Abstract
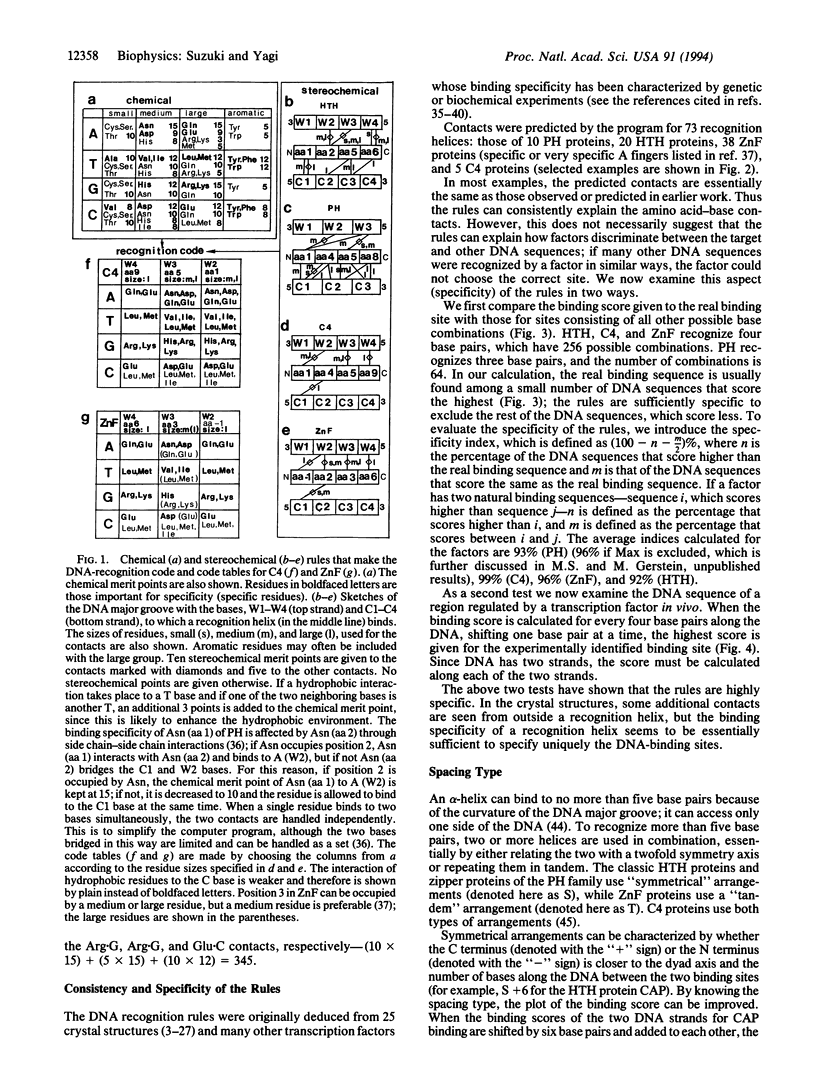
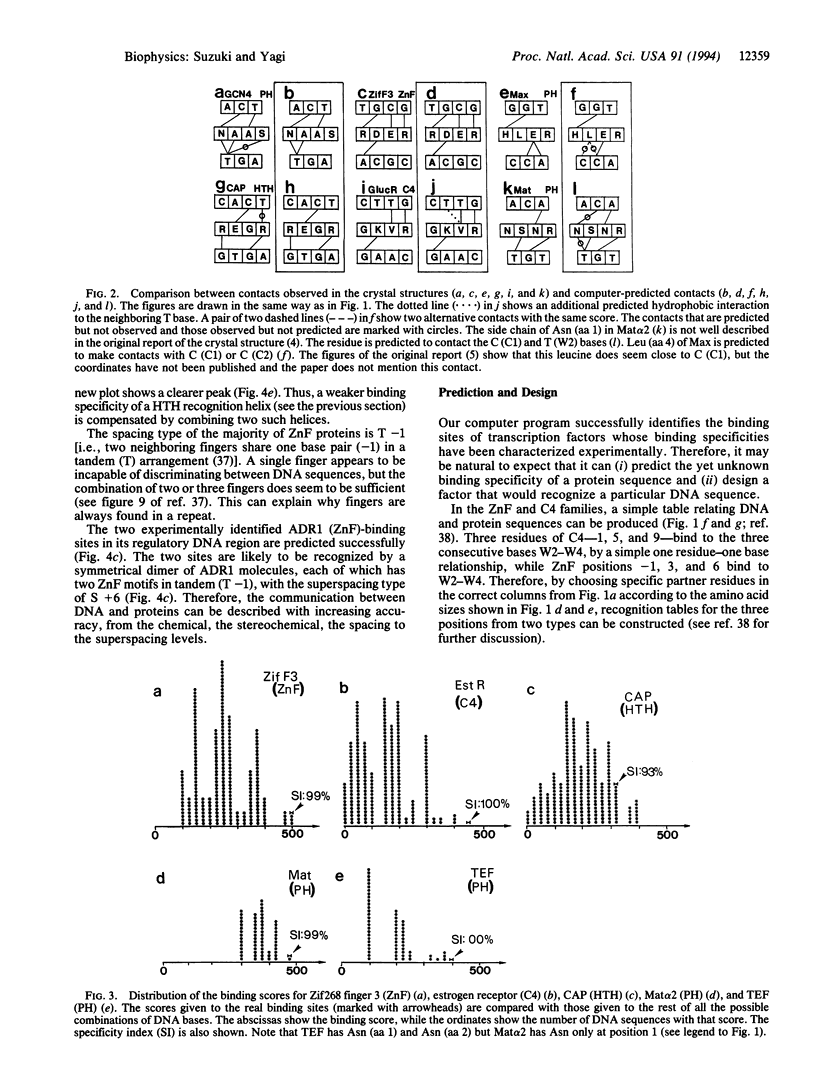
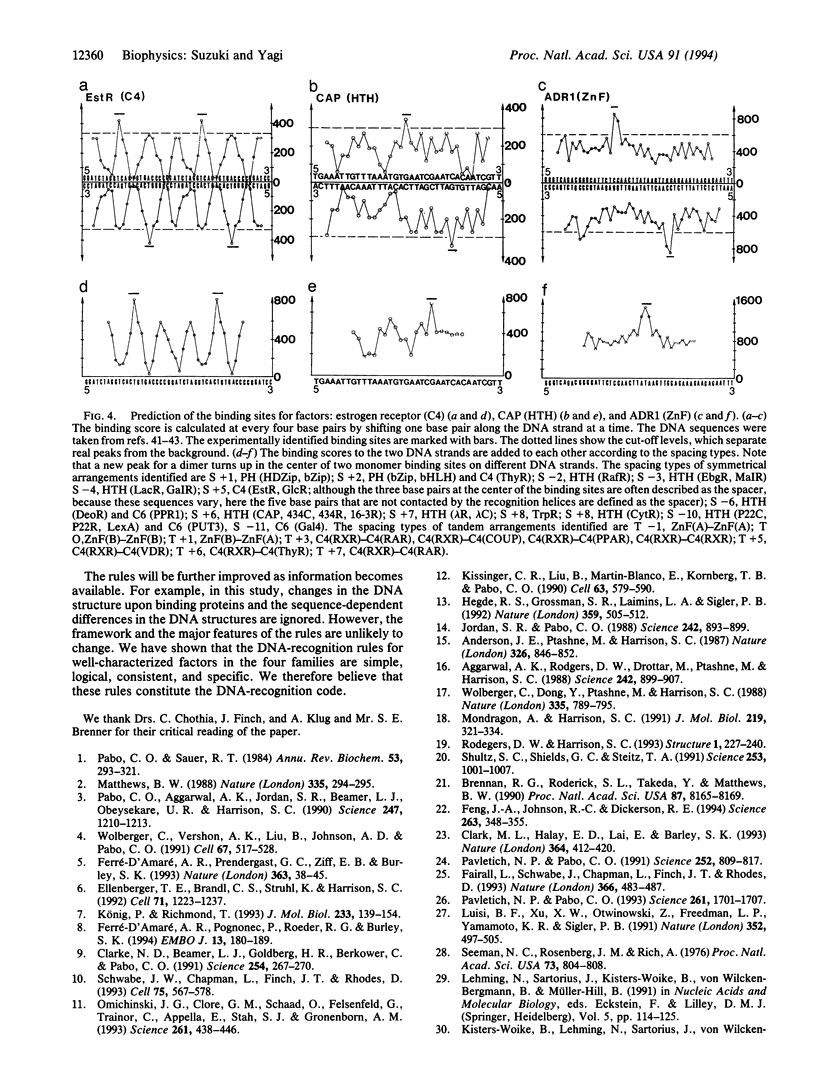
We have previously reported that in four transcription factor families the DNA-recognition rules can be described as (i) chemical rules, which list possible pairings between the 20 amino acid residues and the four DNA bases, and (ii) stereochemical rules, which describe the base and amino acid positions in contact. We have incorporated these rules into a computer program and examined the nature of the rules. Here we conclude that the DNA recognition rules are simple, logical, and consistent. The rules are specific enough to predict DNA-binding characteristics from a protein sequence.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Anderson J. E., Ptashne M., Harrison S. C. Structure of the repressor-operator complex of bacteriophage 434. 1987 Apr 30-May 6Nature. 326(6116):846–852. doi: 10.1038/326846a0. [DOI] [PubMed] [Google Scholar]
- Brennan R. G., Roderick S. L., Takeda Y., Matthews B. W. Protein-DNA conformational changes in the crystal structure of a lambda Cro-operator complex. Proc Natl Acad Sci U S A. 1990 Oct;87(20):8165–8169. doi: 10.1073/pnas.87.20.8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K. L., Halay E. D., Lai E., Burley S. K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993 Jul 29;364(6436):412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Clarke N. D., Beamer L. J., Goldberg H. R., Berkower C., Pabo C. O. The DNA binding arm of lambda repressor: critical contacts from a flexible region. Science. 1991 Oct 11;254(5029):267–270. doi: 10.1126/science.254.5029.267. [DOI] [PubMed] [Google Scholar]
- Deeley M. C., Yanofsky C. Transcription initiation at the tryptophanase promoter of Escherichia coli K-12. J Bacteriol. 1982 Aug;151(2):942–951. doi: 10.1128/jb.151.2.942-951.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjarlais J. R., Berg J. M. Use of a zinc-finger consensus sequence framework and specificity rules to design specific DNA binding proteins. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2256–2260. doi: 10.1073/pnas.90.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger T. E., Brandl C. J., Struhl K., Harrison S. C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992 Dec 24;71(7):1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- Fairall L., Schwabe J. W., Chapman L., Finch J. T., Rhodes D. The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger/DNA recognition. Nature. 1993 Dec 2;366(6454):483–487. doi: 10.1038/366483a0. [DOI] [PubMed] [Google Scholar]
- Feng J. A., Johnson R. C., Dickerson R. E. Hin recombinase bound to DNA: the origin of specificity in major and minor groove interactions. Science. 1994 Jan 21;263(5145):348–355. doi: 10.1126/science.8278807. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A. R., Pognonec P., Roeder R. G., Burley S. K. Structure and function of the b/HLH/Z domain of USF. EMBO J. 1994 Jan 1;13(1):180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D'Amaré A. R., Prendergast G. C., Ziff E. B., Burley S. K. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993 May 6;363(6424):38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- Hegde R. S., Grossman S. R., Laimins L. A., Sigler P. B. Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature. 1992 Oct 8;359(6395):505–512. doi: 10.1038/359505a0. [DOI] [PubMed] [Google Scholar]
- Jordan S. R., Pabo C. O. Structure of the lambda complex at 2.5 A resolution: details of the repressor-operator interactions. Science. 1988 Nov 11;242(4880):893–899. doi: 10.1126/science.3187530. [DOI] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Kisters-Woike B., Lehming N., Sartorius J., von Wilcken-Bergmann B., Müller-Hill B. A model of the lac repressor-operator complex based on physical and genetic data. Eur J Biochem. 1991 Jun 1;198(2):411–419. doi: 10.1111/j.1432-1033.1991.tb16030.x. [DOI] [PubMed] [Google Scholar]
- Klevit R. E. Recognition of DNA by Cys2,His2 zinc fingers. Science. 1991 Sep 20;253(5026):1367–1393. doi: 10.1126/science.1896847. [DOI] [PubMed] [Google Scholar]
- König P., Richmond T. J. The X-ray structure of the GCN4-bZIP bound to ATF/CREB site DNA shows the complex depends on DNA flexibility. J Mol Biol. 1993 Sep 5;233(1):139–154. doi: 10.1006/jmbi.1993.1490. [DOI] [PubMed] [Google Scholar]
- Luisi B. F., Xu W. X., Otwinowski Z., Freedman L. P., Yamamoto K. R., Sigler P. B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991 Aug 8;352(6335):497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- Matthews B. W. Protein-DNA interaction. No code for recognition. Nature. 1988 Sep 22;335(6188):294–295. doi: 10.1038/335294a0. [DOI] [PubMed] [Google Scholar]
- Mondragón A., Harrison S. C. The phage 434 Cro/OR1 complex at 2.5 A resolution. J Mol Biol. 1991 May 20;219(2):321–334. doi: 10.1016/0022-2836(91)90568-q. [DOI] [PubMed] [Google Scholar]
- Omichinski J. G., Clore G. M., Schaad O., Felsenfeld G., Trainor C., Appella E., Stahl S. J., Gronenborn A. M. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science. 1993 Jul 23;261(5120):438–446. doi: 10.1126/science.8332909. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Aggarwal A. K., Jordan S. R., Beamer L. J., Obeysekare U. R., Harrison S. C. Conserved residues make similar contacts in two repressor-operator complexes. Science. 1990 Mar 9;247(4947):1210–1213. doi: 10.1126/science.2315694. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993 Sep 24;261(5129):1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Rodgers D. W., Harrison S. C. The complex between phage 434 repressor DNA-binding domain and operator site OR3: structural differences between consensus and non-consensus half-sites. Structure. 1993 Dec 15;1(4):227–240. doi: 10.1016/0969-2126(93)90012-6. [DOI] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Schwabe J. W., Chapman L., Finch J. T., Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993 Nov 5;75(3):567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler-Tuyns A., Walker P., Martinez E., Mérillat A. M., Givel F., Wahli W. Identification of estrogen-responsive DNA sequences by transient expression experiments in a human breast cancer cell line. Nucleic Acids Res. 1986 Nov 25;14(22):8755–8770. doi: 10.1093/nar/14.22.8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suckow M., von Wilcken-Bergmann B., Müller-Hill B. Identification of three residues in the basic regions of the bZIP proteins GCN4, C/EBP and TAF-1 that are involved in specific DNA binding. EMBO J. 1993 Mar;12(3):1193–1200. doi: 10.1002/j.1460-2075.1993.tb05760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M. Common features in DNA recognition helices of eukaryotic transcription factors. EMBO J. 1993 Aug;12(8):3221–3226. doi: 10.1002/j.1460-2075.1993.tb05991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Gerstein M., Yagi N. Stereochemical basis of DNA recognition by Zn fingers. Nucleic Acids Res. 1994 Aug 25;22(16):3397–3405. doi: 10.1093/nar/22.16.3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Neuhaus D., Gerstein M., Aimoto S. Solution structure of the DNA binding octapeptide repeat of the K10 gene product. Protein Eng. 1994 Apr;7(4):461–470. doi: 10.1093/protein/7.4.461. [DOI] [PubMed] [Google Scholar]
- Thukral S. K., Eisen A., Young E. T. Two monomers of yeast transcription factor ADR1 bind a palindromic sequence symmetrically to activate ADH2 expression. Mol Cell Biol. 1991 Mar;11(3):1566–1577. doi: 10.1128/mcb.11.3.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman J., Harris E., Wilson D., Desplan C. The homeodomain: a new face for the helix-turn-helix? Bioessays. 1992 Mar;14(3):145–150. doi: 10.1002/bies.950140302. [DOI] [PubMed] [Google Scholar]
- Umesono K., Murakami K. K., Thompson C. C., Evans R. M. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991 Jun 28;65(7):1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolberger C., Dong Y. C., Ptashne M., Harrison S. C. Structure of a phage 434 Cro/DNA complex. Nature. 1988 Oct 27;335(6193):789–795. doi: 10.1038/335789a0. [DOI] [PubMed] [Google Scholar]
- Wolberger C., Vershon A. K., Liu B., Johnson A. D., Pabo C. O. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991 Nov 1;67(3):517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]


