Abstract
During microsatellite marker development, researchers must choose from a pool of possible primer pairs to further test in their species of interest. In many cases, the goal is maximizing detectable levels of genetic variation. To guide researchers and determine which markers are associated with higher levels of genetic variation, we conducted a literature review based on 6782 genomic microsatellite markers published from 1997–2012. We examined relationships between heterozygosity (He or Ho) or allele number (A) with the following marker characteristics: repeat type, motif length, motif region, repeat frequency, and microsatellite size. Variation across taxonomic groups was also analyzed. There were significant differences between imperfect and perfect repeat types in A and He. Dinucleotide motifs exhibited significantly higher A, He, and Ho than most other motifs. Repeat frequency and motif region were positively correlated with A, He, and Ho, but correlations with microsatellite size were minimal. Higher taxonomic groups were disproportionately represented in the literature and showed little consistency. In conclusion, researchers should carefully consider marker characteristics so they can be tailored to the desired application. If researchers aim to target high genetic variation, dinucleotide motif lengths with large repeat frequencies may be best.
Keywords: genetic variation, heterozygosity, microsatellite, motif, repeat frequency, taxonomy
For many researchers, microsatellites continue to be the marker of choice for surveys of genetic diversity and structure, as well as paternity analysis and mating system estimates in which codominance is essential. Microsatellites, also known as simple sequence repeats (SSRs) or short tandem repeats (STRs), are typically defined as repeated sequences of one to six bases found throughout the nuclear and plastid genomes of eukaryotes (e.g., Zane et al., 2002; Buschiazzo and Gemmell, 2006; Wheeler et al., 2014). Despite the many benefits of these markers (see Estoup and Angers, 1998; Goldstein and Schlötterer, 1999; Selkoe and Toonen, 2006), a disadvantage in developing genomic markers is that in many cases, microsatellite primers must be developed de novo for a species, especially if primers are not available for testing from closely related species or genera. Even then, primers from related taxa may not be conserved (Rubinsztein et al., 1995; Primmer et al., 1996; Whitton et al., 1997; Morin et al., 1998), often requiring de novo development on a species-by-species basis. Traditional methods of microsatellite marker development involve construction of a genomic library through enrichment for microsatellite repeats, cloning, plasmid isolation, and Sanger sequencing (Zane et al., 2002). Although this process can generate several hundred sequences, only a small subset are usually acceptable for subsequent primer design and evaluation because they must contain a desired repeat, be of the appropriate size (usually 100–300 bp), and have suitable room for primer design in the regions flanking the repeated motif (Squirrell et al., 2003). More recently, next-generation sequencing (NGS) technology has dramatically increased the yield of potential microsatellite primer pairs, generating thousands of individual reads (Ekblom and Galindo, 2011; Hoffman and Nichols, 2011), of which at least 2000 primer pairs may be suitable for further testing (Abdelkrim et al., 2009). Consequently, researchers using either traditional or NGS approaches are eventually faced with an array of primer pairs from which a subset must be selected for further testing in the focal species. How does an investigator decide which primers to choose for further development?
Although there are no commonly accepted criteria for selecting these primer pairs, investigators often choose certain markers based on specific characteristics, which are first described as follows. The nucleotide composition of the repeated sequence is called the motif (Abdelkrim et al., 2009), which can be further described by the motif length, also known as the repeat length (Weber, 1990; Scribner and Pearce, 2000) or sometimes repeat unit (Urquhart et al., 1994) (italicized terms are defined in Appendix 1). The motif length reflects the number of bases in the motif that are repeated [e.g., mononucleotide: (T)n, dinucleotide: (TA)n, trinucleotide: (CGG)n, tetranucleotide: (GAAT)n, pentanucleotide: (GATTC)n, and hexanucleotide: (CCGGTA)n]. The number of times that such a motif appears (n) is known as the repeat frequency or repeat array (Scribner and Pearce, 2000). Multiplying the repeat frequency by the number of base pairs in the motif gives the motif region length or motif size range. These motifs may occur in several different types of arrangements, often referred to as motif type, also known as the motif contiguity (Scribner and Pearce, 2000), repeat pattern, or purity of length (Buschiazzo and Gemmell, 2006). Motif types consist of the following: (1) perfect repeats (Estoup and Angers, 1998; Scribner and Pearce, 2000; Bhargava and Fuentes, 2010), also called simple (Levinson and Gutman, 1987) or pure repeats (Rosenbaum and Deinard, 1998; Buschiazzo and Gemmell, 2006), such as (CA)n or (GTAG)n; (2) compound repeats, which are composed of two or more successive sets of perfect repeats, such as (AT)n(GTC)n (Weber, 1990; Estoup and Angers, 1998; Rosenbaum and Deinard, 1998; Scribner and Pearce, 2000); and (3) interrupted repeats, sometimes called imperfect repeats (Estoup and Angers, 1998; Scribner and Pearce, 2000), which contain an intervening, nonrepeat sequence between two or more perfect or compound repeats, e.g., (TC)nCTAG(CCG)n.
It has been suggested that investigators faced with an array of possible primer pairs should select those associated with dinucleotide repeats over more elaborate motif lengths (tri-, tetra-, or pentanucleotide motifs) to ensure higher levels of genetic variation (Levinson and Gutman, 1987; Grist et al., 1993; Chakraborty et al., 1997; Sup Lee et al., 1999; Ellegren, 2000, 2004). In fact, the majority of microsatellite markers (48–67%) found in many species are dinucleotide repeats, but these are less frequent in coding regions (Li et al., 2002). Trinucleotide and hexanucleotide repeats are thought to be more common in coding regions because they do not cause a frameshift (Toth et al., 2000; Ellegren, 2004). In some cases, AT repeats have been favored over CG repeats as resulting in higher variation (Morgante and Olivieri, 1993). Furthermore, a number of studies point out the importance of using repeats with a minimum repeat frequency (Weber, 1990; Morgante and Olivieri, 1993; Wang et al., 1994). Which, if any, of these suggestions are supported by empirical evidence? In this study, we reviewed more than 6000 published genomic microsatellite markers and their associated genetic diversity values obtained from more than 500 published articles in journals and an associated online database. We focused on genetic diversity in terms of the reported number of alleles (A) and levels of expected and observed heterozygosity (He and Ho). We were interested in the following questions:
1. Are different motif types (perfect vs. imperfect) associated with different levels of genetic variation?
2. Are smaller motif lengths (di-, tri-, etc.) associated with greater levels of genetic variation?
3. Is a higher repeat frequency or larger motif region associated with greater levels of genetic variation?
4. Is there a relationship between fragment size and levels of genetic variation?
5. In utilizing such a unique data set, are certain taxonomic groups disproportionately represented in the microsatellite primer development literature? Are there any trends in levels of genetic variation as revealed by microsatellite markers among these taxonomic groups?
MATERIALS AND METHODS
Database compilation
To analyze genomic microsatellites from all plants, including algae, fungi, and both flowering and nonflowering species, we focused on journals in which such markers are usually reported. The database was constructed from predominantly primer note articles published in Molecular Ecology, Molecular Ecology Notes, Molecular Ecology Resources, and American Journal of Botany (AJB). Data were obtained in one of two ways. In the case of Molecular Ecology and associated publications, microsatellite data were obtained directly for the years 1997 to early 2009 from the Molecular Ecology Resources online database (http://tomato.bio.trinity.edu), where authors are required to submit microsatellite primer information as a condition of publication. In these cases, all entries were screened to contain only plant species. The remainder of Molecular Ecology papers published in 2009–2012 as well as all AJB papers from 1996–2012 were screened manually using the Scopus search engine (Elsevier; http://www.scopus.com) by searching for the keyword “microsatellite*” in titles, abstracts, and key words while excluding the words “Animal,” “Animals,” or “Aves.” All citations from the manually compiled papers were then exported to a database in Mendeley (Mendeley Ltd.; https://www.mendeley.com); the microsatellite primer information and measures of population-level genetic variation (i.e., population screenings) were copied from their appropriate tables within each paper and incorporated into a Microsoft Excel (2007) database together with data previously acquired from the Molecular Ecology Resources online database.
Many studies published more recently have embraced NGS technologies in SSR development, pursuing markers that can be mined from publicly available data sets (e.g., the National Center for Biotechnology Information’s [NCBI] GenBank) of genic regions compiled using expressed sequence tags (ESTs). While this approach can generate thousands of putative markers, they are limited to transcribed regions that are presumably under selection, and may exhibit reduced polymorphism compared with genomic SSRs (Cho et al., 2000; Scott et al., 2000; Eujayl et al., 2001; Rungis et al., 2004; Russell et al., 2004; Chabane et al., 2005; Woodhead et al., 2005; Martin et al., 2010). For these reasons, we limited the database to only include genomic SSRs that are assumed to be under neutral selection. Furthermore, the removal and exclusion of genic SSRs included many agricultural crops, which are traditionally inbred beyond what is expected in natural populations.
In cases in which statistics from multiple populations were reported, we selected the single population with the largest sample size to represent genetic diversity of that study, instead of using mean values calculated across populations. This was done to maintain consistency across papers (e.g., compared to studies with only single population screenings) and to best represent the population-level variation present in the species. In instances where multiple populations with the same sample size were reported, one population was selected at random to include in the database. Although recent studies suggesting ideal characteristics of microsatellite markers have only investigated motifs with a minimum repeat frequency (e.g., >6, >10, or >20; Weber, 1990; Morgante and Olivieri, 1993; Wang et al., 1994, respectively), here we did not discriminate against repeat frequency, so as to incorporate the widest possible breadth and depth of markers developed thus far. We also included both monomorphic and polymorphic markers in the database; monomorphic markers thus served as the baseline for comparison of genetic variability values.
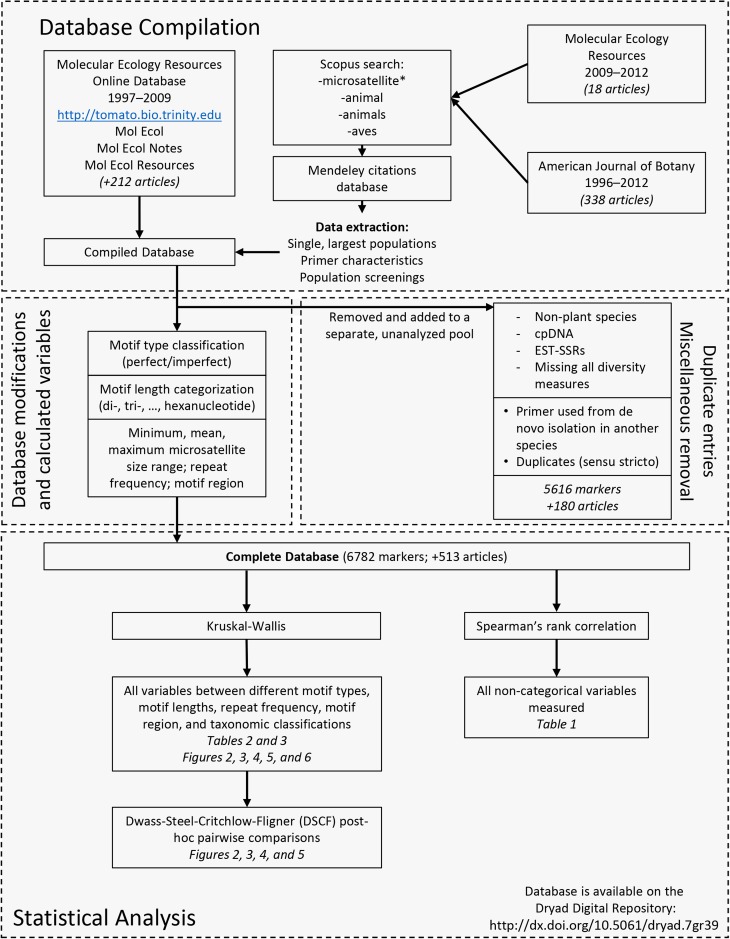
The primer information within the database for each locus consisted of the following information whenever possible: the reported locus name or merit ID (those entries without either of these criteria were assigned a unique number), the primer motif, the number of alleles (A), expected heterozygosity (He), and observed heterozygosity (Ho). We only included data for the species in which the primer was originally designed, as nonspecific primers have shown tendencies to amplify poorly or inconsistently in closely related species (Rubinsztein et al., 1995; Primmer et al., 1996; Whitton et al., 1997; Morin et al., 1998). Within each study, any missing value for a genetic parameter (A, He, or Ho) was represented as a null value but reported zero values were maintained. Figure 1 depicts a schematic workflow of the database compilation.
Fig. 1.
Database compilation/modification workflow. This workflow depicts the general origins, compilations, and modifications of each individual entry into the database with a general “binning” of different variables based on the nature of the measurements and the appropriate statistical analyses used.
Database modifications prior to statistical analysis
Primers were classified by motif type, as either perfect or imperfect based upon the nature or contiguity of the reported repeat motifs. Perfect motifs were simple sequence repeats (SSRs) ranging from two to six nucleotides [e.g., (AC)n or (ATTCGG)n]. Imperfect repeats were classified as compound or interrupted motifs, such as (ATG)n(AC)n or (CT)n(CA)nT(CT)n. The perfect motifs were further classified into different motif lengths, based on the number of nucleotides within the repetitive sequence, from two to six bp. The imperfect motifs, while included in their own separate bin in this study and reported for comparison purposes, were not statistically analyzed in comparison with other SSR motifs because they are more complex in composition and contain greater size ranges. Repeat frequency was extracted when available for each reported marker. The motif region was also calculated for each of the perfect motifs that also included a repeat frequency for each locus. In papers in which a range was reported for the microsatellite size, the minimum, maximum, and mean values of that range were obtained. The lengths of the reported forward and reverse primers (excluding fluorescent label tags) were also calculated. Given that there was very little variation in primer lengths (which only varied within a few base pairs), the data and associated analyses of primer length are not included here (but are available upon request).
Finally, major taxonomic levels from family up to kingdom were incorporated for each species and locus, with a variety of sources used to place taxa accordingly and to update older family classifications as needed (Encyclopedia of Life [http://eol.org/]; ITIS [http://www.itis.gov/]; Stevens, 2001; Mabberley, 2008; The Plant List, 2013; Guiry and Guiry, 2014; Index Fungorum, 2014). A subset of these taxonomic levels were grouped along mono- or dicotyledonous lines and then also in major categories to encapsulate the breadth of divergence in the phylum Tracheophyta. These taxonomic groups include gymnosperms, Nymphaeales, Austrobaileyales, magnoliids, monocots, true eudicots, rosids, and asterids. Mesquite 3.0 build 644 was used to build a cladogram based on the taxonomic tree from the Angiosperm Phylogeny Group (Stevens, 2001).
Duplicate entries
As with any database compilation, duplicate entries had to be addressed. In most cases, straightforward repeated entries were removed. There were, however, a handful of cases of other types of repeated entries that were dealt with individually. Primers were only kept in the database if they included genetic variation parameters for populations of the original species for which they were designed. Therefore, nonspecific primers designed in similarly related species were removed, with the following exceptions. (1) There were three publications (34 entries) in which primer pairs were designed using DNA from two species and the population screening information was provided for distinct populations of each species separately. These were maintained in the database because the primers were effectively species-specific in their design process. (2) In addition, 24 primer pairs revealed multiple loci of amplification in their appropriate species and the population screening information was maintained for each locus as separate primers. (3) Finally, there were 15 entries in which the primer pairs matched but the motif was different. These were all within-species duplicates and were maintained as separate markers. It should be noted that in all cases listed above, alternative permutations, such as reverse complements, were not considered because of the complexity of the data.
The data set with modifications as described above is available on the Dryad Digital Repository (http://dx.doi.org/10.5061/dryad.7gr39; Merritt et al., 2015).
Statistical analysis
SAS/STAT version 9.4 (SAS Institute, Cary, North Carolina, USA) was used to statistically analyze associations across different motif lengths, motif types, microsatellite sizes, repeat frequencies, motif regions, and taxonomic groups with levels of genetic variation, quantified as A, He, and Ho. A Spearman’s rank correlation was used to compare all noncategorical data to identify associations between levels of genetic diversity and microsatellite marker traits. Contingency tables and Fisher’s exact or chi-square tests were used to identify whether certain motif lengths were associated with monomorphic markers. Because the data set exhibited significant deviations from normality in both inspection of quantile-quantile plots and according to the Kolmogorov–Smirnov test, the Kruskal–Wallis test (PROC NPAR1WAY) was used for categorical comparisons. Preliminary analysis used ANOVAs (PROC GLM) with type III sums of squares because this parametric test is generally robust and resistant to deviations from normality; although these tests are not reported here (available upon request), they were in agreement with the results of the Kruskal–Wallis tests.
The posthoc Dwass–Steel–Critchlow–Fligner pairwise comparison (DSCF, a nonparametric equivalent of Tukey’s honest significant difference) test was used to examine differences in levels of genetic variation between each of the groups tested that involve motif lengths.
RESULTS
Trait correlations
A was strongly correlated with He (Spearman’s rank correlation coefficient, rs = 0.835, P < 0.0001; see Table 1) and Ho (rs = 0.530, P < 0.0001). Furthermore, He and Ho were significantly correlated with one another (rs = 0.651, P < 0.0001). There were strong, positive correlations between A and repeat frequency (rs = 0.431, P < 0.0001) and A and motif region (rs = 0.413, P < 0.0001). No significant correlation was found between A and mean microsatellite size (rs = 0.00889, P = 0.467); however, A was inversely correlated with minimum microsatellite size (rs = −0.0769, P < 0.0001) and positively correlated with maximum microsatellite size (rs = 0.127, P < 0.0001). There were no significant correlations with mean microsatellite size and He (rs = −0.0153, P = 0.2226) or Ho (rs = −0.0233, P = 0.0714). A slight but significant inverse correlation was found between minimum microsatellite size and both He (rs = −0.0625, P < 0.0001) and Ho (rs = −0.0581, P < 0.0001); however, maximum microsatellite size was positively correlated with both He (rs = 0.101, P < 0.0001) and Ho (rs = 0.0306, P = 0.0337). Repeat frequency was significantly correlated with He (rs = 0.395, P < 0.0001) and Ho (rs = 0.246, P < 0.0001), but there was no significant correlation with mean microsatellite size (rs = 0.00817, P = 0.5864).
Table 1.
Spearman’s rank correlation matrix comparing levels of genetic variation and marker traits. The upper right side of table contains correlation coefficients (rs) while the bottom left side of the table includes P values with the number of markers included in each pairwise comparison in parentheses. Significant (P < 0.05) correlation coefficients are in bold.
| A | He | Ho | Repeat frequency | Motif region | Mean size | |
| A | – | 0.834 | 0.530 | 0.431 | 0.413 | 0.00889 |
| He | <0.0001 (6336) | – | 0.651 | 0.388 | 0.361 | 0.0153 |
| Ho | <0.0001 (6006) | <0.0001 (5750) | – | 0.246 | 0.213 | −0.0233 |
| Repeat frequency | <0.0001 (4437) | <0.0001 (4250) | <0.0001 (4017) | – | 0.907 | 0.00817 |
| Motif region | <0.0001 (4437) | <0.0001 (4250) | <0.0001 (4017) | <0.0001 (4485) | – | 0.0396 |
| Mean size | 0.4673 (6687) | 0.2226 (6327) | 0.0714 (6016) | 0.5864 (4434) | 0.0084 (4434) | – |
Note: A = number of alleles; He = expected heterozygosity; Ho = observed heterozygosity.
Motif analysis
In analyzing the specific perfect motifs and the imperfect motifs, there were approximately 3061 different motifs reported in the database out of 6782 entries (this estimate refers to unique motifs with differing repeat frequencies and does not take into consideration alternative permutations described below). In the case of the unique dinucleotide repeats, the most abundantly reported motif was GAn (including complementary, reverse, and reverse-complementary permutations: CT, AG, and TC in descending order of frequency), accounting for approximately 34% of all motifs in the data set and 66% of all dinucleotide repeats. The second most abundant dinucleotide motif was CAn (including the reverse, reverse complement, and complementary permutations: AC, GT, and TG) accounting for 15% of all motifs and 30% of all dinucleotide repeats. Of the trinucleotide repeats, the top three most commonly reported were CTTn (including AAG, GAA, and TTC; 31.6% of trinucleotide repeats and 4.01% of all motifs), CAAn (ACC, GTT, and TTG; 11.3% of trinucleotide repeats and 1.43% of all motifs), and GATn (ATC, CTA, and TAG; 6.5% of trinucleotide repeats and 0.826% of all motifs).
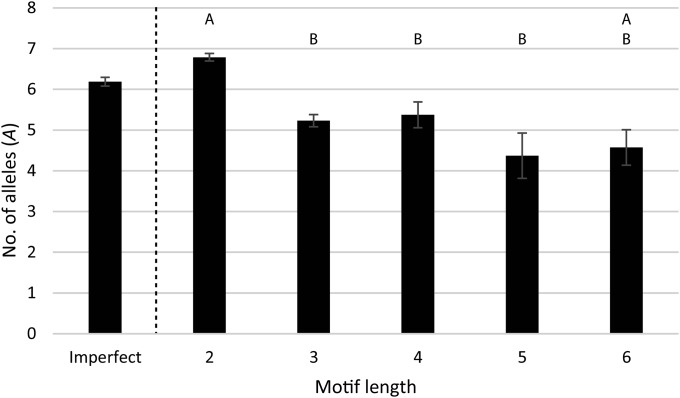
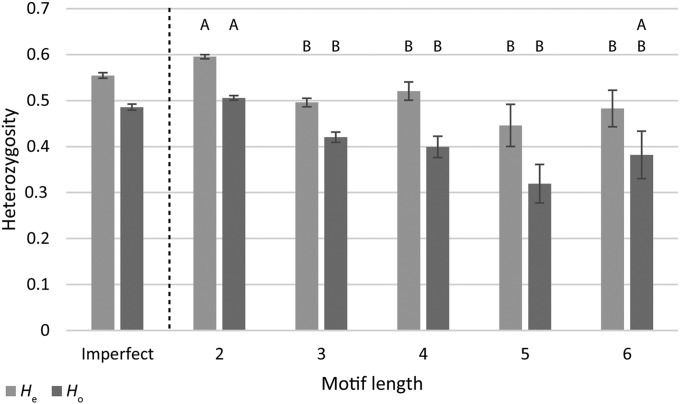
Compared with imperfect motifs, perfect motifs as a group exhibited significantly higher levels of A and He (H = 4.36 and 5.06; P = 0.037 and 0.025, respectively; see Table 2); however, there were no significant differences in Ho (H = 0.04; P = 0.8513). Within perfect motifs, motif lengths differed significantly from one another for A, He, and Ho (H = 107.89, 132.96, and 82.08; P < 0.0001, respectively; see Table 2). The dinucleotide repeat motifs exhibited significantly higher He than any other motif length, and significantly higher A and Ho than the tri-, tetra-, and pentanucleotide repeats (see Table 2, Figs. 2 and 3). Although these significant differences could be a function of the different sample sizes within each motif length group, this is unlikely as the tests incorporate sample size in the calculation.
Table 2.
Marker comparisons of motif types and motif lengths. Each major row includes the mean and number of entries (n) within each category with Kruskal–Wallis statistics (H and the corresponding P value) for comparisons of different major microsatellite characteristic groupings (e.g., motif type and motif length). For each major comparison, levels of genetic variation are included along with repeat frequency, motif region, and mean size. Significant values (P < 0.05) are shown in bold.
| Motif type | Motif lengtha | ||||||
| Variable | Perfect | Imperfect | 2 | 3 | 4 | 5 | 6 |
| A | |||||||
| Mean | 6.43 | 6.19 | 6.81 | 5.23 | 5.37 | 4.39 | 4.57 |
| n | 4595 | 2114 | 3492 | 847 | 185 | 36 | 37 |
| df | 1 | 4 | |||||
| H | 4.36 | 107.89 | |||||
| P value | 0.0369 | <0.0001 | |||||
| He | |||||||
| Mean | 0.575 | 0.554 | 0.598 | 0.495 | 0.520 | 0.461 | 0.473 |
| n | 4398 | 1983 | 3363 | 793 | 174 | 33 | 36 |
| df | 1 | 4 | |||||
| H | 5.06 | 132.96 | |||||
| P value | 0.0245 | <0.0001 | |||||
| Ho | |||||||
| Mean | 0.485 | 0.486 | 0.507 | 0.419 | 0.399 | 0.329 | 0.363 |
| n | 4164 | 1887 | 3193 | 737 | 170 | 34 | 31 |
| df | 1 | 4 | |||||
| H | 0.04 | 82.08 | |||||
| P value | 0.8513 | <0.0001 | |||||
| Repeat frequency | |||||||
| Mean | 13.80 | — | 15.20 | 9.65 | 8.85 | 5.31 | 6.47 |
| n | 4485 | 3427 | 810 | 176 | 36 | 36 | |
| df | 4 | ||||||
| H | 846.38 | ||||||
| P value | <0.0001 | ||||||
| Motif region | |||||||
| Mean | 30.38 | — | 30.40 | 28.96 | 35.41 | 26.53 | 38.83 |
| n | 4485 | 3427 | 810 | 176 | 36 | 36 | |
| df | 4 | ||||||
| H | 28.44 | ||||||
| P value | <0.0001 | ||||||
| Mean sizeb | |||||||
| Mean | 201.25 | 204.50 | 198.03 | 208.00 | 225.45 | 207.79 | 216.03 |
| n | 4592 | 2114 | 3487 | 848 | 186 | 36 | 37 |
| df | 1 | 4 | |||||
| H | 3.91 | 39.56 | |||||
| P value | 0.048 | <0.0001 | |||||
Fig. 2.
Comparison of the number of alleles across different motif lengths. There are significant differences in the motif lengths overall according to the Kruskal–Wallis test; letters above each category depict significantly different groupings based on DSCF posthoc comparisons to further characterize these differences. These groupings indicate that the dinucleotide repeats (2) are significantly different from most other motif lengths. Imperfect motifs are included for side-by-side comparison but were not included in statistical analysis. Error bars represent the standard error of each mean.
Fig. 3.
Comparison of expected and observed levels of heterozygosity across different motif lengths. Letters above each category depict significantly different groupings for either Ho or He, according to DSCF posthoc comparisons. Groupings indicate that dinucleotide repeats (2) are significantly greater than all other motif lengths in He, with the exception of hexanucleotide repeats (6) for Ho. Imperfect motifs are included for side-by-side comparison but were not included in statistical analysis. Error bars represent the standard error of each mean.
Microsatellite characteristics
The mean, minimum, and maximum microsatellite sizes were significantly lower in perfect motif types compared to imperfect motifs (see Table 2). There was a significant difference among the motif lengths in the mean microsatellite size range, with the general trend of size increasing with the number of nucleotides present in the motif (H = 39.6, P < 0.0001; see Table 2). Within perfect motifs, the variation in minimum, mean, and maximum microsatellite sizes was similar across the different motif lengths with respect to magnitude and direction; therefore, only the mean microsatellite size is reported in Table 2.
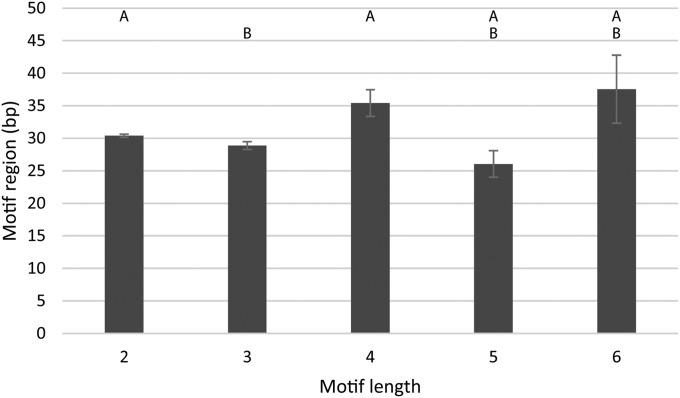
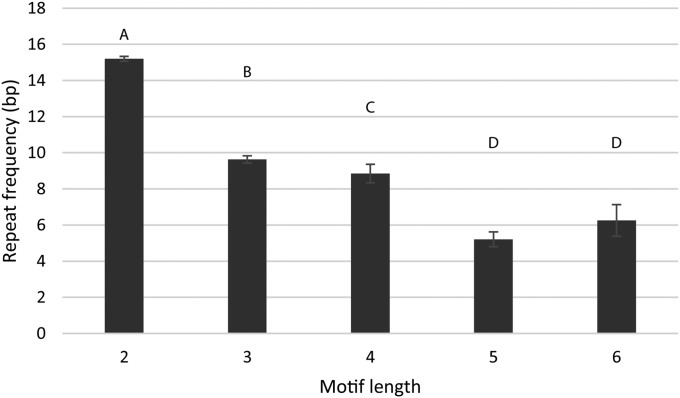
The motif region significantly differed among motif lengths (H = 28.4, P < 0.0001), but there was no consistent trend or relationship across the motif lengths (see Table 2, Fig. 4). Repeat frequencies across the different motif lengths showed very strong significant differences between groups, exhibiting an inverse relationship with the repeat frequency decreasing as motif length increased (H = 846.4, P < 0.0001; see Table 2, Fig. 5).
Fig. 4.
Mean motif region length of different motif lengths. Motif region refers to the total length of the repeated motif size range (e.g., (GCC)6 = 3 bp × 6 = 18). While the lengths of motif regions for each respective motif length may differ significantly from one another, the pattern overall is inconsistent. Error bars represent the standard error of each mean.
Fig. 5.
Mean repeat frequencies of different motif lengths. Repeat frequency refers to the number of times a motif is repeated. Letters above each category depict significantly different groupings according to DSCF posthoc comparisons. There is a significant decrease in repeat frequency as the motif length increases. Error bars represent the standard error of each mean.
Taxonomy
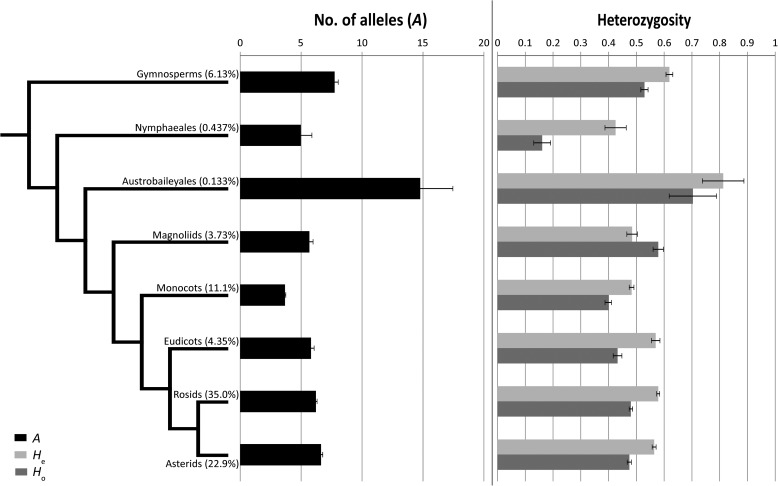
There was no significant difference between monocots and dicots in any of the measures of genetic variation (A, He, and Ho). The monocots did, however, have significantly larger motif regions compared to dicots (H = 17.3, P < 0.0001; see Table 3). Across the different plant taxonomic clades, the gymnosperms exhibited a significantly greater number of alleles than most other plant taxonomic clades, whereas the eudicots, asterids, and rosids had significantly reduced heterozygosity and number of alleles than most of their evolutionarily older counterparts, with the exception of the Nymphaeales (Table 3, Fig. 6).
Table 3.
Group comparisons of major taxonomic clades. Each row includes means and number of entries (n) for each grouping along with Kruskal–Wallis statistics for each comparison made (H and the corresponding P value), including cotyledon type and major taxonomic rankings. Significant (P < 0.05) values are shown in bold.
| Cotyledons | Taxonomic cladesa | ||||||||
| Variable | Dicots | Monocots | Gymnosperms | Nymphaeales | Austrobaileyales | Magnoliids | Eudicots | Rosids | Asterids |
| A | |||||||||
| Mean | 6.33 | 6.49 | 7.75 | 4.97 | 14.78 | 5.68 | 5.81 | 6.21 | 6.63 |
| n | 4374 | 1158 | 416 | 31 | 9 | 270 | 307 | 2447 | 1615 |
| df | 1 | 7 | |||||||
| H | 0.0533 | 54.11 | |||||||
| P value | 0.8175 | <0.0001 | |||||||
| He | |||||||||
| Mean | 0.573 | 0.588 | 0.619 | 0.425 | 0.813 | 0.485 | 0.570 | 0.579 | 0.564 |
| n | 4194 | 1077 | 412 | 29 | 9 | 270 | 280 | 2402 | 1512 |
| df | 1 | 7 | |||||||
| H | 0.968 | 57.69 | |||||||
| P value | 0.3251 | <0.0001 | |||||||
| Ho | |||||||||
| Mean | 0.475 | 0.488 | 0.529 | 0.161 | 0.703 | 0.579 | 0.432 | 0.480 | 0.474 |
| n | 4110 | 1082 | 419 | 29 | 9 | 218 | 298 | 2272 | 1540 |
| df | 1 | 7 | |||||||
| H | 2.07 | 80.65 | |||||||
| P value | 0.1498 | <0.0001 | |||||||
| Repeat frequency | |||||||||
| Mean | 13.5 | 13.8 | 16.0 | 20.2 | 23.8 | 13.7 | 13.8 | 13.5 | 13.5 |
| n | 2860 | 857 | 304 | 13 | 8 | 150 | 185 | 1731 | 942 |
| df | 1 | 7 | |||||||
| H | 1.86 | 65.78 | |||||||
| P value | 0.1770 | <0.0001 | |||||||
| Motif region | |||||||||
| Mean | 29.3 | 31.0 | 34.7 | 40.3 | 47.5 | 29.3 | 29.6 | 29.2 | 29.4 |
| n | 2860 | 857 | 304 | 13 | 8 | 150 | 185 | 1731 | 942 |
| df | 1 | 7 | |||||||
| H | 17.3 | 62.19 | |||||||
| P value | <0.0001 | <0.0001 | |||||||
| Mean size | |||||||||
| Mean | 198.8 | 206.2 | 214.1 | 165.0 | 207.1 | 192.5 | 196.2 | 197.0 | 201.9 |
| n | 4379 | 1157 | 426 | 31 | 9 | 270 | 307 | 2451 | 1616 |
| df | 1 | 7 | |||||||
| H | 1.504 | 28.00 | |||||||
| P value | 0.2201 | 0.0002 | |||||||
Note: A = number of alleles; df = degrees of freedom; He = expected heterozygosity; Ho = observed heterozygosity.
Statistics comparing taxonomic clades include the monocots; however, the means for this group are listed under the Cotyledons column heading.
Fig. 6.
Group comparisons of major taxonomic groups. A (shown in black), He (light gray), and Ho (gray) are shown for each major taxonomic group. The cladogram was drawn with similar taxonomic relationships as those supported by the Angiosperm Phylogeny Group (Stevens, 2001). The percentages in parentheses next to each group name indicate the proportion of entries in the database that comprise each clade. Error bars represent the standard error of each mean.
Even though microsatellite primers tested across genera or more distantly related species had been purposely removed from the data set, there were still 49 entries (24 microsatellite markers) for which the motifs and primer pairs matched across multiple loci. Thirty-eight entries were identified where matching primer sequences (forward and reverse) were found but with differing repeat motifs reported (19 primers total). In all of these instances, they were of the same genera. However, there was one primer with Davidia involucrata Baill. (Nyssaceae) and Hedyotis chrysotricha (Palib.) Merr. (Rubiaceae) where the primer sequences matched, although they did have differing repeat motifs. These matches among primer pairs do not take into account alternative motif permutations.
DISCUSSION
The primary goal of this project was to identify specific characteristics of microsatellites that may aid researchers in choosing effective markers for applications requiring genetic variation, such as quantifying population genetic structure and diversity, estimates of mating systems, and paternity analysis. Now that library development and the isolation of putative microsatellite markers have become relatively straightforward, the remaining challenge in the development process is choosing which markers to further investigate and screen for amplification success and polymorphism. Researchers could better focus their time and effort if they knew specific characteristics of microsatellite markers that are associated with higher levels of genetic variation. Here we generate and use a data set for an empirical review on microsatellite markers that have been developed over the past 18 years, to identify relationships across higher taxa, and conclude with specific recommendations for marker selection.
Markers containing dinucleotide repeats exhibited significantly higher levels of genetic variation in A, He, and Ho than most other motif lengths (Table 2, Figs. 2 and 3). This is consistent with other studies that suggest dinucleotide repeats are generally more variable than other motif lengths, most likely due to the relative ease of mutation via DNA slippage during replication (Levinson and Gutman, 1987; Grist et al., 1993; Strand et al., 1993; Tautz and Schlötterer, 1994; Chakraborty et al., 1997; Sup Lee et al., 1999; Ellegren, 2000, 2004). This slippage is also a potential disadvantage to dinucleotide repeats as it can lead to difficulty in scoring alleles on an electropherogram (i.e., more stutter peaks) compared to larger motif lengths (e.g., Brown et al., 1996). In addition, the differences found here may also be due in part to the very large number of dinucleotide microsatellites reported in the literature relative to all other motif lengths (approximately 73%). Although the overrepresentation of dinucleotides can potentially bias the statistical analysis, the Kruskal–Wallis test incorporates sample size and, therefore, unequal sample sizes should be of minimal concern, especially considering the large number of samples within each motif length (e.g., mean number of entries of all motif lengths = 895). Furthermore, even if investigators have been influenced over the years toward choosing dinucleotide repeat motifs, their popularity may be due in large part to the natural prevalence of dinucleotide repeats throughout plant genome (Brown et al., 1996; Zhao and Ganal, 1996; but see Morgante et al., 2002).
To identify traits conferring greater levels of genetic variation with certain microsatellite markers, primer pairs generating monomorphic loci were included as a baseline for the number of alleles and heterozygosity values. In total, there were only 532 (7.67%) reports of monomorphic markers in the database; this is likely an underrepresentation of the actual proportion in nature because many authors and some journals prefer to exclude monomorphic primers from publication, which biases the data set. Given the importance of monomorphic markers in this and other studies, we recommend that researchers report data from these markers whenever possible. When isolating the more common number of alleles (one to seven), we found that monomorphic markers were more likely to be associated with dinucleotide motif lengths (65.4% of monomorphic primers were dinucleotide) than other motif lengths. However, when put into perspective, the vast majority of microsatellite markers are dinucleotide repeats (72.5%; Fisher’s exact, P < 0.0001), and of these, only 8.67% are monomorphic, compared to 9.6–19.0% of markers possessing three to seven alleles. In addition, some cases of monomorphism may indicate the presence of unseen null alleles, which unfortunately are not always reported in the literature and may be difficult to detect accurately; therefore, null alleles could not be included in our analysis.
Many studies have identified the most abundant repeat motifs reported in the literature (Morgante and Olivieri, 1993; Wang et al., 1994; Brown et al., 1996; Zhao and Ganal, 1996; Zane et al., 2002). Contrary to the suggestion that AT repeats should be preferred (Morgante and Olivieri, 1993), here we found that the dinucleotide motif GAn was the most abundant, both as a unique motif and including (in descending order of frequency) the complement (CTn), reverse (AGn), and reverse complement (TCn). This was similar to previous studies in which GA (or AG) is often reported as one of the most abundant repeat motifs (e.g., Wang et al., 1994; Zane et al., 2002). Furthermore, CTTn was the most abundant trinucleotide repeat, along with the reverse complement (AAGn), complement (GAAn), and reverse (TTCn).
Significantly higher levels of genetic variation (A and He) were found in perfect motif types compared with imperfect motif types (Table 2). This finding corroborates previous suggestions that interrupted motifs reduce stutter and therefore result from mechanisms of mutation (e.g., due to slippage in replication; Richards and Sutherland, 1992, 1994; Pépin et al., 1995; Petes et al., 1997; Rossetto, 2001). It should be noted that the inclusion of compound and interrupted repeats into one (imperfect) category may have skewed the levels of genetic variation detected here for that group of motif types; further study isolating these more-specific motif types may ameliorate the effects of having combined these within a single group. In our study, we were most interested in characteristics of perfect microsatellites; future investigations may wish to focus on subtle variations between compound and interrupted motifs.
The strong correlations between A, He, and Ho in the data set were to be expected, given that they all are measures of genetic diversity. The reduced correlation between A and Ho (rs = 0.530) compared to A and He (rs = 0.835) may be attributed to Ho serving as a direct trait specific to the populations tested as opposed to being a trait of the marker used. The negative correlation between microsatellite motif length with A, He, and Ho strongly supports the suggestion that polymorphism in microsatellite markers is generally found in shorter motif sequences, which are more likely to slip (Grist et al., 1993; Chakraborty et al., 1997; Sup Lee et al., 1999; Ellegren, 2004). The positive correlations detected between repeat frequency or motif region with A, He, and Ho suggest that longer repetitive sequences tend to result in greater polymorphism, regardless of mutation mechanisms (Weber and May, 1989; Valdes et al., 1993; Primmer et al., 1996; Ellegren, 2004). One might expect that microsatellite size range and motif length would be dependent upon one another. However, the lack of a relationship detected here suggests that the associated flanking regions around the motif may play a more important role in determining the size of the overall microsatellite fragment. It should also be noted that microsatellite size incorporates the forward and reverse primer lengths, which are selected by the researcher during primer development. Preliminary analysis, removing respective primer lengths for each microsatellite size range, showed no relationship of primer length with microsatellite size or with levels of heterozygosity. Therefore, primer lengths were included in microsatellite size for the remainder of the study. Although the magnitude of correlations between minimum and maximum microsatellite sizes with A, He, and Ho are low, the directionality (inversely with minimum size and positively maximum size) suggests that greater overall lengths of microsatellite sizes will result in greater levels of genetic variability (e.g., a small minimum size and a large maximum size). This is consistent with the expectation that as the size of the marker increases, so too does the repeat frequency and therefore the number of possible alleles. However, the low magnitude of these correlations suggests that there is a point at which this relationship breaks down.
In this study, we intentionally removed genic (EST) SSRs to focus on genomic markers. While genic markers provide distinct advantages over genomic markers, including cross-species compatibility due to conservation of transcribed regions and a high generation rate at low costs through mining data repositories (Varshney et al., 2005), genomic markers typically provide greater levels of genetic variation (Cho et al., 2000; Scott et al., 2000; Eujayl et al., 2001; Rungis et al., 2004; Russell et al., 2004; Chabane et al., 2005; Woodhead et al., 2005; Martin et al., 2010), present fewer null alleles (Varshney et al., 2005), and are less likely to be subject to direct or artificial selection (in the case of agricultural crops). This is not to say that genomic markers are not without their own drawbacks, including intensive time and resource commitment in isolation, relatively reduced cross-species compatibility, and the inability to inform phenotypic expression. Future investigations could focus on genic markers and examine how their characteristics are associated with measures of genetic variation. This would be helpful as previous studies have suggested that using both genic and genomic markers in concert is a more powerful approach than choosing one marker type over another (e.g., Martin et al., 2010).
Given that our data set included marker and trait information across species and families, we wished to exercise its value by examining variation across higher taxonomic groups. Although there are inconsistent patterns in the variation of microsatellite markers across taxonomic clades, more general trends can be described here. First, there was an underrepresentation of gymnosperms and some extant angiosperms (Nymphaeales and Austrobaileyales) in the literature of reported microsatellite markers. The combined mean number of entries (e.g., microsatellite markers) for the gymnosperms and basal angiosperms was approximately 151, compared to more than 1000 in the angiosperms (monocots, true eudicots, rosids, and asterids), including the magnoliids. This is in part due to the limited number of species (and in some cases, even individuals within species) in these more extant clades, but it also highlights an overall popular interest in angiosperms. Second, the higher genetic variation found in gymnosperms compared to most angiosperm clades may in one sense be attributed to their greater evolutionary age. However, as much as SSRs are thought to form via mutation by expansion due to slippage, there are just as easily contractions of tandemly repeated fragments that reduce the size of SSRs, or gradual elimination of repetitive sequences due to point mutations, given enough time (e.g., into cryptically simple motifs; Tautz et al., 1986; Hancock, 1999). In addition, this variability between gymnosperms and angiosperms may be due to greater genetic admixture among species, especially considering the typical wind-pollination strategies of the gymnosperm clade (Faegri and van der Pijl, 1979) compared to the smaller subset of angiosperms that are wind-pollinated. Although the high genetic diversity reported in Austrobaileyales may reflect its taxonomic age, it is more likely due to a sampling bias as there were only nine markers available for the single species in this clade. In the cases of other taxonomic clades, there may be more subtle, species-specific mechanisms at play. For instance, it has been found in eukaryotes (Chang et al., 2001) and bacteria (LeClerc et al., 1996) that mutations within mismatch repair genes may provide an evolutionary benefit to increased or decreased mutability in gene sets, depending on conferred advantages or disadvantages with such mutations. Species-specific mismatch repair genes in different plant taxa may likewise play a role in the widespread lack of consistent patterns across taxonomic clades.
Related discoveries
Of the 6782 microsatellite markers in the database, there was only one that matched primer pairs (but the motif differed) across different genera and even families, without considering reverse, complementarity, and reverse-complementarity of reported markers. This was between Davidia involucrata (Nyssaceae) and Hedyotis chrysotricha (Rubiaceae). This match is in part due to the primer pair coming from the same laboratory (Du et al., 2012; Yuan et al., 2012), but its rarity also illustrates the limited success of cross-species amplification with genomic markers (Rubinsztein et al., 1995; Primmer et al., 1996; Morin et al., 1998) as compared with genic markers (e.g., Varshney et al., 2005). Ellegren et al. (1995) suggest that while genomic loci may be orthologous among related species, the polymorphism about those loci are not generally conserved as evolutionary age increases.
In this study, we did not differentiate between compound and interrupted motifs because we were more interested in finding characteristics of perfect motifs that may aid investigators in choosing which markers to further pursue during the de novo development phase. However, it is worth noting that interrupted motifs have been suggested as a better choice of marker when comparing more distantly related species as they do show a tendency to mutate at a slower rate compared to more instable tandem repeats of perfect motifs (Rossetto, 2001). Further examination of the characteristics of the various types of imperfect motifs in relation to measures of genetic diversity would be informative.
There was another small number of entries where primer pairs matched—with or without matching motifs—that did not warrant outright removal in the initial steps of compiling the database. All of these were within-species duplicates and occurred in multiple loci. In cases where the motif differed in composition (38 entries total, 19 primer pairs), seven motifs differed in repeat frequency, eight contained entirely different motif configurations, one was a complementary motif of equal length, and the last case consisted of three complementary motifs with differing repeat frequencies. The chances of obtaining such identical primer sequences are very low. For example, forward and reverse primer regions containing the repeated sequence of interest are usually chosen to be 15–30 bp in length by the investigator. The minimum odds of finding this same sequence again are 415–30 or 1 in 1.07e9. Either these primers are not long enough to be unique across the genome or recombination may be the mutagenic force behind these particular markers. NGS technology may be useful in further characterizing the role of recombination and crossing over in microsatellite evolution through identifying like microsatellite markers across multiple loci in genomes.
CONCLUSIONS
In this study, we compiled a large, publicly available data set of characteristics of microsatellite markers published over the past 18 years and showed how these traits are associated with levels of genetic variation. This information can now be used to aid researchers developing new microsatellite markers to conserve their time and resources by choosing the most effective markers for population screening. We also used the data set in a preliminary study to identify trends in levels of genetic variation across major taxonomic groups. While this was only an initial analysis, we encourage further research using this data set to explore levels of genetic variability within and across specific taxonomic families. Other potential uses of the database could include looking for associations between motif lengths and null alleles, or examining differences in markers that are in Hardy–Weinberg equilibrium vs. those that are not; both would necessitate revisiting the original literature to quantify null allele presence or Hardy–Weinberg conformity. In addition, the current study focused on genomic markers, but the workflow described here could also be used to examine analogous characteristics of EST-SSRs. Considering the myriad of EST-SSR resources now available via online databases for nonmodel systems, analyses of EST data would require less time-intensive manual data entry than that described here for genomic markers. Finally, other potential uses of the data set include further exploration of theories into the evolution of repetitive DNA.
Although researchers may benefit from including a variety of different types of microsatellite markers in their genetic investigations, several general conclusions can be drawn from the empirical evidence presented here and in the literature. More generally, attention should be given to comparing genetic variation across studies using different motif lengths, as conclusions may vary due in part to the characteristics of the microsatellites rather than only the natural variation present. We recommend that researchers developing primers for fine-grain analysis of population genetic structure and analysis or mating system estimates should focus on dinucleotide repeats exhibiting a large repeat frequency and wide-ranging overall microsatellite fragment size. When working on a more coarse scale with more distantly diverged (either geographically or temporally) species or taxa, the use of either interrupted repeats or lower repeat frequency perfect motifs may aid in capturing the slower mode and tempo of change while still retaining some degree of relatedness. Microsatellites continue to be important and relevant in a wide variety of studies. Therefore, we recommend that researchers carefully consider the characteristics of the markers that they choose to develop with respect to the types of studies they are intended for, rather than randomly selecting primer pairs to further test in the microsatellite development process.
Appendix 1. Glossary of terms.
Flanking sequence: The nucleotides found immediately on either side of the repeated motif within the microsatellite fragment.
[(forward primer)ACGTGTATATATATATATATATAGAGG(reverse primer)]
Microsatellite marker: The entire sequence containing the forward and reverse primers, the repeated motif of interest, and any intervening flanking sequences. This does not include fluorescent markers used to identify fragments.
[(forward primer)ACGTGTATATATATATATATATAGAGG(reverse primer)]
Motif: Nucleotide composition of the repeated sequence.
[(forward primer)ACGTGTATATATATATATATATAGAGG(reverse primer)]
Motif length: The number of bases within the repeated motif.
[di- (TA)n, tri- (CGG)n, tetra- (GAAT)n, etc.]
Motif type: The arrangement of the repeated motif; these can be perfect [(CA)n or (GTAG)n], compound [(AT)n(GTC)n], or interrupted [(TC)nCTAG(CCG)n]. For our study, compound and interrupted repeat types are known as imperfect.
Motif region: The size range of the repeated motif.
[ACGTGTATATATATATATATATAGAGG = (TA)9 = 2 bp × 9 = 18]
Repeat frequency: The number of times (n) that a motif is repeated within a fragment.
[(TA)9]
LITERATURE CITED
- Abdelkrim J., Robertson B., Stanton J.-A., Gemmell N. 2009. Fast, cost-effective development of species-specific microsatellite markers by genomic sequencing. BioTechniques 46: 185–192. [DOI] [PubMed] [Google Scholar]
- Bhargava A., Fuentes F. F. 2010. Mutational dynamics of microsatellites. Molecular Biotechnology 44: 250–266. [DOI] [PubMed] [Google Scholar]
- Brown S. M., Szewc-McFadden A. K., Kresovich S. 1996. Development and application of simple sequence repeat (SSR) loci for plant genome analysis. In P. P. Javhar , Methods of genome analysis in plants, 147–159. CRC Press, New York, New York, USA. [Google Scholar]
- Buschiazzo E., Gemmell N. J. 2006. The rise, fall and renaissance of microsatellites in eukaryotic genomes. BioEssays 28: 1040–1050. [DOI] [PubMed] [Google Scholar]
- Chabane K., Ablett G. A., Cordeiro G. M., Valkoun J., Henry R. J. 2005. EST versus genomic derived microsatellite markers for genotyping wild and cultivated barley. Genetic Resources and Crop Evolution 52: 903–909. [Google Scholar]
- Chakraborty R., Kimmel M., Stivers D. N., Davison L. J., Deka R. 1997. Relative mutation rates at di-, tri-, and tetranucleotide microsatellite loci. Proceedings of the National Academy of Sciences, USA 94: 1041–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. K., Metzgar D., Wills C., Boland C. 2001. Microsatellites in the eukaryotic DNA mismatch repair genes as modulators of evolutionary mutation rate. Genome Research 11: 1145–1146. [DOI] [PubMed] [Google Scholar]
- Cho Y. G., Ishii T., Temnykh S., Chen X., Lipovich L., McCouch S. R., Park W. D., et al. 2000. Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.). Theoretical and Applied Genetics 100: 713–722. [Google Scholar]
- Du Y.-J., Dai Q.-Y., Zhang L.-Y., Qiu Y.-X., Li J.-H., Fu C.-X. 2012. Development of microsatellite markers for the dove tree, Davidia involucrata (Nyssaceae), a rare endemic from China. American Journal of Botany 99: e206–e209. [DOI] [PubMed] [Google Scholar]
- Ekblom R., Galindo J. 2011. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity 107: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. 2000. Heterogeneous mutation processes in human microsatellite DNA sequences. Nature Genetics 24: 400–402. [DOI] [PubMed] [Google Scholar]
- Ellegren H. 2004. Microsatellites: Simple sequences with complex evolution. Nature Reviews Genetics 5: 435–445. [DOI] [PubMed] [Google Scholar]
- Ellegren H., Primmer C. R., Sheldon B. 1995. Microsatellite evolution: Directionality or bias in locus selection? Nature Genetics 11: 360–362. [DOI] [PubMed] [Google Scholar]
- Estoup A., Angers B. 1998. Microsatellites and minisatellites for molecular ecology: Theoretical and empirical considerations. In G. R. Carvalho , Advances in molecular ecology, 55–85. IOS Press, Burke, Virginia, USA. [Google Scholar]
- Eujayl I., Sorrells M., Baum M., Wolters P., Powell W. 2001. Assessment of genotypic variation among cultivated durum wheat based on EST-SSRS and genomic SSRS. Euphytica 119: 39–43. [Google Scholar]
- Faegri K., van der Pijl L. 1979. The principles of pollination ecology, 3rd ed. Pergamon Press, Oxford, United Kingdom. [Google Scholar]
- Goldstein D. B., Schlotterer C. 1999. Microsatellites: Evolution and applications. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- Grist S. A., Figaira F. A., Morley A. A. 1993. Dinucleotide repeat polymorphisms isolated by the polymerase chain reaction. BioTechniques 15: 304–309. [PubMed] [Google Scholar]
- Guiry M. D., Guiry G. M. 2014. AlgaeBase [online]. World-wide electronic publication, National University of Ireland, Galway, Ireland. Website http://www.algaebase.org [accessed 20 June 2014].
- Hancock J. M. 1999. Microsatellites and other simple sequences: Genomic context and mutational mechanisms. In D. B. Goldstein and C. Schlötterer [eds.], Microsatellites: Evolution and applications, 1–9. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- Hoffman J. I., Nichols H. J. 2011. A novel approach for mining polymorphic microsatellite markers in silico. PLoS One 6: e23283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Index Fungorum. 2014. SIF Index Fungorum [online]. Custodians: The Royal Botanic Gardens Kew represented by the Mycology Section, Landcare Research-NZ, represented by the Mycology Group, and the Institute of Microbiology, Chinese Academy of Science, represented by the State Key Laboratory of Mycology. Website http://indexfungorum.org [accessed 20 June 2014].
- LeClerc J. E., Li B., Payne W. L., Cebula T. A. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274: 1208–1211. [DOI] [PubMed] [Google Scholar]
- Levinson G., Gutman G. A. 1987. Slipped-strand mispairing: A major mechanism for DNA sequences evolution. Molecular Biology and Evolution 4: 203–221. [DOI] [PubMed] [Google Scholar]
- Li Y.-C., Korol A. B., Fahima T., Beiles A., Nevo E. 2002. Microsatellites: Genomic distribution, putative functions and mutational mechanisms: A review. Molecular Ecology 11: 2453–2465. [DOI] [PubMed] [Google Scholar]
- Mabberley D. J. 2008. Mabberley’s plant book, A portable dictionary of plants, their classifications, and uses, 3rd ed. Cambridge University Press, New York, New York, USA. [Google Scholar]
- Martin M., Mattioni C., Cherubini M., Taurchini D., Villani F. 2010. Genetic diversity in European chestnut populations by means of genomic and genic microsatellite markers. Tree Genetics & Genomes 6: 735–744. [Google Scholar]
- Merritt B. J., Culley T. M., Avanesyan A., Stokes R., Brzyski J. 2015. Data from: An empirical review: Characteristics of plant microsatellite markers that confer higher levels of genetic variation. Dryad Digital Repository. http://dx.doi.org/10.5061/dryad.7gr39. [DOI] [PMC free article] [PubMed]
- Morgante M., Olivieri A. M. 1993. PCR-amplified microsatellites as markers in plant genetics. Plant Journal 3: 175–182. [PubMed] [Google Scholar]
- Morgante M., Hanafey M., Powell W. 2002. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nature Genetics 30: 194–200. [DOI] [PubMed] [Google Scholar]
- Morin P. A., Mahboubi P., Wedel S., Rogers J. 1998. Rapid screening and comparison of human microsatellite markers in baboons: Allele size is conserved, but allele number is not. Genomics 53: 12–20. [DOI] [PubMed] [Google Scholar]
- Pépin L., Amigues Y., Lépingle A., Berthier J. L., Bensaid A., Vaiman D. 1995. Sequence conservation of microsatellites between cattle (Bos taurus), goat (Capra hircus) and related species. Examples of use in parentage testing and phylogeny analysis. Heredity 74: 53–61. [DOI] [PubMed] [Google Scholar]
- Petes T. D., Greenwell P. W., Dominska M. 1997. Stabilization of microsatellite sequences by variant repeats in the yeast Saccharomyces cerevisiae. Genetics 146: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Plant List. 2013. The Plant List: Version 1.1 [online]. Website http://www.theplantlist.org [accessed 20 June 2014].
- Primmer C. R., Møller A. P., Ellegren H. 1996. A wide-ranging survey of cross-species amplification in birds. Molecular Ecology 5: 365–378. [DOI] [PubMed] [Google Scholar]
- Richards R. I., Sutherland G. R. 1992. Dynamic mutations: A new class of mutations causing human disease. Cell 70: 709–712. [DOI] [PubMed] [Google Scholar]
- Richards R. I., Sutherland G. R. 1994. Simple repeat DNA is not replicated simply. Nature Genetics 6: 114–116. [DOI] [PubMed] [Google Scholar]
- Rosenbaum H. C., Deinard A. S. 1998. Caution before claim: An overview of microsatellite analysis in ecology and evolutionary biology. In R. DeSalle and B. Schierwater [eds.], Molecular approaches to ecology and evolution, 87–106. Birkhäuser, Boston, Massachusetts, USA. [Google Scholar]
- Rossetto M. 2001. Sourcing of SSR markers from related plant species. In R. Henry , Plant genotyping: The DNA fingerprinting of plants, 211–224. CABI, Wallingford, United Kingdom. [Google Scholar]
- Rubinsztein D. C., Amos W., Leggo J., Goodburn S., Jain S., Li S., Margolis R. L., et al. 1995. Microsatellite evolution—Evidence for directionality and variation in rate between species. Nature Genetics 10: 337–343. [DOI] [PubMed] [Google Scholar]
- Rungis D., Bérubé Y., Zhang J., Ralph S., Ritland C. E., Ellis B. E., Douglas C., et al. 2004. Robust simple sequence repeat markers for spruce (Picea spp.) from expressed sequence tags. Theoretical and Applied Genetics 109: 1283–1294. [DOI] [PubMed] [Google Scholar]
- Russell J., Booth A., Fuller J., Harrower B., Hedley P., Machray G., Powell W. 2004. A comparison of sequence-based polymorphism and haplotype content in transcribed and anonymous regions of the barley genome. Genome 47: 389–398. [DOI] [PubMed] [Google Scholar]
- Scott K. D., Eggler P., Seaton G., Rossetto M., Ablett E. M., Lee L. S., Henry R. J. 2000. Analysis of SSRs derived from grape ESTs. Theoretical and Applied Genetics 100: 723–726. [Google Scholar]
- Scribner K. T., Pearce J. M. 2000. Microsatellites: evolutionary and methodological background and empirical applications at individual, population and phylogenetic levels. In A. J. Baker , Molecular methods in ecology, 235–267. Blackwell, Oxford, United Kingdom. [Google Scholar]
- Selkoe K. A., Toonen R. J. 2006. Microsatellites for ecologists: A practical guide to using and evaluating microsatellite markers. Ecology Letters 9: 615–629. [DOI] [PubMed] [Google Scholar]
- Squirrell J., Hollingsworth M., Woodhead M., Russell J., Lowe A. J., Gibby M., Powell W. 2003. How much effort is required to isolate nuclear microsatellites from plants? Molecular Ecology 12: 1339–1348. [DOI] [PubMed] [Google Scholar]
- Stevens P. F. 2001. Angiosperm Phylogeny Website. Version 12, July 2012 [online]. Website http://www.mobot.org/MOBOT/research/APweb/ [accessed 20 June 2014].
- Strand M., Prolla T. A., Liskay R. M., Peters T. D. 1993. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365: 274–276. [DOI] [PubMed] [Google Scholar]
- Sup Lee J., Hanford M. G., Genova J. L., Farber R. 1999. Relative stabilities of dinucleotide and tetranucleotide repeats in cultured mammalian cells. Human Molecular Genetics 8: 2567–2572. [DOI] [PubMed] [Google Scholar]
- Tautz D., Trick M., Dover G. A. 1986. Cryptic simplicity in DNA is a major source of genetic variation. Nature 322: 652–656. [DOI] [PubMed] [Google Scholar]
- Tautz D., Schlötterer C. 1994. Simple sequences. Current Opinion in Genetics & Development 4: 832–837. [DOI] [PubMed] [Google Scholar]
- Toth G., Gaspari Z., Jurka J. 2000. Microsatellites in different eukaryotic genomes: Survey and analysis. Genome Research 10: 967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart A., Kimpton C. P., Downes T. J., Gill P. 1994. Variation in short tandem repeat sequences: A survey of twelve microsatellite loci for use as forensic identification markers. International Journal of Legal Medicine 107: 13–20. [DOI] [PubMed] [Google Scholar]
- Valdez A. M., Slatkin M., Freimer N. B. 1993. Allele frequencies at microsatellite loci: The stepwise mutation model revisited. Genetics 133: 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney R., Graner A., Sorrells M. E. 2005. Genic microsatellite markers in plants: Features and applications. Trends in Biotechnology 23: 48–55. [DOI] [PubMed] [Google Scholar]
- Wang Z., Weber J. L., Zhong G., Tanksley S. D. 1994. Survey of plant short tandem DNA repeats. Theoretical and Applied Genetics 88: 1–6. [DOI] [PubMed] [Google Scholar]
- Weber J. L. 1990. Informativeness of human (dC-dA)n · (dG-dT)n polymorphisms. Genomics 7: 524–530. [DOI] [PubMed] [Google Scholar]
- Weber J. L., May P. E. 1989. Abundant classes of human DNA polymorphisms which can be typed using the polymerase chain reaction. American Journal of Human Genetics 44: 388–396. [PMC free article] [PubMed] [Google Scholar]
- Wheeler G. L., Dorman H. E., Buchanan A., Challagundla L., Wallace L. E. 2014. A review of the prevalence, utility, and caveats of using chloroplast simple sequence repeats for studies of plant biology. Applications in Plant Sciences 2: 1400059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitton J., Rieseberg L. H., Ungerer M. C. 1997. Microsatellite loci are not conserved across the Asteraceae. Molecular Biology and Evolution 14: 204–209. [DOI] [PubMed] [Google Scholar]
- Woodhead M., Russell J., Squirrell J., Hollingsworth P. M., Mackenzie K., Gibby M., Powell W. 2005. Comparative analysis of population genetic structure in Athyrium distentifolium (Pteridophyta) using AFLPs and SSRs from anonymous and transcribed gene regions. Molecular Ecology 14: 1681–1695. [DOI] [PubMed] [Google Scholar]
- Yuan N., Comes H. P., Mao Y.-R., Qi X.-S., Qiu Y.-X. 2012. Genetic effects of recent habitat fragmentation in the Thousand-Island Lake region of southeast China on the distylous herb Hedyotis chrysotricha (Rubiaceae). American Journal of Botany 99: 1715–1725. [DOI] [PubMed] [Google Scholar]
- Zane L., Bargelloni L., Patarnello T. 2002. Strategies for microsatellite isolation: A review. Molecular Ecology 11: 1–16. [DOI] [PubMed] [Google Scholar]
- Zhao X., Ganal M. W. 1996. Applications of repetitive DNA sequences in plant genome analysis. In Andrew H. Paterson , Genome mapping in plants, 111–125. Academic Press, San Diego, California, USA. [Google Scholar]