Abstract
Carrot (Daucus carota L.) is an important food crop and is useful for studying carotenogenesis due to the quantity and diversity of carotenoids in its roots. Phytoene synthase catalyzes the first committed step in the carotenoid biosynthesis pathway, and its overexpression is the main driving force in the orange phenotype. At present, we lack fundamental knowledge of the role of these genes and their effects on carotenoid accumulation in leaves. In the present study, three backcross inbred lines (BC2S4) with different colored roots derived from a cross between the orange inbred line (Af) and related wild species were used to investigate the role of the duplicated DcPSY genes in root carotenogenesis. Promoter analysis showed that DcPSY genes have diverged substantially in their regulatory sequences after gene duplication. Expression levels of DcPSY1 and DcPSY2 were generally positively correlated with carotenoid content during root development. In mature leaves, total carotenoid content was higher than that in the roots, DcPSY1 expression increased extremely higher than DcPSY2 expression compared with roots, and DcPSY1 was more sensitive than DcPSY2 during leaf de-etiolation under sunlight. These results suggest that DcPSY1 seems to make an important contribution to carotenoid accumulation in the leaves and is important for photosynthesis and photoprotection, but they are not the determining factors of root color. This expands our understanding of the regulation of carotenoid biosynthesis in carrot.
Electronic supplementary material
The online version of this article (doi:10.1007/s11032-014-0163-7) contains supplementary material, which is available to authorized users.
Keywords: Carrot, Backcross inbred lines (BILs), Phytoene synthase gene, Carotenoids accumulation, De-etiolation
Introduction
Carotenoids are a diverse family of red, orange and yellow isoprenoid molecules that serve as accessory pigments in the light-harvesting complex and protect the photosynthetic apparatus against photooxidation (Cazzonelli and Pogson 2010). The plant hormone precursors of abscisic acid (ABA) and strigolactones also belong to this class of molecules (Nambara and Marion-Poll 2005; Umehara et al. 2008; Dun et al. 2009). Carotenoids are indispensable nutrients in the diets of mammals, and α- and β-carotene are particularly important as a source of antioxidants and retinoids, and the precursors of vitamin A (Rao and Rao 2007). Vegetables and fruits are considered to be a good source of bioavailable provitamin A carotenoids (Simon 1997).
Carotenoid biosynthesis is a dynamic process that is localized in plastids in higher plants (Shumskaya et al. 2012). Biosynthetic steps include desaturation, cyclization, hydroxylation and epoxidation (Cazzonelli and Pogson 2010; Ruiz-Solaa and Rodríguez-Concepción 2012). Phytoene synthase (PSY) catalyzes the condensation of two geranylgeranyl diphosphate (GGPP) molecules into one phytoene molecule and is the committal step that diverts carbon flux away from competing pathways and toward carotenoid biosynthesis (Hirschberg 2001; Rodríguez-Villalón et al. 2009). Overexpression of PSY leads to increased accumulation of carotenoids in rice (Oryza sativa) and maize (Zea mays) endosperm (Ye et al. 2000; Aluru et al. 2008; Zhu et al. 2008), canola (Brassica napus) seeds (Shewmaker et al. 1999), tomato (Solanum lycopersicum) fruits (Fraser et al. 2002) and potato (S. tuberosum) tubers (Diretto et al. 2010).
In Arabidopsis, a single PSY gene (AtPSY) regulates phytoene synthesis in all tissues (Ruiz-Solaa and Rodríguez-Concepción 2012), while many plants contain two or more PSY paralogs that have overlapping roles in carotenogenesis in both photosynthetic and non-photosynthetic tissues. In tomato, SlPSY1 is primarily responsible for carotenoid accumulation in flower and fruit, and SlPSY2 performs this function in roots and green tissues (Giorio et al. 2008). In rice and maize, endosperm carotenoid accumulation requires the expression of PSY1, while carotenogenesis in photosynthetic tissues requires the expression of both PSY1 and PSY2 (Gallagher et al. 2004; Li et al. 2008). Drought and salt stress induces carotenogenesis in roots which enhances ABA, and PSY3 is required for this process (Welsch et al. 2008). The regulation of PSY paralogs remains unclear, but allelic variations in PSY could explain carotenogenesis modification in different plant tissues. A delay in lycopene and β-carotene accumulation during tomato fruit ripening was caused by an induced point mutation (P192L) in SlPSY1 (Gady et al. 2012). A single nucleotide polymorphism (SNP) resulting in the A191D mutation in a highly conserved region of MePSY2 enhanced provitamin A levels in cassava roots (Welsch et al. 2010).
Carrot (Daucus carota L. var. sativa, 2n = 2x = 18) is one of the most important sources of dietary carotenoids, being particularly abundant in α- and β-carotene (Simon 1997). Naturally occurring single-locus mutations affecting carotenoid accumulation in carrot roots have been documented, including dominant alleles A (α-carotene accumulation), Io (intense orange xylem), L 1 and L 2 (lycopene accumulation), O (orange xylem), Y, Y 1, and Y 2 (control of differential distribution of α- and β-carotene) as well as recessive alleles y (yellow xylem) and rp (reduced pigmentation) (Umiel and Gabelman 1972; Buishand and Gabelman 1979; Goldman and Breitbach 1996; Simon 2000). The large quantities of diverse carotenoids contribute to the different colors of carrots and are an ideal model for studying carotenoid biosynthesis (Clotault et al. 2008, 2012). A total of 22 putative genes encoding carotenoid biosynthesis enzymes have been mapped in carrot, but none of the root color alleles appear to be located within these genes (Just et al. 2007, 2009; Cavagnaro et al. 2011). Additionally, the high expression of ζ-carotene desaturase (ZDS) and lycopene ε-cyclase (LCYE) might be consistent with the accumulation of lycopene in red cultivars and lutein in yellow cultivars, respectively; however, this hypothesis was not consistent with α- and β-carotene accumulation in orange cultivars (Clotault et al. 2008).
Orange carrots were not widespread until the fifteenth and sixteenth centuries in Europe (Banga 1957; Stolarczyk and Janick, 2011), and recent allelic diversity of SNP data suggests that they arose from selection of yellow cultivars (Banga 1957; Iorizzo et al. 2013). Phytoene synthesis is the limiting step in carotenoid accumulation in carrot roots (Santos et al. 2005). Increased DcPSYs expression was observed in orange carrot roots compared with yellow and white carrots (Bowman et al. 2014). Overexpression of the bacterial Erwinia uredovora PSY gene crtB under the control of a root-specific promoter from yam in wild white carrot cultivar Queen Anne’s Lace (QAL) resulted in increased carotenoid content, which confirmed that PSY expression is the rate-limiting step in the transition from white-to-yellow carrots (Maass et al. 2009). Therefore, it remains unclear whether the high α- and β-carotene content in orange roots is controlled by carotenoid biosynthesis pathway genes or other factors. To uncover the role in carotenoid biosynthesis, knowledge of DcPSY gene structure and functional information concerning the white-to-orange phenotype change would be valuable, as would insight into the regulatory factors that influence DcPSY expression. To date, only limited information has been reported concerning the functional role of carotenoids in photosynthesis and photoprotection in carrot leaves (Stange et al. 2008; Arango et al. 2014; Bowman et al. 2014).
In this study, we determined and analyzed the complete DcPSY1 and DcPSY2 gene sequences, including the promoter regions. An SNP and an InDel marker were, respectively, found to differentiate DcPSY1 and DcPSY2 between the orange inbred line (Af) and related wild species (Ws) and their backcross inbred lines (BILs; BC2S4) with different colored roots. The overlapping roles of the multiple DcPSY genes in the regulation of carotenogenesis in roots and leaves were investigated, and their regulation by light during de-etiolation was also studied.
Materials and methods
Plant materials
The orange inbred line Af was selected and self-pollinated for five generations using the European variety D. carota var. sativa ‘Amsterdam forcing.’ The related wild species D. carota var. carota ‘Songzi’ (Ws) was supplied by the National Mid-term Genebank of Vegetable Genetic Resources, Chinese Academy of Agricultural Sciences, and self-pollinated for three generations. Orange line E2003, yellow line E36101 and pale orange line E02032 were independently selected from the BC1F1 population of (Af × Ws) × Af, backcrossed with Af again, and then self-pollinated for four generations (BC2S4; Fig. S1). Five accession seeds were directly sown in rows at each 5 m2 plot with two agronomic repetitions and had about 300 plants after thinning at the Changping station of the Chinese Academy of Agricultural Sciences on the August 8, 2012.
For gene sequence studies, the roots of the five accessions were randomly sampled from eight plants at the 13th leaf stage. For gene expression and carotenoid accumulation in developmental roots and leaves, the roots of the five accessions were mixed sampled from eight plants with three biological replicates from 0800 to 1000 hours a.m. at the 4th (4 weeks), 5th (5 weeks), 7th (7 weeks), 9th (8 weeks), 11th (10 weeks) and 13th (12 weeks) leaf stages. The leaves of Ws and Af at the 5th and 11th leaf stages were also collected from the eight plants. Both carotenoids and RNA were extracted from the same samples. All samples were frozen in liquid nitrogen and stored at −80 °C until needed.
Cloning of DcPSY1 and DcPSY2
Genomic DNA was extracted from the roots of Ws, Af, E02032, E36101 and E2003 at the 13th leaf stage using a cetyltrimethyl ammonium bromide (CTAB) method modified from Briard et al. (2000). Primers were designed based on the cDNA sequences of DcPSY1 and DcPSY2 (Genebank number: DQ192186 and DQ192187) using Primer Premier 5.0 (http://www.premierbiosoft.com/primerdesign/index.html) as follows: DcPSY1, forward, 5′-TTTCACATTTTTAACAAC-3′, reverse, 5′-TAGAGTAACATAATCCCT-3′; DcPSY2, forward, 5′-GGAAAAAGACAAACCAAA-3′, reverse, 5′-GAGACCATAAGCAACAAG-3′. Amplifications were performed using TranStart FastPfu DNA Polymerase (TransGen Biotech, China) in a MultiGene OptiMax Thermal Cycler (USA). Polymerase chain reaction (PCR) conditions were as follows: 94 °C for 3 min, 35 cycles of 94 °C for 30 s, annealing temperature (45 °C for DcPSY1, 50 °C for DcPSY2) for 30 s, 72 °C for 5 min and a final extension at 72 °C for 10 min. Amplified products were analyzed by 1 % agarose gel electrophoresis, cloned into pESY-Blunt Zero Cloning vector (TransGen Biotech, China) and transformed into Escherichia coli strain JM109 (TransGen Biotech, China). Clones (60 colonies) were picked individually, cultured in 3 ml Luria–Beratni (LB) medium at 37 °C overnight and sequenced at Beijing Genomics Institute, China. Computer analysis of DNA and amino acid sequences was carried out using DNAman software (Lynnonon Biosoft, Canada) and ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html; Thompson et al. 1994).
Cloning of DcPSY promoters using the genomic walking method
Long and accurate (LA) PCR-based genomic walking was performed to clone the DcPSY promoters from Ws and Af using the Genomic Walking Kit (TaKaRa, Japan). Gene-specific primers for cloning the promoters were designed as follows: DcPSY1, p1SP1, 5′-ACCATAGATTGCCCAAACAG-3′; p1SP2, 5′-TACTCAGCACAAACTTCGCC-3′; DcPSY2, p2SP1, 5′-TCTTTTGACCAACGCTGCTT-3′; p2SP2, 5′-TTACCCTTCTCAAGTCTGCCTC-3′. The primary nested PCR products were diluted 1:100 with distilled water for subsequent nested PCR. All manipulations were carried out as described in the manufacturer’s protocol. DNA sequences of the region 1,557 bp upstream of the translational start ATG codon of tomato SlPSY1 and SlPSY2 were obtained from the SOL Genomics Network (http://solgenomics.net), and the corresponding Arabidopsis sequences for AtPSY (At5g17230) were obtained from the P1 clone MKP11 (GeneBank number: AB005238). Maize ZmPSY1 (FJ971252.1), ZmPSY2 (AY325302.1) and ZmPSY3 (DQ372936.1), and rice OsPSY1 (297606578), OsPSY2 (297613623) and OsPSY3 (297610002) sequences were obtained from the NCBI database. Phylogenetic trees were generated using MEGA 5.05 with the neighbor-joining method (Tamura et al. 2011). Putative cis-acting elements within the regions 1,557 bp upstream of SlPSY1, SlPSY2, OsPSY3, DcPSY1 and DcPSY2 were identified by searching PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; Lescot et al. 2002).
UPLC analysis of carotenoids in carrot roots and leaves
Carotenoids were extracted from 200 mg lyophilized roots from the five accessions at the 4th, 5th, 7th, 9th, 11th and 13th leaf stages, and from 100 mg lyophilized leaves from Ws and Af at the 5th and 11th leaf stages using 1 ml of 1:1 acetone:petroleum ether containing 0.1 % butylated hydroxytoluene (BHT). Extracts were dissolved in 2 ml of 1:1 acetone:acetonitrile containing 0.1 % BHT. Carotenoids were separated and analyzed by ultra performance liquid chromatography (Waters, USA). A Waters ACQUITY UPLC BEH C18 Carotenoid column (2.1 mm × 100 mm, 1.7 µm; Waters, USA) was used over 15 min with 1:1 acetone:acetonitrile as the mobile phase. The flow rate was 2 ml/min, the eluent contained 0.01 % BHT to prevent the degradation of carotenoids, and subdued light was used throughout for the same reason. β-Carotene and xanthophylls were identified and quantified based on retention time and peak area compared with authentic standards (Sigma-Aldrich, USA). α-Carotene was calculated according to the β-carotene standard at 450 nm. All extractions and analyses were performed in duplicate. The concentration of each carotenoid was expressed as micrograms per gram dried weight (μg/g DW). Total carotenoid content was estimated by summing the concentrations of α- and β-carotene and xanthophylls. Carotenoid composition pattern was analyzed according to α-, β-carotene and xanthophylls content.
Expression analysis of DcPSY in carrot roots and leaves
Total root RNA from the five accessions at the 4th, 5th, 7th, 9th, 11th and 13th leaf stages and total leaf RNA from Ws and Af at the 5th and 11th leaf stages were extracted using the RNAprep Pure Plant Kit (TianGEN, China). Absorbance at 230, 260 and 280 nm was used for RNA detection and concentration determination using a NanoDrop 2000 (Thermo Scientific, USA). RNA integrity was determined by separation on a 1.2 % agarose gel, and purity was assessed by using the ratio of the absorbance at 260/280 nm and 260/230 nm. Reverse transcription reactions from 1 µg of DNAse I-treated total RNA were carried out using the PrimeScript® RT reagent Kit (TaKaRa, Japan) as described in the instruction manual. Primers for real-time quantitative PCR (qPCR) analysis were designed using Primer Premier 5.0 and verified by BLAST software (http://www.ncbi.nlm.nih.gov/tools/primer-last/index.cgi) according to the DcPSY1 and DcPSY2 sequences. Five housekeeping genes including EF1a (D12709) (Clotault et al. 2008), actin (X17526), tubulin (X16608), ribosomal Protein S10 (AF287338) (Zagon et al. 2010) and ubiquitin (Fuentes et al. 2012) were determined in the developmental roots and leaves according to crossing threshold (C t) values and expression stability by NormFinder (http://www.mdl.dk/publicationsNormFinder.htm). Ubiquitin with the lowest stability value (0.262) was amplified along with the target genes as an endogenous control to normalize expression between different samples. The primers were as follows: DcPSY1, forward, 5′-CTGACACGGTCTCCACATATCC-3′, reverse, 5′-TCCAACTGTTCCAGCAAC GTA-3′; DcPSY2, forward, 5′-GGAACACTACTGATGACCCCA-3′, reverse, 5′-AACTCCCACCTATCCAAAGC-3′; Ubiquitin, forward, 5′-GCTCGAGGACGGCAGAAC-3′, reverse, 5′-CTTGGGCTTGGTGTAGGTCTTC-3′ (Fuentes et al. 2012).
Real-time PCR was performed using a StepOne™ Real-Time PCR System (Applied Biosystems, USA). Reactions contained 2 μl diluted cDNA (100 ng), 0.8 μl of each primer (400 nm), 12.5 μl SYBR Green Master Mix and 0.4 μl ROX Reference Dye (TaKaRa, Japan) in a final volume of 25 μl. Reaction conditions were as follows: 95 °C for 3 min, 40 cycles of 95 °C for 5 s, 56 °C for 30 s and 72 °C for 30 s. The specificity of each primer pair was validated by running a dissociation curve (a single slope and peak was observed for each primer pair). Baseline range and C t values were automatically calculated using StepOne™ Software v. 2.0 (Applied Biosystems, USA). Each sample underwent three biological and three technical repetitions. The fold change in transcript abundance was calculated as 2−∆∆ (Livak and Schmittgen 2001), and the real-time qPCR amplification data were exported into Microsoft Excel 2003. The expression level of each gene in the roots at the 4th leaf stage of Ws was used for calibration. Values are shown as mean ± SD from the replicates of the eight pooled plants.
Correlation analysis of DcPSY transcript levels and carotenoid content
It is important to know whether and when PSY transcript levels correlate with carotenoid accumulation, so that timing can be optimized in breeding and transgenic approaches to enhance carotenoid levels (Li et al. 2008). To relate genotype and root development, the correlation between the transcript levels of the DcPSY genes and the carotenoid content in the five accessions was evaluated. The SAS 9.0 statistical software package was used to calculate the Pearson correlation analysis (r) and test for statistical significance (p). Statistically significant correlation was based on a p value ≤0.05. The correlation analysis method was modified from Li et al. (2008).
DcPSY expression during light-triggered de-etiolation
Transcript levels of several genes encoding carotenogenic enzymes can be upregulated by light, leading to carotenoid accumulation (Li et al. 2008; Stange et al. 2008; Toledo-Ortiz et al. 2010). Expression levels of DcPSY1 and DcPSY2 were quantified to assess their roles in leaf carotenogenesis during de-etiolation under sunlight. Thirty taproots of Ws and Af within 2 cm petioles of the stem apex were grown in the greenhouse at 13–16 °C (night) and 25–28 °C (day) under continuous darkness, respectively. When approximately six leaves had sprouted, plants underwent 8-h light treatment (natural sunlight from 0700 to 1500 hours), 16-h dark treatment (from 1500 to 0700 hours), followed by 8-h light treatment and 8-h darkness treatment. Leaves were sampled every 4 h between 0700 and 2300 hours (three biological replicates), immediately frozen in liquid nitrogen and stored at −80 °C until needed. A bulk of 6–8 individual leaves sampled from three plants for each sampling time was used with three biological replicates. Total RNA extraction, cDNA synthesis and gene expression analysis were carried out as described above.
Results
Variation in DcPSY genes between the orange inbred line and wild species
In order to understand the variation of DcPSY1 and DcPSY2 during breeding, the related wild species D. carota var. carota Ws and the European orange-type inbred line D. carota var. sativa Af were selected to clone the total genomic DNA sequence (DcPSY1: 1,972 bp; DcPSY2: 4,015/4,024 bp) (Fig. 1a). The exon/intron structures of DcPSY1 and DcPSY2 were inferred from the alignment of the genomic sequences to the cDNA coding region (Just et al. 2007). The length of the genomic DNA sequence of DcPSY1 from ATG to TAG was 1,813 bp, which was divided into 5 exons and 4 introns (Fig. 1b). Only an SNP (C66G) was present in the first exon of DcPSY1 between Ws and Af that corresponded to a L22F amino acid substitution in the coding sequence. The length of the genomic DNA sequence of DcPSY2 from ATG to TAG was 3,781/3,790 bp and contained 6 exons and 5 introns (Fig. 1c). The alignment of DcPSY2 showed only a nine-nucleotide InDel sequence (CGCACCAAC) in the third intron between Ws and Af. Three individual offspring (E02032, E36101 and E2003) were randomly selected from the BIL population. The SNP C66G was used to identify the DcPSY1 of E02032 and E36101 from Af and that of E2003 from Ws (Fig. 1b). The InDel marker was used to identify the DcPSY2 of E02032 from Af and that of E36101 and E2003 from Ws (Fig. 1c).
Fig. 1.
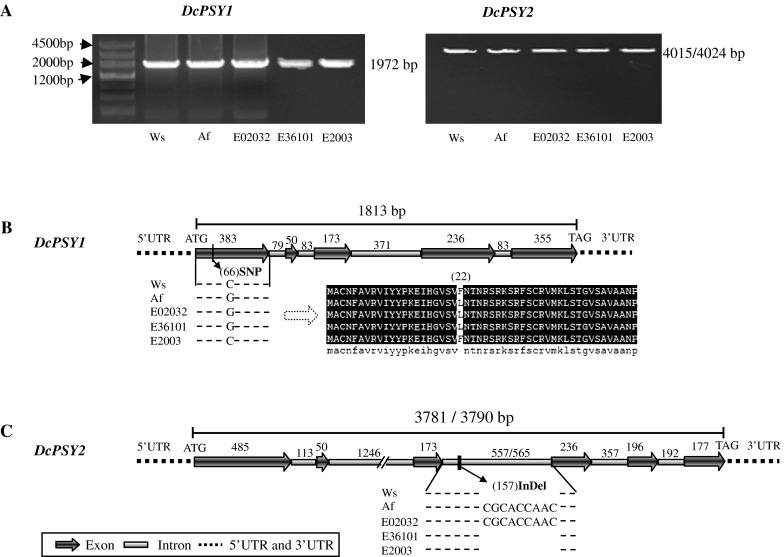
Full-length genomic DNA sequences of DcPSY1 and DcPSY2. a Genomic DNA amplification of DcPSY1 (about 1,972 bp) and DcPSY2 (about 4,015/4,024 bp) from the roots of Ws, Af, E02032, E36101 and E2003. b The genomic DNA sequence of DcPSY1 from ATG to TAG is 1,813 bp and contains 5 exons and 4 introns. The SNP C66G was found in the first exon between Ws and Af, which corresponds to a L22F amino acid change. DcPSY1 of E2003 is from Ws and that of E02032 and E36101 are from Af. c The genomic DNA sequence of DcPSY2 from ATG to TAG is 3,781 and 3,790 bp in Ws and Af, respectively, and contains six exons and five introns. A nine-nucleotide sequence (CGCACCAAC) was found in the third intron between Ws and Af that acts as an InDel marker to differentiate the DcPSY2 of E36101 and E2003 from that of Ws and that of E02032 from that of Af
Divergence in the DcPSY1 and DcPSY2 promoter structures
Promoter studies revealed that cis-regulatory motifs are important in mediating transcriptional regulation of PSY; phytochrome interaction factors (PIFs) directly bind to the promoter of AtPSY and repress expression (Toledo-Ortiz et al. 2010). The putative ABA-response element–coupling element (ABRE–CE) is believed to confer ABA regulation on OsPSY3 and ZmPSY3 (Welsch et al. 2008; Li et al. 2009). It was proposed that the regulatory sequences for most plant genes are within 500 bp (or more conservatively, the length of the target gene) upstream from the start codon (Martin et al. 2010). In this study, DNA sequences that were 1,557 bp upstream from the translation start codons of DcPSY1 and DcPSY2 in Ws and Af were obtained and analyzed for cis-regulatory structures (Fig. 2). This revealed a general lack of similarity for them, but both sequences were the same for Ws and Af indicating that carrot breeding did not affect the promoter region of the DcPSY.
Fig. 2.
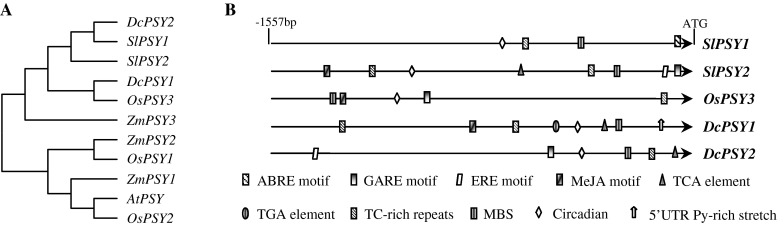
Promoter analysis of PSY genes. a Phylogenetic tree of PSY promoter regions. The promoters of SlPSY1 and SlPSY2 were obtained from the SOL Genomics Network (http://solgenomics.net) and that of AtPSY (At5g17230) was obtained from the P1 clone MKP11. ZmPSY1 (FJ971252.1), ZmPSY2 (AY325302.1), ZmPSY3 (DQ372936.1), OsPSY1 (297606578), OsPSY2 (297613623) and OsPSY3 (297610002) were obtained from the NCBI database. The phylogenetic tree was generated by MEGA 5.05 using the neighbor-joining method (Tamura et al. 2011). b Major putative cis-acting elements in the 5′-untranslated regions of SlPSY1, SlPSY2, OsPSY3, DcPSY1 and DcPSY2. Elements were identified using PlantCare and are shown by different character symbols. Only putative cis-acting elements associated with defense, hormones, circadian regulation and other particular response elements are shown. Motifs positions are relative and not drawn to scale
Phylogenetic analysis showed that the promoter of DcPSY1 grouped with OsPSY3 and that of DcPSY2 grouped with SlPSY1 (Fig. 2a). Consistent with light regulation of carotenoid biosynthesis, multiple light-responsive cis-elements were identified in the promoter regions (Table S1). Ten light-responsive cis-elements (Box 4, GAG motif, BoxII, GATA motif, G-box, I-box, L-box, AAAC motif, ATCT motif and GT1 motif) were found in the DcPSY1 promoter, and five (Box 4, GAG motif, GA motif, AE motif and BoxI) were present in that of DcPSY2. Circadian rhythm-responsive element was found in all analyzed PSY promoters (Fig. 2b). Several hormone response elements were identified with differential occurrence. The ABRE ABA-response and MeJA methyl jasmonic acid-response elements were present in the promoters of SlPSY2, OsPSY3 and DcPSY1, but absent in that of DcPSY2. The GARE GA-response and ERE ethylene-response elements were present in the promoters of SlPSY2 and DcPSY2. The TGA auxin-response element has been shown to interact with carotenoid-derived strigolactone hormones that regulate shoot branching (Hayward et al. 2009), and this element was only found in the DcPSY1 promoter. The TCA salicylic acid-response element was present in the promoter of DcPSY1 and DcPSY2. Some defense/stress-responsiveness elements were found in the promoters, including the MBS MYB-binding site, which is involved in drought inducibility (Yamaguchi-Shinozaki and Shinozaki 1993), and TC-rich repeats were found in the promoters of DcPSY1 and DcPSY2. Notably, a 5′ UTR Py-rich stretch was only present in the DcPSY1 promoter, and this cis-acting element upregulated transcription in the absence of other upstream cis-elements in Lycopersicon esculentum (Daraselia et al. 1996).
Correlation of carotenoid accumulation and DcPSY expression in roots
Control of carotenogenic gene transcription is the primary mechanism by which carotenoid biosynthesis and accumulation are regulated in plants (Kato et al. 2004; Qin et al. 2011). The role of transcriptional regulation of DcPSY genes in carotenoid accumulation in carrot roots remains uncertain. From the 4th to the 13th leaf stages, carotenoid levels in the roots of the five accessions were determined by UPLC. In Ws roots, carotenoid levels remained below the detection threshold during the six leaf stages (Fig. 3a). For α-carotene, the content in Af roots peaked at 831 µg/g DW at the 11th leaf stage before dropping, while in E2003 roots, this peaked at 658 µg/g DW at the 9th leaf stage before decreasing. A small amount of α-carotene (<5 µg/g) was detected in E02032 and E36101 roots. β-Carotene accumulated slowly in Af, E02032 and E2003 up to the 7th leaf stage, and peaked at the 13th leaf stage at 1,098, 811 and 95 µg/g DW, respectively. E36101 contained a small amount of β-carotene (9 µg/g DW) at the 9th leaf stage. For xanthophylls, accumulation was undulating and peaked at 26 µg/g DW in Af during the 13th leaf stage. Total carotenoid accumulation peaked at 1,563 µg/g DW at the 11th leaf stage in Af, and at 1,407 µg/g DW at the 13th leaf stage in E2003. E02032 and E36101 only accumulated levels of 117 and 38 µg/g DW at the 13th and 9th leaf stages, respectively.
Fig. 3.
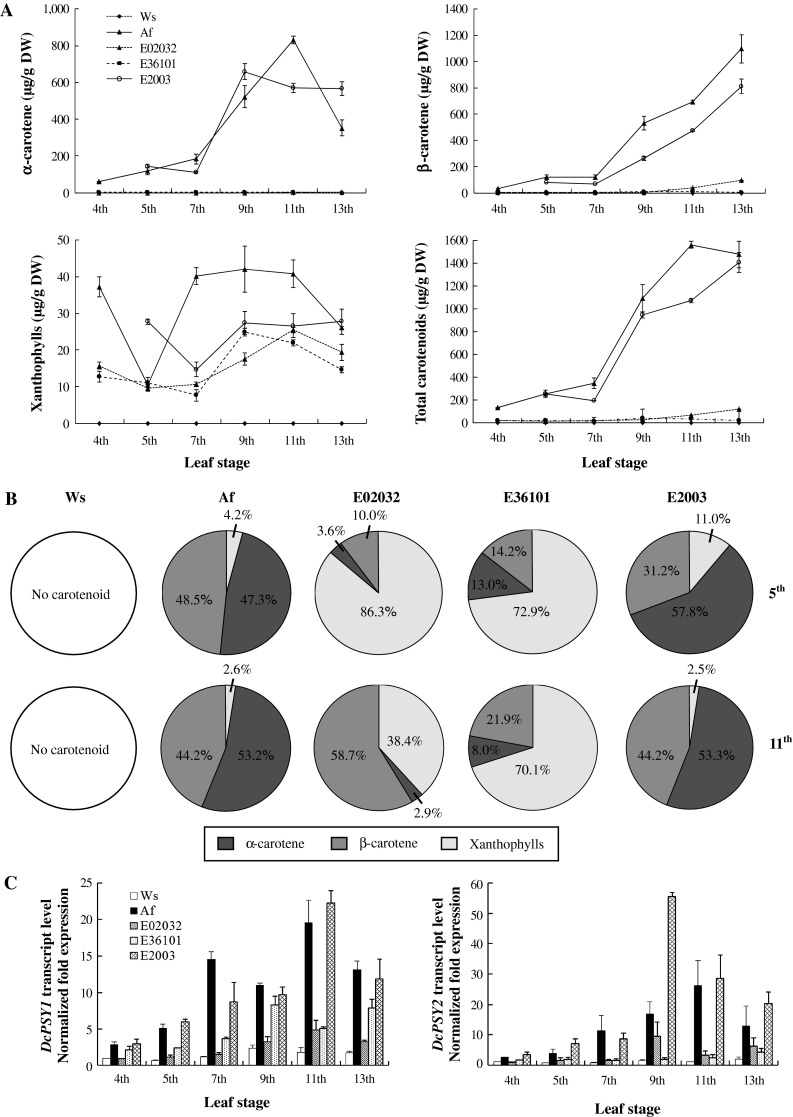
Relationship of carotenoid content and DcPSY gene transcription during carrot root development. a Carotenoid contents were determined by quantitative UPLC. Total carotenoids were calculated by adding α- and β-carotene and xanthophylls. Results are the means of two extraction replicates ± SD. b A comparison of root carotenoid composition patterns among the five accessions was shown at the 5th and 11th leave stage. c DcPSY1 and DcPSY2 transcription levels were determined by real-time qPCR. The values are normalized against ubiquitin transcripts and compared against the expression levels in Ws roots at the 4th leaf stage. Values are the means of three real-time qPCR replicates ±SD
In addition, the carotenoid composition pattern changed during prolonged root development and was different with the exception of Ws (Fig. 3b). The pattern of Af was similar with minor proportion of xanthophylls at the 5th and 11th leaf stages: α-carotene were approximately 47.3 and 53.2 % of total carotenoids, and β-carotene were 48.5 and 44.2 %. The pattern of E2003 turned the same as that of Af at the 11th leaf stage: β-carotene increased from 31.2 to 44.2 %, and α-carotene and xanthophylls decreased from 57.8 to 53.3 % and from 11.0 to 2.5 %, respectively. In E02032, the pattern consisted mostly of xanthophylls (86.3 %) at the 5th leaf stage, and β-carotene (58.7 %) at the 11th leaf stage, and only a minor proportion of α-carotene was present during both stages. In E36101, xanthophylls constituted the major proportion 72.9 and 70.1 % at the two stages, respectively, and levels of β-carotene increased 14.2–21.9 %, but α-carotene decreased from 13.0 to 8.0 %.
DcPSY1 and DcPSY2 were expressed in all five accessions (Fig. 3c). Both transcripts were present at lower levels in Ws, but expression increased 2.4-fold for DcPSY1 and 1.4-fold for DcPSY2 at the 9th leaf stage. In Af, expression levels for both genes peaked around 20-fold and 26-fold above basal levels at the 11th leaf stage. In E2003, DcPSY1 expression was upregulated 22-fold at the 11th leaf stage, while DcPSY2 expression peaked at 55-fold above basal levels at the 9th leaf stage. In E02032, DcPSY1 expression was upregulated fivefold at the 11th leaf stage, while DcPSY2 expression peaked at ninefold at the 9th leaf stage. In E36101, DcPSY1 expression was upregulated eightfold at the 11th leaf stage, while DcPSY2 expression increased slowly during root developments.
The Pearson correlation was used to probe the relationship between DcPSY1 and DcPSY2 expression and carotenoid content (Table S2). In the case of genotype, only expression of DcPSY1 showed significant correlation with α-carotene (r = 0.81, p = 0.05) in Af and xanthophylls (r = 0.89, p = 0.02) in E02032. In root development, expression of DcPSY1 and DcPSY2 generally correlated positively with carotenoid content. The 7th leaf stages were the most highly correlated as follows: α-carotene (DcPSY1: r = 0.99, p = 0.002; DcPSY2: r = 0.99, p = 0.0008), β-carotene (DcPSY1: r = 0.99, p = 0.002; DcPSY2: r = 0.99, p = 0.002) and total carotenoid (DcPSY1: r = 0.99, p = 0.002; DcPSY2: r = 0.98, p = 0.003), respectively.
Correlation of carotenoid accumulation and DcPSY expression in leaves
Carotenoids are abundant in leaves where they play an essential role in photosynthesis and photoprotection. The 5th and 11th leaf stages were selected to examine carotenoid content and DcPSY expression patterns in Ws and Af. The accumulation of α-carotene was significantly different in the two cultivars (Fig. 4a). Levels reached only 7 and 5 µg/g DW in Ws, but were much higher (226 and 223 µg/g DW) in Af at the two stages. The β-carotene content of Ws (509 and 521 µg/g DW) was higher than that of Af (158 and 248 µg/g DW). Xanthophylls and total carotenoid content were similar for Ws and Af leaves; Ws had 1,023 and 1,923 µg/g DW xanthophylls and 1,538 and 2,449 µg/g DW total carotenoid at the two stages, whereas Af had 1,118 and 1,911 µg/g DW xanthophylls and 1,502 and 2,382 µg/g DW total carotenoids, respectively. The composition pattern also showed very different, especially about 0.2 % α-carotene in Ws but 9.3 % in Af at the 11th leaf stage (Fig. 4b).
Fig. 4.
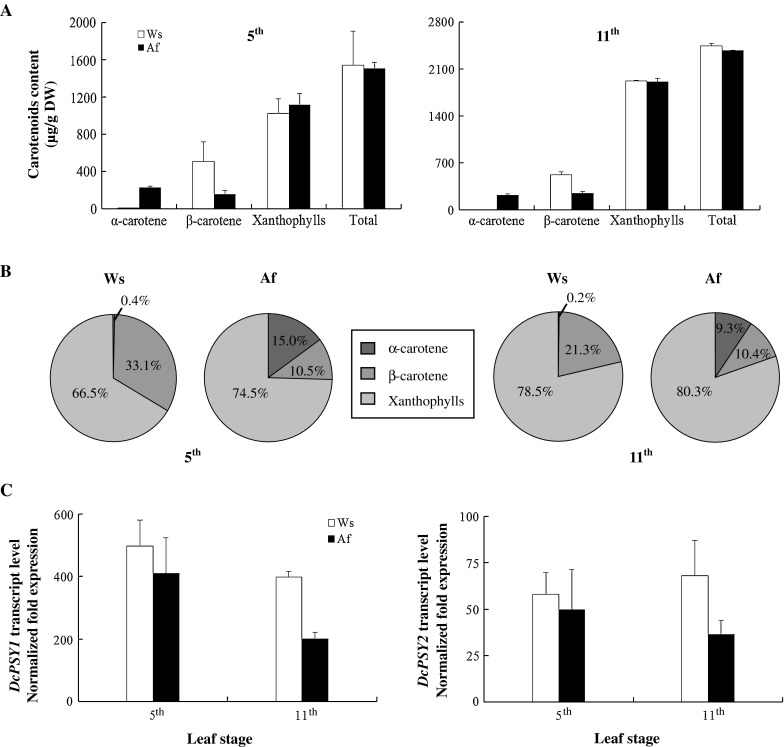
Relationship of carotenoid content and DcPSY gene expression in carrot leaves at the 5th and 11th leaf stages. a α- and β-Carotene, xanthophylls and total carotenoids were compared between the two stages. b Carotenoid composition patterns are shown for Ws and Af leaves. c Relative expression levels of DcPSY1 and DcPSY2 were compared between Ws and Af leaves. Expression levels are normalized against ubiquitin transcripts and compared with expression levels in Ws roots at the 4th leaf stage. Values are the means of three real-time qPCR replicates ±SD
Totally, the expression level of DcPSY1 and DcPSY2 was higher in Ws, especially at the 11th leaf stage (Fig. 4c). From the 5th to the 11th leaf stage, expression of DcPSY1 reduced about 20 % in Ws and 51 % in Af, but that of DcPSY2 increased about 17 % in Ws and decreased about 27 % in Af, respectively. Compared with expression of DcPSY1 and DcPSY2 between the leaves and the roots, DcPSY1 increased 746 times highly than 88 times of DcPSY2 in Ws at the 5th leaf stage, and 80 times than 13 times of DcPSY2 in Af (Figs. 3c, 4c), respectively, which was similar at the 11th leaf stage.
DcPSY expression during leaf de-etiolation under sunlight
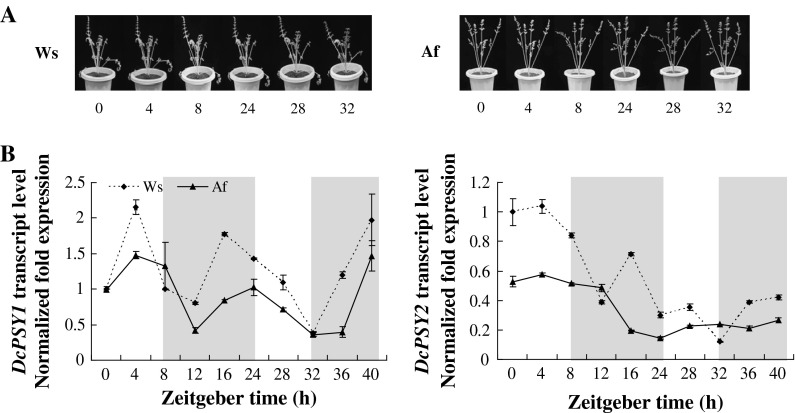
The quantitative and qualitative changes in carotenoid patterns during seedling de-etiolation are associated with concomitant changes in the expression of most genes encoding carotenoid biosynthetic enzymes (Toledo-Ortiz et al. 2010). After 2 days of treatment, the yellow leaves of Ws and Af taproots quickly turned green (Fig. 5a). The expression level of DcPSY1 increased at the beginning of the 4-h sunlight treatment (2.2-fold for Ws, 1.5-fold for Af) and declined in an undulating manner up to 32 h, at which time the expression was 0.4-fold for Ws and Af (Fig. 5b). Expression had returned to peak level by 40 h. In contrast, expression of DcPSY2 was relatively unchanged after initial sunlight treatment and declined in an undulating manner. Expression had recovered slightly by 40 h to 0.4-fold for Ws and 0.3-fold for Af. These results suggested that DcPSY1 is more sensitive than DcPSY2 during leaf de-etiolation.
Fig. 5.
Relative expression levels of DcPSY1 and DcPSY2 during de-etiolation under sunlight. a Overview of leaf color changes under light treatment for 2 days. Af and Ws are shown on the left and right. Photographs were taken during the light treatment hours. b Relative expression levels of DcPSY1 and DcPSY2 under sunlight regulation. Plants were treated with 8-h natural sunlight treatment (from 0700 to 1500 hours), 16-h dark treatment (from 1500 to 0700 hours), followed by 8-h light treatment and 8-h dark treatment. Amplified products are normalized against ubiquitin and calibrated against Ws expression levels during the first 4 h of treatment. Values are mean ± SD from three biological replicates
Discussion
Gene duplication events have profound effects on gene function and regulation (Prince and Pickett 2002). Recent data have revealed that gene duplication and subfunctionalization play a central role in recruitment and regulation of carotenoid accumulation in plants (Gallagher et al. 2004; Giorio et al. 2008; Li et al. 2008). PSY catalyzes the first step in the synthesis of carotenoids and is considered to be both the committed and rate-limiting step (Giorio et al. 2008; Li et al. 2008; Qin et al. 2011). Duplicated PSY genes are present in carrot and share 59 % sequence identity at the amino acid level, which may allow for a finer, more sophisticated cooperative control of carotenoid biosynthesis and accumulation than occurs in plants with a single PSY gene (Clotault et al. 2008). Additionally, DcPSY1 is more closely related to monocot PSYs than it is to other dicot PSY genes (Qin et al. 2011). Gene functional studies like the present work offer new opportunities for α- and β-carotene breeding and transgenic fortification.
The BILs investigated in this study were based on an initial cross of Ws (a white wild phenotype) and Af (an orange inbred line; Fig. S1). Their roots have wide color variability that is mainly dependent on the quantities and types of carotenoid present (Nicolle et al. 2004); however, both parents share a similar genetic background. SNP C66G in the first exon of DcPSY1 and the InDel sequence CGCACCAAC in the third intron of DcPSY2 allowed differentiation of Ws, Af and their offspring (Fig. 1). It is interesting that E02032 with Af DcPSY1 and DcPSY2 had pale orange roots and 117 µg/g DW total carotenoid at the 13th leaf stage, while E36101 with Af DcPSY1 and Ws DcPSY2 had yellow roots and 38 µg/g DW total carotenoid at the 9th leaf stage, and E2003 with Ws DcPSY1 and DcPSY2 had orange roots and 1,407 µg/g DW at the 13th leaf stage (Figs. S1, 3a). However, the carotenoid composition pattern of BILs showed continual changes and some differences compared to Af during prolonged growth (Fig. 3b). These results confirmed that DcPSYs do not determine the carotenoid composition pattern and root color directly, which is consistent with the finding that neither DcPSY1 nor DcPSY2 are colocated with root color loci (Just et al. 2009; Cavagnaro et al. 2011). However, the expression of DcPSY1 and DcPSY2 showed a general positive correlation with carotenoid content in terms of root development experiments (Table S2). Although no carotenoids accumulated in Ws roots, DcPSY1 and DcPSY2 were still expressed (Fig. 3c). This indicated that DcPSYs play a fundamental role in carotenogenesis of the wild and cultivated roots, but the upregulation that leads to carotenoid accumulation in orange roots may be regulated by other factors. Indeed, the substantial favored β-carotene branch pathway was also found in the transgenic wild QAL plants that expressed the yam PSY gene crtB under the control of a root-specific promoter. This plant had yellow roots with 10 % β-carotene of total carotenoid in most lines, but α-carotene was absent (Maass et al. 2009). This result is consistent with observations in other engineered plants, such as canola seeds (Shewmaker et al. 1999), rice (Ye et al. 2000), tomato (Fraser et al. 2002) and potato (Diretto et al. 2010).
The dual role of carotenoids in plants probably explains the presence of multiple paralogs that arose from gene duplication and subsequent specialization in different tissues (Li et al. 2009; Qin et al. 2011). Carrots are regarded as the most important source of carotenoids, and much attention has been focused on carrot carotenoid biosynthesis and genetics (Simon 2000; Just et al. 2007, 2009; Cavagnaro et al. 2011; Clotault et al. 2012; Bowman et al. 2014). However, we still lack fundamental knowledge of the role of these genes in controlling carotenoid accumulation in leaves. Compared with roots, total carotenoid content was higher in mature leaves, reaching 2,449 and 2,382 µg/g DW in Ws and Af leaves at the 11th leaf stage (Fig. 4a), respectively, while Af roots only accumulated 1,563 µg/g DW (Fig. 3a). Xanthophylls were the most abundant carotenoids in Ws and Af leaves (Fig. 4b), which are consistent with the critical role of these pigments in photosynthetic light harvesting and photoprotection (Niyogi et al. 1997). Expression analysis revealed that DcPSY1 transcripts increased extremely higher than that of DcPSY2 in Ws and Af leaves compared with roots (Figs. 3c, 4c). These results suggest that DcPSY1 seems to be more important in carotenoid accumulation in photosynthetic tissues.
The mechanisms of carotenoid accumulation were also different between carrot roots and leaves. Interestingly, Af leaves had 30–40 times higher levels of α-carotene than Ws leaves at the two stages, while the content of β-carotene was comparable (Fig. 4a), which is consistent with the results of Arango et al. (2014). This finding differed from the carotenoid composition observed in other plants (Fig. 4b; Britton 1993). A favored α-carotene branch pathway seems to be altered in the leaves during orange carrot breeding (Arango et al. 2014), which is completely contrary to the findings observed in the β-carotene branch pathway found in engineered plants (Shewmaker et al. 1999; Ye et al. 2000; Fraser et al. 2002; Maass et al. 2009; Diretto et al. 2010). This opinion was supported with more wild, purple, yellow and orange carrot accessions. The results showed that wild, yellow and purple carrot leaves have relatively little α-carotene, but this is higher in all orange accessions (data not shown). Moreover, the mechanism of α-carotene accumulation in the leaf appears to be independent in the root (Figs. 3b, 4b; Arango et al. 2014; Bowman et al. 2014).
The upstream regulatory sequences of DcPSY1 and DcPSY2 have also diverged following gene duplication; the DcPSY1 promoter region clusters with OsPSY3, while the DcPSY2 promoter clusters with SlPSY1 (Fig. 2a). Comparative analysis of the cis-regulatory structures in the duplicated PSY promoters in tomato, carrot and rice indicates regulation by light, circadian rhythms, abiotic stress and hormones (Fig. 2b; Table S1). Since gibberellins share a common biosynthetic precursor (GGPP) with carotenoids (Sun and Kamiya 1994), ABA is synthesized from the cleavage of apocarotenoids (Nambara and Marion-Poll 2005). Therefore, it is important to understand the significance of the ABRE element in the promoter of DcPSY1 and the GARE element in the promoter of DcPSY2 for the regulation of carotenoid accumulation in a metabolic context (Welsch et al. 2008; Li et al. 2009). The auxin TGA-responsive element is only present in the DcPSY1 promoter, despite interaction between auxin and strigolactones in regulating shoot branching (Hayward et al. 2009).
As well as functioning as accessory pigments in photosynthesis, carotenoids are involved in dissipating excess excitation energy from chlorophyll molecules during de-etiolation or under thermal stress, which is a fundamentally important process for preserving the photosynthetic apparatus (Niyogi et al. 1997; Li et al. 2008; Cazzonelli and Pogson 2010). Light has been shown to repress DcPSY1 and DcPSY2 expression in carrot roots (Fuentes et al. 2012), but little information has been reported on the role of sunlight regulation in leaves (Stange et al. 2008). Our system presented an opportunity to address this issue. During leaf de-etiolation, DcPSY1 expression increased at the start of the 4-h sunlight treatment (Fig. 5b), which could increase production of carotenoids to undergo the transition to photosynthetic development (Rodríguez-Villalón et al. 2009; Toledo-Ortiz et al. 2010). In contrast, DcPSY2 expression was relatively unaffected following initial sunlight treatment, but decreased markedly following prolonged treatment. After 2 days of treatment, expression of DcPSY1 recovered to peak levels, but DcPSY2 expression had only recovered slightly. Both Ws and Af yellow leaves turned to a normal green color (Fig. 5a). These results suggest that DcPSY1 is more sensitive and plays a major role in assisting photosynthesis and photoprotection during leaf de-etiolation, which is consistent with the greater number of light-responsive elements and stress-response elements in the promoter (Table S1).
In summary, the wide color variability of the BILs used in this study suggests that DcPSY genes play a fundamental role in carotenogenesis of wild and cultivated carrot roots, but are not the major factors determining root color. DcPSY1 and DcPSY2 expressions were generally positively correlated with carotenoid content in roots, but the factors triggering the increased expression levels in orange roots remain unknown. In mature leaves, total carotenoid content was higher than that in roots, and expression of DcPSY1 increased extremely higher than that of DcPSY2 compared with roots, which indicates that DcPSY1 seems to make a more important contribution to carotenoid accumulation in photosynthetic tissues. The alteration or mutation of the favored α-carotene branch pathway is found in the orange carrot leaves during breeding. The exquisite variation in DcPSY1 expression during de-etiolation suggests a role in assisting photosynthesis and photoprotection. Further studies to identify and characterize additional carotenoid pathway genes and transcription factors are underway using BIL populations. The results are expected to increase our understanding of the regulation of carotenoid biosynthesis and accumulation in carrot, with particular regard to the mutation(s) that affect α-carotene accumulation in leaves.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was partially supported by the Innovation Program of the Chinese Academy of Agricultural Sciences; National Natural Science Foundation of China (Grant Numbers 31101543 and 31272162); the Project from the Ministry of Science and Technology of China (Grant Number 2013BAD01B04). Authors sincerely thank Dr. Philipp W. Simon (Wisconsin University) and Dr. Jung Rong (Nanchang University) for critical review of the manuscript.
References
- Aluru M, Xu Y, Guo R, Wang ZG, Li SS, White W, Wang K, Rodermel S. Generation of transgenic maize with enhanced provitamin A content. J Exp Bot. 2008;59:3551–3562. doi: 10.1093/jxb/ern212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango J, Jourdan M, Geoffriau E, Beyer P, Welsch R (2014) Carotene hydroxylase activity determines the levels of both α-carotene and total carotenoids in orange carrots. Plant Cell 26. doi:10.1105/tpc.113.122127 [DOI] [PMC free article] [PubMed]
- Banga O. The development of the original European carrot material. Euphytica. 1957;6:64–76. [Google Scholar]
- Bowman MJ, Willis DK, Simon PW. Transcript abundance of phytoene synthase 1 and phytoene synthase 2 is associated with natural variation of storage root carotenoid pigmentation in carrot. J Am Soc Hortic Sci. 2014;139:63–68. [Google Scholar]
- Briard M, Le Clerc V, Grzebelus D, Senalik D, Simon PW. Modified protocols for rapid carrot genomic DNA extraction and AFLP™ analysis using silver stain or radioisotopes. Plant Mol Biol Rep. 2000;18:235–241. [Google Scholar]
- Britton G. Carotenoids in chloroplasts pigment-protein complexes. In: Sundqvist C, Ryberg M, editors. Pigment–protein complexes in plastids: synthesis and assembly. San Diego: Academic Press; 1993. pp. 447–483. [Google Scholar]
- Buishand JG, Gabelman WH. Investigations on the inheritance of color and carotenoid content in phloem and xylem of carrot roots (Daucus carota L.) Euphytica. 1979;28:611–632. [Google Scholar]
- Cavagnaro PF, Chung SM, Manin S, Yildiz M, Ali A, Alessandro MS, Iorizzo M, Senalik DA, Simon PW. Microsatellite isolation and marker development in carrot—genomic distribution, linkage mapping, genetic diversity analysis and marker transferability across Apiaceae. BMC Genomics. 2011;12:386–405. doi: 10.1186/1471-2164-12-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli CI, Pogson B. Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010;15:266–274. doi: 10.1016/j.tplants.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Clotault J, Peltier D, Berruyer R, Thomas M, Briard M, Geoffriau E. Expression of carotenoid biosynthesis genes during carrot root development. J Exp Bot. 2008;59:3563–3573. doi: 10.1093/jxb/ern210. [DOI] [PubMed] [Google Scholar]
- Clotault J, Peltier D, Soufflet-Freslon V, Briard M, Geoffriau E. Differential selection on carotenoid biosynthesis genes as a function of gene position in the metabolic pathway: a study on the carrot and dicots. PLoS One. 2012;7:e38724. doi: 10.1371/journal.pone.0038724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daraselia ND, Tarchevskaya S, Narita JO, Daraselia ND, Narita JO. The promoter for tomato 3-hydroxy-3-methylglutaryl coenzyme A reductase gene 2 has unusual regulatory elements that direct high-level expression. Plant Physiol. 1996;112:727–733. doi: 10.1104/pp.112.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diretto G, Al-Babili S, Tavazza R, Scossa F, Papacchioli V, Migliore M, Beyer P, Giuliano G. Transcriptional-metabolic networks in beta-carotene-enriched potato tubers: the long and winding road to the golden phenotype. Plant Physiol. 2010;154:899–912. doi: 10.1104/pp.110.159368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun EA, Brewer PB, Beveridge CA. Strigolactones: discovery of the elusive shoot branching hormone. Trends Plant Sci. 2009;14:364–372. doi: 10.1016/j.tplants.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Fraser PD, Romer S, Shipton CA, Mills PB, Kiano JW, Misawa N, Drake RG, Schuch W, Bramely PM. Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proc Natl Acad Sci USA. 2002;99:1092–1097. doi: 10.1073/pnas.241374598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes P, Pizarro L, Moreno JC, Handford M, Rodriguez-Concepcion M, Stange C. Light-dependent changes in plastid differentiation influence carotenoid gene expression and accumulation in carrot roots. Plant Mol Biol. 2012;79:47–59. doi: 10.1007/s11103-012-9893-2. [DOI] [PubMed] [Google Scholar]
- Gady AL, Vriezen WH, Van de Wal MHBJ, Huang PP, Bovy AG, Visser RGF, Bachem CWB. Induced point mutations in the phytoene synthase 1 gene cause differences in carotenoid content during tomato fruit ripening. Mol Breed. 2012;29:801–812. doi: 10.1007/s11032-011-9591-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher CE, Matthews PD, Li F, Wurtzel ET. Gene duplication in the carotenoid biosynthetic pathway preceded evolution of the grasses. Plant Physiol. 2004;135:1776–1783. doi: 10.1104/pp.104.039818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorio G, Stigliani AL, D’Ambrosio C. Phytoene synthase genes in tomato (Solanum lycopersicum L.)—new data on the structures, the deduced amino acid sequences and the expression patterns. FEBS J. 2008;275:527–535. doi: 10.1111/j.1742-4658.2007.06219.x. [DOI] [PubMed] [Google Scholar]
- Goldman IL, Breitbach DN. Inheritance of a recessive character controlling reduced carotenoid pigmentation in carrot (Daucus carota L.) J Hered. 1996;87:380–382. [Google Scholar]
- Hayward A, Stirnberg P, Beveridge C, Leyser O. Interactions between auxin and strigolactone in shoot branching control. Plant Physiol. 2009;15:400–412. doi: 10.1104/pp.109.137646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg J. Carotenoid biosynthesis in flowering plants. Curr Opin Plant Biol. 2001;4:210–218. doi: 10.1016/s1369-5266(00)00163-1. [DOI] [PubMed] [Google Scholar]
- Iorizzo M, Senalik DA, Ellison SL, Grzebelus D, Cavagnaro PF, Allender C, Brunet J, Spooner DM, Deynze AV, Simon PW. Genetic structure and domestication of carrot (Daucus carota subsp. sativus) (Apiaceae) Am J Bot. 2013;100:930–938. doi: 10.3732/ajb.1300055. [DOI] [PubMed] [Google Scholar]
- Just BJ, Santos CAF, Fonseca MEN, Boiteux LS, Oloizia BB, Simon PW. Carotenoid biosynthesis structural genes in carrot (Daucus carota): isolation, sequence-characterization, single nucleotide polymorphism (SNP) markers and genome mapping. Theor Appl Genet. 2007;114:693–704. doi: 10.1007/s00122-006-0469-x. [DOI] [PubMed] [Google Scholar]
- Just BJ, Santos CAF, Yandell BS, Simon PW. Major QTL for carrot color are positionally associated with carotenoid biosynthetic genes and interact epistatically in a domesticated × wild carrot cross. Theor Appl Genet. 2009;119:1155–1169. doi: 10.1007/s00122-009-1117-z. [DOI] [PubMed] [Google Scholar]
- Kato M, Ikoma Y, Matsumoto H, Sugiura M, Hyodo H, Yano M. Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiol. 2004;134:824–837. doi: 10.1104/pp.103.031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FQ, Vallabhaneni R, Yu J, Rocheford T, Wurtzel ET. The maize phytoene synthase gene family: overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and the thermal stress tolerance. Plant Physiol. 2008;147:1334–1346. doi: 10.1104/pp.108.122119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Tsfadia O, Wurtzel ET. The phytoene synthase gene family in the grasses: subfunctionalization provides tissue-specific control of carotenogenesis. Plant Signal Behav. 2009;4:208–211. doi: 10.4161/psb.4.3.7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maass D, Arango J, Wust F, Beyer P, Welsch R. Carotenoid crystal formation in Arabidopsis and carrot roots caused by increased phytoene synthase protein levels. PLoS One. 2009;4:e6373. doi: 10.1371/journal.pone.0006373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Ellis N, Rook F. Do transcription factors play special roles in adaptive variation? Plant Physiol. 2010;154:506–511. doi: 10.1104/pp.110.161331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- Nicolle C, Simon G, Rock E, Amouroux P, Remesy C. Genetic variability influences carotenoid, vitamin, phenolic, and mineral content in white, yellow, purple, orange, and dark-orange carrot cultivars. J Am Soc Hortic Sci. 2004;129:523–529. [Google Scholar]
- Niyogi KK, Bjorkman O, Grossman AR. The roles of specific xanthophylls in photoprotection. Proc Natl Acad Sci USA. 1997;94:14162–14167. doi: 10.1073/pnas.94.25.14162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- Qin XQ, Coku A, Inoue K, Tian L. Expression, subcellular localization, and cis-regulatory structure of duplicated phytoene synthase genes in melon (Cucumis melo L.) Planta. 2011;234:737–748. doi: 10.1007/s00425-011-1442-8. [DOI] [PubMed] [Google Scholar]
- Rao A, Rao L. Carotenoids and human health. Pharmacol Res. 2007;55:207–216. doi: 10.1016/j.phrs.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Villalón A, Gas E, Rodríguez-Concepción M. Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J. 2009;60:424–435. doi: 10.1111/j.1365-313X.2009.03966.x. [DOI] [PubMed] [Google Scholar]
- Ruiz-Solaa MÁ, Rodríguez-Concepción M (2012) Carotenoid biosynthesis in Arabidopsis: a colorful pathway. The Arabidopsis Book 10:e0158. doi:10.1199/tab.0158 [DOI] [PMC free article] [PubMed]
- Santos CAF, Senalik D, Simon PW. Path analysis suggests phytoene accumulation is the key step limiting the carotenoid pathway in white carrot roots. Genet Mol Biol. 2005;28:287–293. [Google Scholar]
- Shewmaker CK, Sheehy JA, Daley M, Colburn S, Ke DY. Seed-specific overexpression of phytoene synthase: increase in carotenoids and other metabolic effects. Plant J. 1999;20:401–412. doi: 10.1046/j.1365-313x.1999.00611.x. [DOI] [PubMed] [Google Scholar]
- Shumskaya M, Bradbury LMT, Monaco RR, Wurtzel ET. Plastid localization of the key carotenoid enzyme phytoene synthase is altered by isozyme, allelic variation and activity. Plant Cell. 2012;24:3725–3741. doi: 10.1105/tpc.112.104174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon PW. Plant pigments for color and nutrition. HortScience. 1997;32:12–13. [Google Scholar]
- Simon PW. Domestication, historical development, and modern breeding of carrot. Plant Breed Rev. 2000;19:157–190. [Google Scholar]
- Stange C, Fuentes P, Handford M, Pizarro L. Daucus carota as a novel model to evaluate the effect of light on carotenogenic gene expression. Biol Res. 2008;41:289–301. [PubMed] [Google Scholar]
- Stolarczyk J, Janick J. Carrot: history and iconography. Chron Hortic. 2011;51:13–18. [Google Scholar]
- Sun TP, Kamiya Y. The Arabidopsis GAl locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Rodríguez-Concepción M. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc Natl Acad Sci USA. 2010;107:11626–11631. doi: 10.1073/pnas.0914428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, Kyozuka J, Yamaguchi S. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455:195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- Umiel N, Gabelman WH. Inheritance of root color and carotenoid synthesis in carrot, Daucus carota L.: orange vs. red. J Am Soc Hortic Sci. 1972;97:453–460. [Google Scholar]
- Welsch R, Wüst F, Bär C, Al-Babili S, Beyer P. A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol. 2008;147:367–380. doi: 10.1104/pp.108.117028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch R, Arango J, Bar C, Bertha S, Al-Babili S, Beltran J, Chavarriaga P, Ceballos H, Tohme J, Beyer P. Provitamin A accumulation in cassava (Manihot esculenta) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. Plant Cell. 2010;22:3348–3356. doi: 10.1105/tpc.110.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Arabidopsis DNA encoding two desiccation-responsive rd29 genes. Plant Physiol. 1993;101:1119–1120. doi: 10.1104/pp.101.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye XD, Babilli SA, Kloti A, Zhang J, Lucca P, Beyer P, Potrykus I. Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
- Zagon J, Jansen B, Knoppik M, Ehlers A, Kroh LW, Holzhauser T, Vieths S, Broll H. Gene transcription analysis of carrot allergens by relative quantification with single and duplex reverse transcription real-time PCR. Anal Bioanal Chem. 2010;396:483–493. doi: 10.1007/s00216-009-3165-2. [DOI] [PubMed] [Google Scholar]
- Zhu CF, Naqvi S, Breitenbach J, Sandmann G, Christou P, Capell T. Combinatorial genetic transformation generates a library of metabolic phenotypes for the carotenoid pathway in maize. Proc Natl Acad Sci USA. 2008;105:18232–18237. doi: 10.1073/pnas.0809737105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.