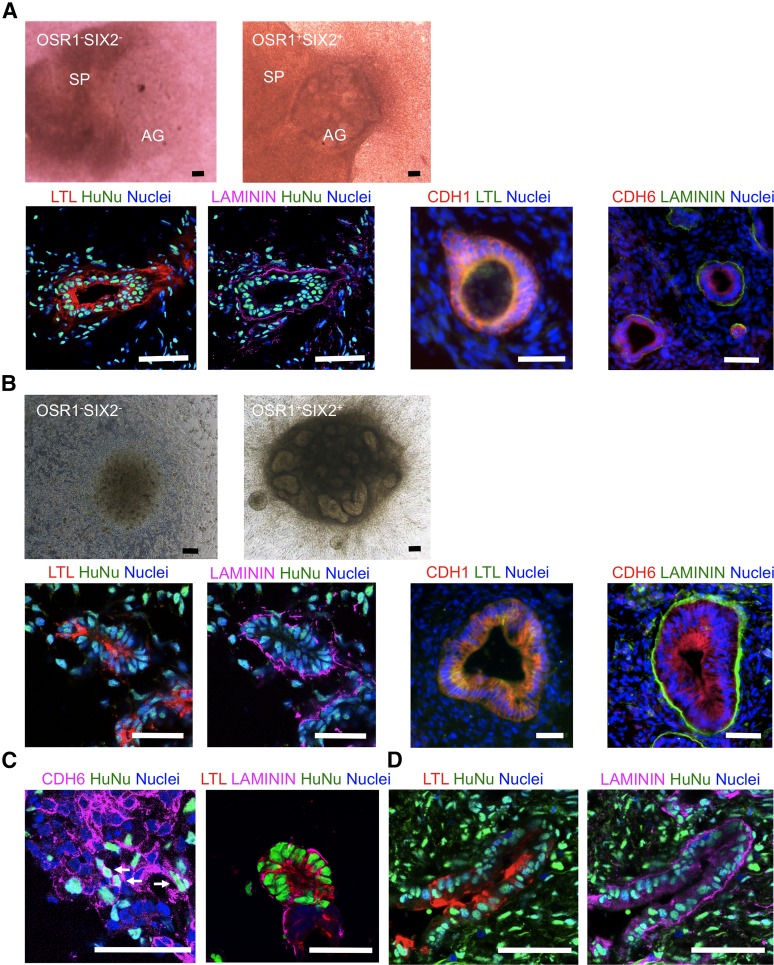
We then considered the pretreatment of OSR1+SIX2+ cells before the cocultures. BMP7 is required for the epithelialization of Six2+ nephron progenitors by Wnt9b [34], and treatment with BIO, a glycogen synthase kinase-3 inhibitor, mimics the action of Wnt9b in the process of nephrogenesis [35]. A previous study demonstrated that multiple factors contained in the conditioned media of UBC lines protect renal progenitors from apoptosis [36, 37]. Therefore, OSR1+SIX2+ cells were treated in UBC-conditioned media supplemented with 50 ng/ml BMP7 and 0.5 μM BIO, which increased the expression of CDH6 (supplemental online Fig. 4). Next, we cocultured the treated OSR1+SIX2+ cell aggregates with mouse embryonic spinal cord tissue in an organ culture setting for 7 days. As a result, the aggregates developed tubular structures that were positive for proximal renal tubule markers, Lotus tetragonolobus lectin (LTL) and CDH6, a marker of polarized epithelia, LAMININ, and an epithelial marker, CDH1, in all 17 organ culture samples examined (Fig. 5A; supplemental online Fig. 5A). The structures consisting of cells positive for glomerular podocyte markers, WT1 and PODOCALYXIN, but not mature podocyte markers, NEPHRIN and PODOCIN, were formed at a low frequency (3 of 17 organ culture samples, 17.6%), suggesting that they might be rather primitive glomerular lineage cells (supplemental online Fig. 5B) [38]. Tubular structures stained with distal tubule markers, such as PAX2, CLC-K2, and CALBINDIN, were not observed (data not shown). In contrast, OSR1−SIX2− cell aggregates did not reconstitute tubular structures and had disappeared by day 7 (Fig. 5A). Similar results were obtained by coculturing the treated OSR1+SIX2+ cell aggregates with NIH3T3 fibroblasts expressing Wnt4, which showed the formation of LTL+LAMININ+ proximal tubule-like structures in all 20 cultures examined and WT1+PODOCALYXIN+ immature glomerulus-like structures in 3 of 20 cultures (15.0%; Fig. 5B).
Figure 5.
Developmental potential of OSR1+SIX2+ renal progenitors to differentiate into renal lineage cells or tissues. (A): The presented macroscopic views show the aggregates of OSR1−SIX2− cells on culture day 5 (upper left) and OSR1+SIX2+ cells on day 7 (upper right) cocultured with E11.5 mouse spinal cord tissue in organ cultures. Lower panels: Immunostaining images of the histological sections of the OSR1+SIX2+ cell aggregates after 7 days of coculture with spinal cord tissue. LTL is a marker of the proximal renal tubules; LAMININ, a marker of the polarized epithelia; CDH1, an epithelial marker; CDH6, an early proximal tubule marker. (B): Coculture experiments with NIH3T3 fibroblasts expressing Wnt4. Macroscopic views of the aggregates of OSR1−SIX2− cells on day 5 (upper left) and OSR1+SIX2+ cells on day 7 (upper right) and immunostaining images of the histological sections of the OSR1+SIX2+ cell aggregates after 7 days of coculture (lower). Note that the lower left two panels in (A) and (B) are of the same sections. (C): Immunostaining images of the organ culture of the OSR1+SIX2+ cells and E11.5 metanephric cells. (D): Immunostaining images of the OSR1+SIX2+ cell aggregates transplanted into the epididymal fat pads of the immunodeficient mice (NOD. CB17-Prkdcscid/J) after 30 days of transplantation. Scale bars = 50 μm. Abbreviations: AG, OSR1−SIX2− or OSR1+SIX2+ cell aggregate; HuNu, human nuclei; LTL, Lotus tetragonolobus lectin; SP, spinal cord.
When we cocultured the treated OSR1+SIX2+ cells with dissociated E11.5 mouse metanephric cells in organ cultures [6, 39], we found some of the human CDH6+ cells integrated into mouse Cdh6+ structures in 5 of 13 samples examined (38.5%), although no C- or S-shaped bodies were clearly identified. The OSR1+SIX2+ cells also contributed to LTL+LAMININ+ proximal tubule-like structures within the mouse metanephric tissues in 7 of 16 samples (43.8%; Fig. 5C). When cocultured with the mouse metanephric cells, the OSR1−SIX2− cells were not integrated into the mouse tissues and instead had formed separated cell aggregates (data not shown). Furthermore, when transplanted into the epididymal fat pads of the immunodeficient mice (NOD.CB17-Prkdcscid/J) [6], the treated OSR1+SIX2+ cells formed LTL+LAMININ+ proximal tubule-like structures in vivo in two of the four grafts examined (Fig. 5D). These results suggest that the OSR1+SIX2+ cells generated from hiPSCs have developmental potential as renal progenitors to mainly form proximal tubule-like structures, although they are not as competent as the nephron progenitors in embryos.
Transplantation of hiPSC-Derived Renal Progenitors Ameliorates Renal Injury in a Mouse Model of AKI
We next examined the therapeutic potential of hiPSC-derived tubulogenic renal progenitors for AKI and transplanted the cellular aggregates of OSR1+SIX2+ cells purified on days 25–28, which were pretreated with UBC-conditioned media, BMP7 and BIO, into the kidney parenchyma of mice with AKI induced by I/R injury just after the injury. Consequently, some OSR1+SIX2+ cells were integrated into the host kidneys and then differentiated into cells positive for a proximal tubular marker LTL within 2 weeks after transplantation (supplemental online Fig. 6). However, the transplantation did not result in any obvious beneficial effects on renal function (data not shown).
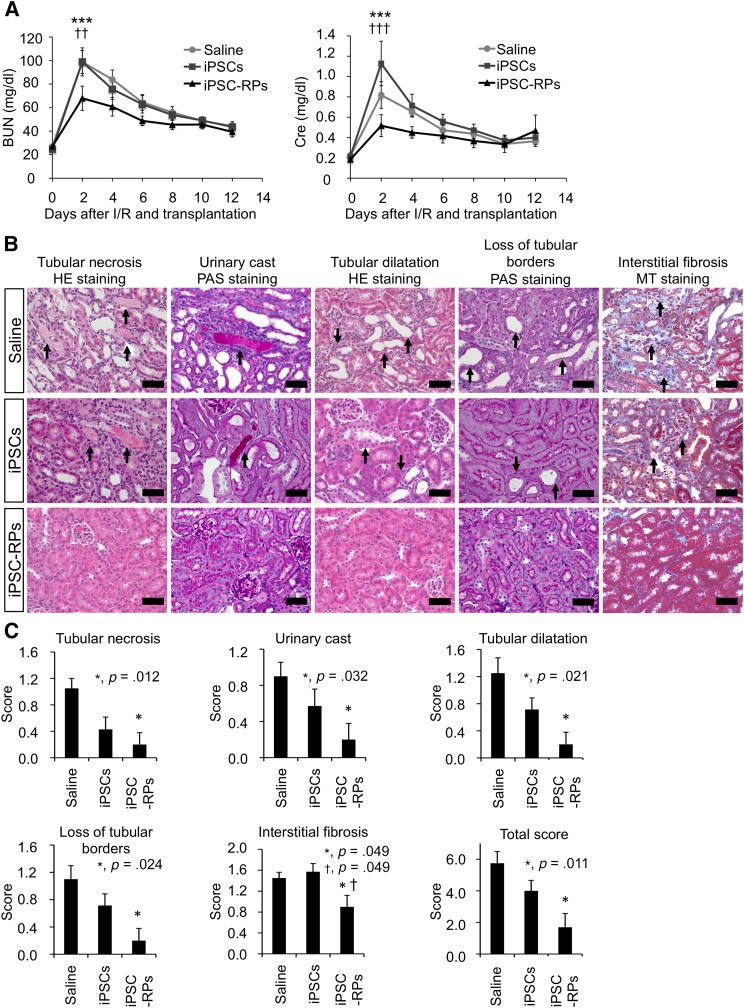
We then considered the therapeutic possibility of the paracrine effects induced by hiPSC-derived renal progenitors for AKI and transplanted the treated OSR1+SIX2+ cell aggregates into the renal subcapsule in the I/R model mice to allow for the administration of a larger number of progenitor cells without causing any additional surgical damage to the host renal tissues. We observed that the transplanted hiPSC-derived renal progenitors did not differentiate into tubular structures nor migrate into the host kidney (supplemental online Fig. 7). However, the renal functional parameter values (BUN and Cre levels) were significantly lower in the mice transplanted with hiPSC-derived renal progenitors than in the control mice treated without cell transplantation (saline) or the mice transplanted with undifferentiated hiPSCs at 2 days after I/R injury (Fig. 6A). Furthermore, histological analyses performed on day 3 after transplantation revealed that the renal parenchymal areas with tubular necrosis, urinary casts, and tubular dilatation were significantly smaller in the mice treated with hiPSC-derived renal progenitors than in the saline group (supplemental online Fig. 8A, 8B). We next compared the therapeutic effects of all four induced cell populations (OSR1−SIX2−, OSR1+SIX2−, OSR1−SIX2+, and OSR1+SIX2+ cells) and found that only the OSR1+SIX2+ cell population significantly reduced both BUN and Cre values on day 2, although the OSR1+SIX2− population decreased only the Cre values on day 8 with statistical significance, which suggests that the therapeutic effects on kidney injury are specific to OSR1+SIX2+ renal progenitors (supplemental online Fig. 9).
Figure 6.
Cell therapy using human iPSC (hiPSC)-derived renal progenitors for the mouse acute kidney (AKI) injury models. (A): The time course analyses of the BUN and plasma Cre levels in the I/R AKI mice that received the renal subcapsular transplantation of hiPSC-derived renal progenitors (n = 6; iPSC-RPs, triangle), undifferentiated hiPSCs (n = 7; iPSCs, square), or saline (n = 11; circle). Statistical significance at the p < .05 level after multiple testing adjustment: ∗∗∗, p < .001 versus saline; ††, p < .01 versus iPSCs; †††, p < .001 versus iPSCs. Least square mean and 95% confidence intervals were estimated according to the mixed effects model for repeated measures. (B): Sections of representative kidney samples from the host mice that received transplantation of iPSC-RPs, iPSCs, or saline were stained with HE, PAS, or MT on day 12 after I/R and transplantation. Representative findings of tubular necrosis, urinary casts, tubular dilatation, loss of tubular borders, and interstitial fibrosis in each treatment group are shown. The arrows indicate the representative area of each finding. Scale bars = 20 μm. (C): Histological scoring of the areas with tubular necrosis, urinary casts, tubular dilatation, loss of tubular borders and interstitial fibrosis and total scores in the host kidneys on day 12 after I/R and transplantation (n = 5 for iPSC-RPs, n = 7 for iPSCs, n = 7 for saline). ∗, p < .05 versus saline; †, p < .05 versus iPSCs. The data are presented as the mean ± SEM in (A) and (C). Abbreviations: BUN, blood urea nitrogen; Cre, creatinine; HE, hematoxylin and eosin; I/R, ischemia/reperfusion; iPSC, induced pluripotent stem cell; iPSC-RP, OSR1+SIX2+ renal progenitor cells; MT, Masson’s trichrome; PAS, periodic acid-Schiff.
Histological analyses performed on day 12 after transplantation revealed that the areas of kidney injury with tubular necrosis, urinary casts, tubular dilatation, loss of tubular borders, and interstitial fibrosis were significantly smaller in the mice treated with hiPSC-derived OSR1+SIX2+ renal progenitors than in the saline group, even after the recovery of the renal function parameters (Fig. 6B, 6C; supplemental online Fig. 10A). Anti-α-smooth muscle actin immunostaining also revealed that the areas of fibrosis were smaller in the mice treated with the OSR1+SIX2+ renal progenitors than in the saline group (supplemental online Fig. 10B). As reported previously, histological changes and fibrosis are important markers indicating the risk of developing late renal failure even when the BUN levels do not differ at the AKI stage [40]. These findings therefore suggest that cell therapy using hiPSC-derived renal progenitors ameliorates the degree of renal tissue damage, including interstitial fibrosis, which indicates the risk of chronic tissue damage.
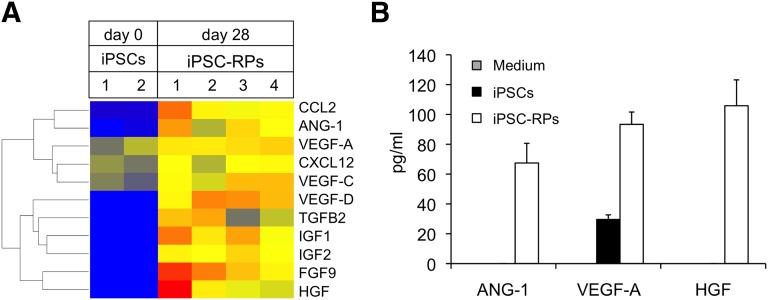
The therapeutic effects of the renal subcapsular transplantation of hiPSC-derived renal progenitors are thus thought to be mainly due to paracrine effects. We identified multiple factors secreted from OSR1+SIX2+ renal progenitors, such as angiopoietin (ANG)-1, vascular endothelial growth factor (VEGF)-A, and hepatocyte growth factor (HGF), that are known to be renoprotective factors produced by mesenchymal stem cells (Fig. 7) [41]. These results suggest that the renal subcapsular transplantation of hiPSC-derived OSR1+SIX2+ renal progenitors could therefore be a potentially effective therapeutic option for AKI.
Figure 7.
OSR1+SIX2+ renal progenitors secrete the growth factors known to be renoprotective factors. (A): The RNA expression of renoprotective factors in the iPSC-RPs compared with that observed in the iPSCs using a microarray analysis. (B): Protein section of renoprotective factors analyzed by the human magnetic luminex screening assay in medium only (medium) and cell culture supernatant of iPSCs and iPSC-RPs. The data from 3 independent experiments are presented as the mean ± SEM (n = 3). Note that the secretion levels of ANG-1, VEGF-A, and HGF in the medium and those of ANG-1 and HGF in the iPSCs were so low that the histogram bars are at the baseline level. Abbreviations: ANG-1, angiopoietin 1; CCL2, C-C motif chemokine 2; CXCL12, C-X-C motif chemokine 12; FGF-9, fibroblast growth factor 9; HGF, hepatocyte growth factor; IGF1, insulin-like growth factor 1; IGF2, insulin-like growth factor 2; iPSCs, inducible pluripotent stem cells; iPSC-RPs, OSR1+SIX2+ renal progenitor cells; TGF-B2, transforming growth factor-β2; VEGF-A, vascular endothelial growth factor A; VEGF-C, vascular endothelial growth factor C; VEGF-D, vascular endothelial growth factor D.
Discussion
In the present study, we established a method to induce the differentiation of hiPSCs into tubulogenic renal progenitors, which subsequently ameliorated renal injury in a mouse model of AKI induced by I/R injury. Although several studies have reported renal progenitor induction from hESCs/iPSCs [14, 32, 42–44], few reports have thus far demonstrated the therapeutic effects of cell therapy using these induced renal cells. Only one recent report showed that hiPSC-derived renal progenitors integrated into the kidney in another type of AKI mouse model induced by cisplatin administration ameliorated renal injury after transplantation via the tail vein [14]. However, it is not clear whether the integration of hiPSC-derived renal progenitors into the host kidney tissues or the trophic effects of factors secreted from the progenitors mainly contribute to the observed therapeutic effects on AKI and whether hiPSC-derived renal progenitors themselves exert these effects, because all hiPSC-derived differentiated cells containing renal progenitors and other cell types were transplanted in that study. In the present study, we have more directly and strictly demonstrated the therapeutic potential of hiPSC-derived renal progenitors by transplanting the purified progenitor population using an OSR1-GFP/SIX2-tdTomato double knock-in hiPSC line and flow cytometry sorting. Hence, the renal progenitors generated in our study might have an advantage for clinical application. The results from both the recent report and the present study suggest the feasibility of cell therapy using hiPSC-derived renal progenitor cells to treat AKI. Although the recent report involved the systemic administration of hiPSC-derived renal progenitors via the tail tip vein, we found that renal subcapsular transplantation, but not parenchymal injection, of hiPSC-derived renal progenitors produced therapeutic benefits in a mouse model of AKI. This observation suggests that the renal subcapsular transplantation of hiPSC-derived renal progenitors might be a suitable method for providing cell therapy against AKI.
In the present study, we have confirmed that hiPSC-derived renal progenitors transplanted into the renal subcapsule exerted therapeutic effects without migrating inside the host kidney, which suggests that some secreted, not cell-bound, factors derived from the progenitor cells working as trophic factors are mainly responsible for the observed therapeutic effects on AKI. Furthermore, we identified multiple growth factors secreted from hiPSC-derived OSR1+SIX2+ cells that have been shown to be renoprotective factors, such as ANG-1, VEGF-A, and HGF (Fig. 7) [41]. However, the systemic administration of the culture supernatant of hiPSC-derived OSR1+SIX2+ cells, which is believed to contain the above trophic factors, via injection into the peritoneal cavity in the AKI mice did not produce a therapeutic effect against kidney injury (supplemental online Fig. 11). When these data are taken together, we assume that some growth factors secreted from the renal progenitors might contribute to the noted therapeutic effects at concentrations higher than a certain therapeutic threshold. It is therefore suggested that the renal subcapsular transplantation of progenitor cells is required to deliver these concentrated factors to the host kidney tissues and thus obtain therapeutic effectiveness. In addition to growth factors, other candidate therapeutic factors secreted from the renal progenitors could include extracellular vesicles/microvesicles, which have been reported to exert therapeutic effects on AKI [45, 46].
In an attempt to elucidate the mechanisms of action of paracrine factors, we also examined the apoptosis and proliferation of cells in the host kidney on day 3 after I/R injury and cell transplantation, when the degree of renal damage had reached a peak, using a terminal deoxynucleotidyl transferase dUTP nick-end labeling assay and proliferating cell nuclear antigen staining, respectively. However, no significant differences were found in the number of apoptotic or proliferating cells between the mouse group transplanted with hiPSC-derived OSR1+SIX2+ progenitor cells and the control group that received saline (supplemental online Fig. 8C). Additional studies are thus needed to elucidate the mechanisms of action underlying the trophic effects of hiPSC-derived renal progenitors, which might also contribute to the development of novel therapeutic drugs for AKI.
However, the hiPSC-derived OSR1+SIX2+ cells generated in the present study had limited developmental competence as renal progenitors, because they differentiated mainly into proximal tubular cells, not glomerular podocytes or distal tubular cells. The renal progenitors induced in other studies showed developmental potential to form glomerulus-like [32] and distal renal tubule-like [32, 42] structure, in addition to the proximal tubules. These differences in developmental competence might derive from the differences in the differentiation protocol.
One of the common principles of previously published differentiation protocols and the current protocol for generating renal progenitors is that they each aim to mimic the developmental process that occurs in embryos, first differentiating ESCs/iPSCs into the primitive streak or mesendoderm, then the IM, followed by eventual nephron progenitor induction, although one report added more developmental steps in their differentiation protocol [32]. When our protocol is compared with previously reported protocols, major differences emerge, including the factors used at the induction step of nephron progenitors. Some reports treated the cells with fibroblast growth factor (FGF)-2 or FGF-9, because these factors have been shown to be expressed in the IM and involved in the formation and maintenance of the metanephric mesenchyme in mouse embryos [14, 32, 42–44, 47–50]. The presence or absence of these factors in the differentiation protocol could lead to differences in the characteristics of the induced renal progenitors.
In contrast, we identified BMP inhibitors and TGF-β isoforms as factors for inducing OSR1+SIX2+ renal progenitors from hiPSCs-derived IM cells by performing an unbiased screening of growth factors and small molecules. These factors were not reported to induce renal progenitors in previous studies [14, 32, 42–44]. However, consistent with our results, in one study, the phosphorylation of Smad1, a molecule downstream of BMP signaling, was strongly detected in the posterior primitive streak at stage 7 and subsequently diminished in the IM at stage 10 in chick embryos, although phosphorylation was also detected in the lateral plate mesoderm. In addition, the Smad1 signal was not activated in the nephrogenic mesenchyme, but rather in the nephric duct at stage 14 [51].
It has been reported that the TGF-β2 secreted by UBC enhances the epithelialization of the mouse metanephric mesenchyme in explant cultures [52]. It has also been suggested that the metanephric mesenchyme changes the composition of the extracellular matrix and induces cellular aggregation in the cap mesenchyme in response to TGF-β signals [53]. Although additional studies are needed to elucidate the exact role of BMP and TGF-β signals in early renal differentiation, these findings and our results suggest the involvement of BMP inhibition and TGF-β activation in the induction of renal progenitors from the IM. Screening for growth factors and small molecules using an unbiased high-throughput method can serve as a powerful tool for identifying novel factors that induce the differentiation of ESCs/iPSCs into renal lineages and elucidating the mechanisms of kidney development.
Additional manipulation and optimization of the differentiation protocol, including the selection of inducing factors, as well as further elucidation of the mechanisms of kidney development, are needed to generate nephron progenitors and renal lineage cells with more potent developmental and/or therapeutic competencies from ESCs/iPSCs.
Conclusion
We have developed a method for inducing the differentiation of hiPSCs into tubulogenic renal progenitors and subsequently demonstrated that the renal subcapsular transplantation of these hiPSC-derived progenitor cells ameliorates renal injury in an AKI mouse model induced by I/R injury. Furthermore, we have demonstrated the therapeutic efficacy of cell therapy with renal cells generated from hiPSCs, suggesting that hiPSC-derived renal lineage cells might be used to develop regenerative therapies for kidney diseases.
Supplementary Material
Acknowledgments
We thank Tomomi Sudo, Nagisa Sakurai, and Yasuhide Masuhara for excellent technical assistance; Erika Moriguchi for excellent secretarial assistance; Katsuhiko Asanuma for the gift of antibodies; Hiroyuki Sakurai, Jonathan Barasch, and Andrew P. McMahon for the gift of cell lines; and Takanori Takebe, Masahiro Enomura, Hiroyuki Koike, Emi Yoshizawa, Hideki Taniguchi, Motoko Yanagita, Satomi Nishijima, Yoh Terada, Kunio Yasunaga, Koji Takakura, Kenichi Suzuki, Hiromu Sato, Kanae Mitsunaga, Akitsu Hotta, Makoto Ikeya, Knut Woltjen, Yasuhiro Yamada, Keisuke Okita, Shinji Masui, Yoshinori Yoshida, Hidetoshi Sakurai, and Kohnosuke Mitani for valuable help and scientific comments. This work was supported by a research grant from the Leading Project of MEXT (to S.Y. and K.O.), the Japan Society for the Promotion of Science through its Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program) (to S.Y. and K.O.), a Grant-in-Aid for Young Scientists (B) (to T. Toyohara), the Japan Science and Technology Agency (JST) through PRESTO (to K.O.), the JST Yamanaka iPS Cell Special Project (to S.Y. and K.O.), the research grant Core Center for iPS Cell Research and Technological Development, Research Center Network for Realization of Regenerative Medicine (to S.Y. and K.O.), and the Uehara Memorial Foundation, Takeda Science Foundation, and iPS Cell Research Fund.
Author Contributions
T. Toyohara: conception/design, collection and/or assembly of data, data analysis and interpretation, manuscript writing; S.-I.M. and Y.Y.: conception/design, collection and/or assembly of data, data analysis and interpretation; S.-I.S.: collection and/or assembly of data, data analysis and interpretation; T.I.: conception/design, collection and/or assembly of data; T. Kawamoto, T. Kasahara, A.H., and H.T.: collection and/or assembly of data; T. Toyoda: conception/design, collection and/or assembly of data; T.A., K.T., N.Y., and S.O.: conception and design; A.S.-O.: collection and/or assembly of data, data analysis and interpretation; Y.S.: data analysis and interpretation; S.Y.: conception/design, financial support, provision of study material or patients; K.O.: conception/design, financial support, provision of study material or patients, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
Y.Y. has uncompensated intellectual property rights as an inventor. N.Y. has compensated employment with Astellas Pharma, Inc. S.Y. has compensated intellectual property rights with iPS Academia Japan and is an uncompensated consultant on the scientific advisory board of iPS Academia Japan. K.O. has compensated intellectual property rights and is an uncompensated consultant on the scientific advisory boards of iPS Portal Inc, Japan; has compensated research funding from Astellas Pharma Inc., Japan; and has uncompensated stock options as founder of iPS Portal Inc., Japan. The other authors indicated no potential conflicts of interest.
References
- 1.Osafune K, Takasato M, Kispert A, et al. Identification of multipotent progenitors in the embryonic mouse kidney by a novel colony-forming assay. Development. 2006;133:151–161. doi: 10.1242/dev.02174. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi A, Valerius MT, Mugford JW, et al. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3:169–181. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mugford JW, Sipilä P, McMahon JA, et al. Osr1 expression demarcates a multi-potent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev Biol. 2008;324:88–98. doi: 10.1016/j.ydbio.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.James RG, Kamei CN, Wang Q, et al. Odd-skipped related 1 is required for development of the metanephric kidney and regulates formation and differentiation of kidney precursor cells. Development. 2006;133:2995–3004. doi: 10.1242/dev.02442. [DOI] [PubMed] [Google Scholar]
- 5.Tena JJ, Neto A, de la Calle-Mustienes E, et al. Odd-skipped genes encode repressors that control kidney development. Dev Biol. 2007;301:518–531. doi: 10.1016/j.ydbio.2006.08.063. [DOI] [PubMed] [Google Scholar]
- 6.Mae S, Shono A, Shiota F, et al. Monitoring and robust induction of nephrogenic intermediate mesoderm from human pluripotent stem cells. Nat Commun. 2013;4:1367. doi: 10.1038/ncomms2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Araoka T, Mae S, Kurose Y, et al. Efficient and rapid induction of human iPSCs/ESCs into nephrogenic intermediate mesoderm using small molecule-based differentiation methods. PLoS One. 2014;9:e84881. doi: 10.1371/journal.pone.0084881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boucher CA, Winchester CL, Hamilton GM, et al. Structure, mapping and expression of the human gene encoding the homeodomain protein, SIX2. Gene. 2000;247:145–151. doi: 10.1016/s0378-1119(00)00105-0. [DOI] [PubMed] [Google Scholar]
- 9.Xu J, Liu H, Park JS, et al. Osr1 acts downstream of and interacts synergistically with Six2 to maintain nephron progenitor cells during kidney organogenesis. Development. 2014;141:1442–1452. doi: 10.1242/dev.103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uchino S, Kellum JA, Bellomo R, et al. Acute renal failure in critically ill patients: A multinational, multicenter study. JAMA. 2005;294:813–818. doi: 10.1001/jama.294.7.813. [DOI] [PubMed] [Google Scholar]
- 11.Chawla LS, Eggers PW, Star RA, et al. Acute kidney injury and chronic kidney disease as interconnected syndromes. N Engl J Med. 2014;371:58–66. doi: 10.1056/NEJMra1214243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prowle JR. Acute kidney injury: An intensivist’s perspective. Pediatr Nephrol. 2014;29:13–21. doi: 10.1007/s00467-013-2411-1. [DOI] [PubMed] [Google Scholar]
- 13.Harari-Steinberg O, Metsuyanim S, Omer D, et al. Identification of human nephron progenitors capable of generation of kidney structures and functional repair of chronic renal disease. EMBO Mol Med. 2013;5:1556–1568. doi: 10.1002/emmm.201201584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imberti B, Tomasoni S, Ciampi O, et al. Renal progenitors derived from human iPSCs engraft and restore function in a mouse model of acute kidney injury. Sci Rep. 2015;5:8826. doi: 10.1038/srep08826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Nakagawa M, Koyanagi M, Tanabe K, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 17.Kajiwara M, Aoi T, Okita K, et al. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:12538–12543. doi: 10.1073/pnas.1209979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okita K, Yamakawa T, Matsumura Y, et al. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells. 2013;31:458–466. doi: 10.1002/stem.1293. [DOI] [PubMed] [Google Scholar]
- 19.Suemori H, Yasuchika K, Hasegawa K, et al. Efficient establishment of human embryonic stem cell lines and long-term maintenance with stable karyotype by enzymatic bulk passage. Biochem Biophys Res Commun. 2006;345:926–932. doi: 10.1016/j.bbrc.2006.04.135. [DOI] [PubMed] [Google Scholar]
- 20.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 21.Susa D, Mitchell JR, Verweij M, et al. Congenital DNA repair deficiency results in protection against renal ischemia reperfusion injury in mice. Aging Cell. 2009;8:192–200. doi: 10.1111/j.1474-9726.2009.00463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, John R, Chen J, et al. IRF-1 promotes inflammation early after ischemic acute kidney injury. J Am Soc Nephrol. 2009;20:1544–1555. doi: 10.1681/ASN.2008080843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Chiara L, Fagoonee S, Ranghino A, et al. Renal cells from spermatogonial germline stem cells protect against kidney injury. J Am Soc Nephrol. 2014;25:316–328. doi: 10.1681/ASN.2013040367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xing L, Cui R, Peng L, et al. Mesenchymal stem cells, not conditioned medium, contribute to kidney repair after ischemia-reperfusion injury. Stem Cell Res Ther. 2014;5:101–112. doi: 10.1186/scrt489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noiri E, Peresleni T, Miller F, et al. In vivo targeting of inducible NO synthase with oligodeoxynucleotides protects rat kidney against ischemia. J Clin Invest. 1996;97:2377–2383. doi: 10.1172/JCI118681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelleher SP, Robinette JB, Miller F, et al. Effect of hemorrhagic reduction in blood pressure on recovery from acute renal failure. Kidney Int. 1987;31:725–730. doi: 10.1038/ki.1987.58. [DOI] [PubMed] [Google Scholar]
- 27.Solez K, Morel-Maroger L, Sraer JD. The morphology of “acute tubular necrosis” in man: Analysis of 57 renal biopsies and a comparison with the glycerol model. Medicine (Baltimore) 1979;58:362–376. [PubMed] [Google Scholar]
- 28.Fitzmaurice G, Laird N, Ware J. Applied Longitudinal Analysis. Hoboken, NJ: Wiley-Interscience; 2004. [Google Scholar]
- 29.Weber S, Taylor JC, Winyard P, et al. SIX2 and BMP4 mutations associate with anomalous kidney development. J Am Soc Nephrol. 2008;19:891–903. doi: 10.1681/ASN.2006111282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao J, Ho JN, Lewis JA, et al. In vivo structure-activity relationship study of dorsomorphin analogues identifies selective VEGF and BMP inhibitors. ACS Chem Biol. 2010;5:245–253. doi: 10.1021/cb9002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osafune K, Caron L, Borowiak M, et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 32.Taguchi A, Kaku Y, Ohmori T, et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell. 2014;14:53–67. doi: 10.1016/j.stem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 33.Kispert A, Vainio S, McMahon AP. Wnt-4 is a mesenchymal signal for epithelial transformation of metanephric mesenchyme in the developing kidney. Development. 1998;125:4225–4234. doi: 10.1242/dev.125.21.4225. [DOI] [PubMed] [Google Scholar]
- 34.Brown AC, Muthukrishnan SD, Guay JA, et al. Role for compartmentalization in nephron progenitor differentiation. Proc Natl Acad Sci USA. 2013;110:4640–4645. doi: 10.1073/pnas.1213971110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park JS, Ma W, O’Brien LL, et al. Six2 and Wnt regulate self-renewal and commitment of nephron progenitors through shared gene regulatory networks. Dev Cell. 2012;23:637–651. doi: 10.1016/j.devcel.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barasch J, Qiao J, McWilliams G, et al. Ureteric bud cells secrete multiple factors, including bFGF, which rescue renal progenitors from apoptosis. Am J Physiol. 1997;273:F757–F767. doi: 10.1152/ajprenal.1997.273.5.F757. [DOI] [PubMed] [Google Scholar]
- 37.Sakurai H, Barros EJ, Tsukamoto T, et al. An in vitro tubulogenesis system using cell lines derived from the embryonic kidney shows dependence on multiple soluble growth factors. Proc Natl Acad Sci USA. 1997;94:6279–6284. doi: 10.1073/pnas.94.12.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawachi H, Abrahamson DR, St John PL, et al. Developmental expression of the nephritogenic antigen of monoclonal antibody 5-1-6. Am J Pathol. 1995;147:823–833. [PMC free article] [PubMed] [Google Scholar]
- 39.Unbekandt M, Davies JA. Dissociation of embryonic kidneys followed by reaggregation allows the formation of renal tissues. Kidney Int. 2010;77:407–416. doi: 10.1038/ki.2009.482. [DOI] [PubMed] [Google Scholar]
- 40.Jiang S, Tang Q, Rong R, et al. Mycophenolate mofetil inhibits macrophage infiltration and kidney fibrosis in long-term ischemia-reperfusion injury. Eur J Pharmacol. 2012;688:56–61. doi: 10.1016/j.ejphar.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Cantaluppi V, Biancone L, Quercia A, et al. Rationale of mesenchymal stem cell therapy in kidney injury. Am J Kidney Dis. 2013;61:300–309. doi: 10.1053/j.ajkd.2012.05.027. [DOI] [PubMed] [Google Scholar]
- 42.Takasato M, Er PX, Becroft M, et al. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat Cell Biol. 2014;16:118–126. doi: 10.1038/ncb2894. [DOI] [PubMed] [Google Scholar]
- 43.Lam AQ, Freedman BS, Morizane R, et al. Rapid and efficient differentiation of human pluripotent stem cells into intermediate mesoderm that forms tubules expressing kidney proximal tubular markers. J Am Soc Nephrol. 2014;25:1211–1225. doi: 10.1681/ASN.2013080831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang M, Han YM. Differentiation of human pluripotent stem cells into nephron progenitor cells in a serum and feeder free system. PLoS One. 2014;9:e94888. doi: 10.1371/journal.pone.0094888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Camussi G, Deregibus MC, Bruno S, et al. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010;78:838–848. doi: 10.1038/ki.2010.278. [DOI] [PubMed] [Google Scholar]
- 46.Cantaluppi V, Gatti S, Medica D, et al. Microvesicles derived from endothelial progenitor cells protect the kidney from ischemia-reperfusion injury by microRNA-dependent reprogramming of resident renal cells. Kidney Int. 2012;82:412–427. doi: 10.1038/ki.2012.105. [DOI] [PubMed] [Google Scholar]
- 47.Barak H, Huh SH, Chen S, et al. FGF9 and FGF20 maintain the stemness of nephron progenitors in mice and man. Dev Cell. 2012;22:1191–1207. doi: 10.1016/j.devcel.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poladia DP, Kish K, Kutay B, et al. Role of fibroblast growth factor receptors 1 and 2 in the metanephric mesenchyme. Dev Biol. 2006;291:325–339. doi: 10.1016/j.ydbio.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 49.Dudley AT, Godin RE, Robertson EJ. Interaction between FGF and BMP signaling pathways regulates development of metanephric mesenchyme. Genes Dev. 1999;13:1601–1613. doi: 10.1101/gad.13.12.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colvin JS, Feldman B, Nadeau JH, et al. Genomic organization and embryonic expression of the mouse fibroblast growth factor 9 gene. Dev Dyn. 1999;216:72–88. doi: 10.1002/(SICI)1097-0177(199909)216:1<72::AID-DVDY9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 51.Faure S, de Santa Barbara P, Roberts DJ, et al. Endogenous patterns of BMP signaling during early chick development. Dev Biol. 2002;244:44–65. doi: 10.1006/dbio.2002.0579. [DOI] [PubMed] [Google Scholar]
- 52.Plisov SY, Yoshino K, Dove LF, et al. TGF beta 2, LIF and FGF2 cooperate to induce nephrogenesis. Development. 2001;128:1045–1057. doi: 10.1242/dev.128.7.1045. [DOI] [PubMed] [Google Scholar]
- 53.Oxburgh L, Chu GC, Michael SK, et al. TGFbeta superfamily signals are required for morphogenesis of the kidney mesenchyme progenitor population. Development. 2004;131:4593–4605. doi: 10.1242/dev.01324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.