Abstract
Oocyte quality is a critical factor limiting the efficiency of assisted reproductive technologies (ART) and pregnancy success in farm animals and humans. ART success is diminished with increased maternal age, suggesting a close link between poor oocyte quality and ovarian-aging. However, the regulation of oocyte quality remains poorly understood. Oocyte quality is functionally linked to ART success because the maternal-to-embryonic transition is dependent on stored maternal factors, which are accumulated in oocytes during oocyte development and growth. The maternal-to-embryonic transition consists of critical developmental processes including maternal RNA depletion and embryonic genome activation. In recent years, key maternal proteins encoded by maternal-effect genes have been determined, primarily using genetically modified mouse models. These proteins are implicated in various aspects of early embryonic development including maternal mRNA degradation, epigenetic reprogramming, signal transduction, protein translation and initiation of embryonic genome activation. Species differences exist in number of cell divisions encompassing the maternal-to-embryonic transition and maternal-effect genes controlling this developmental window. Perturbations of maternal control result in decreased oocyte quality, some of which are associated with ovarian aging.
Keywords: Maternal-effect, Embryo, Reproductive aging, Oocyte quality, Epigenetics, Mitochondria
Introduction
Female fertility, if defined as the percentage of pregnancy to a single insemination, is poor in humans and major domestic animals, such as dairy cows (Spencer 2013). For instance, the conception rate of the first service is only approximately 32% in high-producing cows, which is attributed primarily to the detrimental impacts of lactation on their reproductive system (Sartori et al. 2010). According to the statistics from the Centers for Disease Control and Prevention of the United States (CDC), approximately 10% of women of reproductive age are affected by infertility. Consequently, a growing number of patients resort to assisted reproductive technologies (ART) procedures. Despite much improvement during the last three decades, the average success rate is still poor with 31% pregnant from a single cycle. Of particular note, the success rate is extraordinary low when performed in women of advanced reproductive age (>35 years old), who account for 60% of patients subjected to ART (2012 ART Report from CDC). Such phenomena (usually called reproductive aging), refers to declining fertility with maternal aging and has been well documented in humans and animals (Qiao et al. 2014). For instance, mare fertility decreases between the late teens and early twenties. As mares age further, they become infertile and ovarian activity ceases completely (Carnevale 2008; Altermatt et al. 2009).
The majority of embryo mortality occurs during early developmental stages in various species, such as dairy cows (Sartori et al. 2010), women (Wilcox et al. 1988) and horses (Ginther et al. 1985). In a recent review, Sartori et al. estimated that 37% of embryo deaths occur during the first week after fertilization, primarily resulting from poor oocyte quality in dairy cows (Sartori et al. 2010). In women, growing evidence shows that a large proportion of cases of infertility are attributed to a reduction in oocyte quality, particularly in the case of women of advanced reproductive age (Navot et al. 1991). Indeed, the success rate in ART is significantly higher when women of advanced reproductive age use donor oocytes collected from young women than when using their own eggs (2012 CDC ART Report). It is also noted that in vitro-matured oocytes have lower developmental competence relative to in vivo counterparts (Lonergan et al. 2003; Hansen et al. 2010; Hinrichs 2010). Thus, oocyte quality is a critical component contributing to pregnancy success. Oocyte quality/competence is defined as the ability of oocyte to resume meiosis, cleave and develop to blastocyst stage after fertilization, to implant and develop to term in good health (Sirard et al. 2006).
Specifically, oocyte quality plays an important role in dictating outcome of early embryonic development. This is because the maternal-to-embryonic transition is highly dependent on stored maternal factors, such as subcellular organelles and macromolecules, which are accumulated in oocytes during the course of folliculogenesis and oogenesis. During the maternal-to-embryonic transition, key developmental events occur, including depletion of maternal mRNA transcripts, epigenetic reprogramming/chromatin remodeling, and activation of the newly-formed embryonic genome (Bettegowda et al. 2008). A growing list of maternal proteins encoded by maternal-effect genes have been characterized, primarily using genetically modified mouse models. These factors regulate various aspects of early embryonic development including maternal mRNA degradation, epigenetic modifications/chromatin remodeling, signal transduction, protein translation, onset of embryonic genome activation (EGA), and cell compaction.
In this review, we will summarize the maternal factors that contribute to oocyte developmental competence and functions of maternal-effect genes identified in mammals. We also discuss the relevance of these factors in relation to maternal reproductive aging.
Influences of maternal ovarian follicular environment on oocyte quality
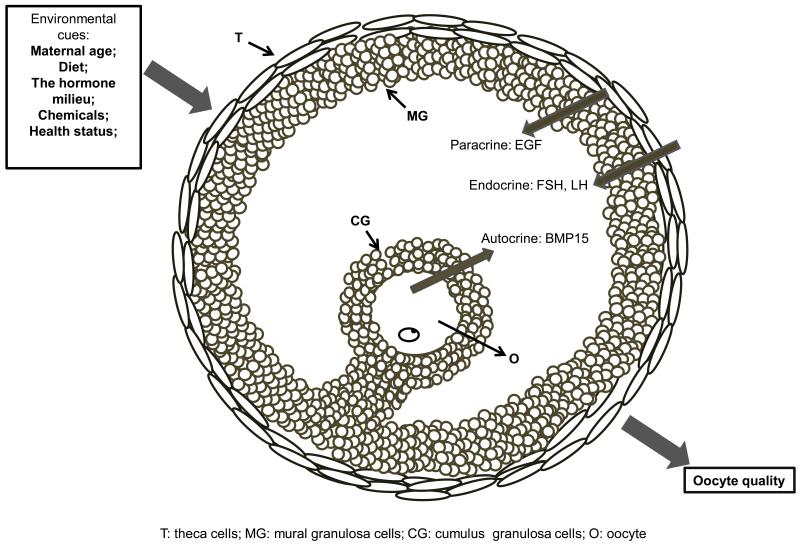
As the largest cell in the body of mammals, the oocyte develops, matures, and obtains developmental competence within the ovarian follicle. Throughout the course of oogenesis, oocyte growth and development is dynamically regulated by various physiological, cellular and molecular events, which are modulated through the ovarian intra-follicular environment (Edson et al. 2009; Hsueh et al. 2014). To some extent, it could be argued that the intra-follicular environment dictates oocyte quality/competence, particularly in the context of the ability to resume meiosis and acquire competent cytoplasm to foster subsequent development after fertilization (Fig. 1).
Fig. 1.
Oocyte acquire developmental competence in ovarian follicles. Environmental cues, such as maternal age, diet, chemicals and health status (e.g. obesity), could impact the intra-follicular environment and thus affect the oocyte developmental competence. A variety of physiological signals (paracrine, endocrine and autocrine) play a critical role in maintaining a proper intra-folliclular environment that nurture a competent oocyte.
The intra-follicular environment is regulated by miscellaneous endocrine, paracrine, and autocrine factors (Fig. 1) (Edson et al. 2009; Hsueh et al. 2014). The endocrine factors (e.g. FSH, LH and E2) are well established in their role in promoting granulosa cell proliferation/differentiation, cumulus expansion, ovulation and final oocyte maturation (Binelli and Murphy 2010). As mediators of endocrine signals, the paracrine factors (EGF, FGF10 etc.) produced from follicular somatic cells are critical for oocyte development and maturation in various species, including mice, horses and cows (Park et al. 2004; Lindbloom et al. 2008; Zhang et al. 2010). Interestingly, recent studies have shown that cell-secreted vesicles consisting of miRNAs and proteins exist in equine ovarian follicular fluid, suggesting a novel form of communication between cells within the follicle (da Silveira et al. 2012). The oocyte also has an active role in regulating follicular development (Su et al. 2009). The bidirectional communication-mediated by gap junctions–between cumulus cells and oocytes is critical for amino acid homeostasis, glycolysis and cholesterol synthesis in oocytes (Su et al. 2009). Two well-characterized TGF-β family members, BMP15 and GDF9, both oocyte-specific proteins, are directly implicated in regulation of meiotic maturation (Wigglesworth et al. 2013), cholesterol synthesis (Su et al. 2008), amino acid transportation (Eppig et al. 2005) and glycolysis (Sugiura et al. 2007), which are utilized to maintain metabolism, growth and maturation of the oocyte. GDF9-null mice are infertile because of developmental arrest of follicles at the primary follicle stage (Dong et al. 1996). BMP15 knockout mice exhibit a defective cumulus phenotype that results in subfertility, while overexpression of BMP15 promotes growth of the ovarian follicle in mice (Yan et al. 2001). Moreover, BMP15 and GDF9 supplementation during in vitro maturation (IVM) improves oocyte developmental competence in cattle (Hussein et al. 2006).
The intra-follicular environment is impacted by a variety of environmental cues such as diet (Fig. 1) (Armstrong et al. 2001; Tilly and Sinclair 2013), the hormonal milieu (Macklon et al. 2006; Altermatt et al. 2012), chemicals (Uzumcu and Zachow 2007; Hunt et al. 2012) and health status [e.g. human polycystic ovary syndrome (Dunaif and Fauser 2013) and equine metabolic syndrome (Sessions-Bresnahan and Carnevale 2014)]. An inappropriate intra-follicular environment could result in poor oocyte quality/competence, which ultimately leads to infertility/subfertility. Dietary restriction causes subfertility or infertility in mice and rats (Selesniemi et al. 2008). In women, low body weight due to dietary restriction is linked to reduced fertility and results in significant changes in gonadotropin hormones to levels similar to those in women with ovarian insufficiency (Bates 1985). Mice on a zinc-deficient diet produced smaller fetuses and are prone to neural tube defects (Tian et al. 2014). In such animals, histone methylation was decreased in oocytes and retained in the maternal pronucleus after fertilization (Tian and Diaz 2013). Furthermore, increased oocyte lipid content and cellular stress are also evident in mouse studies showing poor embryo and fetal development after maternal preconception diabetes or obesity (Luzzo et al. 2012; Wu et al. 2012). The effects of environmental cues on the female reproductive system have been reviewed elsewhere (Uzumcu and Zachow 2007; Wang and Moley 2010; Fleming et al. 2011; Lane et al. 2014). Of particular note, future research on identification of specific maternal factors/oocyte stored factors that are influenced by environmental cues will improve our understanding of the mechanisms underlying the regulation of oocyte competence.
Maternal components contributing to oocyte quality
Spindle
The spindle is a complex subcellular structure with microtubules as fundamental machinery that gathers and segregates chromosome between daughter cells during cell division. Compared with other somatic cell types, oocytes are prone to errors in chromosome segregation that lead to aneuploid embryos/fetuses. The majority of aneuploidies in humans do not result in live offspring, making it a major cause of pregnancy failure. Thus, in human IVF clinics, a significant higher pregnancy rate from single embryo transfer was obtained if embryos are screened for euploidy compared with embryos that assessed only by morphology (Yang et al. 2012). Particularly, aneuploidy is commonly seen in oocytes from women of advanced reproductive age, suggesting it is a major cause of declining fertility with maternal aging (Savva et al. 2010; Kuliev et al. 2011) (Jones and Lane 2013). A similar age-associated increased aneuploidy rate is also observed in mice (Merriman et al. 2012). Growing evidence has shown that most errors in chromosome segregation arise during oocyte meiotic division, with various defects in the meiotic spindle (Howe and FitzHarris 2013; Jones and Lane 2013).
Therefore, much research attention has been drawn to understanding control of the first meiotic division, including microtubule-kinetochore interactions, spindle assembly, and the spindle assembly checkpoint (SAC). Due to the limitations of using human oocytes, most mechanistic research on these topics has used mouse oocytes as a model. Summaries of the control and timing of major events occurring during the metaphase I (MI) phase can be found in recent reviews (Howe and FitzHarris 2013; Jones and Lane 2013; Li and Albertini 2013).
Increased aneuploidy with maternal age has also been seen in mice (Pan et al. 2008; Selesniemi et al. 2011; Merriman et al. 2012), which presents a natural setting in which to search for the molecular causes of maternal age-related aneuploidy. By using this model, microarray analysis indicated that differential oocyte transcript profiles were found between old and young mice (Pan et al. 2008). Furthermore, time-lapse microscopy analysis indicated that there was no difference in the timing of oocytes from old mice entering anaphase of meiosis compared with young mice, suggesting oocyte aging is not likely due to defective SAC (Duncan et al. 2009).
Interestingly, several recent independent studies have reported a dramatic decrease in chromosome-bound cohesion proteins, with weakened chromosome cohesion in oocytes from old mice (Chiang et al. 2010; Lister et al. 2010; Merriman et al. 2012). Specifically, in oocytes from old mice, the distances between sister kinetochores are increased during MI and MII, and abundance of meiotic cohesion protein REC8 is reduced. Live cell imaging followed by immunocytochemical analysis also revealed that the majority of aneuplodies are attributed to weakened centromere cohesion (Chiang et al. 2010).
Despite recent breakthroughs, there is still a dramatic gap in our understanding of molecular mechanisms underlying the regulation of spindle assembly and chromosome segregation in mammalian oocytes. Further studies of these topics will depend on the state-of-the-art live imaging approach to observe detailed events occurring during MI and MII, plus gene-silencing techniques as well as proteomic approaches.
Mitochondria
Well-known as the “powerhouse” in the cell, mitochondria are subcellular organelles with double membranes and are essential for generating the majority of ATP that supplies energy for the body. They are especially important for characteristic energy-consuming events that require adequate energy stores during development such as chromosome segregation and fertilization occurring in oocytes and early embryos (Dumollard et al. 2007). In addition, relevant mitochondrial functions for oocyte/early embryonic development include sequestration and release of calcium, homeostasis of reduction-oxidation, and control of apoptosis.
Mitochondria are maternally inherited and possess their own genome independent of the nuclear genome. Interestingly, the mitochondrial population increases during oocyte maturation followed by an arrest of replication during cleavage stages in early embryos until mitochondrial replication recovers during gastrulation (Dumollard et al. 2007). Therefore, mitochondria in the early embryos originate from the maternal-stored population produced during oogenesis, implying a critical role of maternal mitochondria in early embryonic development; especially DNA of mitochondria is prone to mutations (Tilly and Sinclair 2013).
Dysfunction of mitochondria is linked to a number of human diseases, including obesity-induced diabetes, atherosclerosis and several neurodegenerative disorders (Wallace 2001; Lin and Beal 2006; Di Lisa et al. 2009). Around 1 in 4,000 children born in the United States are afflicted with mitochondrial diseases, which could be attributed to mutations of mitochondrial DNA (Schaefer et al. 2004). In recent years, there is much interest in developing novel ART to prevent transmission of mitochondrial diseases through replacement of mother’s oocyte mitochondria with mitochondria from un-afflicted donors. Pronuclear transfer or spindle transfer can be successfully performed in mice (McGrath and Solter 1983), monkey (Tachibana et al. 2009) and humans (Craven et al. 2010; Paull et al. 2013; Tachibana et al. 2013), and shows promising application in human clinics as a treatment of mitochondrial diseases. However, there are ethical and epigenetic concerns that must be addressed at the same time (Mitalipov and Wolf 2014).
Beside the genetic contribution, mitochondria are also important for early embryogenesis. Abnormal oxidative phosphorylation of mitochondria in mouse oocytes causes a defect in meiotic maturation and fertilization, thus resulting in poor early embryonic development (Van Blerkom et al. 1995). In alignment with this result, increased ATP content in human oocytes is correlated with improved developmental competence of oocytes (Van Blerkom et al. 1995). Microinjection of mitochondria into oocytes leads to reduction of oocyte apoptosis (Perez et al. 2000). It is also believed that insufficient ATP production from oocyte mitochondria is responsible for errors in chromosome segregation (Schon et al. 2000; Eichenlaub-Ritter et al. 2004; Zheng et al. 2007). Several studies have also revealed that mitochondria DNA copy numbers in oocytes and early embryos are correlated with success rates of fertilization and early development (Santos et al. 2006; Spikings et al. 2006). Consistent with this finding, suppressed replication of mitochondria in porcine oocytes during IVM leads to poor early embryonic developmental potential (Spikings et al. 2007). Thus, mitochondrial membrane potential, indicative of mitochondria health, and number of mitochondria may be potential markers of oocyte competence.
Maternally inherited mitochondria are affected by maternal diabetes. Transmission electron microscopy analysis indicates mitochondrial structure of oocytes ovulated from diabetic mice is altered and mitochondrial DNA copy number is increased (Wang et al. 2009). Moreover, the abundance of ATP and metabolites of tricarboxylic acid cycle are reduced in these oocytes, and altered distribution of mitochondria is observed in matured oocytes (Wang et al. 2009). The dysfunctional mitochondria in oocytes from diabetic females may be linked to their poor developmental competence. Indeed, transfer of zygotes from diabetic to non-diabetic foster mothers causes congenital disorders and delayed growth in the offspring, indicating adverse effects of maternal diabetes occur as early as the zygote stage (Wyman et al. 2008).
Mitochondrial dysfunction may contribute to maternal reproductive aging. Previous studies revealed decreases in ATP content, mitochondria numbers and mitochondria DNA copy numbers in aged oocytes, and dramatic morphological changes in oocyte mitochondria occur with aging in various species, including human, rat, pig, cow, mouse and mare (Simsek-Duran et al. 2013; Tilly and Sinclair 2013; Rambags et al. 2014).
Lipids
Lipids are hydrophobic or amphipathic molecules, whose main biological functions include storing energy, cell signaling and as structural components of cell membranes. Fatty acids are one category of lipids that functions as precursors of prostaglandins and cell membrane components. In addition, fatty acids can be stored as triacylglycerides within lipid droplets, which provide a rich source of energy upon demand. Numerous lipid droplets are seen in oocytes from certain species, including cattle, pigs and horses; these to some extent explain the darkness of the oocyte cytoplasm in these species (Dunning et al. 2014). Previous studies have demonstrated darkness of the oocyte cytoplasm is correlated with the oocyte quality. For example, there is a difference in the proportion of oocytes that cleave and develop to the blastocyst stage when bovine oocytes are categorized based on cytoplasm darkness (Jeong et al. 2009). Different darkness of oocyte cytoplasm is also linked with the ratio between saturated and unsaturated fatty acid, with the latter shown to be beneficial for oocyte developmental competence (Kim et al. 2001; Marei et al. 2009; Marei et al. 2010; Lapa et al. 2011).
To date, little direct evidence has been reported regarding the relationship between oocyte lipid content and oocyte competence. However, reduced developmental potential to the blastocyst stage was observed in mice on a diet enriched in n-3 fatty acids for 4 weeks, probably due to corresponding changes in mitochondrial distribution and increases in reactive oxygen species (ROS) levels (Wakefield et al. 2008). Similarly, treatment with a diet enriched in saturated fatty acids results in significant increases in oocyte lipid content, and oocytes exhibit a lipotoxicity response linked with reduced fertilization potential (Wu et al. 2012).
Oocyte lipid content is also affected by the follicular environment, which is–in turn-affected by changes in whole body metabolism. A well-known example to illustrate the association between maternal metabolism and oocyte quality is infertility in high producing cows. It is broadly believed that negative energy balance in post-calving high-producing cows is the primary causes of maternal metabolic disorders that alter the composition of endocrine and biochemical components in ovarian follicles, thus compromising oocyte quality (Thatcher et al. 2006; Thatcher et al. 2011). Numerous studies have attempted to supplement fatty acids to improve fertility in high producing cows (Santos et al. 2008; Thatcher et al. 2011). A diet high in α-linolenic acid leads to increases in α-linolenic acid content in follicular fluid, cumulus-oocyte-complexes and granulosa cells and an increased cleavage rate following in vitro fertilization (IVF) (Zachut et al. 2010).
Impacts of fatty acids on oocyte maturation and competence have also been observed using in vitro embryo production model systems. Supplementation with either stearic or palmitic acid into maturation medium results in defects in cumulus expansion, higher rates of cumulus cell apoptosis and delayed development to MII (Leroy et al. 2005).
Given the importance of lipid metabolism in oocyte developmental competence, our understanding on the key factors (enzymes) involved in this process has been limited. Future research should be directed towards understanding the profiles of regulatory genes related to lipid metabolism and their relevance to oocyte competence. This understanding will shed novel insight on developing strategies to improve oocyte developmental competence in both humans and animals.
Subcortical maternal complex
The subcortical maternal complex (SCMC) is a MDa protein complex that plays an essential role in mouse preimplantation development. Localized in the subcortex of oocytes, the SCMC is assembled during oogenesis (Li et al. 2008a). After fertilization, the SCMC is restricted to the exposed surface of cleaved embryos ultimately making it specifically localized in outside cells of morula and blastocysts (Li et al. 2008a). To date, the SCMC contains a minimum of four maternal proteins: MATER, FLOPED, TLE6 and Filia (Li et al. 2008a). Null mutations of Mater, TLE6 and/or Floped cause developmental arrest during cleavage divisions, and female mutant mice are infertile (Tong et al. 2000; Li et al. 2008a; Yu et al. 2014). In addition, Filia deficiency results in delayed preimplantation development with reduced fecundity, increased aneuploidy and abnormal spindle assembly (Zheng and Dean 2009). Results also indicate MATER, TLE6 and FLOPED are required for the stability of the SCMC (Li et al. 2008a; Zheng and Dean 2009; Yu et al. 2014). Interestingly, one documented role for the SCMC is to regulate the spindle position through cytoplasmic F-actin cytoskeleton (Yu et al. 2014). Further research is needed to explore the complete component identities of the SCMC complex, which will shed novel insights on our understanding of the maternal control of early embryogenesis.
Maternal-effect genes
Maternal-effect genes are genes that when dysfunctional affect the phenotype of offspring regardless of their genotype (Matzuk and Burns 2012). Maternal-effect gene products are especially critical for the early embryonic development after fertilization until massive activation of the embryonic genome occurs. The maternal-effect gene was first described in Drosophila 30 years ago, while the first one in mammals was disclosed in 2000 (Christians et al. 2000; Tong et al. 2000).
Different technical strategies have been used to identity maternal-effect genes in mammals since 2000. The first several maternal-effect genes were discovered in mice using conventional knockout approaches (Christians et al. 2000; Tong et al. 2000; Burns et al. 2003; Wu et al. 2003). To precisely probe the functional roles of maternal genes, for which conventional knockout strategies result in embryonic lethality, conditional knockout paired with RNAi-mediated gene silencing via oocyte injections has been used. A growing number of maternal-effect genes have been found to be essential for early embryogenesis. They are summarized in Table 1.
Table 1.
A list of maternal-effect genes in mammalian early embryonic development (M: mouse; B: bovine)
| Genes | Function | Species | References |
|---|---|---|---|
| Ago2 | Degradation of maternal RNA | M | (Lykke-Andersen et al. 2008) |
| Atg5 | Degradation of maternal protein | M | (Tsukamoto et al. 2008) |
|
Basonucli
n |
Transcriptional factor | M | (Ma et al. 2006) |
| Brg1 | Chromatin remodeling; Embryonic genome activation |
M | (Bultman et al. 2006) |
| Brwd1 | Chromatin remodeling | M | (Philipps et al. 2008) |
| Ctcf | Transcriptional factor; Embryonic genome activation |
M | (Wan et al. 2008) |
| Cdh1 | Cell-cell adhesion during compaction | M | (Larue et al. 1994; Riethmacher et al. 1995; De Vries et al. 2004) |
| Dnmt1 | Maintenance of DNA methylation | M, B | (Hirasawa et al. 2008; Golding et al. 2011) |
| Dnmt3a | De novo methyltransferase | M | (Kaneda et al. 2010) |
| Dnmt3l | De novo methyltransferase | M | (Hata et al. 2002) |
| Dicer1 | Degradation of maternal RNA | M | (Murchison et al. 2007) |
| Dppa3 | Maintenance of DNA methylation | M, B | (Nakamura et al. 2007; Bakhtari and Ross 2014) |
| Filia | Component of SCMC, euploidy maintenance | (Zheng and Dean 2009) | |
| Floped | ND | (Li et al. 2008a) | |
| Fmn2 | Meiotic progression | M | (Leader et al. 2002) |
| H1foo | Replacement of protamines | M | (Becker et al. 2005) |
| H3.3 | Reprogramming; decondensation of chromatin | M | (Lin et al. 2013; Inoue and Zhang 2014; Wen et al. 2014) |
| Hira | H3.3 incorporation; rRNA transcription | M | (Inoue and Zhang 2014; Lin et al. 2014) |
| Hsf1 | Redox homeostasis; mitochondria function | M | (Christians et al. 2000; Bierkamp et al. 2010) |
| Kdm1b | Histone H3 lysine 4 (H3K4) demethylase; de novo DNA methylation |
M | (Ciccone et al. 2009) |
| Mater | Component of SCMC | M, B | (Tong et al. 2000) (Pennetier et al. 2006) |
| mHR6A | Degradation of maternal protein; chromatin remodeling |
M | (Roest et al. 2004) |
| Npm2 | Chromatin remodeling | M | (Burns et al. 2003; Inoue et al. 2011) |
| Padi6 | ND | M | (Yurttas et al. 2008) |
| Pdk1 | ND | M | (Zheng et al. 2010) |
| Pms2 | DNA mismatch Repair | M | (Gurtu et al. 2002) |
| Ring1 | Component of Polycomb protein; chromatin remodeling |
M | (Posfai et al. 2012) |
| Rnf2 | Component of Polycomb protein; chromatin remodeling |
M | (Posfai et al. 2012) |
| SEBOX | ND | M | (Kim et al. 2008) |
| Sox2 | Reprogramming gene expression; | M | (Avilion et al. 2003; Pan and Schultz 2011) |
| Tap73 | Spindle assembly; genomic stability | M | (Tomasini et al. 2008) |
| Tcl1 | ND | M | (Narducci et al. 2002) |
| Tet3 | Active DNA demethylation | M,B | (Gu et al. 2011) |
| Tif1a | Chromatin remodeling | M | (Torres-Padilla and Zernicka-Goetz 2006) |
| Tle6 | ND | M | (Yu et al. 2014) |
| Trim28 | Maintenance of DNA methylation | M | (Messerschmidt et al. 2012) |
| UCH1l | Block polyspermy | M | (Sekiguchi et al. 2006) |
| Zar1 | Early embryonic development | M | (Wu et al. 2003) |
| ZFP36l2 | Degradation of maternal factors | M | (Ramos et al. 2004) |
| Zfp57 | Maintenance of DNA methylation | M | (Li et al. 2008b) |
| JY-1 | ND | B | (Bettegowda et al. 2007; Lee et al. 2014a) |
| Follistatin | ND | B | (Patel et al. 2007; Lee et al. 2009) |
| SMAD4 | ND | B | (Lee et al. 2014b) |
| NOBOX | ND | M, B | (Rajkovic et al. 2004; Tripurani et al. 2011) |
| KPNA7 | Nuclear importing | B | (Tejomurtula et al. 2009) |
Despite the increased understanding of maternal control of early embryonic development in mice through the identification of maternal-effect genes, the information regarding the functional role of maternal genes has been limited in other species, particularly in domestic animals. Due to the practical limitations of using conventional gene knockout strategies in domestic animals, inhibition of mRNA and protein by injection of siRNAs or antibodies is the predominant approach used to determine the functional role of maternal-effect genes (Bettegowda et al. 2007; Tejomurtula et al. 2009; Tripurani et al. 2011; Lee et al. 2014b). We have utilized cattle as a dual-purpose model relevant to both human ART and bovine reproduction with the goal to increase understanding of molecular mediators of oocyte quality and maternal regulation of early embryogenesis. To date, our results support a critical functional role in early embryogenesis in bovine for JY-1 (Bettegowda et al. 2007), Follistatin (Lee et al. 2009), KPNA7 (Tejomurtula et al. 2009), NOBOX (Tripurani et al. 2011) and SMAD4 (Lee et al. 2014c).
The first maternal-effect gene we studied in bovine was identified through analysis of expressed sequence tags (ESTs) from cDNA libraries of bovine oocytes (Yao et al. 2004). Of interest, several ESTs were noted that encode a novel oocyte-expressed transcript: JY-1, whose product is a bovine-specific protein. JY-1 mRNA and protein are present exclusively in the ovary during folliculogenesis. Specifically, JY-1 is exclusively expressed in the oocyte. Gene expression analysis showed levels of JY-1 mRNA are maximal in the germinal vesicle stage oocytes and decline in early embryos to even undetectable levels at 16-cell stage. Microinjection of JY-1 siRNAs into zygotes results in a dramatic decrease in the potential of embryonic development in terms of 8-16 celled embryo and blastocyst rates relative to control embryos (Bettegowda et al. 2007). Addition of exogenous JY-1 protein rescues the development of JY-1-deficient embryos to the blastocyst stage. Moreover, a novel model system whereby JY-1 siRNA was injected into cumulus-enclosed oocytes (germinal vesicle stage) and meiotic arrest maintained for 48 h before IVM, was developed in our lab to examine the effect of reduced JY-1 expression in oocytes on meiotic maturation, cumulus expansion and subsequent development potential after fertilization. Reduction of JY-1 protein in oocytes leads to a dramatic reduction in cumulus expansion, reduced proportion of oocytes developing to MII, and decreased proportions of embryos that developed to the 8-16 cell and blastocyst stages following IVF (Lee et al. 2014a). Effects of oocyte JY-1 depletion on meiotic maturation and cumulus expansion, but not subsequent embryonic development post fertilization, are rescued by supplementation of recombinant JY-1 protein during oocyte culture, supporting a crucial functional role of maternal JY-1 both pre- and post-fertilization (Lee et al. 2014a).
KPNA7 is another bovine maternal-effect gene discovered by analyzing EST sequences from cDNA libraries of oocytes (Yao et al. 2004) and encodes a novel member of the importin α protein family that is involved in the nuclear import of proteins > 40 kDA. KPNA7 mRNA expression is ovary-specific in cattle with high abundance in both immature and mature oocytes and early embryos prior to the 16 cell stage followed by a sharp decrease at the morula and blastocyst stages. KPNA7 deficiency in early embryos led to a reduced developmental potential to the blastocyst stage (Tejomurtula et al. 2009). In addition, compared with other importin α family members, KPNA7 possess a stronger affinity for binding with NPM2 (a maternal-effect nuclear protein), implying a critical role of KPNA7 in the transportation of oocyte-stored nuclear proteins in early embryos (Tejomurtula et al. 2009).
The newborn ovary homeobox encoding gene (NOBOX) is also required for early embryogenesis in cattle (Tripurani et al. 2011). Deficiency of NOBOX mRNA in early embryos results in poor development potential of early embryos to the blastocyst stage, and apparent down-regulation of genes related to signal transduction, cell cycle, and transcriptional regulation during EGA (Tripurani et al. 2011). Furthermore, NOBOX deficiency in early embryos leads to decreased expression of pluripotent genes (Pou5f1/Oct4 and Nanog) and reduced inner cell mass (ICM) cell number (Tripurani et al. 2011). Interestingly, the level of NOBOX mRNA in early embryos is modulated by a particular miRNA (Tripurani et al. 2011).
To further determine differential RNA transcripts associated with poor oocyte competence, we performed microarray analysis using a well-characterized prepubertal model of poor oocyte quality. Results showed follistatin mRNA abundance is positively associated with oocyte competence (Patel et al. 2007). Follistatin increased percentage of embryos cleaving early, percentage of embryos developing to the blastocyst stage and numbers of trophectoderm cells in resulting blastocysts (Lee et al. 2009). Results of comparative experiments in rhesus monkeys also showed positive actions of follistatin treatment on development of IVF embryos to the blastocyst stage and supported translational relevance of results from the bovine model (VandeVoort et al. 2009). Moreover, loss of function studies via microinjection of follistatin siRNAs into zygotes further confirmed the functional role of follistatin in regulating bovine early embryonic development and cell allocation in blastocysts (Lee et al. 2009).
Follistatin selectively binds TGF-β growth factor superfamily ligands, which bind to their receptors and activate SMAD signaling pathways. To increase understanding of the potential mechanism underlying follistatin’s action in early embryos, experiments utilizing a combination of pharmacological and siRNA-mediated inhibition of TGF-β superfamily signaling pathway components in the presence or absence of follistatin treatment were performed. Results to date demonstrated a functional requirement for the common SMAD (SMAD4) and SMAD2/3 (unpublished data) signaling pathways in bovine early embryonic development and suggest stimulatory actions of follistatin on blastocyst rate, but not early cleavage, are blocked when SMAD2/3/4 signaling is inhibited (Lee et al. 2009; Lee et al. 2014c). Collectively, results support a fundamental intrinsic role for follistatin and TGF-β superfamily signaling in bovine early embryonic development.
Relevance to oocyte aging
Altered expression of maternal-effect genes could be associated with reduced oocyte developmental competence characteristic of ovarian aging. Dnmt1o (oocyte-specific DNA methyltransferase 1) and Dnmt1s (somatic form of Dnmt1) mRNA levels are both reduced in oocytes from old animals relative to controls. (Hamatani et al. 2004; Pan et al. 2008). Moreover, nuclear importing of DNMTs is abnormal in oocytes from old animals (Zhang et al. 2011). However, the functional significance is unclear as methylation imprints are maintained for selected imprinted genes, and global methylation level is normal in oocytes from old animals (Lopes et al. 2009). Further research is necessary to clarify how reduced Dnmts affect oocyte quality in models of ovarian aging.
As a maternal-effect gene, p73 is well known for its regulatory role in cell cycle, genome stability and apoptosis (Tomasini et al. 2008). Analysis of p73 expression between oocytes of young women versus women of advanced maternal age shows that TAp73, p73 isoform containing transcription transactivation (TA) domain, levels are reduced in women older than 38 (Guglielmino et al. 2011). Similar results were observed from mouse oocytes. Collectively, defective TAp73 expression in old oocytes could help account for some of the declining fertility associated with maternal aging. Indeed, TAp73-null oocytes exhibit defects in spindle formation and result in poor blastocyst formation in mice (Tomasini et al. 2008).
Mechanisms of maternal control of early embryogenesis
Maternal control of epigenetic reprogramming following fertilization
The fusion of the oocyte and the sperm represents a remarkable moment during which highly differentiated germ cells are reprogrammed to a totipotent status. A variety of developmental events are reset to foster the correct embryonic development, including maintenance of methylation imprints, activation of the embryonic genome and lineage specification. These important developmental events are subject to regulation via epigenetic modifications (DNA methylation, histone modifications etc.)/chromatin remodeling.
Epigenetic modifications are extremely dynamic in early embryos and occur shortly after fertilization, suggesting a critical role in reprogramming the above-mentioned developmental events in early embryos (Fig. 2). For example, active DNA demethylation occurs in the male pronucleus prior to the onset of DNA replication and passive DNA demethylation follows thereafter until the morula stage (Kohli and Zhang 2013). Accordingly, chromatin organization is dramatically different between paternal and maternal pronuclei (Hemberger et al. 2009). This difference is characterized by an asymmetry of DNA demethylation and histone modification patterns (Sarmento et al. 2004; Morgan et al. 2005; Hemberger et al. 2009). Epigenetic reprogramming is believed to resolve this dramatic difference in paternal and maternal chromatin and ensure the transition from differentiated status to totipotency. Due to the transcriptional quiescence in early embryos prior to EGA, proteins (reprogramming factors) stored during oogenesis are believed necessary for the epigenetic reprogramming in early embryos. The powerful ability of oocytes in reprogramming has been demonstrated by the success of somatic cell nuclear transfer (SCNT). Indeed, multiple maternal proteins have been reported to be critical for epigenetic reprogramming.
Fig. 2.
Dynamics of key developmental events occurring during bovine preimplantation development.
Shortly after fertilization and prior to 8-16 cell stage, when embryonic genome activation occurs, early embryonic development relies on maternal-stored factors due to transcriptional silencing during this period. The maternal factors are gradually degraded while the embryonic genome is gradually activated. Genome-wide DNA demethylation occurs in both male and female genomes immediately following fertilization. However, active DNA demethylation–that is independent of DNA replication, exclusively occurs in male genome while passive DNA demethylation, that is dependent on DNA replication, occurs in female genomes. At a certain point between morula and blastocyst stages, de novo DNA methylation is initiated for both male and female genomes. Throughout early embryogenesis, other epigenetic modifications, including histone variants and modifications, are also dynamically reprogrammed. The final goal of the regulation is to reprogram the highly differentiated germ cells (sperm and oocyte) to a totipotent embryo.
Recently, Tet3 has been shown to be a key modulator of active demethylation of paternal DNA after fertilization (Gu et al. 2011). Tet3 is specifically enriched in the paternal pronucleus where conventional DNA methyl groups (5-methylcytosine, 5mc) are oxidized to 5-hydroxylmethylcytosine (5hmc), a novel DNA modification identified recently (Kriaucionis and Heintz 2009; Tahiliani et al. 2009; Inoue and Zhang 2011; Iqbal et al. 2011; Wossidlo et al. 2011). However, Tet3-depleted zygotes fail to exhibit 5mc to 5hmc conversion, indicating a loss of active demethylation of paternal genome. Moreover, reduced fecundity was observed in female mice deficient of Tet3 in the germ line and depletion of Tet3 from oocytes results in reduced reprogramming efficiency in SCNT. These data suggest active demethylation is achieved by DNA hydroxylation catalyzed by Tet3, supporting a critical role for maternal Tet3 in early embryogenesis.
The first protein identified with a documented role in controlling DNA demethylation in zygotes is DPPA3 (Nakamura et al. 2007). DPPA3-null mouse zygotes exhibit active DNA methylation in both paternal and maternal genomes, suggesting its role in prohibiting the maternal genome from active demethylation after fertilization. Additionally, binding of DPPPA3 to H3K9me2-enriched nucleosomes induces a change in chromatin structure. Such change may impact Tet3 affinity for chromatin and causes reduced conversion of 5mc to 5hmc (Nakamura et al. 2007). This role of DPPA3 may be conserved across species as a similar functional role has been documented recently in bovine embryos (Bakhtari and Ross 2014).
Histones are also subject to dramatic reprogramming immediately after fertilization occurs, particularly in terms of histone variants and the post-translational modifications of histone tails (Fig. 2) (Beaujean 2014). It is well known that sperm DNA is highly coated with protamines and requires reorganization at fertilization to ensure successful development of embryos. Maternal histone proteins are critical for completing this reorganization. For example, protamines are rapidly replaced by oocyte-specific linker histone H1Foo (Becker et al. 2005). Recently, several studies also demonstrated that histone variants (H3.3) are crucial for epigenetic regulation/chromatin remodeling which orchestrates gene expression changes in early embryos (Lin et al. 2013; Lin et al. 2014; Wen et al. 2014). Reprogramming of histone post-translational modifications was recently reviewed (Beaujean 2014; Lee et al. 2014d).
H3.3 is one such maternal histone protein important for early embryogenesis. Canonical histone H3 is expressed and deposited onto chromatin during S phase of the cell cycle while the variant H3.3 is found throughout the entire cell cycle. Maternal H3.3 is deposited onto the sperm chromatin shortly after fertilization (1h). Recently, three independent studies revealed an essential role of H3.3 during the process of reprogramming after fertilization in mice (Lin et al. 2013; Inoue and Zhang 2014; Wen et al. 2014). RNAi-mediated knockdown of H3.3 in mouse zygotes results in developmental failure at the morula stage with overly condensed chromosomes, high aneuploidy rate and failure in nucleosome assembly (Lin et al. 2013; Inoue and Zhang 2014), suggesting a critical role of H3.3 in chromatin remodeling in zygotes. Consistent with this idea, Wen et al. showed that reduction of H3.3 from mouse oocytes leads to decreased reprogramming potential and downregulation of key pluripotent genes, which could be rescued by injection of exogenous H3.3 mRNA but not H3.2 using SCNT as a model (Wen et al. 2014). Interestingly, only maternal H3.3 rather than H3.3 from donor somatic cells is responsible for the reprogramming success of SCNT (Wen et al. 2014). The requirement of H3.3 in the reprogramming process may be explained by its role in chromatin remodeling (open/closed chromatin structure) as H3.3 deficiency leads to reduced levels of H3K36me2 and H4K16ac, both markers of open chromatin (Lin et al. 2013).
Hira is a chaperone for H3.3. Hira-mutant females are infertile as embryos generated from such females and wild-type male mice fail to develop beyond zygote stage, suggesting a requirement of maternal Hira in important development events occurring in zygotes (Lin et al. 2014). Consistent with this idea, formation of the maternal pronucleus is blocked after knockdown of Hira in oocytes because of failure in nucleosome assembly in the sperm. Hira also plays a role in maternal chromatin reprogramming since Hira mutant oocytes fail to develop parthenogenetically. Additionally, depletion of Hira causes reduced levels of DNA replication and transcription in both male and female pronuclei.
Embryonic genomic activation
Early embryos gradually develop developmental independence once EGA occurs, a developmental process by which an embryo begins to transcribe its newly formed genome. The onset of EGA varies among species (2 cell in mice, 4-8 cell in human and pig, 8-16 cell in cow and sheep). Proper EGA is critical for normal development (Lee et al. 2014d). Thus, many studies have been performed to determine the mechanism of EGA and gene expression profile changes occurring during the maternal-to-zygotic transition (Lee et al. 2014d). A growing list of maternal factors involved in regulation of EGA have been identified (Table 1). These factors include Brg1 (Bultman et al. 2006), Mater (official name, Nlrp5) (Tong et al. 2000), Padi6 (Yurttas et al. 2008), Hsf1 (Christians et al. 2000), Zar1 (Wu et al. 2003), Npm2 (Burns et al. 2003), Ctcf (Wan et al. 2008), Zfp36l2 (Ramos et al. 2004), Atg5 (Tsukamoto et al. 2008), Ago2 (official name, Eif2c2) (Lykke-Andersen et al. 2008), Basonuclin (Ma et al. 2006), Ring1, Rnf2 (Posfai et al. 2012), and Sox2 (Pan and Schultz 2011). Mutations of any one of the genes encoding these proteins cause developmental arrest at cleavage stages.
The maternal nucleolus is also required for EGA and thus critical for normal early embryonic development in mice (Vogt et al. 2012). In early embryos, the nucleolar precursor bodies (NPBs) undergo dramatic transformation to mature nucleoli. A recent study has shown that LIN28, known for its reprogramming ability in generation of induced pluripotent stem cells (iPSC), is enriched in NPB. Depletion of Lin28 from early embryos results in developmental arrest at 2-4 cell stage, due to the failure of NPB assembly.
Maternal polycomb group (PcG) proteins are crucial for the initiation of EGA (Posfai et al. 2012). PcG proteins are conserved across species with an established role in transcriptional repression. In mammals, PcG proteins have at least two different classes of complexes, PRC1 and PRC2. These complexes are well known for their ability to catalyze mono-ubiquitination of histone H2A and tri-methylation of histone H3 lysine 27. Depletion of maternal Ring1 and Rnf2 from mouse oocytes, both components of PRC1, results in developmental arrest at 2 cell stage after fertilization with wild-type male sperm, suggesting PcG proteins are implicated in the acquisition of oocyte developmental competence during oogenesis by maintaining appropriate chromatin status.
Maternal control of genomic imprinting
Genomic imprinting is an epigenetic process that distinguishes chromosomes inherited from mother and father and results in parent-of-origin-specific gene expression. Central to this process is the creation of differentially methylated regions (DMRs), which exhibit allele-specific epigenetic marks (Plasschaert and Bartolomei 2014). Among these marks, DNA methylation is the most-well-investigated. DMRs are hypomethylated in the expressed parental allele and hypermethylated in the non-expressed parental allele (Plasschaert and Bartolomei 2014). Disruption of DMRs results in loss of imprinting for many imprinted genes and can cause cancer and human defects (Lee and Bartolomei 2013).
Differential epigenetic marks (imprints) are placed on DMR in the germ line and must be maintained throughout development to ensure proper growth and development (Lee and Bartolomei 2013). It is especially critical for maintenance of genomic imprints during preimplantation development because both parental genomes are demethylated while the chromatin is strikingly remodeled. Mechanisms must exist that maintain allele-specific modifications of the DMRs during early development. How these discrete regions are specifically recognized and their specific epigenetic modifications subsequently maintained are poorly understood. It is likely that maternal factors are crucial during preimplantation development to perform this specialized protective role. Indeed, multiple maternal genes have been identified, whose products are involved in protecting imprinted methylation sites during preimplantation development including DPPA3 (Nakamura et al. 2007); ZFP57 (Li et al. 2008b); TRIM28 (Messerschmidt et al. 2012) and DNMT1 (Hirasawa et al. 2008).
Maternal DPPA3 (also known as PGC7 or Stella) is the first maternal protein possessing a protective role in maintenance of methylation imprints. As mentioned above, maternal DPPA3 is required for prohibiting the maternal pronucleus from active demethylation in mouse zygotes and DPPA3 mutants fail to develop beyond the blastocyst stage (Nakamura et al. 2007). Moreover, DPPA3 is also implicated in the protection of imprinted genes with paternally inherited methylation (H19, Rasgrf1). As a result, DPPA3-depletion from oocytes leads to reduced DNA methylation at imprinted genes. Recently, it was found that an underlying histone modification, H3K9me2, is responsible for recruiting DPPA3 to the maternal genome and some paternal imprinting control regions (ICRs), and thus prevents active demethylation by Tet3 (Nakamura et al. 2012).
ZFP57 is a Krüppel-associated box domain (KRAB)–zinc finger protein involved in control of genomic imprinting. During preimplantation development, ZFP57 expression is generally of maternal origin until ZFP57 transcription begins at the blastocyst stage (Zeng et al. 2004). Ablation of both maternal and embryonic ZFP57 lead to embryonic lethality in mice (Li et al. 2008b). Bisulfite sequencing analysis demonstrated that mid-gestation embryos depleted of maternal and embryonic ZFP57 have reduced DNA methylation at both maternal (Snrpn, Peg1, Peg3, Peg5) and paternalimprinted DMRs (Gtl2 ICR) (Li et al. 2008b). The effect of maternal ZFP57 depletion on methylation imprints can be found as early as the blastocyst stage, where ZFP57 depletion results in hypomethylation of Snrpn that can be rescued by embryonic ZFP57 by day 13.5. ZFP57 mutations have also been correlated with abnormal DNA methylation at several imprinted regions in humans (Mackay et al. 2008).
TRIM28 is a scaffold component of a chromatin-modifying complex that mediates transcriptional activity by interaction with KRAB–zinc finger proteins, such as ZFP57, which recognize specific genomic loci (Messerschmidt et al. 2012). TRIM28 is responsible for recruiting epigenetic modifiers that are associated with repressive chromatin (Messerschmidt et al. 2012). Embryonic TRIM28 transcription occurs at the 2-cell stage with the protein detected as early as the 4-cell stage. Maternal TRIM28 deletion results in partial embryonic lethality in mice (Messerschmidt et al. 2012). TRIM28 maternal mutant embryos display loss of methylation imprints at the H19, Snrpn and Gtl2 ICRs but this phenotype varies between embryos (Messerschmidt et al. 2012). H19 imprinted methylation is still reduced in 4- and 8-cell TRIM28 maternal mutant embryos, indicating maternal TRIM28 is required for protection of methylation imprints from passive demethylation.
By comparison, DPPA3 deletion has a global role in protecting the maternal genome while maternal TRIM28 mutation has a specific effect on imprinted genes. This specificity is achieved by interaction of TRIM28 with KRAP ZFP, such as ZFP57, which is expressed in a tissue-specific fashion. Consistent with this idea, TRIM28 and ZFP57 are both enriched at DMRs of imprinted genes, such as H19. Thus, TRIM28 may attract epigenetic modifiers to change chromatin structures to allow transcriptional control. Indeed, multiple epigenetic modifiers have been shown to bind TRIM28 complex, including SETDB1, DNMT1, DNMT3A and DNMT3B (Iyengar and Farnham 2011).
Maternal DNMTs are also critical for the maintenance of methylation imprints during early embryonic development. The DNMT are classified into two categories: de-novo (DNMT3A, 3B, 3L) and maintenance methyltransferse (DNMT1). DNMT1 recognizes hemimethylated DNA, methylating the newly synthesized strand at each replicative cycle. It has been demonstrated that maternal and zygotic DNMT1, but not DNMT3A and 3B, are both required for the maintenance of methylation imprints in preimplantation embryos.
Despite the above information, no universal mechanism has been reported affecting maintenance of methylation imprints of imprinted genes. For example, PGC7 selectively protects the paternally imprinted/methylated DMRs of Rasgrf1 and H19-Igf2 and maternally methylated DMRs of Peg1, Peg3, and Peg10. However, PGC7 is dispensable for the protection of the maternal DMRs of Snrpn and Peg5 and the paternal DMR of IG-DMR1/Meg3. It suggests locus-specific regulation by a cohort of modifiers on methylation imprints.
Cell specification
During mammalian development, the first cell fate decision is clearly made upon the formation of blastocyst (at day 3.5 in mice and 6-7 after insemination in cattle), in which the outside layer of cells is termed trophectoderm (TE) and inside cells named inner cell mass (ICM). TE cells subsequently develop to the majority of placenta. ICM cells will develop into the embryo proper, and are also the source of embryonic stem cells (ESCs). Given specific localization within either ICM (Oct4) or TE (Cdx2), maternal Oct4 and/or Cdx2 were- hypothesized to be critical for the first lineage specification event. However, it was later found that both maternal Oct4 and Cdx2 are dispensable for the first lineage specification (Blij et al. 2012; Frum et al. 2013).
The Hippo-YAP pathway plays an important role in this lineage decision. In outside cells, YAP is localized in the nucleus and induces the TE specific transcription factor Cdx2 by cooperation with TEAD4 while in inside cells YAP is phosphorylated by Lats1/2, Hippo pathway kinases, blocking entry into the nucleus. Recently, a Hippo pathway member upstream of Lats1/2 and YAP, Nf2/Merlin, was shown to be required for YAP phosphorylation in early embryos. Zygotic Nf2 mutant embryos exhibit mild defects in Cdx2 expression and YAP localization while the maternal and zygotic mutants have more severe phenotypes, demonstrating a role of maternal Nf2 in the first lineage decision.
Conclusions and Perspectives
A competent oocyte is a major determinant of pregnancy success, not only providing genetic materials but a variety of functional maternal factors that support early embryogenesis. Despite great progress made towards identification and characterization of the maternal factors, there is still a significant gap in understanding the maternal control of maternal-zygotic-transition, especially in domestic animals, such as cows and horses. Furthermore, there is also a lack of knowledge on the influence of environmental perturbation on the expression and activity of maternal factors. Recent progress in genome sequencing, especially single-cell transcriptomic analysis, will greatly facilitate identification of maternal factors. Furthermore, functional analysis will be convenient with the robust gene engineering tools (e.g. TALEN, CRISPR/Cas9).
Oocyte quality is a dramatic concern for women of advanced age, given a growing number of women that choose to conceive in their later 30s or early 40s. To determine how oocyte quality contributes to poor fertility in women with advanced age, there is a huge interest in effects of maternal age on spindle assembly and chromosome segregation as aneuploidy rate increases as women age. In contrast to this effect on nuclear competence, there is a limited understanding of how cytoplasmic competence/maturation is altered in women with advanced maternal age. Thus, future work should also be directed toward identifying age-associated perturbation in maternal factors. Due to ethical issues and limitations of oocyte numbers in humans, such type of work requires using relevant model such as bovine or equine, which exhibit similarities to the human female reproductive system (Malhi et al. 2007; Carnevale 2008).
Recently, growing attention has been paid to the long-term developmental influences of the use of ART. Aberrant epigenetic regulation is closely associated with low efficiency and the poor outcome of reproductive technologies in both agricultural animals and humans (Grace and Sinclair 2009; El Hajj and Haaf 2013). Evidence demonstrates that aberrant DNA methylation patterns are correlated with the use of reproductive technologies, including superovulation and SCNT (Farin et al. 2006; Market-Velker et al. 2010). Of particular note, superovulation in mouse models induces abnormal imprinted methylation for both paternally and maternally imprinted genes in blastocysts, suggesting maternal-effect genes are affected and are required for maintenance of genomic imprints in early embryos (Market-Velker et al. 2010). However, our understanding of mechanisms of epigenetic reprogramming in early embryos is far from complete. It remains to be identified which maternal factors are disturbed in response to the use of reproductive technologies (e.g. superovulation). Future investigation of the regulation of maternal control on developmental events, such as DNA methylation dynamics, in early embryos will better our understanding of the key factors that contributing to maternal reproductive aging and shed novel insights on further improvements in reproductive technologies in both agricultural animals and humans.
Acknowledgements
The authors’ work presented here was funded by the National Institute of Child Health and Human Development of the National Institutes of Health under award number R01HD072972 and by Michigan State University AgBioResearch.
References
- Altermatt JL, Marolf AJ, Wrigley RH, Carnevale EM. Effects of FSH and LH on ovarian and follicular blood flow, follicular growth and oocyte developmental competence in young and old mares. Anim. Reprod. Sci. 2012;133:191–197. doi: 10.1016/j.anireprosci.2012.06.021. [DOI] [PubMed] [Google Scholar]
- Altermatt JL, Suh TK, Stokes JE, Carnevale EM. Effects of age and equine follicle-stimulating hormone (eFSH) on collection and viability of equine oocytes assessed by morphology and developmental competency after intracytoplasmic sperm injection (ICSI) Reprod. Fertil. Dev. 2009;21:615–623. doi: 10.1071/RD08210. [DOI] [PubMed] [Google Scholar]
- Armstrong DG, McEvoy TG, Baxter G, Robinson JJ, Hogg CO, Woad KJ, Webb R, Sinclair KD. Effect of dietary energy and protein on bovine follicular dynamics and embryo production in vitro: associations with the ovarian insulin-like growth factor system. Biol. Reprod. 2001;64:1624–1632. doi: 10.1095/biolreprod64.6.1624. [DOI] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes. Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhtari A, Ross PJ. DPPA3 prevents cytosine hydroxymethylation of the maternal pronucleus and is required for normal development in bovine embryos. Epigenetics. 2014;9:1271–1279. doi: 10.4161/epi.32087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates GW. Body weight control practice as a cause of infertility. Clin. Obstet. Gynecol. 1985;28:632–644. doi: 10.1097/00003081-198528030-00018. [DOI] [PubMed] [Google Scholar]
- Beaujean N. Histone post-translational modifications in preimplantation mouse embryos and their role in nuclear architecture. Mol. Reprod. Dev. 2014;81:100–112. doi: 10.1002/mrd.22268. [DOI] [PubMed] [Google Scholar]
- Becker M, Becker A, Miyara F, Han Z, Kihara M, Brown DT, Hager GL, Latham K, Adashi EY, Misteli T. Differential in vivo binding dynamics of somatic and oocyte-specific linker histones in oocytes and during ES cell nuclear transfer. Mol. Biol. Cell. 2005;16:3887–3895. doi: 10.1091/mbc.E05-04-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettegowda A, Lee KB, Smith GW. Cytoplasmic and nuclear determinants of the maternal-to-embryonic transition. Reprod. Fertil. Dev. 2008;20:45–53. doi: 10.1071/rd07156. [DOI] [PubMed] [Google Scholar]
- Bettegowda A, Yao J, Sen A, Li Q, Lee KB, Kobayashi Y, Patel OV, Coussens PM, Ireland JJ, Smith GW. JY-1, an oocyte-specific gene, regulates granulosa cell function and early embryonic development in cattle. Proc. Natl. Acad. Sci. 2007;104:17602–17607. doi: 10.1073/pnas.0706383104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierkamp C, Luxey M, Metchat A, Audouard C, Dumollard R, Christians E. Lack of maternal Heat Shock Factor 1 results in multiple cellular and developmental defects, including mitochondrial damage and altered redox homeostasis, and leads to reduced survival of mammalian oocytes and embryos. Dev. Biol. 2010;339:338–353. doi: 10.1016/j.ydbio.2009.12.037. [DOI] [PubMed] [Google Scholar]
- Binelli M, Murphy BD. Coordinated regulation of follicle development by germ and somatic cells. Reprod. Fertil. Dev. 2010;22:1–12. doi: 10.1071/RD09218. [DOI] [PubMed] [Google Scholar]
- Blij S, Frum T, Akyol A, Fearon E, Ralston A. Maternal Cdx2 is dispensable for mouse development. Development. 2012;139:3969–3972. doi: 10.1242/dev.086025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes. Dev. 2006;20:1744–1754. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KH, Viveiros MM, Ren Y, Wang P, DeMayo FJ, Frail DE, Eppig JJ, Matzuk MM. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science. 2003;300:633–636. doi: 10.1126/science.1081813. [DOI] [PubMed] [Google Scholar]
- Carnevale EM. The mare model for follicular maturation and reproductive aging in the woman. Theriogenology. 2008;69:23–30. doi: 10.1016/j.theriogenology.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Chiang T, Duncan FE, Schindler K, Schultz RM, Lampson MA. Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr. Biol. 2010;20:1522–1528. doi: 10.1016/j.cub.2010.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians E, Davis AA, Thomas SD, Benjamin IJ. Maternal effect of Hsf1 on reproductive success. Nature. 2000;407:693–694. doi: 10.1038/35037669. [DOI] [PubMed] [Google Scholar]
- Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461:415–418. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- Craven L, Tuppen HA, Greggains GD, Harbottle SJ, Murphy JL, Cree LM, Murdoch AP, Chinnery PF, Taylor RW, Lightowlers RN, Herbert M, Turnbull DM. Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature. 2010;465:82–85. doi: 10.1038/nature08958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silveira JC, Veeramachaneni DN, Winger QA, Carnevale EM, Bouma GJ. Cell-secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins: a possible new form of cell communication within the ovarian follicle. Biol. Reprod. 2012;86:71. doi: 10.1095/biolreprod.111.093252. [DOI] [PubMed] [Google Scholar]
- De Vries WN, Evsikov AV, Haac BE, Fancher KS, Holbrook AE, Kemler R, Solter D, Knowles BB. Maternal beta-catenin and E-cadherin in mouse development. Development. 2004;131:4435–4445. doi: 10.1242/dev.01316. [DOI] [PubMed] [Google Scholar]
- Di Lisa F, Kaludercic N, Carpi A, Menabo R, Giorgio M. Mitochondria and vascular pathology. Pharmacol. Rep. 2009;61:123–130. doi: 10.1016/s1734-1140(09)70014-3. [DOI] [PubMed] [Google Scholar]
- Dong J, Albertini DF, Nishimori K, Kumar TR, Lu N, Matzuk MM. Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature. 1996;383:531–535. doi: 10.1038/383531a0. [DOI] [PubMed] [Google Scholar]
- Dumollard R, Duchen M, Carroll J. The role of mitochondrial function in the oocyte and embryo. Curr. Top. Dev. Biol. 2007;77:21–49. doi: 10.1016/S0070-2153(06)77002-8. [DOI] [PubMed] [Google Scholar]
- Dunaif A, Fauser BC. Renaming PCOS--a two-state solution. J. Clin. Endocrinol. Metab. 2013;98:4325–4328. doi: 10.1210/jc.2013-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan FE, Chiang T, Schultz RM, Lampson MA. Evidence that a defective spindle assembly checkpoint is not the primary cause of maternal age-associated aneuploidy in mouse eggs. Biol. Reprod. 2009;81:768–776. doi: 10.1095/biolreprod.109.077909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning KR, Russell DL, Robker RL. Lipids and oocyte developmental competence: the role of fatty acids and beta-oxidation. Reproduction. 2014;148:R15–27. doi: 10.1530/REP-13-0251. [DOI] [PubMed] [Google Scholar]
- Edson MA, Nagaraja AK, Matzuk MM. The mammalian ovary from genesis to revelation. Endocr. Rev. 2009;30:624–712. doi: 10.1210/er.2009-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenlaub-Ritter U, Vogt E, Yin H, Gosden R. Spindles, mitochondria and redox potential in ageing oocytes. Reprod. Biomed. Online. 2004;8:45–58. doi: 10.1016/s1472-6483(10)60497-x. [DOI] [PubMed] [Google Scholar]
- El Hajj N, Haaf T. Epigenetic disturbances in in vitro cultured gametes and embryos: implications for human assisted reproduction. Fertil. Steril. 2013;99:632–641. doi: 10.1016/j.fertnstert.2012.12.044. [DOI] [PubMed] [Google Scholar]
- Eppig JJ, Pendola FL, Wigglesworth K, Pendola JK. Mouse oocytes regulate metabolic cooperativity between granulosa cells and oocytes: amino acid transport. Biol. Reprod. 2005;73:351–357. doi: 10.1095/biolreprod.105.041798. [DOI] [PubMed] [Google Scholar]
- Farin PW, Piedrahita JA, Farin CE. Errors in development of fetuses and placentas from in vitro-produced bovine embryos. Theriogenology. 2006;65:178–191. doi: 10.1016/j.theriogenology.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Fleming TP, Lucas ES, Watkins AJ, Eckert JJ. Adaptive responses of the embryo to maternal diet and consequences for post-implantation development. Reprod. Fertil. Dev. 2011;24:35–44. doi: 10.1071/RD11905. [DOI] [PubMed] [Google Scholar]
- Frum T, Halbisen MA, Wang C, Amiri H, Robson P, Ralston A. Oct4 cell-autonomously promotes primitive endoderm development in the mouse blastocyst. Dev. Cell. 2013;25:610–622. doi: 10.1016/j.devcel.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginther OJ, Bergfelt DR, Leith GS, Scraba ST. Embryonic loss in mares: Incidence and ultrasonic morphology. Theriogenology. 1985;24:73–86. doi: 10.1016/0093-691x(85)90213-4. [DOI] [PubMed] [Google Scholar]
- Golding MC, Williamson GL, Stroud TK, Westhusin ME, Long CR. Examination of DNA methyltransferase expression in cloned embryos reveals an essential role for Dnmt1 in bovine development. Mol. Reprod. Dev. 2011;78:306–317. doi: 10.1002/mrd.21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace KS, Sinclair KD. Assisted reproductive technology, epigenetics, and long-term health: a developmental time bomb still ticking. Semin. Reprod. Med. 2009;27:409–416. doi: 10.1055/s-0029-1237429. [DOI] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, Iqbal K, Shi YG, Deng Z, Szabo PE, Pfeifer GP, Li J, Xu GL. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Guglielmino MR, Santonocito M, Vento M, Ragusa M, Barbagallo D, Borzi P, Casciano I, Banelli B, Barbieri O, Astigiano S, Scollo P, Romani M, Purrello M, Di Pietro C. TAp73 is downregulated in oocytes from women of advanced reproductive age. Cell Cycle. 2011;10:3253–3256. doi: 10.4161/cc.10.19.17585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtu VE, Verma S, Grossmann AH, Liskay RM, Skarnes WC, Baker SM. Maternal effect for DNA mismatch repair in the mouse. Genetics. 2002;160:271–277. doi: 10.1093/genetics/160.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatani T, Falco G, Carter MG, Akutsu H, Stagg CA, Sharov AA, Dudekula DB, VanBuren V, Ko MS. Age-associated alteration of gene expression patterns in mouse oocytes. Hum. Mol. Genet. 2004;13:2263–2278. doi: 10.1093/hmg/ddh241. [DOI] [PubMed] [Google Scholar]
- Hansen PJ, Block J, Loureiro B, Bonilla L, Hendricks KE. Effects of gamete source and culture conditions on the competence of in vitro-produced embryos for post-transfer survival in cattle. Reprod. Fertil. Dev. 2010;22:59–66. doi: 10.1071/RD09212. [DOI] [PubMed] [Google Scholar]
- Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nat. Rev. Mol. Cell. Biol. 2009;10:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- Hinrichs K. The equine oocyte: factors affecting meiotic and developmental competence. Mol. Reprod. Dev. 2010;77:651–661. doi: 10.1002/mrd.21186. [DOI] [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes. Dev. 2008;22:1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, FitzHarris G. Recent insights into spindle function in mammalian oocytes and early embryos. Biol. Reprod. 2013;89:71. doi: 10.1095/biolreprod.113.112151. [DOI] [PubMed] [Google Scholar]
- Hsueh AJ, Kawamura K, Cheng Y, Fauser BC. Intraovarian control of early folliculogenesis. Endocr. Rev. 2014:er20141020. doi: 10.1210/er.2014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt PA, Lawson C, Gieske M, Murdoch B, Smith H, Marre A, Hassold T, VandeVoort CA. Bisphenol A alters early oogenesis and follicle formation in the fetal ovary of the rhesus monkey. Proc. Natl. Acad. Sci. 2012;109:17525–17530. doi: 10.1073/pnas.1207854109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein TS, Thompson JG, Gilchrist RB. Oocyte-secreted factors enhance oocyte developmental competence. Dev. Biol. 2006;296:514–521. doi: 10.1016/j.ydbio.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Inoue A, Ogushi S, Saitou M, Suzuki MG, Aoki F. Involvement of mouse nucleoplasmin 2 in the decondensation of sperm chromatin after fertilization. Biol. Reprod. 2011;85:70–77. doi: 10.1095/biolreprod.110.089342. [DOI] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nat. Struct. Mol. Biol. 2014;21:609–616. doi: 10.1038/nsmb.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Farnham PJ. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem. 2011;286:26267–26276. doi: 10.1074/jbc.R111.252569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong WJ, Cho SJ, Lee HS, Deb GK, Lee YS, Kwon TH, Kong IK. Effect of cytoplasmic lipid content on in vitro developmental efficiency of bovine IVP embryos. Theriogenology. 2009;72:584–589. doi: 10.1016/j.theriogenology.2009.04.015. [DOI] [PubMed] [Google Scholar]
- Jones KT, Lane SI. Molecular causes of aneuploidy in mammalian eggs. Development. 2013;140:3719–3730. doi: 10.1242/dev.090589. [DOI] [PubMed] [Google Scholar]
- Kaneda M, Hirasawa R, Chiba H, Okano M, Li E, Sasaki H. Genetic evidence for Dnmt3a-dependent imprinting during oocyte growth obtained by conditional knockout with Zp3-Cre and complete exclusion of Dnmt3b by chimera formation. Genes Cells. 2010 doi: 10.1111/j.1365-2443.2009.01374.x. [DOI] [PubMed] [Google Scholar]
- Kim JY, Kinoshita M, Ohnishi M, Fukui Y. Lipid and fatty acid analysis of fresh and frozen-thawed immature and in vitro matured bovine oocytes. Reproduction. 2001;122:131–138. [PubMed] [Google Scholar]
- Kim KH, Kim EY, Lee KA. SEBOX is essential for early embryogenesis at the two-cell stage in the mouse. Biol. Reprod. 2008;79:1192–1201. doi: 10.1095/biolreprod.108.068478. [DOI] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuliev A, Zlatopolsky Z, Kirillova I, Spivakova J, Cieslak Janzen J. Meiosis errors in over 20,000 oocytes studied in the practice of preimplantation aneuploidy testing. Reprod. Biomed. Online. 2011;22:2–8. doi: 10.1016/j.rbmo.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Lane M, Robker RL, Robertson SA. Parenting from before conception. Science. 2014;345:756–760. doi: 10.1126/science.1254400. [DOI] [PubMed] [Google Scholar]
- Lapa M, Marques CC, Alves SP, Vasques MI, Baptista MC, Carvalhais I, Silva Pereira M, Horta AE, Bessa RJ, Pereira RM. Effect of trans-10 cis-12 conjugated linoleic acid on bovine oocyte competence and fatty acid composition. Reprod. Domest. Anim. 2011;46:904–910. doi: 10.1111/j.1439-0531.2011.01762.x. [DOI] [PubMed] [Google Scholar]
- Larue L, Ohsugi M, Hirchenhain J, Kemler R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc. Natl. Acad. Sci. 1994;91:8263–8267. doi: 10.1073/pnas.91.17.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader B, Lim H, Carabatsos MJ, Harrington A, Ecsedy J, Pellman D, Maas R, Leder P. Formin-2, polyploidy, hypofertility and positioning of the meiotic spindle in mouse oocytes. Nat. Cell Biol. 2002;4:921–928. doi: 10.1038/ncb880. [DOI] [PubMed] [Google Scholar]
- Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Lee KB, Bettegowda A, Wee G, Ireland JJ, Smith GW. Molecular determinants of oocyte competence: potential functional role for maternal (oocyte-derived) follistatin in promoting bovine early embryogenesis. Endocrinology. 2009;150:2463–2471. doi: 10.1210/en.2008-1574. [DOI] [PubMed] [Google Scholar]
- Lee KB, Wee G, Zhang K, Folger JK, Knott JG, Smith GW. Functional role of the bovine oocyte-specific protein JY-1 in meiotic maturation, cumulus expansion, and subsequent embryonic development. Biol. Reprod. 2014a;90:69. doi: 10.1095/biolreprod.113.115071. [DOI] [PubMed] [Google Scholar]
- Lee KB, Zhang K, Folger JK, Knott JG, Smith GW. Evidence Supporting a Functional Requirement of SMAD4 for Bovine Preimplantation Embryonic Development: A Potential Link to Embryotrophic Actions of Follistatin. Biol. Reprod. 2014b;91:62. doi: 10.1095/biolreprod.114.120105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KB, Zhang K, Folger JK, Knott JG, Smith GW. Evidence Supporting a Functional Requirement of SMAD4 for Bovine Preimplantation Embryonic Development: A Potential Link to Embryotropic Actions of Follistatin. Biol. Reprod. 2014c doi: 10.1095/biolreprod.114.120105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Giraldez AJ. Zygotic Genome Activation During the Maternal-to-Zygotic Transition. Annu. Rev. Cell. Dev. Biol. 2014d doi: 10.1146/annurev-cellbio-100913-013027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy JL, Vanholder T, Mateusen B, Christophe A, Opsomer G, de Kruif A, Genicot G, Van Soom A. Non-esterified fatty acids in follicular fluid of dairy cows and their effect on developmental capacity of bovine oocytes in vitro. Reproduction. 2005;130:485–495. doi: 10.1530/rep.1.00735. [DOI] [PubMed] [Google Scholar]
- Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev. Cell. 2008a;15:416–425. doi: 10.1016/j.devcel.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Albertini DF. The road to maturation: somatic cell interaction and self-organization of the mammalian oocyte. Nat. Rev. Mol. Cell. Biol. 2013;14:141–152. doi: 10.1038/nrm3531. [DOI] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell. 2008b;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CJ, Conti M, Ramalho-Santos M. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development. 2013;140:3624–3634. doi: 10.1242/dev.095513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CJ, Koh FM, Wong P, Conti M, Ramalho-Santos M. Hira-mediated h3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev. Cell. 2014;30:268–279. doi: 10.1016/j.devcel.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Lindbloom SM, Farmerie TA, Clay CM, Seidel GE, Jr., Carnevale EM. Potential involvement of EGF-like growth factors and phosphodiesterases in initiation of equine oocyte maturation. Anim. Reprod. Sci. 2008;103:187–192. doi: 10.1016/j.anireprosci.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Lister LM, Kouznetsova A, Hyslop LA, Kalleas D, Pace SL, Barel JC, Nathan A, Floros V, Adelfalk C, Watanabe Y, Jessberger R, Kirkwood TB, Hoog C, Herbert M. Age-related meiotic segregation errors in mammalian oocytes are preceded by depletion of cohesin and Sgo2. Curr. Biol. 2010;20:1511–1521. doi: 10.1016/j.cub.2010.08.023. [DOI] [PubMed] [Google Scholar]
- Lonergan P, Rizos D, Gutierrez-Adan A, Fair T, Boland MP. Oocyte and embryo quality: effect of origin, culture conditions and gene expression patterns. Reprod. Domest. Anim. 2003;38:259–267. doi: 10.1046/j.1439-0531.2003.00437.x. [DOI] [PubMed] [Google Scholar]
- Lopes FL, Fortier AL, Darricarrere N, Chan D, Arnold DR, Trasler JM. Reproductive and epigenetic outcomes associated with aging mouse oocytes. Hum. Mol. Genet. 2009;18:2032–2044. doi: 10.1093/hmg/ddp127. [DOI] [PubMed] [Google Scholar]
- Luzzo KM, Wang Q, Purcell SH, Chi M, Jimenez PT, Grindler N, Schedl T, Moley KH. High fat diet induced developmental defects in the mouse: oocyte meiotic aneuploidy and fetal growth retardation/brain defects. PLoS One. 2012;7:e49217. doi: 10.1371/journal.pone.0049217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen K, Gilchrist MJ, Grabarek JB, Das P, Miska E, Zernicka-Goetz M. Maternal Argonaute 2 is essential for early mouse development at the maternal-zygotic transition. Mol. Biol. Cell. 2008;19:4383–4392. doi: 10.1091/mbc.E08-02-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Zeng F, Schultz RM, Tseng H. Basonuclin: a novel mammalian maternal-effect gene. Development. 2006;133:2053–2062. doi: 10.1242/dev.02371. [DOI] [PubMed] [Google Scholar]
- Mackay DJ, Callaway JL, Marks SM, White HE, Acerini CL, Boonen SE, Dayanikli P, Firth HV, Goodship JA, Haemers AP, Hahnemann JM, Kordonouri O, Masoud AF, Oestergaard E, Storr J, Ellard S, Hattersley AT, Robinson DO, Temple IK. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat. Genet. 2008;40:949–951. doi: 10.1038/ng.187. [DOI] [PubMed] [Google Scholar]
- Macklon NS, Stouffer RL, Giudice LC, Fauser BC. The science behind 25 years of ovarian stimulation for in vitro fertilization. Endocr. Rev. 2006;27:170–207. doi: 10.1210/er.2005-0015. [DOI] [PubMed] [Google Scholar]
- Malhi PS, Adams GP, Mapletoft RJ, Singh J. Oocyte developmental competence in a bovine model of reproductive aging. Reproduction. 2007;134:233–239. doi: 10.1530/REP-07-0021. [DOI] [PubMed] [Google Scholar]
- Marei WF, Wathes DC, Fouladi-Nashta AA. The effect of linolenic Acid on bovine oocyte maturation and development. Biol. Reprod. 2009;81:1064–1072. doi: 10.1095/biolreprod.109.076851. [DOI] [PubMed] [Google Scholar]
- Marei WF, Wathes DC, Fouladi-Nashta AA. Impact of linoleic acid on bovine oocyte maturation and embryo development. Reproduction. 2010;139:979–988. doi: 10.1530/REP-09-0503. [DOI] [PubMed] [Google Scholar]
- Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MR. Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum. Mol. Genet. 2010;19:36–51. doi: 10.1093/hmg/ddp465. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Burns KH. Genetics of mammalian reproduction: modeling the end of the germline. Annu. Rev. Physiol. 2012;74:503–528. doi: 10.1146/annurev-physiol-020911-153248. [DOI] [PubMed] [Google Scholar]
- McGrath J, Solter D. Nuclear transplantation in the mouse embryo by microsurgery and cell fusion. Science. 1983;220:1300–1302. doi: 10.1126/science.6857250. [DOI] [PubMed] [Google Scholar]
- Merriman JA, Jennings PC, McLaughlin EA, Jones KT. Effect of aging on superovulation efficiency, aneuploidy rates, and sister chromatid cohesion in mice aged up to 15 months. Biol. Reprod. 2012;86:49. doi: 10.1095/biolreprod.111.095711. [DOI] [PubMed] [Google Scholar]
- Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335:1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- Mitalipov S, Wolf DP. Clinical and ethical implications of mitochondrial gene transfer. Trends Endocrinol. Metab. 2014;25:5–7. doi: 10.1016/j.tem.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 2005;14(Spec No 1):R47–58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes. Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, Tanaka S, Shiota K, Nakano T. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Liu YJ, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, Nakano T. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature. 2012;486:415–419. doi: 10.1038/nature11093. [DOI] [PubMed] [Google Scholar]
- Narducci MG, Fiorenza MT, Kang SM, Bevilacqua A, Di Giacomo M, Remotti D, Picchio MC, Fidanza V, Cooper MD, Croce CM, Mangia F, Russo G. TCL1 participates in early embryonic development and is overexpressed in human seminomas. Proc. Natl. Acad. Sci. 2002;99:11712–11717. doi: 10.1073/pnas.182412399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navot D, Bergh PA, Williams MA, Garrisi GJ, Guzman I, Sandler B, Grunfeld L. Poor oocyte quality rather than implantation failure as a cause of age-related decline in female fertility. Lancet. 1991;337:1375–1377. doi: 10.1016/0140-6736(91)93060-m. [DOI] [PubMed] [Google Scholar]
- Pan H, Ma P, Zhu W, Schultz RM. Age-associated increase in aneuploidy and changes in gene expression in mouse eggs. Dev. Biol. 2008;316:397–407. doi: 10.1016/j.ydbio.2008.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Schultz RM. Sox2 modulates reprogramming of gene expression in two-cell mouse embryos. Biol. Reprod. 2011;85:409–416. doi: 10.1095/biolreprod.111.090886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Su YQ, Ariga M, Law E, Jin SL, Conti M. EGF-like growth factors as mediators of LH action in the ovulatory follicle. Science. 2004;303:682–684. doi: 10.1126/science.1092463. [DOI] [PubMed] [Google Scholar]
- Patel OV, Bettegowda A, Ireland JJ, Coussens PM, Lonergan P, Smith GW. Functional genomics studies of oocyte competence: evidence that reduced transcript abundance for follistatin is associated with poor developmental competence of bovine oocytes. Reproduction. 2007;133:95–106. doi: 10.1530/rep.1.01123. [DOI] [PubMed] [Google Scholar]
- Paull D, Emmanuele V, Weiss KA, Treff N, Stewart L, Hua H, Zimmer M, Kahler DJ, Goland RS, Noggle SA, Prosser R, Hirano M, Sauer MV, Egli D. Nuclear genome transfer in human oocytes eliminates mitochondrial DNA variants. Nature. 2013;493:632–637. doi: 10.1038/nature11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennetier S, Perreau C, Uzbekova S, Thelie A, Delaleu B, Mermillod P, Dalbies-Tran R. MATER protein expression and intracellular localization throughout folliculogenesis and preimplantation embryo development in the bovine. BMC Dev. Biol. 2006;6 doi: 10.1186/1471-213X-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez GI, Trbovich AM, Gosden RG, Tilly JL. Mitochondria and the death of oocytes. Nature. 2000;403:500–501. doi: 10.1038/35000651. [DOI] [PubMed] [Google Scholar]
- Philipps DL, Wigglesworth K, Hartford SA, Sun F, Pattabiraman S, Schimenti K, Handel M, Eppig JJ, Schimenti JC. The dual bromodomain and WD repeat-containing mouse protein BRWD1 is required for normal spermiogenesis and the oocyte-embryo transition. Dev. Biol. 2008;317:72–82. doi: 10.1016/j.ydbio.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasschaert RN, Bartolomei MS. Genomic imprinting in development, growth, behavior and stem cells. Development. 2014;141:1805–1813. doi: 10.1242/dev.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai E, Kunzmann R, Brochard V, Salvaing J, Cabuy E, Roloff TC, Liu Z, Tardat M, van Lohuizen M, Vidal M, Beaujean N, Peters AH. Polycomb function during oogenesis is required for mouse embryonic development. Genes. Dev. 2012;26:920–932. doi: 10.1101/gad.188094.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J, Wang ZB, Feng HL, Miao YL, Wang Q, Yu Y, Wei YC, Yan J, Wang WH, Shen W, Sun SC, Schatten H, Sun QY. The root of reduced fertility in aged women and possible therapentic options: current status and future perspects. Mol Aspects Med. 2014;38:54–85. doi: 10.1016/j.mam.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Rajkovic A, Pangas SA, Ballow D, Suzumori N, Matzuk MM. NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science. 2004;305:1157–1159. doi: 10.1126/science.1099755. [DOI] [PubMed] [Google Scholar]
- Rambags BP, van Boxtel DC, Tharasanit T, Lenstra JA, Colenbrander B, Stout TA. Advancing maternal age predisposes to mitochondrial damage and loss during maturation of equine oocytes in vitro. Theriogenology. 2014;81:959–965. doi: 10.1016/j.theriogenology.2014.01.020. [DOI] [PubMed] [Google Scholar]
- Ramos SB, Stumpo DJ, Kennington EA, Phillips RS, Bock CB, Ribeiro-Neto F, Blackshear PJ. The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development. 2004;131:4883–4893. doi: 10.1242/dev.01336. [DOI] [PubMed] [Google Scholar]
- Riethmacher D, Brinkmann V, Birchmeier C. A targeted mutation in the mouse E-cadherin gene results in defective preimplantation development. Proc. Natl. Acad. Sci. 1995;92:855–859. doi: 10.1073/pnas.92.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roest HP, Baarends WM, de Wit J, van Klaveren JW, Wassenaar E, Hoogerbrugge JW, van Cappellen WA, Hoeijmakers JH, Grootegoed JA. The ubiquitin-conjugating DNA repair enzyme HR6A is a maternal factor essential for early embryonic development in mice. Mol. Cell. Biol. 2004;24:5485–5495. doi: 10.1128/MCB.24.12.5485-5495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JE, Bilby TR, Thatcher WW, Staples CR, Silvestre FT. Long chain fatty acids of diet as factors influencing reproduction in cattle. Reprod. Domest. Anim. 2008;43(Suppl 2):23–30. doi: 10.1111/j.1439-0531.2008.01139.x. [DOI] [PubMed] [Google Scholar]
- Santos TA, El Shourbagy S, St John JC. Mitochondrial content reflects oocyte variability and fertilization outcome. Fertil. Steril. 2006;85:584–591. doi: 10.1016/j.fertnstert.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Sarmento OF, Digilio LC, Wang YM, Perlin J, Herr JC, Allis CD, Coonrod SA. Dynamic alterations of specific histone modifications during early murine development. J. Cell Sci. 2004;117:4449–4459. doi: 10.1242/jcs.01328. [DOI] [PubMed] [Google Scholar]
- Sartori R, Bastos MR, Wiltbank MC. Factors affecting fertilisation and early embryo quality in single- and superovulated dairy cattle. Reprod. Fertil. Dev. 2010;22:151–158. doi: 10.1071/RD09221. [DOI] [PubMed] [Google Scholar]
- Savva GM, Walker K, Morris JK. The maternal age-specific live birth prevalence of trisomies 13 and 18 compared to trisomy 21 (Down syndrome) Prenatal Diagn. 2010;30:57–64. doi: 10.1002/pd.2403. [DOI] [PubMed] [Google Scholar]
- Schaefer AM, Taylor RW, Turnbull DM, Chinnery PF. The epidemiology of mitochondrial disorders--past, present and future. Biochim Biophys Acta. 2004;1659:115–120. doi: 10.1016/j.bbabio.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Schon EA, Kim SH, Ferreira JC, Magalhaes P, Grace M, Warburton D, Gross SJ. Chromosomal non-disjunction in human oocytes: is there a mitochondrial connection? Hum Reprod. 2000;15(Suppl 2):160–172. doi: 10.1093/humrep/15.suppl_2.160. [DOI] [PubMed] [Google Scholar]
- Sekiguchi S, Kwon J, Yoshida E, Hamasaki H, Ichinose S, Hideshima M, Kuraoka M, Takahashi A, Ishii Y, Kyuwa S, Wada K, Yoshikawa Y. Localization of ubiquitin C-terminal hydrolase L1 in mouse ova and its function in the plasma membrane to block polyspermy. Am J Pathol. 2006;169:1722–1729. doi: 10.2352/ajpath.2006.060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selesniemi K, Lee HJ, Muhlhauser A, Tilly JL. Prevention of maternal aging-associated oocyte aneuploidy and meiotic spindle defects in mice by dietary and genetic strategies. Proc. Natl. Acad. Sci. 2011;108:12319–12324. doi: 10.1073/pnas.1018793108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selesniemi K, Lee HJ, Tilly JL. Moderate caloric restriction initiated in rodents during adulthood sustains function of the female reproductive axis into advanced chronological age. Aging Cell. 2008;7:622–629. doi: 10.1111/j.1474-9726.2008.00409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions-Bresnahan DR, Carnevale EM. The effect of equine metabolic syndrome on the ovarian follicular environment. J. Anim. Sci. 2014;92:1485–1494. doi: 10.2527/jas.2013-7275. [DOI] [PubMed] [Google Scholar]
- Simsek-Duran F, Li F, Ford W, Swanson RJ, Jones HW, Jr., Castora FJ. Age-associated metabolic and morphologic changes in mitochondria of individual mouse and hamster oocytes. PLoS One. 2013;8:e64955. doi: 10.1371/journal.pone.0064955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirard MA, Richard F, Blondin P, Robert C. Contribution of the oocyte to embryo quality. Theriogenology. 2006;65:126–136. doi: 10.1016/j.theriogenology.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Spencer TE. Early pregnancy: Concepts, challenges, and potential solutions. Animal Frontiers. 2013;3:48–55. [Google Scholar]
- Spikings EC, Alderson J, St John JC. Transmission of mitochondrial DNA following assisted reproduction and nuclear transfer. Hum. Reprod. Update. 2006;12:401–415. doi: 10.1093/humupd/dml011. [DOI] [PubMed] [Google Scholar]
- Spikings EC, Alderson J, St John JC. Regulated mitochondrial DNA replication during oocyte maturation is essential for successful porcine embryonic development. Biol. Reprod. 2007;76:327–335. doi: 10.1095/biolreprod.106.054536. [DOI] [PubMed] [Google Scholar]
- Su YQ, Sugiura K, Eppig JJ. Mouse oocyte control of granulosa cell development and function: paracrine regulation of cumulus cell metabolism. Semin. Reprod. Med. 2009;27:32–42. doi: 10.1055/s-0028-1108008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YQ, Sugiura K, Wigglesworth K, O’Brien MJ, Affourtit JP, Pangas SA, Matzuk MM, Eppig JJ. Oocyte regulation of metabolic cooperativity between mouse cumulus cells and oocytes: BMP15 and GDF9 control cholesterol biosynthesis in cumulus cells. Development. 2008;135:111–121. doi: 10.1242/dev.009068. [DOI] [PubMed] [Google Scholar]
- Sugiura K, Su YQ, Diaz FJ, Pangas SA, Sharma S, Wigglesworth K, O’Brien MJ, Matzuk MM, Shimasaki S, Eppig JJ. Oocyte-derived BMP15 and FGFs cooperate to promote glycolysis in cumulus cells. Development. 2007;134:2593–2603. doi: 10.1242/dev.006882. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Amato P, Sparman M, Woodward J, Sanchis DM, Ma H, Gutierrez NM, Tippner-Hedges R, Kang E, Lee HS, Ramsey C, Masterson K, Battaglia D, Lee D, Wu D, Jensen J, Patton P, Gokhale S, Stouffer R, Mitalipov S. Towards germline gene therapy of inherited mitochondrial diseases. Nature. 2013;493:627–631. doi: 10.1038/nature11647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sparman M, Sritanaudomchai H, Ma H, Clepper L, Woodward J, Li Y, Ramsey C, Kolotushkina O, Mitalipov S. Mitochondrial gene replacement in primate offspring and embryonic stem cells. Nature. 2009;461:367–372. doi: 10.1038/nature08368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejomurtula J, Lee KB, Tripurani SK, Smith GW, Yao J. Role of importin alpha8, a new member of the importin alpha family of nuclear transport proteins, in early embryonic development in cattle. Biol. Reprod. 2009;81:333–342. doi: 10.1095/biolreprod.109.077396. [DOI] [PubMed] [Google Scholar]
- Thatcher W, Santos JE, Staples CR. Dietary manipulations to improve embryonic survival in cattle. Theriogenology. 2011;76:1619–1631. doi: 10.1016/j.theriogenology.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Thatcher WW, Bilby TR, Bartolome JA, Silvestre F, Staples CR, Santos JE. Strategies for improving fertility in the modern dairy cow. Theriogenology. 2006;65:30–44. doi: 10.1016/j.theriogenology.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Tian X, Anthony K, Neuberger T, Diaz FJ. Preconception zinc deficiency disrupts postimplantation fetal and placental development in mice. Biol. Reprod. 2014;90:83. doi: 10.1095/biolreprod.113.113910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Diaz FJ. Acute dietary zinc deficiency before conception compromises oocyte epigenetic programming and disrupts embryonic development. Dev. Biol. 2013;376:51–61. doi: 10.1016/j.ydbio.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly JL, Sinclair DA. Germline energetics, aging, and female infertility. Cell Metab. 2013;17:838–850. doi: 10.1016/j.cmet.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, Khan F, Itie-Youten A, Wakeham A, Tsao MS, Iovanna JL, Squire J, Jurisica I, Kaplan D, Melino G, Jurisicova A, Mak TW. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes. Dev. 2008;22:2677–2691. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong ZB, Gold L, Pfeifer KE, Dorward H, Lee E, Bondy CA, Dean J, Nelson LM. Mater, a maternal effect gene required for early embryonic development in mice. Nat. Genet. 2000;26:267–268. doi: 10.1038/81547. [DOI] [PubMed] [Google Scholar]
- Torres-Padilla ME, Zernicka-Goetz M. Role of TIF1alpha as a modulator of embryonic transcription in the mouse zygote. J Cell Biol. 2006;174:329–338. doi: 10.1083/jcb.200603146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripurani SK, Lee KB, Wang L, Wee G, Smith GW, Lee YS, Latham KE, Yao J. A novel functional role for the oocyte-specific transcription factor newborn ovary homeobox (NOBOX) during early embryonic development in cattle. Endocrinology. 2011;152:1013–1023. doi: 10.1210/en.2010-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto S, Kuma A, Mizushima N. The role of autophagy during the oocyte-to-embryo transition. Autophagy. 2008;4:1076–1078. doi: 10.4161/auto.7065. [DOI] [PubMed] [Google Scholar]
- Uzumcu M, Zachow R. Developmental exposure to environmental endocrine disruptors: consequences within the ovary and on female reproductive function. Reprod. Toxicol. 2007;23:337–352. doi: 10.1016/j.reprotox.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Blerkom J, Davis PW, Lee J. ATP content of human oocytes and developmental potential and outcome after in-vitro fertilization and embryo transfer. Hum Reprod. 1995;10:415–424. doi: 10.1093/oxfordjournals.humrep.a135954. [DOI] [PubMed] [Google Scholar]
- VandeVoort CA, Mtango NR, Lee YS, Smith GW, Latham KE. Differential effects of follistatin on nonhuman primate oocyte maturation and pre-implantation embryo development in vitro. Biol. Reprod. 2009;81:1139–1146. doi: 10.1095/biolreprod.109.077198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt EJ, Meglicki M, Hartung KI, Borsuk E, Behr R. Importance of the pluripotency factor LIN28 in the mammalian nucleolus during early embryonic development. Development. 2012;139:4514–4523. doi: 10.1242/dev.083279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield SL, Lane M, Schulz SJ, Hebart ML, Thompson JG, Mitchell M. Maternal supply of omega-3 polyunsaturated fatty acids alter mechanisms involved in oocyte and early embryo development in the mouse. Am J Physiol Endocrinol Metab. 2008;294:E425–434. doi: 10.1152/ajpendo.00409.2007. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm for degenerative diseases and ageing. Novartis Found Symp. 2001;235:247–263. doi: 10.1002/0470868694.ch20. discussion 263-246. [DOI] [PubMed] [Google Scholar]
- Wan LB, Pan H, Hannenhalli S, Cheng Y, Ma J, Fedoriw A, Lobanenkov V, Latham KE, Schultz RM, Bartolomei MS. Maternal depletion of CTCF reveals multiple functions during oocyte and preimplantation embryo development. Development. 2008;135:2729–2738. doi: 10.1242/dev.024539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Moley KH. Maternal diabetes and oocyte quality. Mitochondrion. 2010;10:403–410. doi: 10.1016/j.mito.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Ratchford AM, Chi MM, Schoeller E, Frolova A, Schedl T, Moley KH. Maternal diabetes causes mitochondrial dysfunction and meiotic defects in murine oocytes. Mol. Endocrinol. 2009;23:1603–1612. doi: 10.1210/me.2009-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen D, Banaszynski LA, Liu Y, Geng F, Noh KM, Xiang J, Elemento O, Rosenwaks Z, Allis CD, Rafii S. Histone variant H3.3 is an essential maternal factor for oocyte reprogramming. Proc. Natl. Acad. Sci. 2014;111:7325–7330. doi: 10.1073/pnas.1406389111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigglesworth K, Lee KB, O’Brien MJ, Peng J, Matzuk MM, Eppig JJ. Bidirectional communication between oocytes and ovarian follicular somatic cells is required for meiotic arrest of mammalian oocytes. Proc. Natl. Acad. Sci. 2013;110:E3723–3729. doi: 10.1073/pnas.1314829110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox AJ, Weinberg CR, O’Connor JF, Baird DD, Schlatterer JP, Canfield RE, Armstrong EG, Nisula BC. Incidence of early loss of pregnancy. N Engl J Med. 1988;319:189–194. doi: 10.1056/NEJM198807283190401. [DOI] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2 doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- Wu LL, Russell DL, Norman RJ, Robker RL. Endoplasmic reticulum (ER) stress in cumulus-oocyte complexes impairs pentraxin-3 secretion, mitochondrial membrane potential (DeltaPsi m), and embryo development. Mol. Endocrinol. 2012;26:562–573. doi: 10.1210/me.2011-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Viveiros MM, Eppig JJ, Bai Y, Fitzpatrick SL, Matzuk MM. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat. Genet. 2003;33:187–191. doi: 10.1038/ng1079. [DOI] [PubMed] [Google Scholar]
- Wyman A, Pinto AB, Sheridan R, Moley KH. One-cell zygote transfer from diabetic to nondiabetic mouse results in congenital malformations and growth retardation in offspring. Endocrinology. 2008;149:466–469. doi: 10.1210/en.2007-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Wang P, DeMayo J, DeMayo FJ, Elvin JA, Carino C, Prasad SV, Skinner SS, Dunbar BS, Dube JL, Celeste AJ, Matzuk MM. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol. Endocrinol. 2001;15:854–866. doi: 10.1210/mend.15.6.0662. [DOI] [PubMed] [Google Scholar]
- Yang Z, Liu J, Collins GS, Salem SA, Liu X, Lyle SS, Peck AC, Sills ES, Salem RD. Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: results from a randomized pilot study. Mol Cytogenet. 2012;5:24. doi: 10.1186/1755-8166-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Ren X, Ireland JJ, Coussens PM, Smith TP, Smith GW. Generation of a bovine oocyte cDNA library and microarray: resources for identification of genes important for follicular development and early embryogenesis. Physiol. Genomics. 2004;19:84–92. doi: 10.1152/physiolgenomics.00123.2004. [DOI] [PubMed] [Google Scholar]
- Yu XJ, Yi Z, Gao Z, Qin D, Zhai Y, Chen X, Ou-Yang Y, Wang ZB, Zheng P, Zhu MS, Wang H, Sun QY, Dean J, Li L. The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics. Nat Commun. 2014;5:4887. doi: 10.1038/ncomms5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurttas P, Vitale AM, Fitzhenry RJ, Cohen-Gould L, Wu W, Gossen JA, Coonrod SA. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development. 2008;135:2627–2636. doi: 10.1242/dev.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachut M, Dekel I, Lehrer H, Arieli A, Arav A, Livshitz L, Yakoby S, Moallem U. Effects of dietary fats differing in n-6:n-3 ratio fed to high-yielding dairy cows on fatty acid composition of ovarian compartments, follicular status, and oocyte quality. J. Dairy Sci. 2010;93:529–545. doi: 10.3168/jds.2009-2167. [DOI] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev. Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Zhang K, Hansen PJ, Ealy AD. Fibroblast growth factor 10 enhances bovine oocyte maturation and developmental competence in vitro. Reproduction. 2010;140:815–826. doi: 10.1530/REP-10-0190. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lu DY, Ma WY, Li Y. Age-related changes in the localization of DNA methyltransferases during meiotic maturation in mouse oocytes. Fertil. Steril. 2011;95:1531–1534. e1531. doi: 10.1016/j.fertnstert.2010.06.050. [DOI] [PubMed] [Google Scholar]
- Zheng P, Dean J. Role of Filia, a maternal effect gene, in maintaining euploidy during cleavage-stage mouse embryogenesis. Proc. Natl. Acad. Sci. 2009;106:7473–7478. doi: 10.1073/pnas.0900519106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Vassena R, Latham KE. Effects of in vitro oocyte maturation and embryo culture on the expression of glucose transporters, glucose metabolism and insulin signaling genes in rhesus monkey oocytes and preimplantation embryos. Mol. Human Reprod. 2007;13:361–371. doi: 10.1093/molehr/gam014. [DOI] [PubMed] [Google Scholar]
- Zheng W, Gorre N, Shen Y, Noda T, Ogawa W, Lundin E, Liu K. Maternal phosphatidylinositol 3-kinase signalling is crucial for embryonic genome activation and preimplantation embryogenesis. EMBO Rep. 2010;11:890–895. doi: 10.1038/embor.2010.144. [DOI] [PMC free article] [PubMed] [Google Scholar]