Background: A mutation, L166P, in DJ-1, is linked to Parkinson disease.
Results: The Sdj1-L169P fission yeast orthologue of DJ1-L166P is misfolded, associated with chaperones, and degraded via two ubiquitin-proteasome dependent pathways.
Conclusion: Sdj1-L169P is subject to a two-step degradation pathway.
Significance: Mapping the degradation pathways for misfolded proteins is important for our basic understanding of protein quality control in health and disease.
Keywords: chaperone; Parkinson disease; Parkinson disease (autosomal recessive, early onset) 7 (PARK7); proteasome; proteostasis; ubiquitin
Abstract
A mutation, L166P, in the cytosolic protein, PARK7/DJ-1, causes protein misfolding and is linked to Parkinson disease. Here, we identify the fission yeast protein Sdj1 as the orthologue of DJ-1 and calculate by in silico saturation mutagenesis the effects of point mutants on its structural stability. We also map the degradation pathways for Sdj1-L169P, the fission yeast orthologue of the disease-causing DJ-1 L166P protein. Sdj1-L169P forms inclusions, which are enriched for the Hsp104 disaggregase. Hsp104 and Hsp70-type chaperones are required for efficient degradation of Sdj1-L169P. This also depends on the ribosome-associated E3 ligase Ltn1 and its co-factor Rqc1. Although Hsp104 is absolutely required for proteasomal degradation of Sdj1-L169P aggregates, the degradation of already aggregated Sdj1-L169P occurs independently of Ltn1 and Rqc1. Thus, our data point to soluble Sdj1-L169P being targeted early by Ltn1 and Rqc1. The fraction of Sdj1-L169P that escapes this first inspection then forms aggregates that are subsequently cleared via an Hsp104- and proteasome-dependent pathway.
Introduction
Cells are frequently challenged by environmental and physiological stress conditions, which can lead to protein misfolding. Also in the absence of stress, protein misfolding may occur as a result of mutation or defects in synthesis, intracellular trafficking, or association with other macromolecules. If such partially denatured proteins are not efficiently eliminated, they can form toxic intracellular aggregates. To prevent accumulation of such toxic species, nature has evolved elaborate quality control mechanisms. The two main strategies employed seek either to shield the misfolded proteins from aggregation and refold them to the native state by the assistance of molecular chaperones, or, if the native state is unachievable, target the clients for proteasomal degradation or autophagy (1–3). Faults in either system can lead to a build-up of toxic protein species, which has been associated with a variety of severe human diseases, in particular neurodegenerative disorders (4).
In eukaryotic cells, most intracellular proteins are degraded via the ubiquitin-proteasome system. This system depends on a cascade of three types of enzymes termed E1, E2, and E3 that conjugate ubiquitin to specific target proteins (5). Subsequently, proteins marked with polyubiquitin are degraded by the 26S proteasome, a large proteolytic particle found in the nucleus and cytosol of all eukaryotic cells (6). The E3 ubiquitin-protein ligases are the main factors that determine the specificity of ubiquitin conjugation, and thus ultimately control protein degradation. In regard to the degradation of misfolded proteins, it is vital that substrate selection is both sufficiently broad to include a wide variety of substrates, and highly specific, so that only misfolded proteins are targeted (7).
Studies, primarily in yeast, have been pivotal for our current understanding of how misfolded proteins are targeted for degradation. Thus, misfolded secretory proteins are recognized by endoplasmic reticulum (ER)5 luminal chaperones and targeted for so-called ER-associated degradation via the E3 ubiquitin-protein ligases, Hrd1 and Doa10 (8), whereas misfolded cytosolic and nuclear proteins are primarily recognized by Hsp70-type chaperones and ubiquitylated by various cytosolic and/or nuclear E3s such as CHIP, Ubr1, Ubr11, and San1 (3, 7, 9–11). More recently, it was found that ribosomes are equipped with E3 ligases, which carry out co-translational quality control (12). These include the E3 ubiquitin ligase Ltn1, which upon ribosome stalling ubiquitylates nascent proteins to signal their degradation (13, 14). Mutants in lister, the mouse orthologue of yeast LTN1, exhibit profound early-onset and progressive neurological dysfunction (15), suggesting that early quality control is an important mechanism for ensuring proper clearance of aberrant proteins. Studying protein quality control pathways requires appropriate substrates, and to determine how misfolded proteins are targeted, it is essential that the degradation pathways for more misfolded proteins are mapped.
The PARK7/DJ-1 gene is well studied due to its link to monogenic Parkinson disease (16). The encoded DJ-1 protein belongs to the DJ-1/ThiJ/PfpI protein superfamily, which includes proteases, chaperones, kinases, catalases, and transcriptional regulators (17). The DJ-1 protein is a homodimer, where each monomer contains a single domain that adopts a flavodoxin-like helix-strand-helix sandwich (18). The C-terminal α8-helix, which together with the α7-helix is involved in dimerization, is conserved in all DJ-1 orthologues and distinguishes the DJ-1 protein family from many related protein families, including the PfpI and Hsp31 families (19). Several mutations in the DJ-1 gene, all of which are predicted to lead to loss of DJ-1 function, either through deletion or destabilizing mutations, have been connected with Parkinson disease (16). One of the most extensively studied DJ-1 polymorphisms is the L166P point mutation (20) that is located in helix α7. This DJ-1 variant is less structured and unable to form dimers even with wild type DJ-1 (18, 21). In addition, DJ-1-L166P is prone to aggregate in vitro (21, 22), and is rapidly degraded via an unknown mechanism (21, 23).
Here, we map the degradation pathway for the DJ-1-L166P orthologue in the fission yeast Schizosaccharomyces pombe (19). In the following, we will refer to this protein as Sdj1, for S. pombe DJ-1. We show that the fission yeast Sdj1 point mutant is misfolded, fails to dimerize, and aggregates in large cytosolic inclusions. In vivo, the protein is rapidly degraded by the proteasome through a pathway that depends on the Hsp70-type chaperone Ssa2, the Hsp104 disaggregase, and the ribosome-associated E3 ubiquitin-protein ligase Ltn1. Although deletion of any of these proteins leads to accumulation of aggregated Sdj1, aggregate clearance occurs independently of Ltn1, suggesting that Ltn1 regulates degradation of some Sdj1 prior to aggregate formation. In conclusion, our results support a model in which the fission yeast DJ-1 mutant is cleared by a two-step proteasomal degradation pathway. Thus, initially misfolded Sdj1 is regulated by Ltn1 and Rqc1. The fraction that escapes this first inspection then continues to form cytosolic aggregates, which are cleared via an Hsp104- and proteasome-dependent pathway.
Experimental Procedures
S. pombe Strains and Techniques
Fission yeast strains used in this study (Table 1) are derivatives of wild type heterothallic strains 972h− and 975h+. Some strains were purchased from Bioneer (24), others were generously provided by Dr. Jeremy S. Hyams, Dr. Colin Gordon, Dr. Paul Russell, Dr. Mitsuhiro Yanagida, Dr. Snezhana Oliferenko, Dr. Iva M. Tolic-Nørrelykke, and Dr. Shao-Win Wang. Standard genetic methods and media were used and S. pombe transformations were performed using lithium acetate (25). PCR mutagenesis was performed as described (26).
TABLE 1.
Fission yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| Wild type | leu1–32 ura4-D18 | Laboratory stock |
| nas6Δ | nas6::G418 leu1–32 ura4-D18 | 24 |
| atg1Δ | atg1::G418 leu1–32 ura4-D18 | 24 |
| ubr1Δ | ubr1::G418 leu1–32 ura4-D18 | 67 |
| ubr11Δ | ubr11::G418 leu1–32 ura4-D18 | 67 |
| san1Δ | san1::G418 leu1–32 ura4-D18 | 24 |
| hul5Δ | hul5::G418 leu1–32 ura4-D18 | 24 |
| hrd1Δ | hrd1::G418 leu1–32 ura4-D18 | 24 |
| doa10Δ | doa10::G418 leu1–32 ura4-D18 | 24 |
| ltn1Δ | ltn1::G418 leu1–32 ura4-D18 | 24 |
| ufd4Δ | ufd4::G418 leu1–32 ura4-D18 | 24 |
| pub1Δ | pub1::G418 leu1–32 ura4-D18 | 24 |
| pub2Δ | pub2::G418 leu1–32 ura4-D18 | 24 |
| pub3Δ | pub3::G418 leu1–32 ura4-D18 | 24 |
| dma1Δ | dma1::G418 leu1–32 ura4-D18 | 24 |
| hel2Δ | hel2::G418 leu1–32 ura4-D18 | 24 |
| rqc1Δ | rqc1::G418 leu1–32 ura4-D18 | 24 |
| ltn1Δhsp104Δ | ltn1::G418 hsp104::NAT leu1–32 ura4-D18 | This study |
| ssa1Δ | ssa1::G418 leu1–32 ura4-D18 | 24 |
| ssa2Δ | ssa2::G418 leu1–32 ura4-D18 | 65 |
| sks2Δ | sks2::G418 leu1–32 ura4-D18 | 24 |
| hsp104Δ | hsp104::NAT leu1–32 ura4-D18 | This study |
| cox4-RFP | leu1–32::nmt1::coxIVDsRFP (leu1+) ura4-D18 | J. Hyams |
| pabp-mCherry | Pabp::pabp-mCherry (G418) ura4-D18 | 38 |
| hsp104-mCherry | hsp104::hsp104-mCherry (G418) ura4-D18 | 39 |
Plasmids and PCR
To generate Sdj1 (SPAC22E12.03c) constructs, full-length sdj1+ cDNAs were amplified and inserted into pDONR221 (Invitrogen). To generate the Sdj1 L169P variant, site-directed mutagenesis was performed on this plasmid using QuikChange (Stratagene) as described by the manufacturer. For expression in S. pombe, the inserts from the pDONR221 vectors were transferred to the pDUAL vector system containing a C-terminal YFH (YFP, FLAG, and His6) tag (27) using Gateway cloning technology (Invitrogen). For expression of recombinant protein in Escherichia coli, the inserts from the pDONR221 vectors were transferred to the pDEST15 and pDEST17 (Invitrogen) expression vectors, using Gateway cloning technology (Invitrogen).
Binding Assays
The GST fusion proteins were expressed in E. coli BL21 (DE3) or E. coli Rosetta (DE3) and bound to glutathione-Sepharose 4 beads (GE Healthcare) as described by the manufacturer. The protein/bead ratio was 1 mg/ml. Binding experiments were carried out using 20 μl of beads in 1 ml of cleared (13,000 × g for 30 min) 50% extract from E. coli expressing His6-tagged Sdj1. The extracts were prepared by sonication in buffer A (25 mm Tris-HCl, pH 7.5, 50 mm NaCl, 10% glycerol, 0.1% Triton X-100, 2 mm DTT, 1 mm PMSF, and Complete protease inhibitors (Roche Applied Science)). After 4–16 h of tumbling at 4 °C, the beads were washed 4 times in 10 ml of buffer A and resuspended in 30 μl of SDS sample buffer. 20 μl of the samples were analyzed on 12% SDS gels and subjected to Western blotting.
Immunoprecipitations were performed from 50-ml cultures in mid-exponential phase. Cells were lysed using glass beads in 1 volume of buffer B (25 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10% glycerol, 1 mm PMSF and Complete protease inhibitors (Roche Applied Science)) and cleared by centrifugation (13,000 × g for 30 min). The soluble fraction was tumbled end-over-end for 4 h at 4 °C with 10 μl of GFP-trap (Chromotek) beads. The beads were then washed 4 times in 10 ml of buffer B and resuspended in 30 μl of SDS sample buffer. 20 μl of the samples were separated on 12% SDS gels and subjected to Western blot analysis.
Antibodies
The antisera, used in Western blots, were 5A5 mouse monoclonal anti-Hsp70 (Abcam), mouse monoclonal anti-His6 (Qiagen), TAT1 mouse monoclonal anti-tubulin (Abcam), and goat anti-GST (Sigma). Secondary antibodies were purchased from DAKO Cytomation.
Growth Assays
Growth assays on solid media were performed essentially as described (28). Briefly, the S. pombe strains to be assayed were grown to an A600 nm of 0.4–0.8. The cells were then diluted in media to an A600 nm of exactly 0.40. Serial 5-fold dilutions of this culture were prepared before 5 μl of each dilution was spotted onto solid EMM2 media plates and incubated at the indicated temperature until colonies formed.
Fluorescence Microscopy
For microscopy, S. pombe cells were grown to mid-exponential phase. Samples of 5 ml were collected, and the cells were washed by centrifugation (800 × g for 2 min) in sterile-filtered PBS. The cells were fixed with 6% formaldehyde in PBS for 30 min and washed extensively with PBS. The fixed cells were mounted on coverslips using fluoromount (Sigma) and analyzed in a fluorescence microscope (Zeiss AxioImager Z1) and CCD camera (Hamamatsu ORCA-ER).
Aggregate quantification was performed using a hemocytometer. For each sample 150–250 cells were counted.
Mass Spectroscopy
For the mass spectroscopy analyses, 50-ml cultures, expressing Sdj1-YFH, Sdj1-L169P-YFH, or as a control, YFH, were grown to mid-exponential phase. The cells were harvested by centrifugation (800 × g for 2 min) and washed in buffer B (50 mm HEPES/KOH, pH 7.5, 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% sodium deoxycholate) supplemented with 1 mm PMSF and Complete EDTA-free protease inhibitors (Roche Applied Science). The cells were then resuspended in 1 ml of buffer B with inhibitors and carefully dripped into liquid nitrogen. The frozen cell aggregates were then broken with a MM400 ball mill (Retsch) and the extracts were cleared twice by centrifugation (4000 rpm, 10 min). The YFH-tagged proteins were isolated using GFP-trap beads (Chromotek) and washed extensively in buffer B with protease inhibitors as above. Finally, the isolated protein-proteins interactomes were analyzed by mass spectrometry as described previously (29).
Circular Dichroism Spectroscopy
Far-UV circular dichroism spectra were recorded on 5 μm Sdj1 in 10 mm NaH2PO4, pH 7.0, at room temperature on a Jasco J-810 Spectropolarimeter in a 1-mm Quartz SUPRASIL cuvette (Hellma). A total of 15 scans were accumulated from 250 to 190 nm, and a buffer spectrum was recorded identically and subtracted. The scanning speed was 20 nm/min, response time 1 s, and data pitch 0.1 nm. The final spectra were processed by FFT filtering and converted into mean residue ellipticity values.
Protein Degradation Assays
The degradation of Sdj1 and Sdj1 variants was followed in cycloheximide-treated cultures by electrophoresis and blotting as described previously (30). Blots were quantified using the Un-Scan-It software (Silk Scientific). Bortezomib was purchased from LC Laboratories.
Real-time PCR
Total RNA was purified by wild type and ltn1Δ cells using hot phenol as described previously (31), converted to cDNA using Transcriptor First Strand cDNA synthesis kit (Roche Applied Science), and analyzed by real-time PCR using Brilliant SYBR Green QPCR Master Mix (Stratagene) on an MX4000 machine (Stratagene). Actin (act1+) was used for normalization.
In Silico Saturation Mutagenesis with FoldX
The FoldX energy function version 3.1 was used to estimate the free-energy change upon mutations of Sdj1 (32). FoldX is an empirical energy function that employs an all-atom representation of the protein and it has been tested on a dataset of more than 1000 sequence variants from more than 20 different proteins. FoldX calculations were carried out for each of the monomer structures included in the PDB entry 4QYT (33) to assess the reproducibility of the results, and the average is reported here. The RepairPDB function of FoldX was first applied to the wild type structures, during which we also replaced the oxidized (cysteine-sulfinate) Cys-111 present in the crystal structure with its standard thiol form. The known high structural similarity between the reduced and oxidized forms of the human protein (33) suggests that this is a valid approach.
Structures for saturation mutagenesis were generated using an in-house Python program that allows for the introduction of all 19 possible point mutants at each position of the protein using multithreading calculations. The BuildModel function of FoldX was employed and five independent runs were carried out. The results were then averaged and the standard deviation calculated. The mutational free energies were predicted consistently in the different runs using the different starting structures with standard deviations less than 0.8 kcal/mol, which should be considered together with the typical prediction error of FoldX that is also about 0.8 kcal/mol (34).
Results
Sdj1 Is the Fission Orthologue of Human DJ-1
In a database search for proteins involved in stress response and proteostasis, we noticed that the S. pombe SPAC22E12.03c protein was highly similar to the human Parkinson disease protein PARK7/DJ-1. Sequence alignments showed that SPAC22E12.03c is 28% identical and 56% similar to human DJ-1 (Fig. 1A). This similarity is distributed evenly throughout the DJ-1 sequence, and also includes residues in the α8-helix (Fig. 1A) that distinguishes the DJ-1 protein family from related protein families (19). In addition, both the putative active site cysteine residue Cys-106 (C111 in fission yeast) and the disease-relevant Leu-166 (Leu-169 in fission yeast) are conserved (Fig. 1A). We therefore named the SPAC22E12.03c protein Sdj1 for S. pombe DJ-1. Budding yeast, on the other hand, only contains a member of the related Hsp31 family that constitutes another branch of the DJ-1/ThiJ/PfpI superfamily (35, 36).
FIGURE 1.
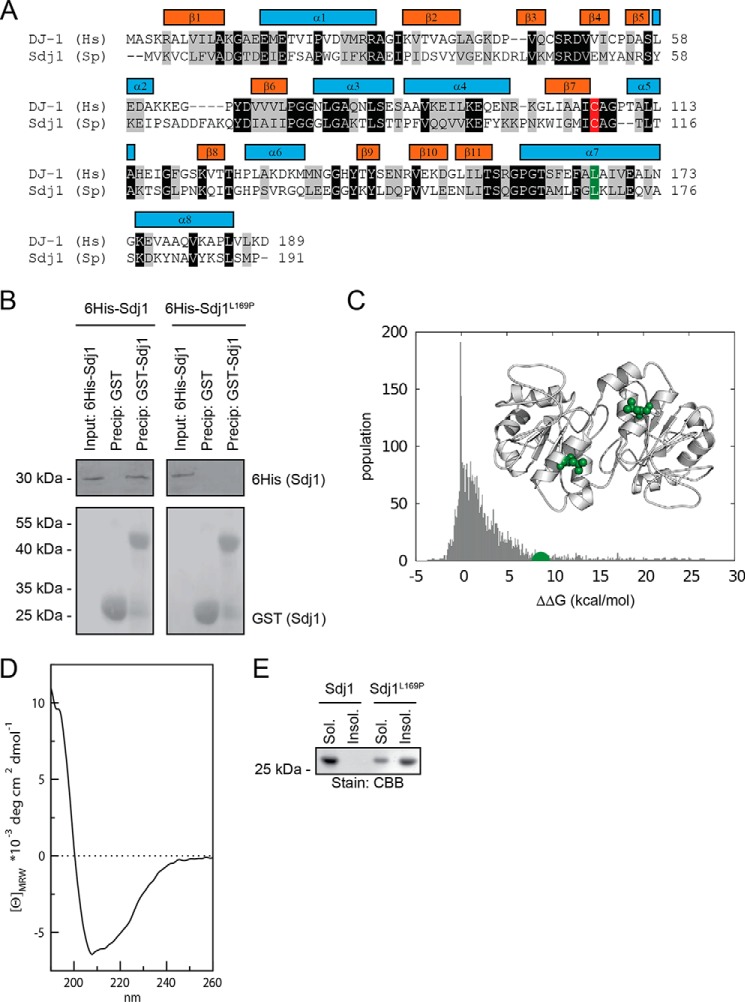
Sdj1 is a fission yeast homolog of DJ-1 and is induced by stress. A, ClustalW sequence alignment of human (Hs) DJ-1 and S. pombe (Sp) SPAC22E12.03c (Sdj1). Identical and homologous residues have been shaded black and gray, respectively. The secondary structure elements of DJ-1 are indicated by colored bars. Note that both the putative active site cysteine residue Cys-106 (Cys-111 in fission yeast) (marked in red) and the disease relevant Leu-166 (L169 in fission yeast) (marked in green) are conserved. B, GST and GST-tagged Sdj1 were used in pulldown experiments with purified His6-tagged Sdj1 or Sdj1-L169P. The precipitated material was analyzed by SDS-PAGE and blotting for the His6-tagged Sdj1 variants (upper panel) or, as a control for even loading, GST (lower panel). C, the plot shows the changes in thermodynamic stability upon all the possible single-site mutations of Sdj1 calculated by FoldX. The value of L169P is marked (green). Note that most Sdj1 mutations are not predicted to affect the Sdj1 structure (values near 0 kcal/mol), very few are predicted to structurally stabilize Sdj1 (negative ΔΔG values), whereas the L169P mutation (green) should destabilize the Sdj1 structure (positive ΔΔG value). The inset shows structure of the Sdj1 homodimer (Protein Data Bank code 4QYT) with the position of Leu-169 (green) marked. D, far-UV circular dichroism spectrum of wild type recombinant Sdj1 purified from E. coli. The maximum at 195 nm, and minimum at 208 nm with the broad shoulder at 222 nm, suggest that the protein is folded and highly α-helical. E, purified wild type Sdj1 and Sdj1-L169P were separated into soluble and insoluble fractions by high-speed centrifugation before the fractions were resolved SDS-PAGE and stained with Coomassie Brilliant Blue (CBB).
The Sdj1-L169P Protein Is Unable to Dimerize and Misfolds
Before studying the Sdj1-L169P degradation mechanism, we first performed a number of experiments to test if S. pombe Sdj1 and Sdj1-L169P behave similarly to human DJ-1 and DJ-1 L166P, respectively.
The disease-associated L166P point mutation induces severe structural distortion in the human DJ-1 protein, which makes the protein unable to form homodimers (21, 22). To test if the corresponding S. pombe mutation, L169P, has similar structural consequences for Sdj1, we generated recombinant His6- and GST-tagged versions of Sdj1 and Sdj1-L169P in E. coli. Glutathione-Sepharose beads loaded with GST-tagged Sdj1, or as a control, GST, were then used for co-precipitation experiments with His6-tagged Sdj1 or Sdj1-L169P. As expected, we found that GST-tagged Sdj1 was able to co-precipitate wild type His6-tagged Sdj1, but not His6-tagged Sdj1-L169P (Fig. 1B). In agreement with this, we observe that the Leu-169 residue is located in the proximity of the subunit-subunit interface in the published crystal structure of Sdj1 (33) (Fig. 1C).
To obtain further indication that Sdj1-L169P is indeed structurally destabilized, we predicted the effects induced upon mutations on Sdj1 stability by FoldX, using the published crystal structure (33) as a starting structure for the calculations. In particular, we performed in silico saturation mutagenesis, introducing all possible single site mutations into the wild type Sdj1 sequence. Using FoldX, we then predicted the change in thermodynamic folding stability with respect to the wild type protein (ΔΔG) (supplemental File S1). The distribution of the resulting ΔΔG values revealed, as expected, that most Sdj1 mutations only moderately affect the thermodynamic stability of Sdj1, i.e. that many ΔΔG values are relatively close to 0 kcal/mol (Fig. 1C). The L169P mutation was, however, predicted to destabilize Sdj1 strongly (∼8 kcal/mol) (Fig. 1C).
To further study whether Sdj-L169P was misfolded, we purified recombinant Sdj1 and Sdj1-L169P from E. coli and analyzed them by circular dichroism spectroscopy. Although the spectrum of wild type Sdj1 suggested that Sdj1, as expected from the known crystal structure, is a folded, helical protein (Fig. 1D), the purified Sdj1-L169P appeared insoluble at the concentrations required for these analyses. Thus, when separating Sdj1 or Sdj1-L169P into soluble and insoluble fractions by high speed centrifugation, Sdj1 was present only in the soluble fraction, whereas Sdj1-L169P was almost exclusively found in the insoluble fraction, suggesting that Sdj1-L169P is misfolded and forms insoluble aggregates (Fig. 1E).
To further substantiate that Sdj1-L169P like DJ1-L166P is misfolded, we analyzed the subcellular localization of Sdj1 and Sdj1-L169P in S. pombe cells. In human cells DJ-1 is localized to the cytosol and nucleus (23), and under stress conditions enriched at mitochondria (37). Sdj1, carrying a C-terminal YFP, FLAG, and His6 (YFH) tag, was evenly distributed throughout the cytosol and nucleus, and did not markedly co-localize with the mitochondrial marker protein, Cox4 (Fig. 2A). This localization was not affected by stress conditions (not shown). However, the Sdj1-L169P mutant aggregated in distinct cytosolic inclusions (Fig. 2A). The aggregates were visible in ∼14% of the cells and each positive cell contained on average only 1–2 of these inclusions.
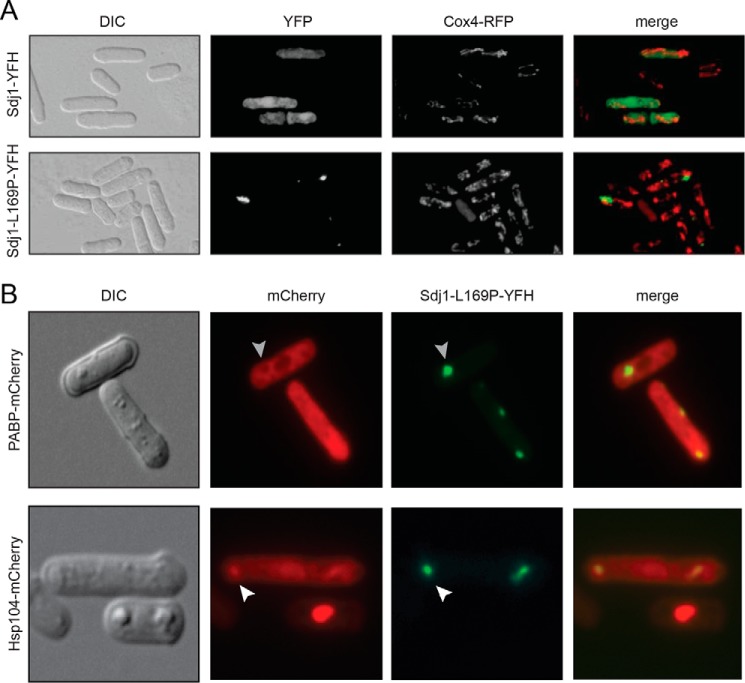
FIGURE 2.
Sdj1-L169P aggregates with Hsp104. A, wild type cells expressing Cox4-RFP and Sdj1-YFH (upper panels) or Sdj1-L169P-YFH (lower panels) were fixed and analyzed by DIC microscopy or fluorescence microscopy as indicated. Note that Sdj1 is spread throughout the cell, whereas Sdj1-L169P forms intracellular aggregates. No co-localization with the mitochondrial Cox4 protein was observed. B, wild type cells expressing mCherry-tagged PABP (left panels) or Hsp104 (right panels) and Sdj1-L169P-YFH (lower panels) were fixed and analyzed by DIC microscopy or fluorescence microscopy as indicated. Note that the PABP signal is excluded from Sdj1-L169P aggregates (gray arrowhead), whereas the Hsp104 and Sdj1-L169P signals overlap (white arrowheads).
PABP is a marker of stress granules, RNA-enriched granules that are formed in response to certain stress conditions (38), whereas Hsp104 is a large molecular chaperone involved in protein quality control and disaggregation of various cytosolic protein aggregates (39). To further examine the nature of the observed Sdj1-L169P aggregates, the localization of Sdj1-L169P was analyzed in strains expressing mCherry-labeled PABP (Pab1) or Hsp104. We did not observe any stress granule formation in cells expressing Sdj1-L169P (Fig. 2B), and hence did not observe PABP enrichment in association with Sdj1-L169P inclusions (Fig. 2B). On the contrary, it appeared that the PABP-mCherry signal was excluded from larger Sdj1-L169P aggregates (Fig. 2B), suggesting that the Sdj1-L169P aggregates were not stress granules. Contrasting this, we observed an enrichment of mCherry-labeled Hsp104 in association with Sdj1-L169P inclusions (Fig. 2B), supporting that these structures contain misfolded and aggregated Sdj1-L169P.
Sdj1-L169P Is Rapidly Degraded via the Ubiquitin-Proteasome Pathway
Having established that Sdj1-L169P, like human DJ1-L166P, is misfolded, we next analyzed the turnover of Sdj1-L169P protein in fission yeast cells.
First, we observed that the steady-state level of Sdj1-L169P was reduced compared with wild type Sdj1 (Fig. 3A). In cultures where protein synthesis was blocked by addition of cycloheximide, we found that wild type Sdj1 appeared stable, whereas the Sdj1-L169P protein was rapidly degraded (Fig. 3B). However, when the proteasome inhibitor bortezomib was added to the culture, the degradation was blocked (Fig. 3B), suggesting that the reduced Sdj1-L169P steady-state level was primarily caused by proteasomal degradation of the protein. To further study if Sdj1-L169P degradation occurred via the proteasome, and not by autophagy, the degradation kinetics of Sdj1-L169P was followed in nas6Δ and atg1Δ mutants, where proteasomal degradation and autophagy, respectively, have been compromised by mutation. Indeed, Sdj1-L169P was more stable in cells deleted for nas6, but not in atg1-deleted cells (Fig. 3C). By quantifying the degradation we found that whereas Sdj1-L169P had a half-life of ∼1 h in wild type and atg1Δ cells, the half-life in the nas6Δ strain was longer than the time course of the experiment (i.e. >3 h) (Fig. 3D).
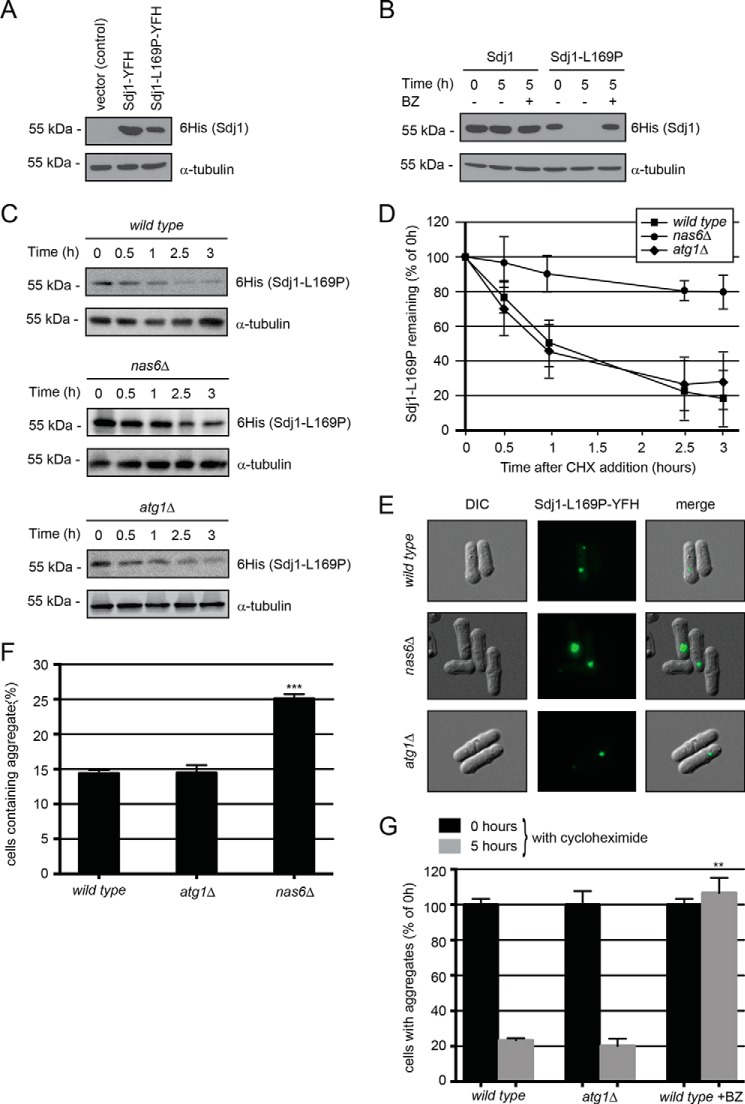
FIGURE 3.
Sdj1-L169P is degraded by the ubiquitin-proteasome system. A, the steady-state levels of Sdj1-YFH, Sdj1-C111A-YFH, and Sdj1-L169P-YFH were compared by SDS-PAGE and Western blotting using antibodies to His6 (to detect the YFH tag on the Sdj1 variants) and as a loading control α-tubulin. B, the amount of Sdj1-YFH or Sdj1-L169P-YFH was followed in cultures treated with cycloheximide (CHX) for 5 h. To some cultures, 1 mm of the proteasome inhibitor bortezomib (BZ) was also added. Equal loading was checked using antibodies to tubulin. C, the amount of Sdj1-L169P-YFH was followed in wild type, nas6Δ, and atg1Δ strains treated with cycloheximide. Equal loading was checked using antibodies to tubulin. D, quantification of degradation experiments as in C. Wild type, filled square; nas6Δ, filled circle; atg1Δ, filled diamond. The error bars indicate the S.E. ± mean (n = 4). E, fluorescence micrographs showing the Sdj1-L169P containing aggregates in the indicated genetic backgrounds. Note the larger aggregates in the nas6Δ strain. F, the number of the Sdj1-L169P-YFH aggregate containing cells was determined by fluorescence microscopy for the indicated strains. The error bars indicate the S.E. ± mean (n = 3, ***, p < 0.001, Student's t test). G, the stability of Sdj1-L169P aggregates in the indicated strains was determined by counting the number of cells containing aggregates in cultures treated with cycloheximide. The number of aggregate containing cells at 0 h was normalized to 100%. The error bars indicate the S.E. ± mean (S.E.) (n = 3, ***, p < 0.001, Student's t test). Before normalization the data were: wild type (0 h), 14.38 ± 0.47 (S.E.); wild type (5 h), 3.35 ± 0.18; atg1Δ (0 h), 14.45 ± 1.11; atg1Δ (5 h), 2.91 ± 0.60; wild type + BZ (0 h), 14.38 ± 0.47; wild type + BZ (5 h), 15.32 ± 1.24.
In accordance with this, the Sdj1-L169P aggregates were also considerably larger in nas6Δ cells compared with the atg1Δ and wild type strains (Fig. 3E), and a higher fraction of nas6Δ cells contained aggregates (25% in nas6Δ cells versus 14% in wild type cells) (Fig. 3F). Finally, we examined the in vivo stability of the Sdj1-L169P aggregates. After 5 h of cycloheximide treatment, the number of cells containing Sdj1-L169P aggregates was reduced to about 20% of the initial amount in wild type and atg1Δ cells (Fig. 3G). However, in the presence of bortezomib, the aggregates appeared stable (Fig. 3G), again confirming that Sdj1-L169P degradation occurs via the ubiquitin-proteasome system.
Sdj1-L169P Associates with the Ssa1 and Ssa2 Hsp70 Chaperones
Because, E3s and substrate proteins form transient enzyme-substrate complexes, substrate-specific E3s can in some cases be identified through their binding to the substrate (40). Therefore, as a starting point for identifying components involved in the degradation of Sdj1-L169P, we compared the protein-protein interactomes of Sdj1 and Sdj1-L169P by quantitative mass spectrometry. To this end, quadruplicates of wild type S. pombe cells expressing YFH-tagged Sdj1, Sdj1-L169P, or as a control, YFH, were grown to late exponential phase and whole cell lysates were prepared. The tagged proteins were isolated by immunoprecipitation and the co-precipitated proteins were identified by liquid chromatography coupled to high-resolution mass spectrometry as previously described (29). As expected, the immunoprecipitated Sdj1 protein was enriched in the precipitated material, along with 144 interaction partners, which were not found in the YFH control. All the identified Sdj1-interacting proteins are listed in supplemental File S2. Sorting the Sdj1 interaction partners according to their gene ontology annotations for biological processes revealed a strong enrichment of translation-related proteins. Thus, 41% of the Sdj1 interacting proteins compared with 11% of the entire fission yeast proteome (supplemental File S2) were gene ontology annotated to translation. In accordance with this, DJ-1 has previously been shown to bind and regulate certain mRNAs (41). In addition, the E. coli DJ-1 orthologue, Yaj1, has also been shown to associate with ribosomes (42, 43).
Interestingly, a few proteins were found to specifically interact with Sdj1-L169P (supplemental File S2). Among these were Ssa1 and Ssa2, two highly similar cytosolic Hsp70-type chaperones. To independently validate the data from mass spectrometry, we subsequently performed immunoprecipitation experiments with wild type Sdj1 and Sdj1-L169P. Indeed, we found that Hsp70 (Ssa1 and Ssa2) was specifically associated with Sdj1-L169P and not wild type Sdj1 (Fig. 4A).
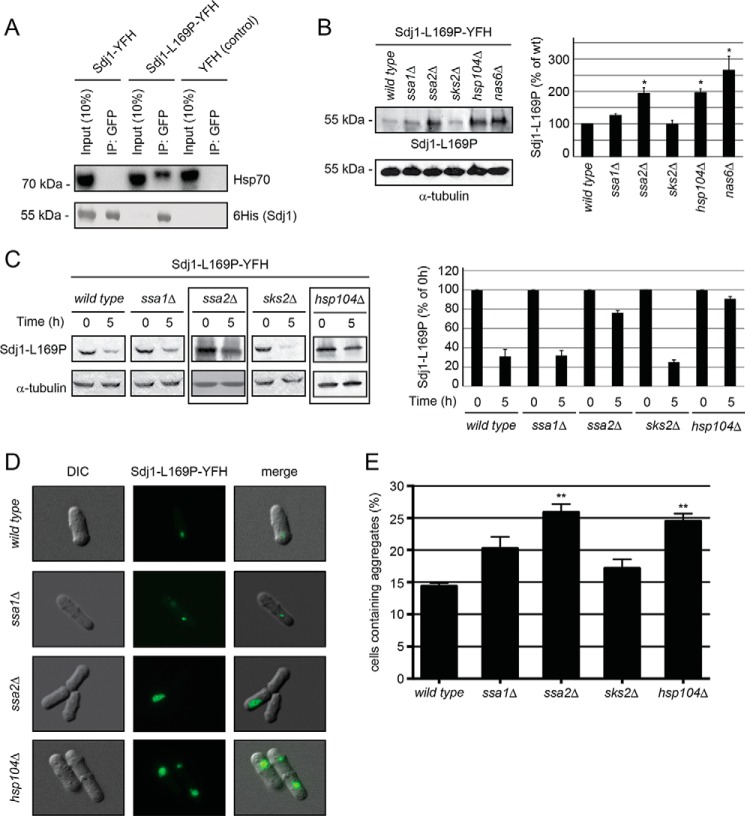
FIGURE 4.
Sdj1-L169P degradation depends on chaperones. A, immunoprecipitates (IP) from wild type S. pombe cells expressing Sdj1-YFH, Sdj1-L169P-YFH, and as a control, YFH. After adjusting the loading, the precipitated material was resolved by SDS-PAGE and analyzed by Western blotting, using antibodies to Hsp70 (upper panel) and His6 (lower panel). B, left panel, the steady-state level of Sdj1-L169P-YFH in the indicated strains was determined by Western blotting of whole cell lysates. Tubulin served as a loading control. Right panel, bar diagrams showing quantification of Western blots normalized to the wild type control. The error bars indicate the S.E. ± mean (n = 3, *, p < 0.05, Student's t test). C, left panel, the amount of Sdj1-L169P-YFH was followed in the indicated chaperone mutant strains treated with cycloheximide. Equal loading was checked using antibodies to tubulin. Note that Sdj1-L169P is stabilized in the ssa2Δ and hsp104Δ strains (boxed). Right panel, bar diagrams showing quantification of Western blots normalized to the signal at 0 h. The error bars indicate the S.E. ± mean (n = 3). D, fluorescence micrographs showing the Sdj1-L169P containing aggregates in the indicated genetic backgrounds. Note the larger aggregates in the ssa2Δ and hsp104Δ strains. E, the number of the Sdj1-L169P-YFH aggregate containing cells was determined by fluorescence microscopy for the indicated strains. The error bars indicate the S.E. ± mean (n = 3, ***, p < 0.001, Student's t test). DIC, differential interference contrast.
Sdj1-L169P Degradation Depends on the Hsp104 and Hsp70 Chaperones
Because we observed that Sdj1-L169P associated with both the Ssa1 and Ssa2 Hsp70-type chaperones and co-localized with the Hsp104 disaggregase, we tested if the steady-state level of Sdj1-L169P was affected in strains where these or other chaperones were compromised by mutation. Indeed, the Sdj1-L169P level was increased in ssa1, ssa2, and hsp104 null mutants, with the most marked effect observed in the ssa2Δ and hsp104Δ strains (Fig. 4B). Accordingly, Ssa2 and Hsp104 were required for efficient Sdj1-L169P degradation, whereas no clear change in the stability of the protein was observed in the ssa1 null mutant (Fig. 4C). The degradation did, however, not rely on the related Hsp70 chaperone Sks2 (Fig. 4, B and C). The Sdj1-L169P aggregates also appeared significantly larger in the ssa2 and hsp104 null mutants, compared with wild type cells (Fig. 4D). Furthermore, when quantifying the fraction of cells containing Sdj1-L169P aggregates, significantly more aggregates were detected in ssa2Δ and hsp104Δ cells (Fig. 4E). We therefore conclude that the Ssa2 Hsp70-type chaperone and the Hsp104 chaperone are involved in chaperone-dependent degradation of Sdj1-L169P.
Degradation of Sdj1-L169P Depends on the E3 Ubiquitin Ligase Ltn1
Although we were hoping that the proteomic analyses would reveal which E3 ubiquitin-protein ligase was responsible for targeting Sdj1-L169P for degradation, such enzyme-substrate complexes are often transient, and our dataset unfortunately did not provide us with any clues as to which E3 was involved in the degradation.
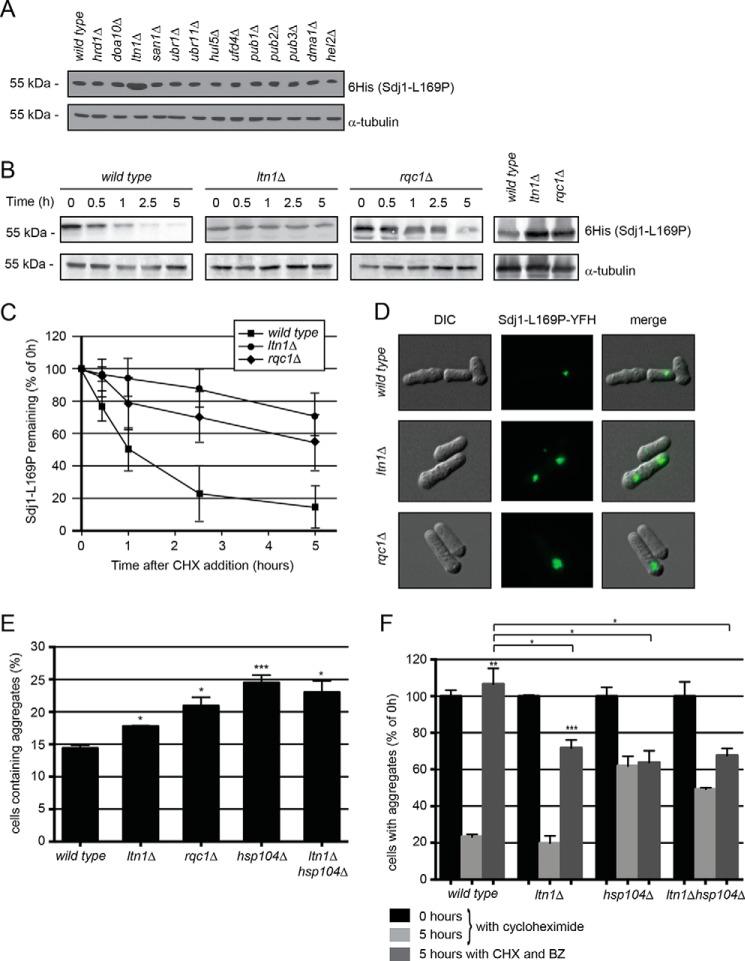
The S. pombe genome encodes roughly 100 different E3 ubiquitin-protein ligases, so to identify the E3 targeting Sdj1-L169P for degradation, we hypothesized that the relevant E3 would likely be specific for misfolded proteins (7) and/or be induced by stress conditions. These parameters led us to select 13 different E3 candidates from available transcriptomic data (44) and literature survey: Hrd1, Doa10, Ltn1, San1, Ubr1, Ubr11, Hul5, Ufd4, Pub1, Pub2, Pub3, Dma1, and Hel2. We then compared the steady-state level of Sdj1-L169P in null mutants of each of these E3s. Surprisingly, only in the ltn1Δ strain did we observe an increased level of Sdj1-L169P (Fig. 5A).
FIGURE 5.
The E3 Ltn1 targets nascent Sdj1-L169P for degradation. A, the steady-state level of Sdj1-L169P-YFH in the indicated strains was determined by Western blotting of whole cell lysates. Tubulin served as a loading control. B, the amount of Sdj1-L169P-YFH was followed in the wild type, ltn1Δ, and rqc1Δ strains treated with cycloheximide (CHX). Equal loading was checked using antibodies to tubulin. C, quantification of degradation experiments as in B. Wild type, filled square; ltn1Δ, filled circle; rqc1Δ, filled diamond. The error bars indicate the S.E. ± mean (n = 4). D, fluorescence micrographs showing the Sdj1-L169P containing aggregates in the indicated genetic backgrounds. Note the larger aggregates in the ltn1Δ and rqc1Δ strains. E, the number of the Sdj1-L169P-YFH aggregate containing cells was determined by fluorescence microscopy for the indicated strains. The error bars indicate the S.E. ± mean (n = 3, *, p > 0.5; ***, p < 0.001, Student's t test). F, the stability of Sdj1-L169P aggregates in the indicated strains was determined by counting the number of cells containing aggregates in cultures treated with cycloheximide. To some cultures (dark gray) the proteasome inhibitor bortezomib (BZ) was also added. The number of aggregate containing cells at 0 h was normalized to 100%. The error bars indicate the S.E. ± mean (n = 3, **, p < 0.01, Student's t test). Before normalization the data were: wild type (0 h), 14.38 ± 0.47 (S.E.); wild type (5 h), 3.35 ± 0.18; wild type + BZ (5 h), 15.32 ± 1.24; ltn1Δ (0 h), 17.77 ± 0.10 (S.E.); ltn1Δ (5 h), 3.52 ± 0.70; ltn1Δ + BZ (5 h), 12.74 ± 0.78; hsp104Δ (0 h), 24.49 ± 1.18; hsp104Δ (5 h), 15.15 ± 1.28; hsp104Δ + BZ (5 h), 15.62 ± 1.56; ltn1Δhsp104Δ (0 h), 23.01 ± 1.79; ltn1Δhsp104Δ (5 h), 11.33 ± 0.19; ltn1Δhsp104Δ + BZ (5 h), 15.54 ± 0.91.
Studies in budding yeast have shown that Ltn1 (also known as Rkr1) is involved in ribosome-associated quality control in cooperation with the proteins Rqc1 and Tae2, forming the ribosome quality control complex (45) (46). To verify the involvement of this complex in the degradation of Sdj1-L169P, we followed the Sdj1-L169P degradation in ltn1 and rqc1 null mutants. Indeed, Sdj1-L169P was more stable in both ltn1 and rqc1 deletion strains (Fig. 5B). Although degradation was still observed in the ltn1Δ and rqc1Δ strains, Sdj1-L169P was significantly stabilized in these mutants compared with the wild type control (Fig. 5C). Accordingly, the steady-state level of Sdj1-L169P was also increased in the rqc1Δ strain (Fig. 5B). Furthermore, in these strains the Sdj1-L169P aggregates were enlarged (Fig. 5D) and more abundant than in the wild type strain (Fig. 5E), although the effect was not as dramatic as for the hsp104Δ and ssa2Δ mutants (Fig. 5E), and was not further increased in an hsp104Δltn1Δ double mutant (Fig. 5E).
Because Ltn1 and Rqc1 are ribosome-associated proteins involved in quality control of nascent proteins, it is unlikely that they regulate degradation of already aggregated Sdj1-L169P. Accordingly, when studying the stability of the formed Sdj1-L169P aggregates in these strains, we observed that they were cleared as efficiently as in the wild type strain both in the mutant lacking ltn1 (Fig. 5F) and in the rqc1 null mutant (not shown). This shows that the accumulation of Sdj1-L169P, observed in these strains, is not a consequence of reduced aggregate degradation, which suggests that Ltn1 regulates Sdj1-L169P degradation upstream of aggregate formation.
In contrast to this, the degradation of the Sdj1-L169P aggregates was severely inhibited in the hsp104 deletion strain (Fig. 5F). Furthermore, the residual aggregate degradation, observed in the hsp104 deletion strain, was not mediated by the proteasome (Fig. 5F), showing that proteasomal degradation of Sdj1-L169P aggregates is entirely dependent on the Hsp104 chaperone, and that this aggregate degradation pathway cannot be rescued by non-proteasomal pathways. Also in the ltn1 null cells aggregate degradation was no longer exclusively mediated by the proteasome but partially occurred through a non-proteasomal pathway (Fig. 5F), presumably by autophagy. This suggests that cells deprived of Ltn1 or Hsp104-mediated proteasomal pathways may utilize autophagy as a back-up system for disposing of this substrate.
Because aggregate degradation does not rely on Ltn1 (Fig. 5F), another E3 must function in targeting aggregated Sdj1-L169P for proteasomal degradation. In an attempt to identify this E3, we followed aggregate degradation in the remaining 12 E3 mutants after treating with cycloheximide. However, in no single deletion mutant were the aggregates significantly stabilized or more abundant than in the wild type (not shown), indicating that either the relevant E3 was one not included in our selection, or possibly that multiple E3s collaborate in aggregate clearance.
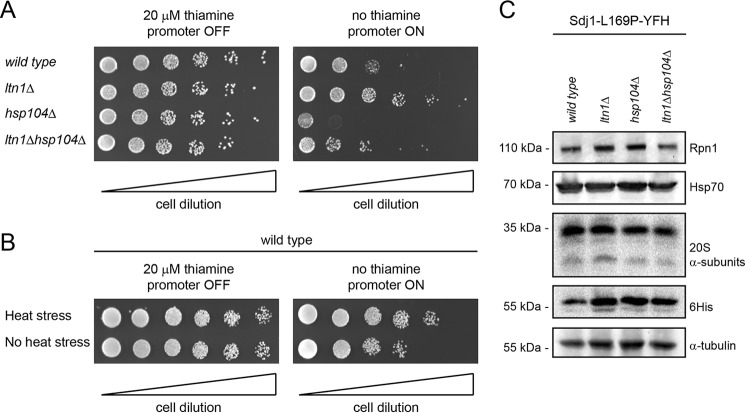
Sdj1-L169P Inhibits Growth of Wild Type and hsp104Δ, but Not of ltn1Δ Strains
It has been well documented that the pathogenesis of protein misfolding disorders is related to the toxicity of the aberrant protein species (47). To test how Sdj1-L169P expression affected the yeast cells, we performed growth assays on strains expressing Sdj1-L169P under control of the thiamine-regulated nmt1 promoter. Here, expression of Sdj1-L169P significantly inhibited the growth of wild type cells (Fig. 6A). This effect was aggravated in mutants lacking Hsp104, but was, surprisingly, suppressed in the ltn1Δ strain (Fig. 6A). Expression of Sdj1-L169P in mutants lacking both Ltn1 and Hsp104 resulted in an intermediate phenotype that appeared more like the situation in the wild type strain (Fig. 6A). We speculated that the Sdj1-L169P-resistant phenotype of ltn1Δ cells could be a result of the cells triggering a stress response pathway that increases their tolerance for misfolded protein. To test this hypothesis, we repeated the growth assays using cells that were subjected to a 45-min heat shock at 40 °C prior to plating. Indeed, this heat shock treatment led to a slightly increased tolerance to Sdj1-L169P (Fig. 6B), indicating that the resistance of ltn1Δ cells to Sdj1-L169P expression is caused by an up-regulated stress response, or that heat treatment or ltn1 deletion leads to Sdj1-L169P being more rapidly deposited in aggregates, which are generally considered less toxic than soluble misfolded proteins.
FIGURE 6.
Sdj1-L169P expression is toxic to wild type and hsp104Δ cells. A, growth comparison on solid media of the indicated strains transformed with an nmt1 thiamine-regulated expression construct for Sdj1-L169P. Note that expression of Sdj1-L169P (right panel) inhibits cell growth for the wild type, hsp104Δ, and ltn1Δhsp104Δ strains, but not for the ltn1Δ strain. B, growth comparison on solid media of wild type cells transformed with an nmt1 thiamine-regulated expression construct for Sdj1-L169P either subjected to heat shock prior to plating or untreated. Note that Sdj1-L169P expressing cells grow better when they are heat shocked. C, the steady-state levels of the indicated proteins were determined by blotting of whole cell extracts from wild type, ltn1Δ, hsp104Δ, and ltn1Δhsp104Δ strains. Tubulin served as a loading control.
To determine whether the ltn1Δ cells were stressed, we performed mRNA quantification of ltn1Δ wild type cells by real-time PCR. This did, however, not reveal any significant change in the level of the ER-stress activated bip1+ gene, and only a modest 2.8 ± 0.5-fold (S.E.) increase of the Hsf1-responsive hsp104+ gene in the ltn1Δ mutant relative to wild type. In addition, the Sdj1-L169P-resistant phenotype of ltn1Δ cells was unchanged in cells grown in the presence of the proteasomal inhibitor bortezomib (not shown). Furthermore, no up-regulation of proteasomal subunits or Hsp70 chaperones was observed in these cells (Fig. 6C), suggesting that the increased tolerance of ltn1Δ cells is independent of proteasome or Hsp70 abundance.
Discussion
Because the accumulation of misfolded proteins has been linked to a variety of human diseases (48), the quality control mechanisms that govern their turnover have been the subject to intense investigation (3, 49). Most of these studies have focused on post-translational quality control mechanisms that handle the degradation of already misfolded and aggregated proteins. However, in recent years it has become clear that protein quality control is exerted already during synthesis (50). Thus, in human cells up to about 15% of newly synthesized proteins are co-translationally ubiquitylated (51).
In the present work we have mapped the degradation pathway for Sdj1-L169P, the fission yeast homologue of the human DJ1-L166P protein involved in Parkinson disease. Like human DJ1-L166P, yeast Sdj1-L169P appeared misfolded both in vivo and in vitro. Our FoldX calculations predict that Sdj1-L169P is thermodynamically destabilized by ∼8 kcal/mol. None of the other residues linked to disease in human DJ-1 are preserved in S. pombe Sdj1, but for the DJ-1 residues Leu-10, Met-26, and Asp-149 (16), residues with similar chemical properties are found in Sdj1 (Val-8, Phe-24, and Glu-152, respectively). In our calculations, the disease-causing Sdj1 mutations V8P, F24I, and E152A would lead to structural destabilization with ΔΔG values of 6.3, 3.7, and 2.5 kcal/mol, respectively. Hence, at least in the human DJ-1-L10P mutant we would predict is also a substrate of a quality control pathway.
Our data on fission yeast Sdj1-L169P support a model where the mutant protein is cleared by a two-step quality control system. First, and perhaps already during translation, the ribosome-associated Ltn1 and Rqc1 regulate the Sdj1-L169P level. The Sdj1-L169P protein that escapes during this early point then forms cytosolic aggregates that are subsequently solubilized and degraded by an Hsp104-dependent proteasomal pathway. Because Ltn1 and Rqc1 are significantly less abundant in cells than ribosomes (52), it is likely that some Sdj1-L169P slips past this ribosomal quality control system and is subsequently degraded by other mechanisms.
Ltn1 has primarily been recognized as an E3 ligase dedicated to the degradation of products resulting from the translation of aberrant non-stop mRNAs (13, 45, 53), and was found not to influence the degradation of a quality control substrate (13). Our results suggest that Ltn1 not only targets nascent chains of aberrant mRNAs, but also may play a role in targeting other misfolded aggregation-prone proteins. This is in line with studies showing that co-translational ubiquitylation is increased in response to stress conditions that facilitate protein misfolding (51), and that the main characteristic of nascent polypeptides undergoing co-translational ubiquitylation is enrichment of properties that enhance aggregation and hinder folding (54). Furthermore, in budding yeast, another misfolded substrate was shown to accumulate in various E3 mutant strains, including an ltn1 deletion strain (55). We speculate that also the association between Sdj1 and the ribosome may bring misfolded Sdj1 into proximity with the Ltn1 ubiquitin ligase, thereby further increasing the likelihood that the protein will be recognized as a target.
Ltn1 has previously been reported to function redundantly with the less described E3 ligase Hel2 in co-translational ubiquitylation (54). However, because the degradation of Sdj1-L169P was unaffected in a hel2 deletion strain, our results suggest that these ubiquitin ligases must differ to some extent in their function.
That Sdj1-L169P appears only partly stabilized in ltn1Δ and rqc1Δ strains, when evaluated by blotting procedures, contradicts the observation that the aggregates are as efficiently eliminated in these strains as in wild type cells. Possibly, this discrepancy is caused by an increased stabilization of soluble Sdj1-L169P in these strains, because the whole cell lysates used for blotting will contain both soluble and aggregated Sdj1-L169P. Unfortunately, our attempts to distinguish between these two subpopulations of Sdj1-L169P have not been successful.
In wild type cells, the Sdj1-L169P aggregates were degraded exclusively by the proteasome, which requires the aggregates to be resolubilized prior to degradation (6). Accordingly, proteasomal aggregate degradation was entirely dependent on the Hsp104 chaperone, which is well known to mediate the disaggregation of misfolded substrates in yeast cells (56, 57), but intriguingly no Hsp104 orthologues are found in higher eukaryotes. Surprisingly, aggregate re-solubilization did not occur in the presence of the proteasomal inhibitor bortezomib, indicating that disaggregation and proteasomal degradation are tightly coupled processes.
Deletion of either the ssa1 or ssa2 chaperones resulted in accumulation of Sdj-L169P although the effect was markedly higher in the ssa2 null strain. Although Hsp70 chaperones are well known mediators of chaperone-assisted degradation (3, 7), it is difficult to fully distinguish a direct role for these chaperones in mediating Sdj1-L169P degradation from the likely increased tendency of the protein to aggregate in the absence of a major cytosolic Hsp70 chaperone. Nonetheless, Hsp70-type chaperones have previously been shown to be vital for Hsp104 function (57, 58), and were also shown to associate with translating ribosomes (59), suggesting that Ssa1 and Ssa2 may potentially be involved in both Hsp104- and Ltn1-mediated pathways. Ssa1 and Ssa2 are 95% identical proteins and it is therefore surprising that Ssa1 is not able to compensate for the loss of Ssa2. This suggests functional variation between the two chaperones despite their similarity. Functional differences between budding yeast Ssa1 and Ssa2 have been noted before (60–62), but may in our case simply be explained by the fact that Ssa2 is much more abundant in S. pombe than Ssa1 (52).
Wild type cells, expressing Sdj1-L169P, only display a relatively mild growth phenotype, whereas hsp104Δ cells, expressing Sdj1-L169P, display severe growth retardation. This correlates with the increased Sdj1-L169P accumulation observed in the hsp104 null strain and is in accordance with budding yeast studies in which Hsp104 was seen to induce tolerance to various stress conditions that induce protein misfolding and aggregation (63, 64). Recently, it was shown in budding yeast that the ribosomal quality control activity is regulated by a stress signaling pathway, but also itself propels a signal to the stress-activated transcription factor Hsf1 (46). As all the components of the ribosomal quality control system are conserved, this might also be the case in fission yeast and may explain why heat shock treatment or deletion of ltn1 leads to an Sdj1-L169P-resistant phenotype. However, our mRNA quantification of hsp104, a known Hsf1 target (65), only revealed a modest increase in the ltn1Δ strain. This suggests that perhaps the Sdj1-L169P-resistant phenotype observed after heat treatment or in the ltn1 deletion is rather caused by Sdj1-L169P being more rapidly deposited in aggregates, which are normally considered less toxic than soluble misfolded proteins. Thus, perhaps aberrant translation products present in the ltn1Δ cells indirectly affect Sdj1-L169P solubility and degradation.
Interestingly, human DJ-1 was itself recently shown to regulate the activity of the 20S proteasome (66), and presumably wild type Sdj1 functions in a similar manner in fission yeast. However, as the experiments presented here were all performed in an sdj1+ wild type genetic background, and Sdj1-L169P fails to interact with Sdj1, such a function of Sdj1 is unlikely to be relevant for our analyses on the degradation of misfolded Sdj1-L169P.
In conclusion, we describe a novel two-step quality control pathway using Sdj1-L169P as a model substrate. The requirement of two sequentially acting pathways represents a novel concept in protein quality control and demonstrates the importance of rapid and efficient clearance of misfolded and aggregation-prone substrates.
Author Contributions
S. G. M., I. B. L., F. K., and R. H. P. conceived and designed the experiments. S. G. M., I. B. L., E. G. P., C. T. M., E. P., and B. B. K. performed the experiments. S. G. M., I. B. L., K. L. L., M. L. N., F. K., R. H. P. analyzed the data. S. G. M. and R. H. P. wrote the paper.
Supplementary Material
Acknowledgments
We thank Camilla S. Jensen for help in the early stages of the work, and Anne-Marie Bonde-Lauridsen for expert technical assistance. In addition we thank Dr. Minoru Yoshida, Dr. Jeremy S. Hyams, Dr. Colin Gordon, Dr. Mitsuhiro Yanagida, Dr. Snezhana Oliferenko, Dr. Iva M. Tolic-Nørrelykke, and Dr. Shao-Win Wang for reagents, and Dr. Klavs B. Hendil, Dr. Olaf Nielsen, and Dr. Christian Holmberg for helpful discussions and comments on the manuscript. The authors declare that they have no conflicts of interest with the contents of this article.
This work was supported in part by grants from the Danish Council for Independent Research (Natural Sciences), The Novo Nordisk Foundation, The Danish Cancer Society, and the Lundbeck Foundation (to R. H.-P., B. B .K., and K. L.-L.).

This article contains supplemental Files S1 and S2.
- ER
- endoplasmic reticulum.
References
- 1. Hartl F. U., Hayer-Hartl M. (2009) Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16, 574–581 [DOI] [PubMed] [Google Scholar]
- 2. Hartl F. U., Bracher A., Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 [DOI] [PubMed] [Google Scholar]
- 3. Kettern N., Dreiseidler M., Tawo R., Höhfeld J. (2010) Chaperone-assisted degradation: multiple paths to destruction. Biol. Chem. 391, 481–489 [DOI] [PubMed] [Google Scholar]
- 4. Ross C. A., Poirier M. A. (2004) Protein aggregation and neurodegenerative disease. Nat. Med. 10, S10–S17 [DOI] [PubMed] [Google Scholar]
- 5. Pickart C. M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 6. Finley D. (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kriegenburg F., Ellgaard L., Hartmann-Petersen R. (2012) Molecular chaperones in targeting misfolded proteins for ubiquitin-dependent degradation. FEBS J. 279, 532–542 [DOI] [PubMed] [Google Scholar]
- 8. Vembar S. S., Brodsky J. L. (2008) One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell. Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gardner R. G., Nelson Z. W., Gottschling D. E. (2005) Degradation-mediated protein quality control in the nucleus. Cell 120, 803–815 [DOI] [PubMed] [Google Scholar]
- 10. Guerriero C. J., Weiberth K. F., Brodsky J. L. (2013) Hsp70 targets a cytoplasmic quality control substrate to the San1p ubiquitin ligase. J. Biol. Chem. 288, 18506–18520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kriegenburg F., Jakopec V., Poulsen E. G., Nielsen S. V., Roguev A., Krogan N., Gordon C., Fleig U., Hartmann-Petersen R. (2014) A chaperone-assisted degradation pathway targets kinetochore proteins to ensure genome stability. PLoS Genet. 10, e1004140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang F., Canadeo L. A., Huibregtse J. M. (2015) Ubiquitination of newly synthesized proteins at the ribosome. Biochimie 114, 127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bengtson M. H., Joazeiro C. A. (2010) Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467, 470–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lyumkis D., Oliveira dos Passos D., Tahara E. B., Webb K., Bennett E. J., Vinterbo S., Potter C. S., Carragher B., Joazeiro C. A. (2014) Structural basis for translational surveillance by the large ribosomal subunit-associated protein quality control complex. Proc. Natl. Acad. Sci. U.S.A. 111, 15981–15986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chu J., Hong N. A., Masuda C. A., Jenkins B. V., Nelms K. A., Goodnow C. C., Glynne R. J., Wu H., Masliah E., Joazeiro C. A., Kay S. A. (2009) A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 106, 2097–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Corti O., Lesage S., Brice A. (2011) What genetics tells us about the causes and mechanisms of Parkinson's disease. Physiol. Rev. 91, 1161–1218 [DOI] [PubMed] [Google Scholar]
- 17. Bandyopadhyay S., Cookson M. R. (2004) Evolutionary and functional relationships within the DJ1 superfamily. BMC Evol. Biol. 4, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anderson P. C., Daggett V. (2008) Molecular basis for the structural instability of human DJ-1 induced by the L166P mutation associated with Parkinson's disease. Biochemistry 47, 9380–9393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lucas J. I., Marín I. (2007) A new evolutionary paradigm for the Parkinson disease gene DJ-1. Mol. Biol. Evol. 24, 551–561 [DOI] [PubMed] [Google Scholar]
- 20. Bonifati V., Rizzu P., Squitieri F., Krieger E., Vanacore N., van Swieten J. C., Brice A., van Duijn C. M., Oostra B., Meco G., Heutink P. (2003) DJ-1(PARK7), a novel gene for autosomal recessive, early onset parkinsonism. Neurol. Sci. 24, 159–160 [DOI] [PubMed] [Google Scholar]
- 21. Olzmann J. A., Brown K., Wilkinson K. D., Rees H. D., Huai Q., Ke H., Levey A. I., Li L., Chin L. S. (2004) Familial Parkinson's disease-associated L166P mutation disrupts DJ-1 protein folding and function. J. Biol. Chem. 279, 8506–8515 [DOI] [PubMed] [Google Scholar]
- 22. Baulac S., LaVoie M. J., Strahle J., Schlossmacher M. G., Xia W. (2004) Dimerization of Parkinson's disease-causing DJ-1 and formation of high molecular weight complexes in human brain. Mol. Cell. Neurosci. 27, 236–246 [DOI] [PubMed] [Google Scholar]
- 23. Miller D. W., Ahmad R., Hague S., Baptista M. J., Canet-Aviles R., McLendon C., Carter D. M., Zhu P. P., Stadler J., Chandran J., Klinefelter G. R., Blackstone C., Cookson M. R. (2003) L166P mutant DJ-1, causative for recessive Parkinson's disease, is degraded through the ubiquitin-proteasome system. J. Biol. Chem. 278, 36588–36595 [DOI] [PubMed] [Google Scholar]
- 24. Kim D. U., Hayles J., Kim D., Wood V., Park H. O., Won M., Yoo H. S., Duhig T., Nam M., Palmer G., Han S., Jeffery L., Baek S. T., Lee H., Shim Y. S., Lee M., Kim L., Heo K. S., Noh E. J., Lee A. R., Jang Y. J., Chung K. S., Choi S. J., Park J. Y., Park Y., Kim H. M., Park S. K., Park H. J., Kang E. J., Kim H. B., Kang H. S., Park H. M., Kim K., Song K., Song K. B., Nurse P., Hoe K. L. (2010) Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 28, 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moreno S., Klar A., Nurse P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823 [DOI] [PubMed] [Google Scholar]
- 26. Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 [DOI] [PubMed] [Google Scholar]
- 27. Matsuyama A., Shirai A., Yashiroda Y., Kamata A., Horinouchi S., Yoshida M. (2004) pDUAL, a multipurpose, multicopy vector capable of chromosomal integration in fission yeast. Yeast 21, 1289–1305 [DOI] [PubMed] [Google Scholar]
- 28. Andersen K. M., Jensen C., Kriegenburg F., Lauridsen A. M., Gordon C., Hartmann-Petersen R. (2011) Txl1 and Txc1 are co-factors of the 26S proteasome in fission yeast. Antioxid. Redox. Signal. 14, 1601–1608 [DOI] [PubMed] [Google Scholar]
- 29. Poulsen J. W., Madsen C. T., Young C., Poulsen F. M., Nielsen M. L. (2013) Using guanidine-hydrochloride for fast and efficient protein digestion and single-step affinity-purification mass spectrometry. J. Proteome. Res. 12, 1020–1030 [DOI] [PubMed] [Google Scholar]
- 30. Hartmann-Petersen R., Wallace M., Hofmann K., Koch G., Johnsen A. H., Hendil K. B., Gordon C. (2004) The Ubx2 and Ubx3 cofactors direct Cdc48 activity to proteolytic and nonproteolytic ubiquitin-dependent processes. Curr. Biol. 14, 824–828 [DOI] [PubMed] [Google Scholar]
- 31. Lyne R., Burns G., Mata J., Penkett C. J., Rustici G., Chen D., Langford C., Vetrie D., Bähler J. (2003) Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC. Genomics 4, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. (2005) The FoldX web server: an online force field. Nucleic Acids Res. 33, W382–W388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Madzelan P., Labunska T., Wilson M. A. (2012) Influence of peptide dipoles and hydrogen bonds on reactive cysteine pKa values in fission yeast DJ-1. FEBS J. 279, 4111–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guerois R., Nielsen J. E., Serrano L. (2002) Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 320, 369–387 [DOI] [PubMed] [Google Scholar]
- 35. Wilson M. A., St Amour C. V., Collins J. L., Ringe D., Petsko G. A. (2004) The 1.8-A resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: a member of the DJ-1/ThiJ/PfpI superfamily. Proc. Natl. Acad. Sci. U.S.A. 101, 1531–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Graille M., Quevillon-Cheruel S., Leulliot N., Zhou C. Z., Li de la Sierra Gallay I., Jacquamet L., Ferrer J. L., Liger D., Poupon A., Janin J., van Tilbeurgh H. (2004) Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family. Structure 12, 839–847 [DOI] [PubMed] [Google Scholar]
- 37. Canet-Avilés R. M., Wilson M. A., Miller D. W., Ahmad R., McLendon C., Bandyopadhyay S., Baptista M. J., Ringe D., Petsko G. A., Cookson M. R. (2004) The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. U.S.A. 101, 9103–9108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang C. Y., Wen W. L., Nilsson D., Sunnerhagen P., Chang T. H., Wang S. W. (2012) Analysis of stress granule assembly in Schizosaccharomyces pombe. RNA. 18, 694–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Coelho M., Dereli A., Haese A., Kühn S., Malinovska L., DeSantis M. E., Shorter J., Alberti S., Gross T., Tolić-Norrelykke I. M. (2013) Fission yeast does not age under favorable conditions, but does so after stress. Curr. Biol. 23, 1844–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Poulsen E. G., Steinhauer C., Lees M., Lauridsen A. M., Ellgaard L., Hartmann-Petersen R. (2012) HUWE1 and TRIP12 collaborate in degradation of ubiquitin-fusion proteins and misframed ubiquitin. PLoS One 7, e50548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van der Brug M. P., Blackinton J., Chandran J., Hao L. Y., Lal A., Mazan-Mamczarz K., Martindale J., Xie C., Ahmad R., Thomas K. J., Beilina A., Gibbs J. R., Ding J., Myers A. J., Zhan M., Cai H., Bonini N. M., Gorospe M., Cookson M. R. (2008) RNA binding activity of the recessive parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc. Natl. Acad. Sci. U.S.A. 105, 10244–10249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kthiri F., Gautier V., Le H. T., Prère M. F., Fayet O., Malki A., Landoulsi A., Richarme G. (2010) Translational defects in a mutant deficient in YajL, the bacterial homolog of the parkinsonism-associated protein DJ-1. J. Bacteriol. 192, 6302–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Le H. T., Gautier V., Kthiri F., Malki A., Messaoudi N., Mihoub M., Landoulsi A., An Y. J., Cha S. S., Richarme G. (2012) YajL, prokaryotic homolog of parkinsonism-associated protein DJ-1, functions as a covalent chaperone for thiol proteome. J. Biol. Chem. 287, 5861–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen D., Toone W. M., Mata J., Lyne R., Burns G., Kivinen K., Brazma A., Jones N., Bähler J. (2003) Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14, 214–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Defenouillère Q., Yao Y., Mouaikel J., Namane A., Galopier A., Decourty L., Doyen A., Malabat C., Saveanu C., Jacquier A., Fromont-Racine M. (2013) Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc. Natl. Acad. Sci. U.S.A. 110, 5046–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C. C., Li G. W., Zhou S., King D., Shen P. S., Weibezahn J., Dunn J. G., Rouskin S., Inada T., Frost A., Weissman J. S. (2012) A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Herczenik E., Gebbink M. F. (2008) Molecular and cellular aspects of protein misfolding and disease. FASEB J. 22, 2115–2133 [DOI] [PubMed] [Google Scholar]
- 48. Kim Y. E., Hipp M. S., Bracher A., Hayer-Hartl M., Hartl F. U. (2013) Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82, 323–355 [DOI] [PubMed] [Google Scholar]
- 49. Amm I., Sommer T., Wolf D. H. (2014) Protein quality control and elimination of protein waste: the role of the ubiquitin-proteasome system. Biochim. Biophys. Acta 1843, 182–196 [DOI] [PubMed] [Google Scholar]
- 50. Pechmann S., Willmund F., Frydman J. (2013) The ribosome as a hub for protein quality control. Mol. Cell 49, 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang F., Durfee L. A., Huibregtse J. M. (2013) A cotranslational ubiquitination pathway for quality control of misfolded proteins. Mol. Cell 50, 368–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marguerat S., Schmidt A., Codlin S., Chen W., Aebersold R., Bähler J. (2012) Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151, 671–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shao S., von der Malsburg K., Hegde R. S. (2013) Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol. Cell 50, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Duttler S., Pechmann S., Frydman J. (2013) Principles of cotranslational ubiquitination and quality control at the ribosome. Mol. Cell 50, 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Theodoraki M. A., Nillegoda N. B., Saini J., Caplan A. J. (2012) A network of ubiquitin ligases is important for the dynamics of misfolded protein aggregates in yeast. J. Biol. Chem. 287, 23911–23922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Parsell D. A., Kowal A. S., Singer M. A., Lindquist S. (1994) Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372, 475–478 [DOI] [PubMed] [Google Scholar]
- 57. Glover J. R., Lindquist S. (1998) Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 [DOI] [PubMed] [Google Scholar]
- 58. Lee J., Kim J. H., Biter A. B., Sielaff B., Lee S., Tsai F. T. (2013) Heat shock protein (Hsp) 70 is an activator of the Hsp104 motor. Proc. Natl. Acad. Sci. U.S.A. 110, 8513–8518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Horton L. E., James P., Craig E. A., Hensold J. O. (2001) The yeast hsp70 homologue Ssa is required for translation and interacts with Sis1 and Pab1 on translating ribosomes. J. Biol. Chem. 276, 14426–14433 [DOI] [PubMed] [Google Scholar]
- 60. Brown C. R., McCann J. A., Chiang H. L. (2000) The heat shock protein Ssa2p is required for import of fructose-1,6-bisphosphatase into Vid vesicles. J. Cell Biol. 150, 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sharma D., Martineau C. N., Le Dall M. T., Reidy M., Masison D. C., Kabani M. (2009) Function of SSA subfamily of Hsp70 within and across species varies widely in complementing Saccharomyces cerevisiae cell growth and prion propagation. PLoS One 4, e6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sharma D., Masison D. C. (2011) Single methyl group determines prion propagation and protein degradation activities of yeast heat shock protein (Hsp)-70 chaperones Ssa1p and Ssa2p. Proc. Natl. Acad. Sci. U.S.A. 108, 13665–13670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sanchez Y., Lindquist S. L. (1990) HSP104 required for induced thermotolerance. Science 248, 1112–1115 [DOI] [PubMed] [Google Scholar]
- 64. Sanchez Y., Taulien J., Borkovich K. A., Lindquist S. (1992) Hsp104 is required for tolerance to many forms of stress. EMBO J. 11, 2357–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vjestica A., Zhang D., Liu J., Oliferenko S. (2013) Hsp70-Hsp40 chaperone complex functions in controlling polarized growth by repressing Hsf1-driven heat stress-associated transcription. PLoS Genet. 9, e1003886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Moscovitz O., Ben-Nissan G., Fainer I., Pollack D., Mizrachi L., Sharon M. (2015) The Parkinson's-associated protein DJ-1 regulates the 20S proteasome. Nat. Commun. 6, 6609. [DOI] [PubMed] [Google Scholar]
- 67. Penney M., Samejima I., Wilkinson C. R., McInerny C. J., Mathiassen S. G., Wallace M., Toda T., Hartmann-Petersen R., Gordon C. (2012) Fission yeast 26S proteasome mutants are multi-drug resistant due to stabilization of the Pap1 transcription factor. PLoS One 7, e50796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.