Abstract
In the yeast Saccharomyces cerevisiae, distinct regions of the 1.2-kb telomerase RNA (TLC1) bind to the catalytic subunit Est2p and to accessory proteins. In particular, a bulged stem structure binds the essential regulatory subunit Est1p. We now show that the Est1p-binding domain of the RNA can be moved to three distant locations with retention of telomerase function in vivo. We present the Est1p relocation experiment in the context of a working model for the secondary structure of the entire TLC1 RNA, based on thermodynamic considerations and comparative analysis of sequences from four species. The model for TLC1 has three long quasihelical arms that bind the Ku, Est1p, and Sm proteins. These arms emanate from a central catalytic core that contains the template and Est2p-binding region. Deletion mutagenesis provides evidence that the Sm arm exists in vivo and can be shortened by 42 predicted base pairs with retention of function; therefore, precise positioning of Sm proteins, like Est1p, is not required within telomerase. In the best-studied ribonucleoprotein enzyme, the ribosome, the RNAs have specific three-dimensional structures that orient the functional elements. In the case of yeast telomerase, we propose that the RNA serves a very different function, providing a flexible tether for the protein subunits.
Telomeric DNA consists of repeated sequences that serve as binding sites for double- and single-stranded DNA-binding proteins, important for maintaining proper structure and function of the chromosome end (1). The repeat sequence of telomeric DNA is established by the ribonucleoprotein (RNP) enzyme telomerase (2, 3), which uses a portion of its RNA subunit as a template for a reverse transcription reaction catalyzed by telomerase reverse transcriptase (TERT; encoded by the EST2 gene in yeast) (4).
In addition to binding TERT, budding yeast telomerase RNA binds accessory proteins (see Table 2, which is published as supporting information on the PNAS web site) (4, 5). A stem–loop structure binds to the Ku heterodimer, which regulates telomerase action at broken chromosome ends in addition to natural telomeres (6). The essential telomerase subunit Est1p, which is also found in humans and fission yeast (7–9), binds a bulged stem element in the TLC1 telomerase RNA (10). Est1p may target telomerase to the telomere and/or activate telomerase once bound there; it is not required for in vitro telomerase activity (11–13). Finally, the TLC1 RNA binds to the Sm proteins, known to bind to a consensus sequence RAU4–6GR (R = purine) in small nuclear RNP RNAs (14). Binding of Sm proteins is required for efficient biogenesis of TLC1 (15), whereas maturation of telomerase RNA in other eukaryotes involves different pathways (5).
In contrast to the conserved catalytic subunit TERT, telomerase RNA components are highly variable in both sequence and size. A phylogenetically supported secondary structure model for the ≈450-nt vertebrate telomerase RNA has been proposed on the basis of sequences from 32 different species, including fish, amphibians, reptiles, birds, and mammals (16). For the budding yeasts, however, the problem of deducing secondary structure has been confounded by multiple factors: the strikingly rapid divergence in sequence of the telomerase RNAs, their large size, and the unavailability of many sequences. Although the Kluyveromyces sequences have provided some phylogenetic information used to generate schematic models of two possible conformations of this telomerase RNA (17), no detailed model for any entire budding yeast telomerase RNA has yet been proposed.
Most of the Saccharomyces cerevisiae telomerase RNA, outside a region containing the template and Est2p- and Est1p-binding sites, is dispensable for function (18). This finding correlates with the fact that the vast majority of yeast telomerase RNA sequence is changing extraordinarily rapidly through evolution. These genetic and phylogenetic data have suggested that yeast telomerase RNA may serve a novel role for an RNA in a ribonucleoprotein complex, functioning primarily as a flexible tether or “scaffold” for proteins in the telomerase RNP (18, 19). Here we test this hypothesis directly by examining the requirement for relative positioning of telomerase proteins within the complex. We propose that telomerase RNA has a central core that carries out its enzymatic function, plus three flexible RNA arms that bind the Est1p, Ku, and Sm proteins.
Methods
mfold Analysis. TLC1 RNA sequences were submitted to the mfold version 3.1 web server (www.bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi), which calculates RNA folding on the basis of experimentally determined free energies (Fig. 4, which is published as supporting information on the PNAS web site) (20, 21). The default RNA-folding parameters on the web site were used, except the window parameter was set at 7 to obtain additional possible structures with higher global folding energies. This set of parameters yielded 67 possible folds, and the first, last, and every fifth predicted structure were surveyed to assess prevalence of substructures.
Phylogenetic Comparison and alifold Analysis. S. cerevisiae, Saccharomyces paradoxus, Saccharomyces mikatae, and Saccharomyces bayanus TLC1 RNAs were aligned by using clustalw (www.ebi.ac.uk/clustalw) with subsequent manual refinement (Fig. 5, which is published as supporting information on the PNAS web site). This alignment was then used as input for alifold software (http://rna.tbi.univie.ac.at/cgi-bin/alifold.cgi). The output of possible covarying nucleotides from alifold was then analyzed manually, comparing the most credible covarying nucleotides against the mfold and the alignment. Nearly all of the most credible covarying nucleotides identified by alifold were present in the mfold model. However, certain other highly credible covarying nucleotides that were not paired in the mfold lowest-energy structure were then forced (F) to pair (numbering refers to positions within S. cerevisiae TLC1 sequence; see sequence alignment shown in Fig. 5): F 158 447 1, F 121 828 1, F 128 822 1, F 237 268 1, F 275 342 1, F 209 363 1, F 295 326 1, F 960 1115 1, F 963 1106 1, F 74 875 1, F 82 869 1, F 148 460 1, and F 152 454 1, where the 1 in each entry indicates that only one base pair is forced. The final, phylogenetically improved structure is shown in Fig. 1 (see also Fig. 6, which is published as supporting information on the PNAS web site).
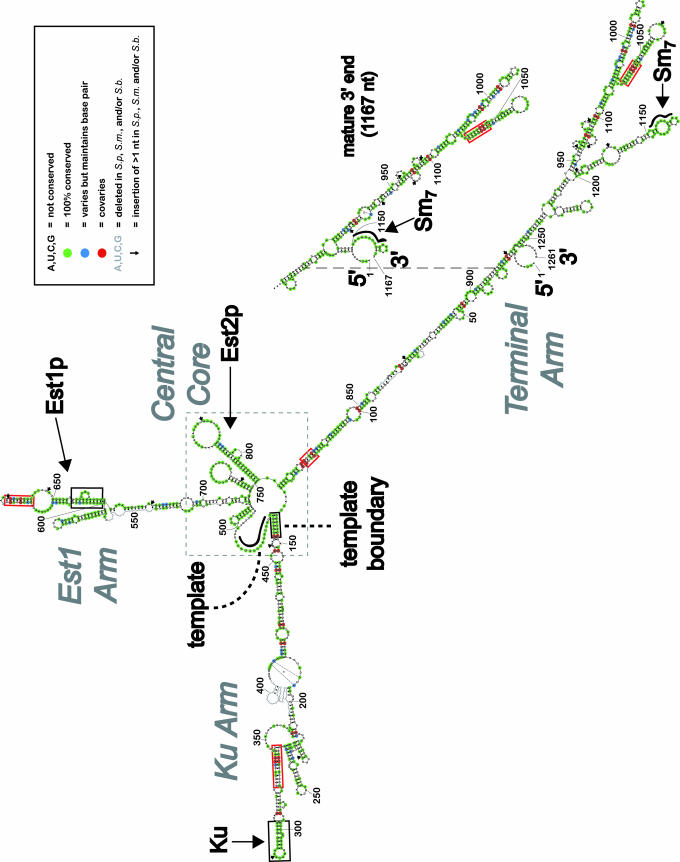
Fig. 1.
A working model of S. cerevisiae telomerase RNA secondary structure. Two forms of TLC1 with different 3′ ends (≈1,261 and 1,167 nt) are hypothesized to be precursor and mature forms, respectively (15, 32, 33). Phylogenetic data for each nucleotide were derived from alignment of TLC1 from S. cerevisiae, S. paradoxus (S.p.), S. mikatae (S.m.), and S. bayanus (S.b.) (Fig. 5). The template used for reverse transcription and the Sm7 complex binding site are known to be single-stranded RNA regions (thick black contoured lines). Previously identified secondary structure elements that bind Est1p or Ku are framed in black boxes. RNA elements important for telomerase RNA template function are identified by dashed black lines. Red boxes highlight regions of the RNA that are strongly supported by phylogenetic analysis. A dashed gray box indicates the central core region containing the template and Est2p catalytic subunit-binding region.
Targeted Mutagenesis of TLC1. Mutations in TLC1 were generated with DNA primers harboring the desired mutant sequence, which were then used for PCR. Amplified products were subcloned into vector pSD107 (a CEN/TRP1 vector containing TLC1 with endogenous promoter and terminator sequences) (15). All clones were sequenced.
Plasmids and Yeast. Strain TCy43 (MATa ura3-53 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 VR::ADE2–TEL adh4::URA3–TEL Δtlc1::LEU2 rad52::LEU2 pTLC1-LYS2-CEN) was used for all experiments (15) and is a Δtlc1 mutant harboring wild-type TLC1 on a plasmid marked with LYS2. For the plasmid shuffle, constructs expressing TLC1 mutants [based on the TLC1WT plasmid pSD107 (15), a pRS314 derivative (CEN/ARS TRP1) (22)] were transformed into TCy43 (grown in -lysine medium) and incubated on -tryptophan -lysine plates at 30°C. Colonies were then restreaked to -tryptophan +α-aminoadipate, which counterselects for the LYS2 plasmid harboring wild-type TLC1, and grown at 23°C. The cells expressing mutant RNAs were then tested for phenotypes after restreaking to -tryptophan medium and growing at 30°C. Mutants that supported yeast growth for 10 restreaks were considered to be functional.
TLC1 expression plasmids with mutations in the terminal arm are as follows: pDZ107 [TLC1 Δ22–102::(CG)5], pDZ108 [TLC1 Δ846–914::(CG)5], and pDZ109 (TLC1 Δ22–102::(CG)5 Δ846–914::(CG)5]. The mutants harboring a repositioned Est1p-binding site are as follows: pDZ110 (TLC1WT + 524–704 at 1033), pDZ111 (TLC1WT + 524–704 at 220), pDZ112 (TLC1WT + 524–704 at 450), pDZ113 (TLC1 bulgeΔ + 524–704 at 1033), pDZ114 (TLC1 bulgeΔ + 524–704 at 220), and pDZ115 (TLC1 bulgeΔ + 524–704 at 450). Controls for the repositioning of the Est1p-binding site are pDZ123 (TLC1WT + 524–704 bulgeΔ at 1033) and pDZ124 (TLC1 bulgeΔ + 524–704 bulgeΔ at 1033). bulgeΔ, deletion of nucleotides 660–664, prevents Est1p binding.
Results
A Working Model of S. cerevisiae Telomerase RNA Secondary Structure. We began to model the secondary structure of TLC1 RNA by using mfold to predict energetically favorable RNA conformations (20). This software, which is based on experimentally determined thermodynamic parameters for RNA folding (21), cannot be expected to fully succeed in predicting the secondary structure for such a large RNA. Nonetheless, we were encouraged that mfold correctly predicted the existence of all five known TLC1 elements (listed in Table 2; see Fig. 4 for the most energetically favorable prediction, ΔG° = -338.3 kcal/mol). The Ku hairpin structure, which differed just slightly from the reported binding element (23), was present in 11 of 14 mfold predictions that were within 3.5% of the free energy of the energetically most favorable. The bulged stem reported to bind Est1p (10) was present in 13 of 14. The template boundary helix, which is important for correct template usage and is formed by a long-range pairing (24), was present in 11 of 14. The template was predicted to be single-stranded (an essential feature for this portion of the RNA to be free for reverse transcription) in 12 of 14 of these lowest-energy structure predictions. The Sm-binding RNA consensus sequence, which is bound by a ring of the seven Sm proteins (25, 26), was predicted by mfold to be predominantly single-stranded, existing on one side of a well conserved internal loop near the tip of an RNA arm. In addition, the 5′ and 3′ ends of the TLC1 were always predicted to be closely juxtaposed, which is a common property of natural RNA structures, including tRNAs, rRNAs, RNase P, and signal recognition particle (SRP) RNA.
Thus, the mfold prediction of secondary structure for TLC1 RNA appears to be worth further scrutiny. Overall, the model suggests that the Ku, Est1, and Sm protein-binding sites exist at the ends of three prominent, largely helical “arms” of the RNA, extending from a central core. Intriguingly, both the template and the Est2p catalytic subunit binding region (nucleotides 728–864) (18) reside at the central core, despite being ≈300 nucleotides away in primary sequence.
Refining the Model on the Basis of Comparative Analysis with Other Saccharomyces TLC1 RNAs. Kluyveromyces budding yeast telomerase RNAs are significantly different in sequence from Saccharomyces TLC1, making them useful for studying some conserved elements (10, 17) but essentially useless for beginning to deduce overall RNA secondary structure. Therefore, with the aim of comparing less divergent telomerase RNAs, we acquired sequences for the region predicted to contain the TLC1 gene in S. paradoxus, S. mikatae, and S. bayanus from Manolis Kellis and Eric Lander (Massachusetts Institute of Technology, Cambridge) (27). As expected, sequences from these organisms in the ≈2 kb between the PDX3 and CSG2 genes on the right arm of chromosome II contained likely orthologs of S. cerevisiae TLC1: the central 14 of 16 template nucleotides (3) were 100% conserved and high sequence identity was identified in the well characterized functional regions, as well as other locations (Fig. 5). Interestingly, the telomerase RNA sequences have apparently diverged significantly among the four Saccharomyces species, as alignment of TLC1 sequences showed only 43% identity (Table 1). In contrast, alignments performed in identical fashion on likely orthologs of other coding and noncoding RNAs from these same four species yielded high sequence identity (82–100%).
Table 1. Telomerase RNA sequence is less conserved than that of other RNAs among four Saccharomyces species.
| RNA | Size, nt | Sequence identity, % (all four species) |
|---|---|---|
| Telomerase RNA | 1,261 | 43 |
| 5S rRNA | 121 | 100* |
| 18S rRNA | 1,800 | 99* |
| Actin mRNA (ORF) | 1,128 | 92† |
| RNase P RNA | 369 | 84* |
| U1 small nuclear RNA | 371‡ | 82 |
Nonprotein coding RNA sequences are unannotated in the database
The four actin protein sequences show 100% identity
Alignment is for the 5′-most 371 of 568 nt
The Saccharomyces telomerase RNA sequences were inspected for covarying nucleotides that would support the existence of conserved RNA helices. Identification of such covariation was facilitated by using alifold, a computer program that identifies credible covariation events on the basis of aligned sequences from different species and the most energetically favored folding predictions (28–30). Most of the 24 identified covarying nucleotide pairs were near stretches of 100% conserved nucleotides, where important structure and function are most likely to exist. Where alifold identified highly credible covariation events that were not already present in the mfold model, the covarying nucleotides were then forced to pair by constraining mfold. This combination of thermodynamic and phylogenetic predictions led to the working model that we propose for S. cerevisiae TLC1 in Fig. 1 (see also Fig. 6). Based on the criterion that two covariation events “prove” the existence of a particular helix, four novel helices are thus supported by covariation in the proposed model (red boxes in Fig. 1).
Telomerase Functions When Est1p Is Tethered to the RNA at Diverse Positions. The yeast telomerase RNA must bind Est1p to function in vivo (10). To test whether Est1p must bind in a specific orientation within telomerase, we repositioned its binding site in the RNA to diverse locations and assessed telomerase function in vivo. We took advantage of a plasmid expressing a bulge deletion RNA that cannot bind Est1p (10) and then reintroduced a 180-nt wild-type Est1p-binding site at three unnatural locations (Fig. 2A). We chose positions 220, 450, and 1033 because they were in nonessential regions of the RNA (18) that were also not well conserved. For all of the designed constructs, mfold indicated that TLC1 RNA would fold as it does in wild type, with the inserted Est1p site predicted to be appended to an otherwise unperturbed RNA structure (data not shown).
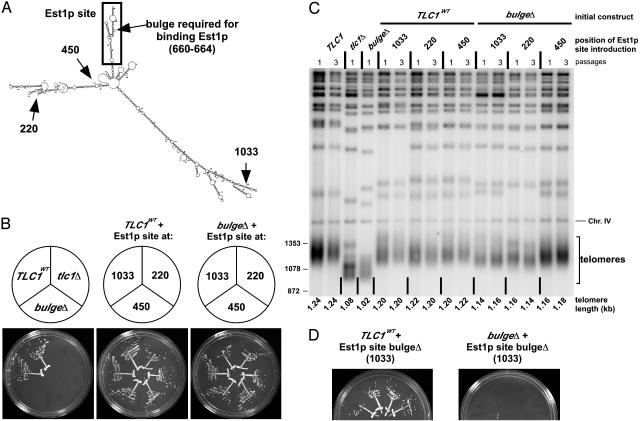
Fig. 2.
Relocation of the Est1p-binding site is tolerated by telomerase. (A) Schematic showing position of the bulged stem required for binding Est1p (arrow) (10) as well as positions to which the Est1p-binding site (boxed region) was relocated. (B) Growth of cells with the Est1p-binding site moved to three unnatural positions after ≈100 generations. Two independent isolates are shown for each TLC1 clone. (C) Southern blot of telomeric DNA from cells harboring TLC1 RNA with repositioned Est1p-binding site. Genomic DNA was digested with XhoI and probed for telomere repeats. A fragment of chromosome IV (Chr. IV) was identified by a second probe and served as an internal control for relative mobility, used to more accurately quantify length of the smallest telomeric restriction fragments (telomeres). Numbers to left indicate size markers (bp). (D) Relocated Est1p site without a bulge does not rescue Est1p-binding defective RNA. Four independent isolates are shown.
These TLC1 RNAs harboring relocated Est1p-binding sites were all capable of supporting cell growth (Fig. 2B). Furthermore, the telomere lengths supported by these RNA constructs were similar to wild type as well as respective wild-type control RNAs with the corresponding Est1p-binding site insertion (Fig. 2C). To test the possibility that insertion of Est1p-binding sequences at these locations somehow rescued the activity of the mutated Est1p-binding site at the endogenous location, we inserted bulge-deleted versions of the Est1p site at the same three locations. As expected, the bulge-deleted Est1p sites could not restore telomerase function when introduced at any of these three locations (Fig. 2D Right and data not shown). Furthermore, the same RNA sequences had no apparent effect on growth when inserted into wild-type TLC1 (Fig. 2D Left and data not shown), demonstrating that the inability of these inserted sequences to rescue the Est1p-binding-defective RNA was not due to a negative secondary effect on telomerase. Thus, Est1p can be tethered to TLC1 RNA at diverse positions and still provide telomerase function in vivo. Although we present this result in the context of the secondary structure model, this conclusion is model-independent.
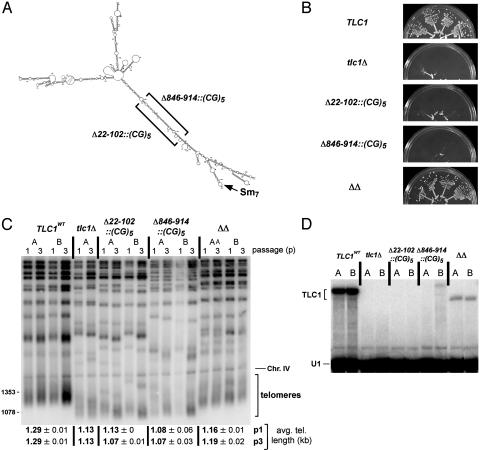
Testing the Existence and Length Requirement of the Terminal Arm. We tested the existence and length requirement of the longest unbranched section of the terminal arm, which contains the 5′ and 3′ ends as well as the Sm-binding site. Nucleotides 22–102 and 846–914 were each replaced by the sequence (CG)5 (Fig. 3A), and the ability of each RNA to support telomerase activity in vivo was assessed. Replacement of either 22–102 or 846–914 with (CG)5 caused senescence (Fig. 3B). This result correlates with mfold modeling of these mutant TLC1 RNAs: in both cases, the predicted structure of the Est2p-binding region (and sometimes other regions) differed significantly from that observed in wild-type TLC1. If Est2p is no longer able to bind to telomerase RNA, the enzyme will not function (18). In contrast, when the Δ22–102::(CG)5 and Δ846–914::(CG)5 mutations were present in the same RNA, cells exhibited wild-type growth (Fig. 3B) and telomere length was restored to nearly wild-type levels (ΔΔ, Fig. 3C). TLC1 RNA levels for the compensatory mutant were 11% of wild-type levels (ΔΔ, Fig. 3D), whereas the RNA levels of the two senescent single mutants were essentially undetectable, probably because these RNAs did not support RNP formation. Thus, the data support the existence of the proposed terminal arm of TLC1, because combining two deleterious single mutations would be unlikely to restore function if the two regions where (CG)5 sequences were introduced were not paired to each other in the structure. Furthermore, the length requirement for the arm is not fixed, but rather the arm can be shortened without significant loss of function. This finding also means that the Sm-binding site need not be held at a fixed distance from the central core in the RNA secondary structure, but can still function when moved closer.
Fig. 3.
Testing the existence and requirement of the terminal arm. (A) Schematic showing the nucleotide replacements in each single mutant and the double mutant. Nucleotides 22–102 were replaced with the sequence (CG)5, predicted to pair in the double mutant (ΔΔ) with the (CG)5 that replaced nucleotides 846–914 on the other side of the predicted terminal arm. (B) Growth of four independent isolates of each mutant is shown after ≈100 generations. (C) Length of telomeric DNA from helix replacement mutants. Numbers to left indicate size markers (bp). (D) Northern blot showing expression of TLC1 RNA in terminal arm mutants. A second probe for U1 small nuclear RNA was used as an internal control for loading and relative mobility. Cells were grown to stationary phase before being harvested.
Discussion
For the case of the essential regulatory subunit Est1p, we have shown that multiple structural rearrangements of its binding site within telomerase RNA provide function. Repositioning of RNA sequence sufficient for binding Est1p (nucleotides 524–704), including the nucleotides shown to form a required bulged stem (10), to position 220, 450, or 1033 is tolerated by telomerase in vivo. In addition, truncation of the terminal arm of the RNA “reels in” the Sm site with respect to the central core in the secondary structure model, yet is still compatible with Sm function. These results suggest that the overall structure of telomerase is at least somewhat flexible and that telomerase RNA tethers Est1p and the Sm proteins to the RNP rather than positioning them precisely within a highly structured complex.
The hypothesis that yeast telomerase RNA provides a flexible scaffold for protein subunits arose naturally from previous observations. The bulk of yeast telomerase RNA sequence is changing rapidly during evolution (ref. 17 and this study) and most of TLC1 RNA is dispensable for function (18). These findings suggested that only a few discrete RNA structures in TLC1 might be required for its function and that the rest of the RNA serves primarily to tether these elements together.
The working secondary structure model that we propose fits nicely with this hypothesis; accessory protein binding sites exist at the tips of three long quasihelical arms, which project from a central catalytic core. A high degree of nucleotide conservation (more the exception than the rule for TLC1) exists in these regions of TLC1 RNA that are known to bind proteins. Sequence covariation and conservation also suggest that the RNA stems around nucleotides 1000–1050 may serve as a site for binding protein(s). Other conserved structures in TLC1, such as the base of the terminal arm (nucleotides 100–136 paired with 815–850), probably play RNA-specific roles important for the overall folding of telomerase RNA. The structure model provides an explanation for the earlier result that deletion of nucleotides 101–138 interferes with Est2p binding (18); these nucleotides now appear to be just across the central loop from the main Est2p-binding structure (A. Chappell and V. Lundblad, personal communication). Finally, most of the regions where TLC1 RNA sequence has changed rapidly reside in the middle of the arms proposed by our initial model. These regions are the most difficult to model and may be poorly conserved because the structure is relatively unimportant. Thus, phylogenetic analysis, which requires conserved structure for success, may not be applicable for deducing the true structure of these segments of S. cerevisiae TLC1. However, it is also likely that with more sequences and improvements in RNA structure prediction, modeling of at least some of these vast regions will be significantly improved.
How does the S. cerevisiae telomerase RNA model compare with those for ciliates and vertebrates? It is tempting to speculate that the proposed central core of TLC1 is analogous to the ciliate RNA and to the template/pseudoknot domains of the vertebrate RNAs. Such a conserved core with yeast-specific functional appendages has previously been described for U1 small nuclear RNA (31). However, such comparisons for telomerase RNA are premature, since, for example, it is not even clear whether the Kluyveromyces RNA (17) forms an overall structure analogous to that of Saccharomyces.
In conclusion, we have proposed a working secondary structure for the rapidly evolving S. cerevisiae telomerase RNA. The model has already been useful in guiding genetic experiments to test the length requirement of the prominent terminal arm as well as engineering repositioned Est1p-binding sites. On the basis of these experiments we propose that budding yeast telomerase RNA serves a previously unrecognized function for an RNA in an RNP, acting as a flexible scaffold for protein subunits. The mode by which TLC1 achieves its function is flexible, in the sense that dramatic repositioning of the essential Est1p site is accommodated. Additionally, our results suggest that the RNA may have structural flexibility, such that the telomerase RNP, in contrast to the ribosome, is a rather loosely ordered complex of RNA and protein subunits.
Supplementary Material
Acknowledgments
We thank Anita Seto, April Livengood, Art Zaug, Quentin Vicens, Stefan Aigner, and Feng Guo. We thank Andy Chappell, Vicki Lundblad, Manolis Kellis, and Eric Lander for sharing information before publication and Anita Seto, Quentin Vicens, Michael Rosbash, and David Shore for helpful suggestions on the manuscript. D.C.Z. is an associate of the Howard Hughes Medical Institute. This work was supported by National Institutes of Health Grant GM28039.
Abbreviations: RNP, ribonucleoprotein; TERT, telomerase reverse transcriptase.
References
- 1.Blackburn, E. H. (2001) Cell 106, 661-673. [DOI] [PubMed] [Google Scholar]
- 2.Greider, C. W. & Blackburn, E. H. (1989) Nature 337, 331-337. [DOI] [PubMed] [Google Scholar]
- 3.Singer, M. S. & Gottschling, D. E. (1994) Science 266, 404-409. [DOI] [PubMed] [Google Scholar]
- 4.Lingner, J., Hughes, T. R., Shevchenko, A., Mann, M., Lundblad, V. & Cech, T. R. (1997) Science 276, 561-567. [DOI] [PubMed] [Google Scholar]
- 5.Cech, T. R. (2004) Cell 116, 273-279. [DOI] [PubMed] [Google Scholar]
- 6.Stellwagen, A. E., Haimberger, Z. W., Veatch, J. R. & Gottschling, D. E. (2003) Genes Dev. 17, 2384-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lundblad, V. & Szostak, J. W. (1989) Cell 57, 633-643. [DOI] [PubMed] [Google Scholar]
- 8.Beernink, H. T., Miller, K., Deshpande, A., Bucher, P. & Cooper, J. P. (2003) Curr. Biol. 13, 575-580. [DOI] [PubMed] [Google Scholar]
- 9.Reichenbach, P., Hoss, M., Azzalin, C. M., Nabholz, M., Bucher, P. & Lingner, J. (2003) Curr. Biol. 13, 568-574. [DOI] [PubMed] [Google Scholar]
- 10.Seto, A. G., Livengood, A. J., Tzfati, Y., Blackburn, E. & Cech, T. R. (2002) Genes Dev. 16, 2800-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lingner, J., Cech, T. R., Hughes, T. R. & Lundblad, V. (1997) Proc. Natl. Acad. Sci. USA 94, 11190-11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lundblad, V. (2003) Curr. Biol. 13, R439-R441. [DOI] [PubMed] [Google Scholar]
- 13.Taggart, A. K. & Zakian, V. A. (2003) Curr. Opin. Cell Biol. 15, 275-280. [DOI] [PubMed] [Google Scholar]
- 14.Will, C. L. & Lührmann, R. (2001) Curr. Opin. Cell Biol. 13, 290-301. [DOI] [PubMed] [Google Scholar]
- 15.Seto, A. G., Zaug, A. J., Sobel, S. G., Wolin, S. L. & Cech, T. R. (1999) Nature 401, 177-180. [DOI] [PubMed] [Google Scholar]
- 16.Chen, J. L., Blasco, M. A. & Greider, C. W. (2000) Cell 100, 503-514. [DOI] [PubMed] [Google Scholar]
- 17.Tzfati, Y., Knight, Z., Roy, J. & Blackburn, E. H. (2003) Genes Dev. 17, 1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livengood, A. J., Zaug, A. J. & Cech, T. R. (2002) Mol. Cell. Biol. 22, 2366-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans, S. K. & Lundblad, V. (2002) Genetics 162, 1101-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuker, M. (2003) Nucleic Acids Res. 31, 3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathews, D. H., Sabina, J., Zuker, M. & Turner, D. H. (1999) J. Mol. Biol. 288, 911-940. [DOI] [PubMed] [Google Scholar]
- 22.Sikorski, R. S. & Hieter, P. (1989) Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterson, S. E., Stellwagen, A. E., Diede, S. J., Singer, M. S., Haimberger, Z. W., Johnson, C. O., Tzoneva, M. & Gottschling, D. E. (2001) Nat. Genet 27, 64-67. [DOI] [PubMed] [Google Scholar]
- 24.Seto, A. G., Umansky, K., Tzfati, Y., Zaug, A. J., Blackburn, E. H. & Cech, T. R. (2003) RNA 9, 1323-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kambach, C., Walke, S., Young, R., Avis, J. M., de la Fortelle, E., Raker, V. A., Lührmann, R., Li, J. & Nagai, K. (1999) Cell 96, 375-387. [DOI] [PubMed] [Google Scholar]
- 26.Thore, S., Mayer, C., Sauter, C., Weeks, S. & Suck, D. (2003) J. Biol. Chem. 278, 1239-1247. [DOI] [PubMed] [Google Scholar]
- 27.Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E. S. (2003) Nature 423, 241-254. [DOI] [PubMed] [Google Scholar]
- 28.Hofacker, I. L., Fekete, M., Flamm, C., Huynen, M. A., Rauscher, S., Stolorz, P. E. & Stadler, P. F. (1998) Nucleic Acids Res. 26, 3825-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hofacker, I. L. (2003) Nucleic Acids Res. 31, 3429-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofacker, I. L., Fekete, M. & Stadler, P. F. (2002) J. Mol. Biol. 319, 1059-1066. [DOI] [PubMed] [Google Scholar]
- 31.Kretzner, L., Krol, A. & Rosbash, M. (1990) Proc. Natl. Acad. Sci. USA 87, 851-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bosoy, D., Peng, Y., Mian, I. S. & Lue, N. F. (2003) J. Biol. Chem. 278, 3882-3890. [DOI] [PubMed] [Google Scholar]
- 33.Chapon, C., Cech, T. R. & Zaug, A. J. (1997) RNA 3, 1337-1351. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.