Abstract
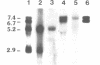
The neural cell adhesion molecule (NCAM) exists in at least three different protein isoforms which are selectively expressed by different cell types and at different stages of development. They are encoded by four to five different transcripts that are derived from a single gene. Here we report the exon--intron structure of the 3' part of the mouse NCAM gene. This region contains six exons. The 5' exon is constitutively expressed in all four prominent size classes of NCAM mRNAs detected in the mouse brain. The second exon contains the poly(A) addition sites for the two smaller mRNAs of 5.2 and 2.9 kb which differ in the length of their 3' non-coding regions and seem both to encode NCAM-120. This second exon is absent in the largest 7.4 kb transcript which encodes NCAM-180; in the 6.7 kb mRNA, which appears to code for NCAM-140, the second and the fifth exon have been spliced out. This data explains how the prominent four transcripts and three protein isoforms of mouse NCAM are generated from a single gene. The alternatively spliced fifth exon is surrounded by inverted repeats potentially capable of secondary structure formation, that may sequester this exon in a loop.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barthels D., Santoni M. J., Wille W., Ruppert C., Chaix J. C., Hirsch M. R., Fontecilla-Camps J. C., Goridis C. Isolation and nucleotide sequence of mouse NCAM cDNA that codes for a Mr 79,000 polypeptide without a membrane-spanning region. EMBO J. 1987 Apr;6(4):907–914. doi: 10.1002/j.1460-2075.1987.tb04837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Andreadis A., Nadal-Ginard B. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Nadal-Ginard B. Complete nucleotide sequence of the fast skeletal troponin T gene. Alternatively spliced exons exhibit unusual interspecies divergence. J Mol Biol. 1986 Apr 5;188(3):313–324. doi: 10.1016/0022-2836(86)90157-9. [DOI] [PubMed] [Google Scholar]
- Cathala G., Savouret J. F., Mendez B., West B. L., Karin M., Martial J. A., Baxter J. D. A method for isolation of intact, translationally active ribonucleic acid. DNA. 1983;2(4):329–335. doi: 10.1089/dna.1983.2.329. [DOI] [PubMed] [Google Scholar]
- Covault J., Merlie J. P., Goridis C., Sanes J. R. Molecular forms of N-CAM and its RNA in developing and denervated skeletal muscle. J Cell Biol. 1986 Mar;102(3):731–739. doi: 10.1083/jcb.102.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham B. A., Hemperly J. J., Murray B. A., Prediger E. A., Brackenbury R., Edelman G. M. Neural cell adhesion molecule: structure, immunoglobulin-like domains, cell surface modulation, and alternative RNA splicing. Science. 1987 May 15;236(4803):799–806. doi: 10.1126/science.3576199. [DOI] [PubMed] [Google Scholar]
- Cunningham B. A., Hoffman S., Rutishauser U., Hemperly J. J., Edelman G. M. Molecular topography of the neural cell adhesion molecule N-CAM: surface orientation and location of sialic acid-rich and binding regions. Proc Natl Acad Sci U S A. 1983 May;80(10):3116–3120. doi: 10.1073/pnas.80.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Eustachio P., Owens G. C., Edelman G. M., Cunningham B. A. Chromosomal location of the gene encoding the neural cell adhesion molecule (N-CAM) in the mouse. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7631–7635. doi: 10.1073/pnas.82.22.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson G., Gower H. J., Barton C. H., Prentice H. M., Elsom V. L., Moore S. E., Cox R. D., Quinn C., Putt W., Walsh F. S. Human muscle neural cell adhesion molecule (N-CAM): identification of a muscle-specific sequence in the extracellular domain. Cell. 1987 Sep 25;50(7):1119–1130. doi: 10.1016/0092-8674(87)90178-4. [DOI] [PubMed] [Google Scholar]
- Edelman G. M. Cell adhesion molecules in the regulation of animal form and tissue pattern. Annu Rev Cell Biol. 1986;2:81–116. doi: 10.1146/annurev.cb.02.110186.000501. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Gennarini G., Hirn M., Deagostini-Bazin H., Goridis C. Studies on the transmembrane disposition of the neural cell adhesion molecule N-CAM. The use of liposome-inserted radioiodinated N-CAM to study its transbilayer orientation. Eur J Biochem. 1984 Jul 2;142(1):65–73. doi: 10.1111/j.1432-1033.1984.tb08251.x. [DOI] [PubMed] [Google Scholar]
- Gennarini G., Hirsch M. R., He H. T., Hirn M., Finne J., Goridis C. Differential expression of mouse neural cell-adhesion molecule (N-CAM) mRNA species during brain development and in neural cell lines. J Neurosci. 1986 Jul;6(7):1983–1990. doi: 10.1523/JNEUROSCI.06-07-01983.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goridis C., Hirn M., Santoni M. J., Gennarini G., Deagostini-Bazin H., Jordan B. R., Kiefer M., Steinmetz M. Isolation of mouse N-CAM-related cDNA: detection and cloning using monoclonal antibodies. EMBO J. 1985 Mar;4(3):631–635. doi: 10.1002/j.1460-2075.1985.tb03676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H. T., Barbet J., Chaix J. C., Goridis C. Phosphatidylinositol is involved in the membrane attachment of NCAM-120, the smallest component of the neural cell adhesion molecule. EMBO J. 1986 Oct;5(10):2489–2494. doi: 10.1002/j.1460-2075.1986.tb04526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H. T., Finne J., Goridis C. Biosynthesis, membrane association, and release of N-CAM-120, a phosphatidylinositol-linked form of the neural cell adhesion molecule. J Cell Biol. 1987 Dec;105(6 Pt 1):2489–2500. doi: 10.1083/jcb.105.6.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemperly J. J., Edelman G. M., Cunningham B. A. cDNA clones of the neural cell adhesion molecule (N-CAM) lacking a membrane-spanning region consistent with evidence for membrane attachment via a phosphatidylinositol intermediate. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9822–9826. doi: 10.1073/pnas.83.24.9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemperly J. J., Murray B. A., Edelman G. M., Cunningham B. A. Sequence of a cDNA clone encoding the polysialic acid-rich and cytoplasmic domains of the neural cell adhesion molecule N-CAM. Proc Natl Acad Sci U S A. 1986 May;83(9):3037–3041. doi: 10.1073/pnas.83.9.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C. C. Homocopolymer sequences in the spacer of a sea urchin histone gene repeat are sensitive to S1 nuclease. Nature. 1982 Feb 25;295(5851):714–716. doi: 10.1038/295714a0. [DOI] [PubMed] [Google Scholar]
- Kornblihtt A. R., Umezawa K., Vibe-Pedersen K., Baralle F. E. Primary structure of human fibronectin: differential splicing may generate at least 10 polypeptides from a single gene. EMBO J. 1985 Jul;4(7):1755–1759. doi: 10.1002/j.1460-2075.1985.tb03847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S. E., Thompson J., Kirkness V., Dickson J. G., Walsh F. S. Skeletal muscle neural cell adhesion molecule (N-CAM): changes in protein and mRNA species during myogenesis of muscle cell lines. J Cell Biol. 1987 Sep;105(3):1377–1386. doi: 10.1083/jcb.105.3.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan B. A., Johnson W. A., Hirsh J. Regulated splicing produces different forms of dopa decarboxylase in the central nervous system and hypoderm of Drosophila melanogaster. EMBO J. 1986 Dec 1;5(12):3335–3342. doi: 10.1002/j.1460-2075.1986.tb04648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B. A., Hemperly J. J., Prediger E. A., Edelman G. M., Cunningham B. A. Alternatively spliced mRNAs code for different polypeptide chains of the chicken neural cell adhesion molecule (N-CAM). J Cell Biol. 1986 Jan;102(1):189–193. doi: 10.1083/jcb.102.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C., Mattei M. G., Mattei J. F., Santoni M. J., Goridis C., Jordan B. R. Localization of the human NCAM gene to band q23 of chromosome 11: the third gene coding for a cell interaction molecule mapped to the distal portion of the long arm of chromosome 11. J Cell Biol. 1986 Mar;102(3):711–715. doi: 10.1083/jcb.102.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybroe O., Albrechtsen M., Dahlin J., Linnemann D., Lyles J. M., Møller C. J., Bock E. Biosynthesis of the neural cell adhesion molecule: characterization of polypeptide C. J Cell Biol. 1985 Dec;101(6):2310–2315. doi: 10.1083/jcb.101.6.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odermatt E., Tamkun J. W., Hynes R. O. Repeating modular structure of the fibronectin gene: relationship to protein structure and subunit variation. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6571–6575. doi: 10.1073/pnas.82.19.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens G. C., Edelman G. M., Cunningham B. A. Organization of the neural cell adhesion molecule (N-CAM) gene: alternative exon usage as the basis for different membrane-associated domains. Proc Natl Acad Sci U S A. 1987 Jan;84(1):294–298. doi: 10.1073/pnas.84.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollerberg E. G., Sadoul R., Goridis C., Schachner M. Selective expression of the 180-kD component of the neural cell adhesion molecule N-CAM during development. J Cell Biol. 1985 Nov;101(5 Pt 1):1921–1929. doi: 10.1083/jcb.101.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollerberg G. E., Burridge K., Krebs K. E., Goodman S. R., Schachner M. The 180-kD component of the neural cell adhesion molecule N-CAM is involved in cell-cell contacts and cytoskeleton-membrane interactions. Cell Tissue Res. 1987 Oct;250(1):227–236. doi: 10.1007/BF00214676. [DOI] [PubMed] [Google Scholar]
- Prentice H. M., Moore S. E., Dickson J. G., Doherty P., Walsh F. S. Nerve growth factor-induced changes in neural cell adhesion molecule (N-CAM) in PC12 cells. EMBO J. 1987 Jul;6(7):1859–1863. doi: 10.1002/j.1460-2075.1987.tb02444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld M. G., Amara S. G., Evans R. M. Alternative RNA processing: determining neuronal phenotype. Science. 1984 Sep 21;225(4668):1315–1320. doi: 10.1126/science.6089345. [DOI] [PubMed] [Google Scholar]
- Ruppert C., Goldowitz D., Wille W. Proto-oncogene c-myc is expressed in cerebellar neurons at different developmental stages. EMBO J. 1986 Aug;5(8):1897–1901. doi: 10.1002/j.1460-2075.1986.tb04442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B., Greene J. M., Green M. R. Cryptic branch point activation allows accurate in vitro splicing of human beta-globin intron mutants. Cell. 1985 Jul;41(3):833–844. doi: 10.1016/s0092-8674(85)80064-7. [DOI] [PubMed] [Google Scholar]
- Sadoul K., Meyer A., Low M. G., Schachner M. Release of the 120 kDa component of the mouse neural cell adhesion molecule N-CAM from cell surfaces by phosphatidylinositol-specific phospholipase C. Neurosci Lett. 1986 Dec 23;72(3):341–346. doi: 10.1016/0304-3940(86)90538-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni M. J., Barthels D., Barbas J. A., Hirsch M. R., Steinmetz M., Goridis C., Wille W. Analysis of cDNA clones that code for the transmembrane forms of the mouse neural cell adhesion molecule (NCAM) and are generated by alternative RNA splicing. Nucleic Acids Res. 1987 Nov 11;15(21):8621–8641. doi: 10.1093/nar/15.21.8621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Small S. J., Shull G. E., Santoni M. J., Akeson R. Identification of a cDNA clone that contains the complete coding sequence for a 140-kD rat NCAM polypeptide. J Cell Biol. 1987 Nov;105(5):2335–2345. doi: 10.1083/jcb.105.5.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- Solnick D., Lee S. I. Amount of RNA secondary structure required to induce an alternative splice. Mol Cell Biol. 1987 Sep;7(9):3194–3198. doi: 10.1128/mcb.7.9.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Wharton K. A., Johansen K. M., Xu T., Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985 Dec;43(3 Pt 2):567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]