Abstract
Amyotrophic lateral sclerosis is a rapidly progressing neurodegenerative disease associated with protein misfolding and aggregation. Most cases are characterized by TDP-43 positive inclusions, while a minority of familial ALS cases are instead FUS and SOD1 positive respectively. Cells can generate inclusions of variable type including previously characterized aggresomes, IPOD or JUNQ structures depending on the misfolded protein. SOD1 invariably forms JUNQ inclusions but it remains unclear whether other ALS protein aggregates arise as one of these previously described inclusion types or form unique structures. Here we show that FUS variably partitioned to IPOD, JUNQ or alternate structures, contain a mobile fraction, were not microtubule dependent and initially did not contain ubiquitin. TDP-43 inclusions formed in a microtubule independent manner, did not contain a mobile fraction but variably colocalized to JUNQ inclusions and another alternate structure. We conclude that the RNA binding proteins TDP-43 and FUS do not consistently fit the currently characterised inclusion models suggesting that cells have a larger repertoire for generating inclusions than currently thought, and imply that toxicity in ALS does not stem from a particular aggregation process or aggregate structure.
Amyotrophic Lateral Sclerosis (ALS) is characterised by the progressive and selective death of upper and lower motor neurones in the motor cortex and spinal cord. This leads to loss of muscle control, muscle atrophy and invariably death, generally within 3–5 years of diagnosis. The cause(s) of most cases of ALS (sporadic ALS; sALS) remain undefined, however, approximately 5–10% of cases are inherited (familial ALS; fALS). Mutations in genes now known to cause ALS include SOD11, FUS/TLS2,3, VAPB4, TARDBP5,6, OPTN7, VCP8, SQSTM19, UBQLN210, PFN111, MATR312 and hexanucleotide expansions in C9ORF7213.
In common with other neurodegenerative diseases such as Alzheimer’s disease, Creutzfeldt-Jakob disease, Parkinson’s disease, Huntington’s disease14, there is growing evidence that a correlation exists between protein aggregate formation and neuronal loss in ALS spinal cord15,16,17,18,19. Indeed, the progression of disease has been proposed to be due to propagation of protein misfolding and aggregation20. Inclusions associated with neurodegenerative disease are made up of insoluble proteinaceous material that consist mainly of one main constituent14, but also are composed of a variety of proteins, including molecular chaperones21,22 and the proteasome23. It is unclear how these additional components become part of the inclusions, but at least some may be there due to the functional interaction, such as chaperone and degradation machinery, with misfolded proteins in the aggregates24. In Huntington’s disease, huntingtin with expanded poly glutamine repeats (polyQ) predominates in neuronal inclusions. In Parkinson’s disease, inclusions known as Lewy bodies are thought to invariably consist mainly of α-synuclein. Ubiquitin positive round and skein-like inclusions in most cases of ALS are immunoreactive for TDP-4325,26, the exceptions are SOD1 and FUS familial cases of ALS which are associated with SOD1 and FUS positive inclusions respectively2,3,27.
More generally, previous work has identified common structures created during aggregation of a range of proteins, such as soluble oligomeric aggregates, that can be toxic when applied to cells28. However, in ALS pathology it remains unclear if the inclusions, or other aggregate form, are themselves toxic or if they are a symptom of proteotoxicity, reflecting a cellular response/adaptation to misfolded protein. Within the cellular milieu, protein aggregates or inclusions can be generated by several distinct pathways. Originally, cell driven, microtubule dependant juxtanuclear inclusions of CFTR ensheathed by vimentin were discovered and termed aggresomes29. These may be related to the juxtanuclear quality control compartment (referred to as JUNQ30) later observed in yeast and mammalian cells. JUNQ-like inclusions are enriched in ubiquitylated proteins, proteasome subunits, chaperones such as Hsp70 and have a mobile fraction30,31. In addition, an insoluble protein deposit (IPOD) compartment has been observed in yeast and mammalian cells and is a dense, immobile compartment consisting of insoluble protein aggregates that (at least initially) are not ubiquitinated30,32. While postmortem studies have identified ALS inclusions that are either TDP-43, FUS or SOD1 immunoreactive, a thorough characterisation and comparison of how these inclusions arise has not been performed. While misfolded mutant SOD1 is consistently associated with JUNQ-like inclusions as reported previously30,33, we now report for the first time that while FUS and TDP-43 can form iPOD and/or JUNQ-like inclusions respectively they can also partition to inclusions that are distinct from SOD1 inclusions, distinct from each other, and from other described inclusions types. These data may suggest that cells have a broader array of pathways for generating proteinacious inclusions than originally thought, and imply that toxicity in ALS does not stem from a particular aggregation process or aggregate structure.
Results
Distinct partitioning of ALS associated mutant SOD1 from TDP-43wt aggregates
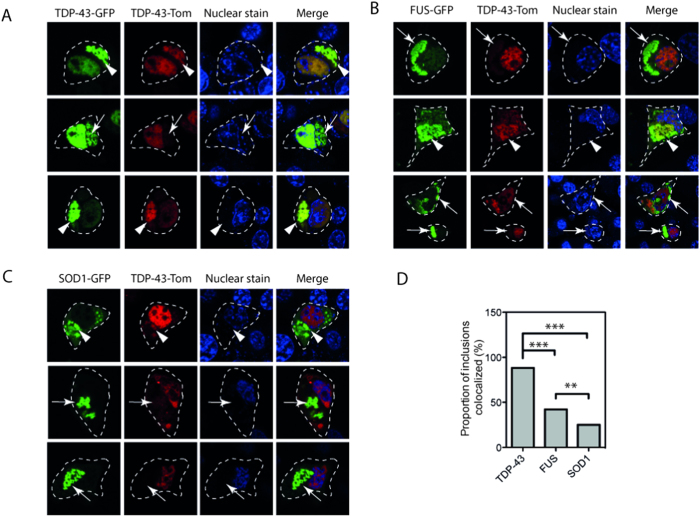
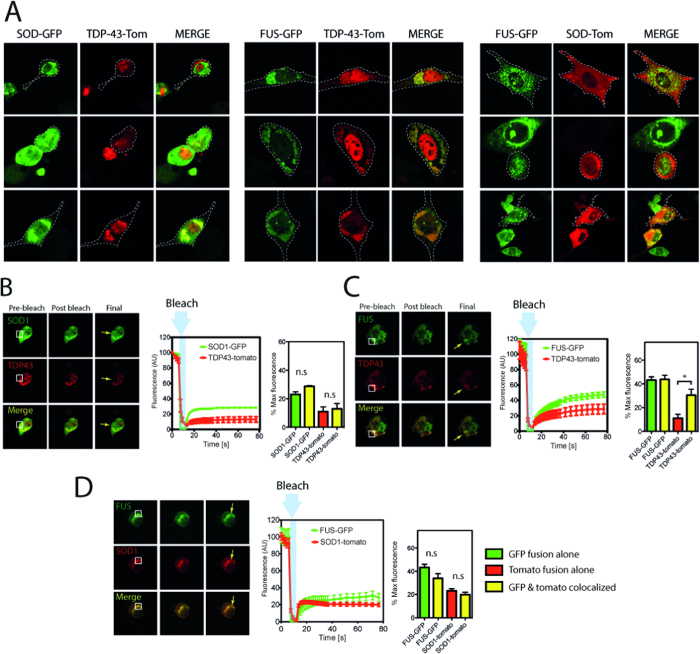
Given that many ALS inclusions contain TDP-43wt, we sought to examine whether overexpression of mutant TDP-43, FUS or SOD1 would induce TDP-43 aggregation and if so whether the aggregates accumulate in distinct cellular compartments. We used TDP-43wt-tomato34, which under basal conditions remains predominantly in the nucleus, as a reporter for TDP-43 recruitment into aggregates. Aggregated TDP-43 has previously been shown to recruit TDP-43wt into cytoplasmic inclusions35. Consistent with this, we observe that co-expression of TDP-43M337V-GFP and TDP-43wt-tomato produces cytosolic inclusions (defined as fluorescent foci > 2 μm) positive for both mutant and TDP-43wt in approximately 88% of cells expressing both constructs that contain inclusions (Fig. 1A,D; white arrow heads). However, while the bulk of TDP-43M337V is retained in the cytosol in inclusions, co-localization of TDP-43wt with these inclusions did not always deplete the nucleus of TDP-43wt. Indeed, in some instances inclusion formation of TDP-43M337V did not recruit TDP-43wt (Fig.1A, white arrows).
Figure 1. Aggregated mutant TDP-43 and FUS recruit TDP-43wt.
NSC-34 cells were transiently transfected with TDP-43wt-tomato and examined by confocal microscopy after 48 hours. (A) Mutant TDP-43M337V-GFP coexpressed with TDP-43wt-tomato. (B) Mutant FUSR495X co-expressed with TDP-43wt-tomato. (C) SOD1A4V co-transfected with TDP-43wt tomato. White arrow heads indicate areas of colocalization of GFP and tomato tagged protein. White arrows indicate distinct inclusion structures containing one tagged protein only. (D) Proportion of cells with aggregates where both mutant TDP-43, FUS and SOD1 colocalize with TDP-43wt. Data is combined from 3 independent experiments, Z-tests were used to compare differences in proportions **p < 0.01, ***p < 0.001.
Although FUS can be found associated with ubiquitin positive inclusions in sALS36, FUS positive inclusions in mutant FUS fALS cases are not TDP-43 positive37. In addition, it has been suggested that FUS and TDP-43 may have different aggregation pathways in cells38. In the current study, expression of FUSR495X -GFP lead to inclusions that when co-expressed with TDP-43wt-tomato resulted in co-aggregation of mutant FUS and TDP-43wt (Fig. 1B,C, white arrow heads; ~ 42% of cells containing inclusions of FUSR495X and TDP-43wt). This suggests that at least in some cases, and under the conditions used here, FUS recruits TDP-43 to the same type of aggregates in neuron-like cells. Similar to expression of mutant TDP-43, not all FUSR495X inclusions recruited TDP-43wt (Fig. 1B, white arrows).
In sporadic ALS the co-localization of SOD1 with TDP-43 positive inclusions is rare, and SOD1 fALS cases are characterized by inclusions positive for SOD1 but negative for TDP-4339. Consistent with these data, our cell model of overexpression of SOD1A4V rarely resulted in the aggregation of both SOD1A4V and TDP-43wt (Fig. 1C; 1 occurrence in every 140 cells containing SOD1). However, in the event that TDP-43 aggregation occurred in the presence of SOD1A4V aggregates ~ 25% of these inclusions colocalized. Notably, there was a significant difference between the proportion of cells containing colocalization of TDP-43wt in cells containing aggregates of TDP-43M337V, FUSR495X, SOD1A4V and TDP-43wt (z-test; n = 73, 165, 79 respectively; p < 0.01) suggesting different mechanisms of aggregation in each case.
Given the predominant co-aggregation of mutant TDP-43 and FUS but not SOD1 with TDP-43wt, we examined the potential role of RNA in the construction of these inclusions. Firstly, we used actinomycin D to supress RNA synthesis (Fig. S1C) to determine if reducing overall RNA levels would alter inclusion formation. Treatment of cells with actinomycin D overnight did not significantly change the number of inclusions formed in cells expressing TDP-43M337V, FUSR495X or SOD1A4V (Fig. S1). Next we lysed cells containing inclusions and then treated lysates with RNase. We reasoned that if TDP-43 and FUS were bound to inclusions at least partly via interactions with RNA then treatment would disrupt inclusions. In the absence of RNase inclusions were trapped in our filter trap assay (Fig. S1D,E). Similar levels of TDP-43M337V, FUSR495X, and SOD1A4V were found trapped regardless of RNase treatment (Fig. S1D,E), suggesting inclusion formation was independent of RNA.
Microtubule disruption affects SOD1 inclusion formation but not TDP-43 and FUS
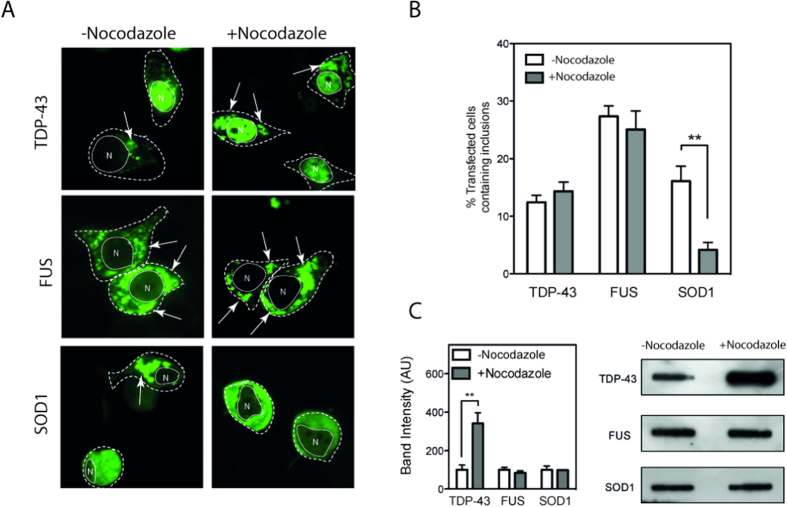
Previous work indicates that microtubule disruption promotes TDP-43 inclusions40 and halts the formation of a very specific FUS inclusion type formed by the 1–359 FUS fragment41. Similarly, aggresomes, IPOD and JUNQ structures are suppressed when microtubules are disrupted29,30. To compare the effects of destabilization of microtubules on ALS inclusions we treated NSC-34 cells transfected with mutant FUS, TDP-43 or SOD1 fused to GFP with microtubule destabilizer nocodazole. We observed large cytoplasmic inclusions that are cloud or sponge-like in appearance (referred to as conglomerate in ALS pathology) in approximately 15% of SOD1A4V -GFP transfected cells (Fig. 2A,B) but after nocodazole treatment this was significantly reduced (p < 0.01; Student’s t-test) to fewer than 5% of transfected cells had large inclusions. The remaining cells did not appear to have smaller granular foci as observed in other aggregating systems such as occurs in the Htt42 model but remained in a diffuse pattern of fluorescence. This is consistent with previous observations of mutant SOD1 forming JUNQ-like aggresomes that are microtubule dependent43. In contrast, NSC-34 cells transfected with either TDP-43M337V or FUSR495X showed no significant difference in percentage of inclusions present regardless of nocodazole treatment (Fig. 2A,B). This suggests that neither FUS nor TDP-43 aggregate in a manner consistent with aggresomes, IPOD or JUNQ structures. However, it must be noted that although there is evidence that IPOD–like inclusions require intact microtubules to aggregate, evidence also exists to show that polyQ-104 protein aggregates similarly in the presence or absence of nocodazole44. To complement image analysis we also performed filter trap assays to examine any difference in total trappable aggregated protein under the same conditions. We found that although the number of SOD1 inclusions decreased upon nocodazole treatment (Fig. 2B, p < 0.05; Student’s t-test), total aggregation measured by filter trap did not (Fig. 2C). Similarly, FUS aggregation did not change after nocodazole treatment. In contrast, total trappable TDP-43 aggregates significantly increased upon nocodazole treatment (Fig. 2C; p < 0.01; Student’s t-test).
Figure 2. Microtubule destabilization prevents SOD1 inclusions but not TDP-43 and FUS inclusions.
NSC-34 cells were transiently transfected with mutant TDP-43, FUS, or SOD1-GFP and after 24 hours incubated with or without 33 μM nocodazole. (A) Confocal images of cells expressing GFP fusion proteins in the presence or absence of nocodazole. Dotted white line represents cell outline, solid white line represents nuclear outline obtained from transmission image (denoted N), white arrows indicate inclusions. (B) Quantification of the proportion of cells with inclusions in transfected cells. At least 6 fields of view from each timepoint were counted (minimum 30 cells per field) and scored. Experiments were performed 3 times and bar charts represent mean and standard deviation. ** indicates p < 0.01 (C) After treatment with nocodazole cells were lysed and lysates were used in filter trap assays. Trapped material represents aggregates. Western blotting of resulting filter trap assay and quantification using imageJ. Experiments were performed 3 times and bar charts represent mean and standard deviation. **indicates p < 0.01.
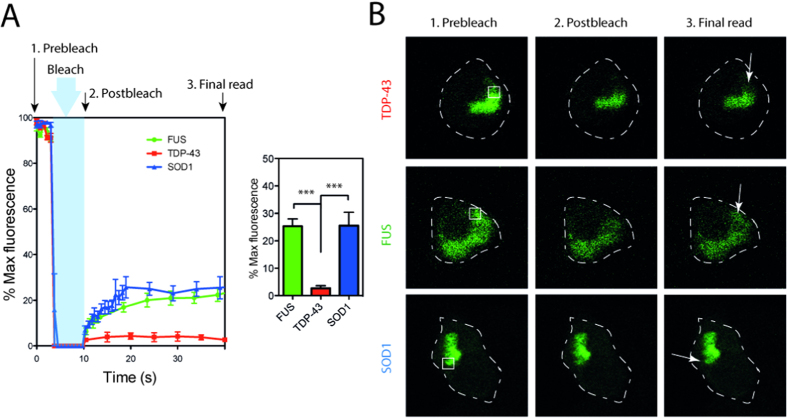
TDP-43 aggregates are immobile
IPOD structures are thought to be predominantly immobile, while JUNQ and aggresome structures contain a mobile fraction31,45. Here we used fluorescence recovery after photo bleaching (FRAP) to examine the mobility of molecules within ALS inclusions. Using a high powered laser pulse we bleached a region of GFP positive inclusions that corresponded to < 25% of any given inclusion and then recorded the subsequent fluorescence over 40 s. We found that there was minimal fluorescence recovery after bleaching TDP-43M337V inclusions (Fig. 3A,B) confirming recent observations46. In contrast, our data suggests that both SOD1 and FUS inclusions recover approximately 25% of their original fluorescence, significantly more recovery than TDP-43 (Tukey’s multiple comparison test; p < 0.001), suggesting a mobile fraction or porous structure within the inclusions (Fig. 3A,B). The recovery patterns of each of the aggregates was similar to that of the soluble protein tested in both wt and mutant expressing cells (Fig. S2). Therefore we cannot rule out the possibility that the protein species diffusing in to the inclusion are soluble species from outside of the aggregate and do not represent diffusion from within the aggregate itself.
Figure 3. SOD1 and FUS inclusions contain a mobile fraction.
FRAP analysis of inclusions formed in NSC-34 cells expressing mutant TDP-43, FUS or SOD1-GFP. (A) Mean fluorescence intensity (from within the ROI) plotted over time. Prebleach intensity was recorded, and recovery was recorded for up to 40 s. Results are means and standard deviation from n = 10. Mean fluorescence at 40 s is quantified in histogram. ***indicates p < 0.001. (B) Representative confocal images of prebleach, post bleach and recovery endpoint. ROI is marked in white, timing of image relative to data collection is marked in panel A.
FUS but not TDP-43 and SOD1 co-aggregate with IPOD substrate Huntingtinex146Q
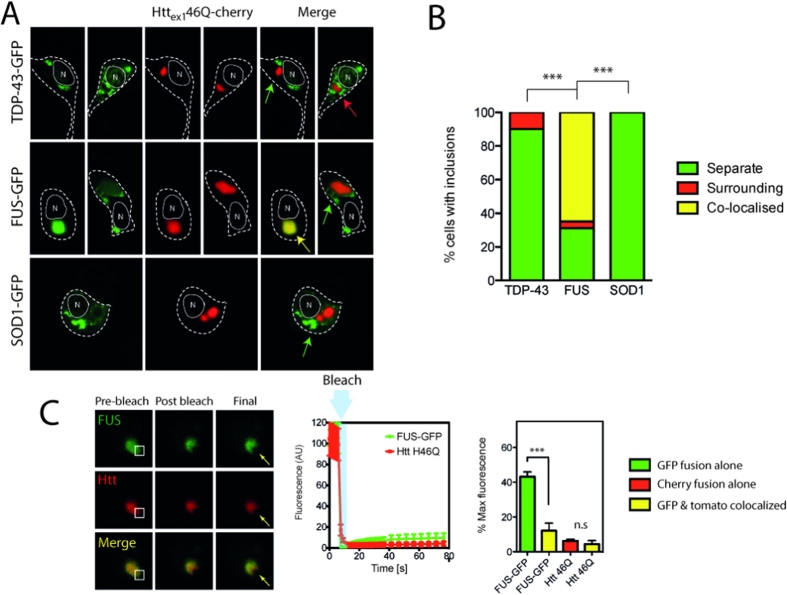
Proteins that aggregate in to insoluble deposits can be recruited in to the same IPOD structure in cells regardless of their amino acid sequence31,33. To test whether ALS mutants would form IPOD-like structures we co-transfected NSC-34 cells with IPOD substrate Huntingtinex146Q fused to cherry (Httex146Q-cherry) and either TDP-43M337V, FUSR495X, or SOD1A4V GFP fusions. Expression of Httex146Q consistently generated a round IPOD-like inclusion in the cytoplasm or nuclei in NSC-34 cells (Fig. 4A). Consistent with previous work, co-expression of Httex146Q-cherry with mutant SOD1 produced inclusions of SOD1 distinct from Httex146Q-cherry inclusions in 100% of cells expressing both proteins (Fig. 4A,B)30,31,33. When TDP-43M337V was co-expressed with Httex146Q-cherry, inclusions were predominantly separate with no co-localization (Fig. 4A green arrow). However, in ~ 10% of cells containing aggregates of both TDP-43 and Htt the TDP-43M337V aggregates were tightly packed around the dense Htt core (Fig. 4A red arrow). In contrast, a statistically significant increase in FUS aggregates, compared to TDP-43 and SOD1, (n = 46; compared to SOD1 (n = 42) and TDP-43 (n = 45); z-test, p < 0.001) co-localized to Httex146Q-cherry IPOD-like structures (Fig. 4A yellow arrow). The remaining FUS aggregates were found to be similar to those observed for TDP-43 with separate inclusions, with a small percentage of those tightly packed around the Httex146Q-cherry core (Fig. 4B).
Figure 4. SOD1 and TDP-43 inclusions are distinct from iPOD.
Cells were transfected with Httex146Q-mcherry and either mutant TDP-43, FUS or SOD1-GFP. The cells were imaged 48 h after transfection (A) and then the number of inclusions colocalized or surrounding the Htt iPOD quantified (B). Green arrows indicate distinct and separate inclusions, red arrows indicate GFP tagged inclusions surrounding Htt inclusions, yellow arrows indicate colocalization. A minimum of 100 cells with inclusions were counted in each case and the experiment was repeated n = 2. A z-test of proportions was conducted on combined dataset and ***denotes p < 0.001. (C) FRAP measurements were performed on Htt-FUS positive inclusions. White box indicates ROI, arrow indicates bleached area. Mean fluorescence intensity was followed for 80 s and data shown are means and standard deviations n = 6. Histogram is data from FUS and Htt colocalized at 80 s compared to that of either FUS-GFP or Htt-RFP alone. ***indicates p < 0.001.
Given that we had found that FUS aggregates contained a mobile fraction, and that previous work had demonstrated that IPOD structures contain immobile aggregates, we used FRAP to investigate if FUS aggregates remained mobile in IPOD structures. When a region of an inclusion that contained both FUS and Httex146Q-cherry was bleached we found that neither FUS nor Httex146Q-cherry fluorescence recovered significantly (Fig. 4C), indicating that FUS was mostly immobile in IPOD structures. Under the same conditions, FUS-only aggregates recovered around 40% of the bleached fluorescence (Fig. S3C) and Httex146Q-cherry was immobile with no fluorescence recovery (Fig. S3A). These results indicate a significant decrease in recovery of FUS fluorescence in the presence of Htt aggregates (Fig. 4C; p < 0.001; Student’s t-test). In the cases where colocalization of Htt and FUS occurred, we observed a pattern of fluorescence in which ~ 80% of total FUS fluorescence and ~ 90% of total Htt was consistently associated with the dual labelled inclusions (Fig. S4A).
TDP-43 and FUS variably co-aggregate with SOD1A4V JUNQ
SOD1 is a well-characterized JUNQ substrate30,31,33. To test whether TDP-43 or FUS might be JUNQ substrates we co-transfected TDP-43M337V or FUSR495X with SOD1A4V fusion proteins. In cells that contained inclusions of both TDP-43 and SOD1 we observed that inclusions were distinct 50% of the time, the remaining cells contained inclusions that were positive for both TDP-43 and SOD1 (Fig. 5A). In the cases where colocalization occurred, we observed a pattern of fluorescence in which ~ 80% of total SOD1 fluorescence was consistently associated with the dual labelled inclusions (Fig. S4B). In contrast, the amount of TDP-43 present in these inclusions was significantly lower in comparison to SOD1 (p < 0.001; Student’s t-test) and ranged from ~ 50% to ~ 70% (Fig. S4B). This suggested to us that under certain specific conditions TDP-43 could be a JUNQ substrate. Similarly, while FUS did not form round dot like inclusions that were observed when co-expressed with Httex146Q-cherry, there was co-localization with SOD1A4V inclusions in approximately 30% of cells containing inclusions of both SOD1 and FUS (Fig. 5A). In the cases where there was inclusion colocalization ~ 80% of the total cellular SOD1 and FUS was present in the inclusion suggesting an active co-aggregation process (Fig. S4C). When TDP-43M337V and FUSR495X were aggregated in the same cell around 85% of the inclusions were positive for both TDP-43 and FUS (Fig. 5A). In a manner similar to that of SOD1 we observed that colocalized inclusions consistently contained ~ 70% of total cellular FUS, these same inclusions variably contained a significantly lower amount (p < 0.05, Student’s t-test), ~ 50% of the TDP-43 signal (Fig. S4D), consistent with TDP-43 aggregation being a secondary process.
Figure 5. Mutant TDP-43 can co-aggregate to JUNQ like SOD1 positive structures.
NSC-34 cells were cotransfected with combinations of mutant SOD1, TDP-43 and FUS and imaged after 48 h (A). Mutant TDP-43 could be found colocalized with SOD1 in some cases. (B) Mutant SOD1 and TDP-43 positive inclusions were analyzed using FRAP. White box indicates ROI, arrow indicates bleached area. Mean fluorescence intensity was followed for 80 s and data shown are means and standard deviations n = 6. Histogram is data from SOD1 and TDP-43 colocalized at 80 s compared to that of either SOD1-GFP or TDP-43-tomato alone. (C) Mutant FUS and TDP-43 positive inclusions were analyzed using FRAP. White box indicates ROI, arrow indicates bleached area. Mean fluorescence intensity was followed for 80 s and data shown are means and standard deviations n = 6. Histogram is data from FUS and TDP-43 colocalized at 80 s compared to that of either FUS-GFP or TDP-43-tomato alone. *indicates p < 0.05. (D) Mutant FUS and SOD1 positive inclusions were analyzed using FRAP. White box indicates ROI, arrow indicates bleached area. Mean fluorescence intensity was followed for 80 s and data shown are means and standard deviations n = 9. Histogram is data from FUS and SOD1 colocalized at 80 s compared to that of either FUS-GFP or SOD1-tomato alone.
Since we had observed TDP-43 inclusions to be immobile and a significant proportion of TDP-43 inclusions co-localized with SOD1 inclusions we performed FRAP to examine the mobility of these proteins in dual labeled inclusions. When bleached with 488 nm laser, TDP-43M337V remained immobile while SOD1A4V-GFP signal partially diffused back in to the bleached area—consistent with a previously observed mobile fraction (Fig. 5B). In contrast, FRAP measurements on inclusions that contained both FUSR495X and TDP-43M337V indicated that when TDP-43 was recruited in to FUS positive inclusions a proportion of the TDP-43 signal was recovered, consistent with a mobile fraction (Fig. 5C). This is consistent with TDP-43 being a secondary aggregator in these inclusions. When inclusions contained both FUSR495X and SOD1A4V both FUS and SOD1 retained a diffusible fraction (Fig. 5D).
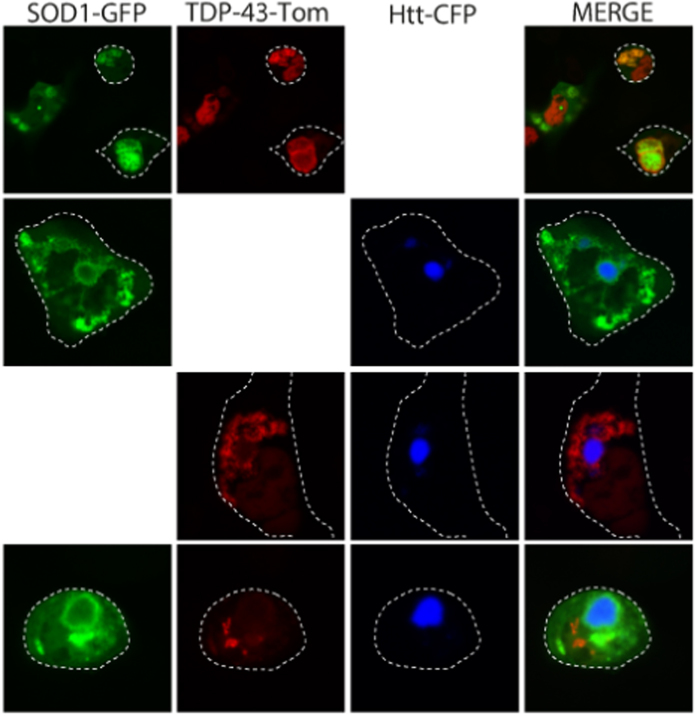
Given the different structural properties of TDP-43 and SOD1 inclusions, but their partial overlap when co-expressed, we performed triple transfections of TDP-43M337V-tomato, SOD1A4V -GFP and Httex146Q-Cerulean fluorescent protein. In this scenario, TDP-43 co-localized with SOD1A4V aggregates in some instances, but never co-localized with Httex146Q-Cerulean inclusions (Fig. 6). In some cases the cells were able to compartmentalize all three inclusions from one another. In these cases many of the TDP-43 inclusions appeared to be in the nucleus or very close to the nuclear envelope (Supplementary Material).
Figure 6. Triple transfection reveals 3 distinct inclusions.
NSC-34 cells were cotransfected with combinations of SOD1, TDP-43 and Htt including a triple transfection of all three plasmids and imaged after 48 h.
FUS and TDP-43 inclusions are ubiquitylated late compared to SOD1 inclusions
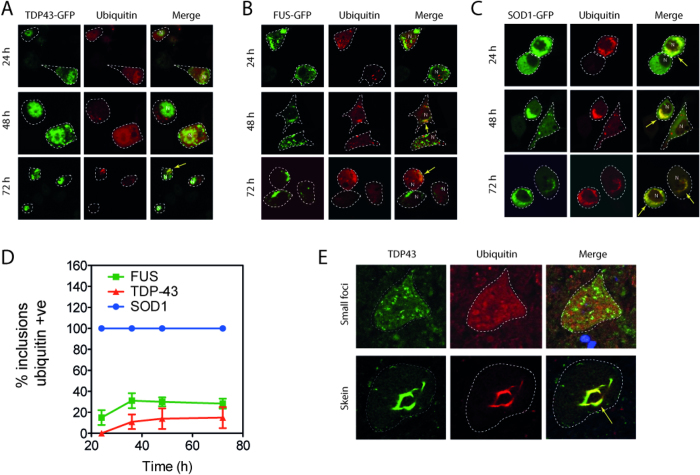
Ubiquitin positive inclusions are a hallmark of ALS, however the mechanism and dynamics of ubiquitylation and inclusion formation cannot be determined from postmortem tissue. Recent work on Huntingtin systems shows that although ubiquitylated Htt positive inclusions are present in post mortem tissue, in cell culture systems ubiquitin co-localizes to inclusions relatively late32. To compare the association of ubiquitin with ALS inclusions we initially co-transfected NSC-34 cells with mRFP-ubiquitin and SOD1A4V, TDP-43M337V or FUSR495X GFP fusions. SOD1A4V inclusions were always observed to co-localize with ubiquitin starting at our earliest observations 24 hours after transfection (Fig. 7C,D). In contrast, TDP-43M337V and FUSR495X did not always co-localize with ubiquitin with around 15 and 30% of inclusions respectively becoming positive for mRFP-ubiquitin after 72 h (Fig. 7A,B,D). To confirm this was not an artefact of the co-expression with mRFP-ubiquitin fusion we performed ubiquitin- immunostaining on cells expressing mutant FUS, TDP-43 and SOD1-GFP alone and observed similar ubiquitin co-localization patterns at 48–72 hours (Fig. S5).
Figure 7. Ubiquitin associates with TDP-43 and FUS inclusions late compared to SOD1.
NSC-34 cells were transiently transfected with mutant TDP-43, FUS, or SOD1-GFP with mRFP-ubiquitin and examined by confocal microscopy at various times after transfection (A–C). Quantification of proportion of inclusions in dual transfected cells that contain both GFP fusion proteins and ubiquitin (D). 8 fields of view from each timepoint were counted (minimum 30 cells per field) and scored. Experiment was performed in triplicate and is representative of 2 independent experiments. (E) Human ALS post mortem tissue was stained for both TDP-43 and ubiquitin. 40 inclusions were imaged across two cases of sporadic ALS. Representative images of small foci (not colocalizing with ubiquitin) and large skeins that colocalize to ubiquitin staining are shown.
Our results in cell culture are seemingly incongruous with those from post mortem ALS spinal cord sections that suggest that ubiquitin is a near universal marker of TDP-43 inclusions. To examine the relationship between TDP-43 and ubiquitin in human ALS we stained human ALS post mortem spinal cord sections and observed that 38 out of 40 TDP-43 inclusions (from 2 SALS cases) co-localized with ubiquitin. Strikingly, the 2 out of 40 inclusions that did not co-localize with ubiquitin (Fig. 7E) were smaller and more punctate (similar to the inclusions seen in our cell models after 48 h), while the ubiquitin-positive inclusions were generally extremely large skein-like inclusions (Fig. 7E). Interestingly, we rarely observed skein-like inclusions in our cell culture models, suggesting that skeins may develop over a long period of time.
Given the late redistribution of ubiquitin we tested whether the ubiquitin negative inclusions might co-localize to autophagy marker LC3. We observed that while both TDP-43 and FUS inclusions were not co-localized with LC3 they were found adjacent to LC3 positive foci. In contrast, there was no observable relationship between SOD1 inclusions and LC3 (Fig. S6).
Discussion
The role of protein aggregates in the pathology of a range of neurodegenerative diseases such as Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease has gained increasing support14,47. Generally in these cases there is one dominant pathogenic protein or peptide, such as Aβ, tau, α-synuclein, and huntingtin that misfolds and aggregates potentially causing cellular damage along the way. ALS is also associated with protein misfolding and aggregation, however although all cases are associated with ubiquitin-positive inclusions, the make up of these inclusions varies depending on their location in the CNS and the underlying genetic profile of the patient. Given that protein aggregation is a unifying feature of ALS, we hypothesized that the underlying process of aggregate formation, and the compartment to which the aggregates are driven would be similar, even though the individual proteins involved may differ. If so, this hypothesis may provide common therapeutic strategies across all ALS variants.
We investigated three fALS-associated genes which encode proteins previously observed to accumulate in ubiquitylated inclusions in postmortem tissue. While TDP-43 positive inclusions are observed in sALS and TDP-43 fALS cases, the inclusions in SOD1 and FUS associated fALS are TDP-43 negative, and SOD1 and FUS positive respectively. Our results confirm that SOD1 is a JUNQ-like aggregate compartment substrate30,31,33. Expression of mutant SOD1 results in the microtubule-dependent appearance of large juxtanuclear conglomerate inclusions that always co-localize with ubiquitin (Table 1). As observed in post mortem tissue, the co-aggregation of SOD1 and TDP-43 is relatively rare in cells expressing both mutant SOD1 and TDP-43wt.
Table 1. Summary of features of the inclusions of TDP-43M337V, FUSR495X and SOD1A4V.
| TDP-43 | FUS | SOD1 | |
|---|---|---|---|
| Recruits TDP-43WT | ✓ (88%*) | ✓ (42%*) | ✓ (25%*) |
| Microtubule dependent | × | × | ✓ |
| Contain a mobile fraction | × | ✓ | ✓ |
| Co-aggregate with Httex146Q | × | ✓ | × |
| Co-aggregate with JUNQ substrate SOD1 | ✓50%* | ✓30%* | ✓ |
| Co-localisation with ubiquitin | ✓Late | ✓Late | ✓Early |
*expressed as a percentage of cells containing both types of inclusions.
In contrast, TDP-43 and FUS form distinct, microtubule-independent inclusions that do not initially associate with ubiquitin in cell culture. In addition, TDP-43 and FUS inclusions are also structurally distinct from each other, as TDP-43 inclusions are immobile while FUS inclusions contain a mobile fraction (see Table 1). Neither of these inclusions fit the current definition of known aggregate compartments, including aggresomes, IPOD and JUNQ, suggesting the engagement of distinct protein quality control elements to assemble the inclusions. This may be an inherent property of the solution states of the proteins in question given we observed that the diffusion properties of non-aggregated FUS and TDP-43 were vastly different to that of SOD1. FRAP measurements of SOD1 indicate it has a rapidly diffusing element, consistent with previous work suggesting it is predominantly dimeric in cells, with mutations promoting monomer formation33. In contrast, FRAP measurements of non-aggregated TDP-43 and FUS (mutant and wt) indicate a very slowly diffusing species consistent with very large complexes, likely with mRNA or in the case of nuclear protein, DNA. The inherent property of TDP-43 and FUS to form functional aggregates may underlie the aggregation of these slowly diffusing species in a manner similar to stress granule formation48. Removal of stress granules is, at least in part, regulated by autophagy pathways49, and dysfunction of autophagy results in TDP-43 inclusion formation46. This contrasts the UPS-dependent pathways involved in misfolded SOD1 degradation50 and may partially explain the differences in the way SOD1, FUS and TDP-43 inclusions are formed.
While ubiquitin is thought to be universally present in TDP-43 inclusions, data from our models indicate that ubiquitin joins the inclusions late in the process. This is reminiscent of Htt inclusions32, and may represent ubiquitylation for degradation via autophagy rather than for degradation by the proteasome. This is consistent with the presence of autophagosomes in close proximity to the surface of inclusions in ALS post mortem tissue51. Interestingly, both K48 and K63 linked ubiquitin chains, directing proteins to the proteasome and autophagy pathways respectively, have been shown to localize to TDP-43 inclusions46 suggesting that autophagic processes may be active at these sites. While our model shows a minority of cells containing TDP-43 inclusions that co-localize with ubiquitin, most TDP-43 positive inclusions in human ALS spinal cord contain ubiquitin. However, our work shows that smaller, presumably early-stage aggregates do not overlap with ubiquitin staining in ALS post-mortem spinal motor neurons, consistent with our cell model and the concept that TDP-43 is ubiquitylated late in the aggregation process.
Specific proteins are consistently sequestered to very specific aggregation compartments in the cell. For example, proteins containing polyQ expansions have been shown to reliably deposit in IPOD-like inclusions in mammalian cells30,31,33. In addition, SOD1 and proteins with polyA expansions are consistently found in JUNQ-like aggregate compartments in cells30,31,33. Interestingly, when SOD1 is modified by fusion to a polyQ repeat it can be directed to the IPOD compartment, but without this artificial modification it consistently appears in the JUNQ compartment31. We confirm that while SOD1 is consistent in its cellular aggregation patterns, TDP-43 and FUS are not, displaying variable aggregation structures presumably via distinct aggregation pathways. We observed that TDP-43 could aggregate distinctly from SOD1, Htt and FUS in our cell models, and it could also co-aggregate with SOD1 and FUS. Interestingly, in these cases TDP-43 aggregates had altered diffusion properties in FUS inclusions but not in SOD1 inclusions, suggesting a co-aggregation process with mutant SOD1 but a secondary aggregation process in the case of mutant FUS. Our analysis showing a trend for a larger percentage of total FUS in these inclusions compared to that of TDP-43 supports this. The work presented here is also consistent with human post mortem tissue where TDP-43 does not accumulate with Htt in a case study of coexisting Huntington’s disease with ALS52 (although both polyQ repeats and TDP inclusions were present) and is not found in mutant SOD1 inclusions27. Similarly, FUS could co-aggregate with Htt, TDP-43 and SOD1 or it could form distinct FUS inclusions. When localized with Htt, FUS altered its diffusion properties suggesting in this case FUS aggregation was a secondary process. FUS has been found to accumulate with polyQ aggregates53 and with TDP-43 aggregates in human post mortem tissue36. This inconsistent behavior may arise due to the fact that both TDP-43 and FUS are extremely aggregation prone and may easily be pushed down several aggregation pathways within cells. Regardless, these RNA binding proteins also form inclusions that do not fit within the definitions of IPOD or JUNQ. We propose that a specific mechanism exists in cells to deal with aggregating RNA binding proteins, which we name RNA Interactor Specific Compartments/Inclusions (RISCI).
The work presented here suggests that large inclusions formed by ALS proteins are RNA independent, consistent with the formation of inclusions resulting from inappropriate misfolding related interactions rather than native interactions with RNA binding partners. The RNA binding proteins TDP-43 and FUS are intrinsically aggregation prone38,54, but while mutations in TDP-43 associated with familial ALS accelerate the rate at which it aggregates this is not the case for FUS. It is thought that the aggregation propensity of these proteins is due to the so-called prion-domain present in many RNA binding proteins55. In addition, mutations in SOD1 alter the protein stability and aggregation propensity in a way that correlates with disease progression in SOD1 associated fALS56. This intrinsic instability in these ALS proteins requires that the cell maintain protein homeostasis, either by investing in folding machinery to ensure these proteins maintain their native structure or degradation machinery to remove non-functional and misfolded protein. In general, if a cell is unable to maintain protein homeostasis it can actively partition misfolded proteins in specific ways given the individual proteins solution state, ubiquitylation state, and cellular compartment. In the current work we observe that the structures formed by TDP-43 and FUS are different, suggesting distinct pathways of inclusion formation. This is consistent with previous demonstration of fundamental differences in FUS and TDP-43 aggregation properties and a lack of overlap between genetic modifiers of FUS and TDP-43 toxicity in yeast models38,57. One possible major difference is the recent discovery that FUS forms porous hydrogels48 to which TDP-43 can bind; this is consistent with our work presented here that shows that FUS inclusions in cells have a diffusible component and that TDP-43 can be recruited in to these diffusible structures. While our work did not examine the relationship between aggregation and cellular dysfunction there are convincing arguments that suggest FUS and TDP-43 mutations that result in aggregates can cause either loss or gain of function58. Mutations in FUS can lead to inappropriate phosphorylation of RNA polymerase II59, loss of nuclear Gems60, and mutant FUS does not rescue from endogenous FUS knock down in Zebrafish61 consistent with toxicity from loss of function. In contrast, evidence exists to demonstrate that FUS does not need to be cleared from the nucleus to induce toxicity associated with cytoplasmic accumulation58 consistent with a toxic gain of function. Knock-down of zebrafish TDP-43 gene leads to a phenotype similar to that of expression of mutant human TDP-43, and can be rescued by expression of the wild type but not mutant human mutant TDP-43 consistent with at least a partial loss of function, while overexpression in yeast leads to aggregation and cellular dysfunction suggesting a toxic gain of function of aggregated TDP-4357. The data presented in this paper demonstrating that TDP-43, FUS and SOD1 can form structurally distinct aggregates in the cell combined with the fact that TDP-43 and FUS have very different functions to that of SOD1 but result in the same disease phenotype is consistent with a unified model of toxic gain of function ALS protein aggregates. It is tempting to speculate that one potential mechanism that could explain why distinct aggregates could be similarly detrimental to cellular function is the recruitment of other proteins (e.g. ubiquitin, optineurin, ubiquilin2) that are critical for cellular function. However, we cannot rule out that the loss of nuclear TDP-43 and FUS in to aggregates leads to a toxic loss of function. In reality the detrimental effects of mutations in these genes is likely to be a complex combination of both loss and gain of function.
Several mechanisms for forming inclusions in mammalian cells have been described, including IPOD, JUNQ30, aggresomes62, aggresome like structures (ALIS)63, and ER associated degradation-associated vesicles64. The almost ubiquitous nature of these types of inclusions in neurodegenerative disease, and the diverse and specific mechanisms that underpin their formation, reveals their importance to cellular protein quality control and human pathology. It is likely that these represent just a fraction of the pathways for aggregation in cells and further characterization of these processes is an important avenue for further investigation.
In conclusion, we observe that TDP-43 and FUS can form inclusions that do not fit the current definitions for known aggregation processes in cells and at least in some cases aggregate into RNA interactor specific compartments/inclusions (RISCI). However, they do not consistently aggregate in to one kind of inclusion type. Further understanding of the active processes promoting the formation of aggregate structures may help explain the apparent pathological heterogeneity in ALS that results in the formation of round ubiquitylated inclusion, skeins, Lewy body-like hyaline inclusions, and basophilic inclusions.
Materials and Methods
Plasmids
The pEGFP-N1 vectors containing human SOD1wt, SOD1A4V and G93A SOD1 were generated as described65. Expression vector pCMV6-AC-GFP containing human TDP-43 and FUS was obtained from OriGene (USA). SOD1-tomato constructs were generated by replacing the GFP sequences in the GFP-tagged constructs65 with the tomato red fluorescent protein. TDP-43wt-tomato (Addgene plasmid 28205, provided by Zuoshang Xu34), mRFP-Ubiquitin (Addgene plasmid 11935, provided by Nico Dantuma66) and pmRFP-LC3 (Addgene plasmid 21075, Provided by Tamotsu Yoshimori67) constructs were acquired from Addgene (USA). The vectors describing Httex1(46Q) fusions (C-terminally) to mCherry and Cerulean were generated as described33,68.
Cell Culture and Transfection
Neuroblastoma x Spinal cord hybrid NSC-34 cells69 were maintained in Dulbecco’s modified Eagles Medium (DMEM) supplemented with 10% foetal bovine serum (FBS, Bovogen Biologicals, Australia). Cells were kept at 37 °C in a humidified incubator with 5% atmospheric CO2. For confocal microscopy, cells were grown on 13 mm round coverslips in 24-well plates or on 4 or 8-well chambered coverglass (Nalge Nunc International, USA). Cells were grown in 6-well plates for cell lysate experiments.
Cells were transfected using Lipofectamine 2000 (Invitrogen, USA) according to manufacturer’s instructions with 0.5 μg DNA per well for a 24-well plate or 4-well chambered coverglass, 0.2 μg DNA per well for 8-well chambered coverglass and 2 μg DNA per well for 6-well plates. For co-transfections and triple transfections, the amount of DNA was divided equally between constructs. The DNA: Lipofectamine ratio used was 1:5 (w/v). Media was refreshed 5 h after transfection. For disruption of microtubules, cells were incubated with 33 μM nocadazole (Sigma-Aldrich, USA) overnight (~ 18 h) before analysis 48 h post-transfection.
Cell Lysis
Transfected NSC-34 cells in 6-well plates (+/− nocadazole) were harvested 48 h post-transfection with trypsin/EDTA (Gibco, USA). Cells were washed with phosphate buffered saline (PBS) before being pelleted by centrifugation (21000 × g for 45 s at 4 °C). Cell pellets were resuspended in ~ 50 μl lysis buffer (600 mM KCl, 20 mM Tris-Cl (pH 7.8), 20% (v/v) glycerol, Complete® Protease Inhibitor Cocktail (Roche, Switzerland)) and subjected to three freeze/thaw cycles in liquid nitrogen. To digest DNA, 250 units of DNase (Roche) was added to each lysate and incubated at 37 °C for 30 min. Lysates were then centrifuged at 300 × g for 5 min to pellet any large debris before the protein concentration was determined by BCA Assay.
Filter Trap Assay
Cell lysates with a total protein concentration of 100 μg were adjusted to 200 μl with a final sodium dodecyl sulphate (SDS) concentration of 2% in PBS. Lysates were filtered through a 0.2 μm cellulose acetate membrane (Whatman, UK) pre-equilibrated in 1% SDS in PBS using a Bio-Dot SF microfiltration unit (Bio-Rad, USA) under vacuum. Large aggregates are trapped in the membrane, while soluble material passes through70. The membrane was blocked in heat denatured casein (HDC) for 1 h at 37 °C before probing for aggregates with either monoclonal mouse anti-GFP-HRP conjugated antibody (1:1000 in HDC, OriGene—TA150043) overnight at 4 °C or polyclonal rabbit anti-GFP (1:10 000 in HDC, Abcam, UK—ab290) overnight at 4 °C followed by anti-rabbit IgG-HRP conjugated antibody (1:2000 in HDC, Bio-Rad—170–6515) for 1 h at 37 °C.
Laser Scanning Confocal Microscopy
Imaging of NSC-34 co-transfections was performed 24–72 hours post transfection. NSC-34 cells grown on coverslips were transfected with GFP-tagged mutant SOD1, TDP-43 and FUS and fixed for 20 min at room temperature (RT) with 4% paraformaldehyde (PFA) (Merck Millipore, USA) in PBS 48 h post transfection. Cells were permeabilised in 1% tritonX-100 (TX-100) in PBS for 30 min on ice before blocking for 30 min at RT with 5% FBS, 1% BSA 0.3% TX-100 in PBS. Cells were incubated with primary antibodies (see below) overnight at 4 °C followed by Alexa Fluor®-conjugated secondary antibodies reactive to mouse or rabbit IgG for 1 h on ice. All antibodies were diluted in 1% BSA, 0.1% TX-100 in PBS and cells were washed with PBS between each incubation step. Following staining, coverslips were mounted onto slides using anti-fadent mounting medium (Citifluor, UK). All cells were imaged using a Leica TCS SP5 Confocal microscope.
Antibodies
The following primary antibodies were used for cell staining experiments: monoclonal mouse anti-ubiquitin (25 μg/ml, ab7254), monoclonal mouse anti-sequestosome 1/p62 (10 μg/ml, ab56416), monoclonal mouse anti-ubiquilin 2 (1 μg/ml, ab190283) and polyclonal rabbit anti-optineurin (12 μg/ml, ab23666). Rabbit polyclonal IgG (ab171870), purified mouse IgG2a kappa (Immunology Consultants Laboratory Inc, USA—RS-90G2A) and mouse IgG1 monoclonal antibody (Chemicon, Australia—MABC002) were used as isotype controls. All antibodies were obtained from Abcam unless otherwise specified.
FRAP
FRAP was performed on transfected NSC-34 cells 48 h post transfection using the LAS AF FRAP Application Wizard on the Leica TCS SP5 Confocal Microscope. Images were acquired using the 63 × objective with two line averages and a scan speed of 700 Hz. Five pre-bleach images were acquired over 7.5 s with the 488 nm laser set at 20% power. The region of interest (ROI) was then bleached using the ‘zoom in ROI’ method over four frames of 1.5 s at 100% laser power. Recovery was monitored for over 1 min with the laser power set back at 20%.
Immunostaining of post mortem tissue
Dual immunofluorescence was performed on spinal cord tissue from two SALS cases using a rabbit polyclonal anti-TDP-43 antibody (Proteintech, 10782-2-AP) and mouse monoclonal anti-ubiquitin antibody (Chemicon, MAB1510). Five μm spinal cord sections were deparaffinised with xylene, and rehydrated with a descending series of diluted ethanol and water. Antigens were retrieved by heating sections in 10 mM citrate buffer (pH 6.0). Non-specific background was blocked with 1% bovine serum albumin (BSA) (Sigma Aldrich). Sections were incubated at 4 °C overnight with the primary antibodies (1:1000 for anti-TDP-43 and 1:100 anti-ubiquitin), followed by incubation with secondary antibodies (anti-rabbit conjugated with Alexa Fluor 488 and secondary anti-mouse conjugated with Alexa Fluor 555; Life Technologies) for 1 hour at room temperature. Slides were coverslipped using the Prolong Gold antifade reagent.
Confocal fluorescence imaging was performed using a Leica DM6000 upright laser scanning confocal microscope with Leica application suite advanced fluorescence software. Images were acquired with a 40 × oil immersion objective. Images were acquired using sequential mode to avoid crosstalk between two dyes.
Additional Information
How to cite this article: Farrawell, N. E. et al. Distinct partitioning of ALS associated TDP-43, FUS and SOD1 mutants into cellular inclusions. Sci. Rep. 5, 13416; doi: 10.1038/srep13416 (2015).
Supplementary Material
Acknowledgments
JJY is supported by the ARC in the form of a Discovery Early Career award, NHMRC project grant 1003032, and an MNDRIA grant in aid. DMH is supported by an ARC Future Fellowship, an ARC Discovery grant and NHMRC project grants. DNS is supported by NHMRC project grants. Tissues were received from the New South Wales Brain Banks and Queensland Brain Bank (Australian Brain Bank Network). IPB is supported by an MND Australia Leadership Grant.
Footnotes
Author Contributions J.J.Y. conceptualized, designed, performed and supervised the experiments, analyzed the data and wrote the paper. N.E.F. designed and performed experiments, analyzed data and wrote the paper. I.A.L.S., S.T.W. and I.P.B. performed experiments and analyzed data. D.M.H. and D.N.S. contributed to experimental design, contributed resources and to writing/editing of the manuscript.
References
- Rosen D. R. et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62 (1993). [DOI] [PubMed] [Google Scholar]
- Kwiatkowski T. J. Jr et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323, 1205–1208 (2009). [DOI] [PubMed] [Google Scholar]
- Vance C. et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323, 1208–1211 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura A. L. et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. American Journal of Human Genetics 75, 822–831 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E. et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nature Genetics 40, 572–574 (2008). [DOI] [PubMed] [Google Scholar]
- Sreedharan J. et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama H. et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature 465, 223–226 (2010). [DOI] [PubMed] [Google Scholar]
- Johnson J. O. et al. Exome Sequencing Reveals VCP Mutations as a Cause of Familial ALS. Neuron 68, 857–864 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S. L., Majcher V., Searle M. S. & Layfield R. SQSTM1 mutations—Bridging Paget disease of bone and ALS/FTLD. Experimental Cell Research 325, 27–37 (2014). [DOI] [PubMed] [Google Scholar]
- Deng H. X. et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477, 211–215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. H. et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature 488, 499–503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. O. et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci 17, 664–666 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F. & Dobson C. M. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 75, 333–366 (2006). [DOI] [PubMed] [Google Scholar]
- Giordana M. T. et al. TDP-43 redistribution is an early event in sporadic amyotrophic lateral sclerosis. Brain Pathol 20, 351–360 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettschneider J. et al. TDP-43 pathology and neuronal loss in amyotrophic lateral sclerosis spinal cord. Acta neuropathologica 128, 423–437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticozzi N., Ratti A. & Silani V. Protein aggregation and defective RNA metabolism as mechanisms for motor neuron damage. CNS and Neurological Disorders—Drug Targets 9, 285–296 (2010). [DOI] [PubMed] [Google Scholar]
- Leigh P. N. et al. Ubiquitin-immunoreactive intraneuronal inclusions in amyotrophic lateral sclerosis. Morphology, distribution, and specificity. Brain 114, 775–788 (1991). [DOI] [PubMed] [Google Scholar]
- Strong M. J., Kesavapany S. & Pant H. C. The pathobiology of amyotrophic lateral sclerosis: A proteinopathy? Journal of Neuropathology and Experimental Neurology 64, 649–664 (2005). [DOI] [PubMed] [Google Scholar]
- Grad L. I. et al. Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc Natl Acad Sci USA 111, 3620–3625 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman M. Y. & Goldberg A. L. Cellular Defences against unfolded proteins: A cell biologist thinks about neurodegenerative diseases. Neuron 29, 15–32 (2001). [DOI] [PubMed] [Google Scholar]
- Yerbury J. J. & Kumita J. R. Protein chemistry of amyloid fibrils and chaperones: Implications for amyloid formation and disease. Current Chemical Biology 4, 89–98 (2010). [Google Scholar]
- Huang Q. & Figueiredo-Pereira M. E. Ubiquitin/proteasome pathway impairment in neurodegeneration: Therapeutic implications. Apoptosis 15, 1292–1311 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerbury J. J. et al. The small heat shock proteins alphaB-crystallin and Hsp27 suppress SOD1 aggregation in vitro. Cell stress & chaperones 18, 251–257 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong L. K., Neumann M., Sampathu D. M., Lee V. M. Y. & Trojanowski J. Q. TDP-43 proteinopathy: The neuropathology underlying major forms of sporadic and familial frontotemporal lobar degeneration and motor neuron disease. Acta Neuropathologica 114, 63–70 (2007). [DOI] [PubMed] [Google Scholar]
- Liscic R. M., Grinberg L. T., Zidar J., Gitcho M. A. & Cairns N. J. ALS and FTLD: Two faces of TDP-43 proteinopathy. European Journal of Neurology 15, 772–780 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie I. R. A. et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Annals of Neurology 61, 427–434 (2007). [DOI] [PubMed] [Google Scholar]
- Bolognesi B. et al. ANS binding reveals common features of cytotoxic amyloid species. ACS chemical biology 5, 735–740 (2010). [DOI] [PubMed] [Google Scholar]
- Johnston J. A., Ward C. L. & Kopito R. R. Aggresomes: a cellular response to misfolded proteins. The Journal of cell biology 143, 1883–1898 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D., Kopito R. & Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature 454, 1088–1095 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg S. J. et al. Compartmentalization of superoxide dismutase 1 (SOD1G93A) aggregates determines their toxicity. Proceedings of the National Academy of Sciences of the United States of America 109, 15811–15816 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp M. S. et al. Indirect inhibition of 26S proteasome activity in a cellular model of Huntington’s disease. The Journal of cell biology 196, 573–587 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polling S. et al. Misfolded polyglutamine, polyalanine, and superoxide dismutase 1 aggregate via distinct pathways in the cell. J Biol Chem 289, 6669–6680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. et al. The C-terminal TDP-43 fragments have a high aggregation propensity and harm neurons by a dominant-negative mechanism. PloS one 5, e15878 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che M. X., Jiang Y. J., Xie Y. Y., Jiang L. L. & Hu H. Y. Aggregation of the 35-kDa fragment of TDP-43 causes formation of cytoplasmic inclusions and alteration of RNA processing. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 25, 2344–2353 (2011). [DOI] [PubMed] [Google Scholar]
- Deng H. X. et al. FUS-immunoreactive inclusions are a common feature in sporadic and non-SOD1 familial amyotrophic lateral sclerosis. Annals of Neurology 67, 739–748 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair I. P. et al. FUS mutations in amyotrophic lateral sclerosis: clinical, pathological, neurophysiological and genetic analysis. Journal of neurology, neurosurgery, and psychiatry 81, 639–645 (2010). [DOI] [PubMed] [Google Scholar]
- Sun Z. et al. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol 9, e1000614 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S. et al. TDP-43 is consistently co-localized with ubiquitinated inclusions in sporadic and Guam amyotrophic lateral sclerosis but not in familial amyotrophic lateral sclerosis with and without SOD1 mutations. Neuropathology : official journal of the Japanese Society of Neuropathology 29, 672–683 (2009). [DOI] [PubMed] [Google Scholar]
- Pesiridis G. S., Tripathy K., Tanik S., Trojanowski J. Q. & Lee V. M. A “two-hit” hypothesis for inclusion formation by carboxyl-terminal fragments of TDP-43 protein linked to RNA depletion and impaired microtubule-dependent transport. J Biol Chem 286, 18845–18855 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelkovnikova T. A., Robinson H. K., Connor-Robson N. & Buchman V. L. Recruitment into stress granules prevents irreversible aggregation of FUS protein mislocalized to the cytoplasm. Cell Cycle 12, 3194–3202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski P. J., Ning K., D’Souza-Schorey C. & Fields S. Requirement of an intact microtubule cytoskeleton for aggregation and inclusion body formation by a mutant huntingtin fragment. Proc Natl Acad Sci USA 99, 727–732 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J. A., Dalton M. J., Gurney M. E. & Kopito R. R. Formation of high molecular weight complexes of mutant Cu, Zn-superoxide dismutase in a mouse model for familial amyotrophic lateral sclerosis. Proceedings of the National Academy of Sciences of the United States of America 97, 12571–12576 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meriin A. B. et al. Intracellular aggregation of polypeptides with expanded polyglutamine domain is stimulated by stress-activated kinase MEKK1. J Cell Biol 153, 851–864 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Gedalya T. et al. Cyclosporin-A-induced prion protein aggresomes are dynamic quality-control cellular compartments. J Cell Sci 124, 1891–1902 (2011). [DOI] [PubMed] [Google Scholar]
- Scotter E. L. et al. Differential roles of the ubiquitin proteasome system and autophagy in the clearance of soluble and aggregated TDP-43 species. J Cell Sci 127, 1263–1278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles T. P., Vendruscolo M. & Dobson C. M. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol 15, 384–396 (2014). [DOI] [PubMed] [Google Scholar]
- Kato M. et al. Cell-free Formation of RNA Granules: Low Complexity Sequence Domains Form Dynamic Fibers within Hydrogels. Cell 149, 753–767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J. R., Kolaitis R. M., Taylor J. P. & Parker R. Eukaryotic Stress Granules Are Cleared by Autophagy and Cdc48/VCP Function. Cell 153, 1461–1474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa J. et al. Dorfin Ubiquitylates Mutant SOD1 and Prevents Mutant SOD1-mediated Neurotoxicity. Journal of Biological Chemistry 277, 36793–36798 (2002). [DOI] [PubMed] [Google Scholar]
- Arima K. et al. Immunohistochemical and ultrastructural characterization of ubiquitinated eosinophilic fibrillary neuronal inclusions in sporadic amyotrophic lateral sclerosis Acta Neuropathologica 96, 75–85 (1998). [DOI] [PubMed] [Google Scholar]
- Tada M. et al. Coexistence of Huntington’s disease and amyotrophic lateral sclerosis: a clinicopathological study. Acta Neuropathologica 124, 749–760 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi H., Koyano S., Susuki Y., Nukina N. & Kuroiwa Y. The RNA-binding protein FUS/TLS is a common aggregate-interacting protein in polyglutamine diseases. Neuroscience Research 66, 131–133 (2010). [DOI] [PubMed] [Google Scholar]
- Johnson B. S. et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. Journal of Biological Chemistry 284, 20329–20339 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J. et al. Analysis of hnRNPA1, A2/B1, and A3 genes in patients with amyotrophic lateral sclerosis. Nature 495, 467–473 (2013).23455423 [Google Scholar]
- Wang Q., Johnson J. L., Agar N. Y. R. & Agar J. N. Protein aggregation and protein instability govern familial amyotrophic lateral sclerosis patient survival. PLoS Biology 6, 1508–1526 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. S., McCaffery J. M., Lindquist S. & Gitler A. D. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci USA 105, 6439–6444 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday G. et al. Mechanisms of disease in frontotemporal lobar degeneration: gain of function versus loss of function effects. Acta Neuropathol 124, 373–382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J. C. et al. FUS is sequestered in nuclear aggregates in ALS patient fibroblasts. Mol Biol Cell 25, 2571–2578 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuiji H. et al. Spliceosome integrity is defective in the motor neuron diseases ALS and SMA. EMBO molecular medicine 5, 221–234 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E. et al. FUS and TARDBP but not SOD1 interact in genetic models of amyotrophic lateral sclerosis. PLoS genetics 7, e1002214 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito R. Aggresomes, inclusion bodies and protein aggregation. Trends in Cell Biology 10, 524–530 (2000). [DOI] [PubMed] [Google Scholar]
- Szeto J. et al. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy 2, 189–199 (2006). [DOI] [PubMed] [Google Scholar]
- Kamhi-Nesher S. et al. A Novel Quality Control Compartment Derived from the Endoplasmic Reticulum. Molecular Biology of the Cell 12, 1711–1723 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B. J. et al. Impaired extracellular secretion of mutant superoxide dismutase 1 associates with neurotoxicity in familial amyotrophic lateral sclerosis. J Neurosci 25, 108–117 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S. et al. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes and Development 20, 1343–1352 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Noda T. & Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 (2007). [DOI] [PubMed] [Google Scholar]
- Ramdzan Y. M. et al. Conformation sensors that distinguish monomeric proteins from oligomers in live cells. Chemistry & biology 17, 371–379 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashman N. R. et al. Neuroblastoma x spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev Dyn 194, 209–221, 10.1002/aja.1001940306 (1992). [DOI] [PubMed] [Google Scholar]
- Wanker E. E. et al. Membrane filter assay for detection of amyloid-like polyglutamine-containing protein aggregates. Methods in Enzymology 309, 375–386 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.