Summary
Inflammasomes are oligomeric signaling complexes that promote caspase activation and maturation of proinflammatory cytokines. Structural and biophysical studies have shed light on the mechanisms of nucleic acid recognition and signaling complex assembly involving the AIM2 (absent in myeloma 2) and IFI16 (γ-interferon-inducible protein 16) inflammasomes. However, our understanding of the mechanisms of the NLRP3 (nucleotide-binding oligomerization-like receptor family, pyrin domain-containing protein 3) activation, either by nucleic acids or by other reported stimuli, has remained elusive. Exciting recent progress on the filament formation by the ASC (apoptosis-associated speck-like protein containing a caspase recruitment domain) pyrin domain and the IFI16–double stranded DNA complex has established that the formation of higher order polymers is one of the general mechanisms for signaling platform assembly in innate immune system. The paradigm-changing discovery of the extracellular function of the NLRP3–ASC inflammasome has opened the door for therapeutic targeting the inflammasome filament formation for various clinical conditions. Future characterization of the canonical and non-canonical inflammasome complexes will undoubtedly reveal more surprises on their structure and function and enrich our understanding of the molecular mechanisms of ligand recognition, activation, and regulation.
Keywords: inflammasome, nucleic acid recognition, filament formation, extracellular function
Introduction
Inflammasome is a multi-protein signaling complex that mediates the activation of caspase-1 and maturation of proinflammatory cytokines such as interleukin-1β (IL-1β) and IL-18 (1, 2). Such oligomeric complexes consist of the receptor/sensor molecules, the adapter ASC (apoptosis-associated speck-like protein containing a caspase recruitment domain), and procaspase-1. All of the inflammasome receptors/sensors contain at least one of the signaling domains that belong to the death domain superfamily. The death domain superfamily members adopt globular folds of anti-parallel six helix bundles with polarized charge distribution on their surface (3). Among the four families of the death domain (DD), death effector domain, caspase recruiting domain (CARD), and pyrin domain (PYD), the latter two are involved in the assembly of the inflammasomes through homotypic protein–protein interactions. Several inflammasome receptors/sensors have been reported in the literature, including the PYHIN family members AIM2 (absent in melanoma 2) (4–7) and IFI16 (interferon-inducible protein 16) (8, 9), and the NOD (nucleotide-binding oligomerization)-like receptor (NLR) family members NLRP1 (1), NLRP3 (10), NLRC4 (11–14), NLRP6 (15, 16), NLRP7 (17), and NLRP12 (18). In addition to the above ‘canonical’ inflammasome, recent identification of the non-canonical inflammasome involving the recognition of intracellular LPS by caspase-4/5/11 illustrated a completely new mode of caspase activation and broadened our perspective on innate immune recognition (19–21). Analogous to the regulation of the NLRP3 inflammasome activation by interferon-inducible guanylate binding protein 5 (GBP5) (22), several GBP proteins are involved in the regulation of the non-canonical inflammasome pathway and execution of pyroptosis, perhaps through inducing the lysis of bacteria-containing vacuoles (23, 24).
A diverse array of stimuli has been reported to activate the inflammasomes. However, not all of these have been demonstrated to physically interact with the receptor or sensor molecules per se. AIM2 (4–6) and IFI16 (8) have been shown to form inflammasomes that respond to DNA molecules in the cytosol and nucleus, respectively. The mechanisms of such sequence-independent DNA recognition have been characterized by structural and biophysical studies (25–28). Among the various microbial and sterile stimuli that activate the NLRP3 inflammasome, nucleic acids such as bacterial RNA (29), viral RNA (30), oxidized mitochondrial DNA (31), bacterial RNA:DNA hybrids (32), bacterial mRNA, tRNA, and rRNA (33), RNA from group B Streptococcus associated with lysosomal components (34), dsRNA concomitant with membrane permeabilization and potassium efflux (35), and dsRNA associated DHX33 helicase that is ubiquitinated by tripartite motif 33 (TRIM33) (36, 37) have been shown to induce NLRP3 activation. At present, the underlying mechanisms have not been defined, but it is likely that NLRP3 responds to these different forms of nucleic acids indirectly through other proteins such as the RNA helicases. In addition, Sha et al. (33) reported that human macrophages respond to bacterial mRNA, tRNA, and rRNA through IL-1β and IL-18 production, whereas murine macrophages only recognize bacterial mRNA. The mechanism of such distinct recognition is not well understood, but these observations do suggest species differences in inflammasome response to nucleic acids.
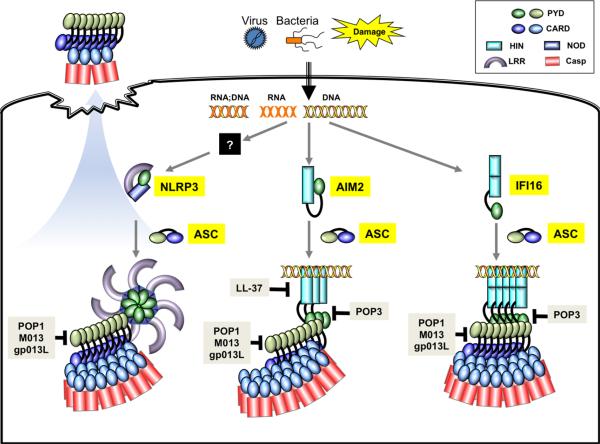
Two important aspects of the nucleic acid-recognizing inflammasomes are of particular interests. First, neither AIM2 nor IFI16 contains an oligomerization domain, in contrast to the ATP-dependent oligomerization NOD domain among the NLR family members. Therefore, a long piece of dsDNA serves as both the ligand and the oligomerization platform for both receptors (Fig. 1). Secondly, because nucleic acids are universal genetic materials, pathological stimulation of these nucleic acid-recognizing inflammasomes by self-DNA could potentially lead to autoimmune and autoinflammatory disorders. In fact, aberrant immune responses involving AIM2 or IFI16 have been suggested to contribute to lupus pathogenesis (38), and IFI16 was implicated in the pathophysiology of Sjögren's syndrome (39) and systemic sclerosis (40). Furthermore, elevated AIM2 expression has been linked to cytosolic DNA recognition in keratinocytes from patients with psoriasis lesion (41). Interestingly, this study also identified the antimicrobial peptide LL-37 as an antagonist for AIM2 activation in keratinocytes, perhaps through the sequestration of cytosolic DNA from the AIM2 receptor and subsequent reduction in proinflammatory cytokines production. This is in contrast to the roles of LL-37 in promoting DNA sensing by TLR9 in plasmacytoid dendritic cells (42) and keratinocytes (43), which lead to enhanced type I interferon production. Clearly distinct subcellular location of the different DNA receptors impacts their regulation by the LL-37 peptide. It would be important to investigate whether other positively charged antimicrobial peptides also play a role in the regulation of nucleic acid-sensing pathways.
Fig. 1. Cartoon presentation of the ligand recognition, assembly, and regulation of nucleic acid-sensing AIM2, IFI16, and NLRP3 inflammasomes.
Even though NLRP3 has been found to be important for sensing of RNA:DNA hybrid and RNA, it is currently not clear whether it directly associates with these nucleic acids or functions through other binding partners. AIM2 resides in an autoinhibited state in the absence of dsDNA binding, whereas the conformation of the DNA-free IFI16 appears to be more extended. Similar to AIM2 and NLRC4, NLRP3 is presumed to be in an autoinhibited conformation in the resting state, in which its leucine-rich repeat (LRR) domain interacts with the NOD and PYD domains. Activation of AIM2 and NLRP3 leads to the formation of filaments involving the adapter ASC, whereas the IFI16–DNA complex has been shown to adopt filament structure in the absence of ASC. Upon pyroptotic cell death, large specks composed mostly of polymerized ASC are released into the extracellular environment to perpetuate inflammation. A number of host and microbe-encoded proteins or peptide have been shown to modulate inflammasome function through the regulation of ligand binding or PYD–PYD interactions. Keys to the individual domains are shown on the upper right corner.
In this review, I summarize recent findings on DNA recognition, inflammasome assembly and regulation for AIM2 and IFI16, as well as the extracellular function of the inflammasomes. I also provide a perspective on future studies of the canonical and non-canonical inflammasomes.
AIM2 and IFI16 initiate immune responses to dsDNA
The AIM2 protein was first identified in a tumor suppressor screening as an interferon γ-inducible protein, whose expression pattern is correlated with reversion of tumorigenesis (44). However, AIM2 by itself was not sufficient to reverse tumor growth but may collaborate with other tumor suppressors (45). AIM2 was re-discovered in 2009 by several groups simultaneously using proteomics and bioinformatics approaches as a novel dsDNA sensor in the cytosol. AIM2 assembles an inflammasome complex with the adapter ASC and procaspase-1 to induce the maturation and secretion of proinflammatory cytokines (4–7). These studies also revealed that AIM2 initiates inflammatory responses to double stranded (ds) DNA but not single stranded (ss) DNA or RNA, consistent with its role in immune responses to vaccinia virus, mouse cytomegalovirus, human papillomavirus, Fransicella tularensis, and Listeria monocytogenes (46–52), but not Sendai virus (46). The AIM2-mediated immune response is not specific for viral or bacterial DNA: synthetic DNA such as poly (dA:dT), DNA of apparent random sequences, or host DNA could all activate the AIM2 inflammasome. The lack of sequence specificity in DNA recognition establishes that AIM2 functions as a bona fide pattern recognition receptor that can respond to threats from diverse pathogens or microbes. Nevertheless, this also suggests that it may play a role in certain autoimmune pathologies involving DNA. The HIN 200 (hematopoietic interferon-inducible nuclear proteins with a 200-amino-acid repeat) domain at the C-terminus of AIM2 was identified as the primary DNA recognition site, based on previous sequence analysis of the HIN domains that consist of two OB (oligonucleotide/oligosaccharide-binding) folds (53).
IFI16 was reported to be targeted by antinuclear antibodies in lupus patients but not in healthy individuals (54), but a mechanistic connection between the function of IFI16 and the pathogenesis of lupus remains to be established. In the cytosolic compartment, IFI16 recognizes DNA from vaccinia virus and herpes simplex virus type 1 and activates the stimulator of interferon genes (STING) pathway for inter-feron production (9). IFI16 also functions as a DNA sensor in the nucleus during infection of endothelial cells by Kaposi's sarcoma-associated herpesvirus (KSHV) (8). Upon binding to nuclear DNA, which may originate from the KSHV genome, IFI16 associates with the adapter ASC and procaspase-1 initially in the nucleus, and redistributes to the cytoplasm through unknown mechanisms. This was correlated with the induction of IL-1β during KSHV infection that is dependent on IFI16 and ASC. IFI16 mediates immune responses to not only DNA virus but also lentiviruses such as HIV-1. Several groups have demonstrated that IFI16 activates the STING pathway and inflammasome formation during HIV-1 replication (55–57). This recognition of lentivirus is dependent on the reverse-transcribed HIV-1 ssDNA that adopts stem-loop structures, likely mimetics of dsDNA (55). Importantly, in macrophages, the activation of the IFI16–STING pathway in the cytosol serves to control HIV infection, whereas in CD4+ T cells, the stimulation of the IFI16 inflammasome leads to pyroptotic cell death that may account for the majority of the CD4+ T-cell depletion, which drives the devastating pathophysiology in acquired immune deficiency syndrome (AIDS). Surprisingly, in the cytosolic compartment of T cells, IFI16 colocalizes with microbial DNA and STING but does not induce interferon production (58). Understanding the mechanisms of this cell type-specific immune response to DNA, particularly the potential blockade of the IFI16–STING pathway in CD4+ T cells will be important future areas of research.
Mechanism of dsDNA recognition by the AIM2 and IFI16 HIN domains
The mechanisms of the sequence-independent DNA recognition have been elucidated through structural and biophysical studies (25–28), which demonstrated that the AIM2 HIN domain binds the dsDNA sugar phosphate backbone using a highly positively charged concave surface spanning its OB1 fold, linker, and OB2 fold. The few hydrogen bonds observed between the HIN domain and the DNA bases in the crystals are considered ‘sequence-neutral’ interactions that often involve water molecules and therefore could be accommodated by any of the four DNA bases. The N-ter-mini of the HIN domains are located distal to the DNA-binding surface, which may facilitate the recruitment of and assembly with the adapter ASC through the N-terminal PYD domains. The non-specific binding to the DNA backbone is also permissive to minor variations of the protein–DNA interface upon tilting and sliding of the HIN domain along the dsDNA. This adaptability may facilitate the recruitment of multiple copies of the AIM2 receptor to a long stretch of dsDNA to assemble oligomeric signaling complexes with the adapters and effector enzymes.
Initial structural characterization of the IFI16 HIN domains revealed their tandem OB folds and suggested their association with tumor suppressor p53 (59), in agreement with previous reports that IFI16 promoted cellular senescence, was downregulated in various cancers and tumor cell lines (60, 61), and was required for maximal p53 activation (62). However, it was not clear how each of the IFI16 HIN domains specifically interact with different regions of p53, and how such interactions promote the tumor suppressor activities of p53. Structural studies of the IFI16 HINb domain in complex with dsDNA demonstrated its ability to non-specifically associate with DNA in a similar manner as the AIM2 HIN domain. The positively charged concave surface of the HINb domain interacts with the DNA backbone, albeit with smaller interface and lower affinity compared with the AIM2 HIN domain (25).
Rings, disks, and filaments: assembly of the inflammasome complexes
The similarity between the inflammasome and apoptosome was recognized early on, based on similar domain structures of the inflammasome receptors/sensors and the apoptosome protein Apaf-1, as well as the fact that both are signaling platforms evolved for the activation of caspases (63, 64). It was proposed that an inflammasome complex could adopt a symmetrical ring-like structure resembling the seven- or eightfold symmetric apoptosome (65–67). This was partially confirmed by electron microscopy studies of the recombinantly reconstituted NLRP1 inflammasome that displayed ring-like structures with five- or sevenfold symmetry (68). In contrast, the NAIP5–NLRC4 inflammasome purified from cells transfected with NAIP5, NLRC4, and flagellin constructs showed disk-like complexes with 11 or 12 protomers (69). Notably, the sizes of these two inflammasomes vary, with the NLRP1 inflammasome ring possessing an outer diameter of approximately 13 nm (68), and the NAIP5–NLRC4 inflammasome a diameter of approximately 28 nm (69). The exact ratio of NAIP5 versus NLRC4 in the inflammasome complex was not clear, but it was suggested that NLRC4 was overstoichiometric to NAIP5. The latter study also reported that NLRC4 forms elongated rod-like structures of approximately 25 nm in width and 30–200 nm in length, which may represent non-physiological multimers formed by the overexpressed NLRC4 that undergoes spontaneous activations.
The inflammasome adapter ASC has been shown to form single large supramolecular specks of approximately 2 lm in diameter in activated macrophages (70, 71). A lot of efforts have since been focused on understanding the protein interactions that mediate such giant assembly. Using nuclear magnetic resonance spectroscopy and modeling methods, Vajjhala et al. (72) identified three different surfaces on the PYD domain from pyrin that bind two surfaces on the ASC PYD domain. It was however not clear how one of the ASC PYD surface could engage two different surfaces at pyrin with comparable affinities, and whether trimerization of pyrin is required for the pyrin–ASC assembly.
Two recent studies using electron microscopy reported a major breakthrough on the structural characterization of large ASC clusters (73, 74). Surprisingly, the ASC PYD domain was shown to form elongated filament structures instead of rings or disks. The filament is hollow in the center with an inner and outer diameter of 2 nm and 9 nm, respectively (73). Formation of the ASC PYD filaments was inefficient by itself, but was significantly enhanced by the AIM2 PYD, the AIM2–DNA complex, and the NLRP3 PYD–NOD, consistent with the essential roles of the receptor–adapter PYD–PYD interactions during inflammasome formation. Both AIM2 and NLRP3 locate at the ends of the ASC filaments in substoichiometric amounts compared with the ASC PYD, and ATP enhanced the ASC filament formation involving NLRP3 (73). Interestingly, electron microscopy images of the NLRP3 PYD–NOD sample also showed disk-like structures of approximately 20 nm diameter as well as filaments. It is currently unclear if these structures bear resemblance to the electron microscopy images of the NLRP1 and NAIP5–NLRC4 inflammasomes. The elegant electron microscopy structure of the helical ASC PYD cluster near atomic resolution illustrates asymmetric type I, II, and III interfaces similar to previous reports of the DD complexes (75, 76), but the details of the domain–domain interactions show significant differences due to the shift in orientations of the individual domains and the structural differences between DD and PYD domains. Consistent with previous findings, electrostatic interactions play an important role among all three interfaces and high salt buffers disrupt the ASC filament formation. Analyses of the reconstituted AIM2 inflammasome and endogenous NLRP3 inflammasome suggest that the receptor PYD domains nucleate the ASC PYD polymerization, which in turn nucleates the caspase-1 filament formation through CARD–CARD interactions. In both cases, caspase-1 is overstoichiometric to ASC, which is in turn overstoichiometric to the receptor molecules, in agreement with the observation of inflammasome specks from patients that contain significantly more ASC molecules than NLRP3 (see below) (77). The PYD and CARD domain-mediated filaments may be the building blocks of the macroscopic puncta or speck structures observed in cells containing activated inflammasomes.
Indeed, macroscopic filament formation has also been observed for IFI16 in complex with dsDNA using negative stain electron microscopy (78). Such filaments have a width of 20–25 nm and require the presence of the PYD domain in the context of the full-length IFI16, as well as a cysteinespecific cross-linker to preserve the integrity of the IFI16–DNA complex. The minimal length of dsDNA is 60 bp and optimal length is 150 bp, generally consistent with previous observation of the minimal dsDNA length for optimal inter-feron production via IFI16 stimulation (9, 25). Biophysical analysis of the IFI16–dsDNA interaction revealed that the IFI16 PYD domain mediates cooperative DNA binding, and mutating the predicted type I PYD–PYD interface residues essentially eliminated such cooperativity. It was proposed that the cooperative DNA binding by the IFI16 oligomers may have evolved for nucleic acid sensing in the nucleus: it facilitates immune responses to longer pieces of microbial dsDNA that permits cooperative assembly of the IFI16 oligomers, while at the same time suppresses the association of IFI16 with abundant short self-DNA fragments that is not conducive to the formation of stable IFI16–DNA filaments required for signaling (78).
Extracellular inflammasome specks are danger signals that propagate inflammation
Inflammasomes have traditionally been viewed as macromolecular signaling platforms that reside and function in the cytosolic compartment. Two ground-breaking studies have changed this paradigm. Franklin (79) and Baroja-Mazo (77) reported that inflammasome specks or particles are present in the extracellular environment, perhaps through the release of intracellular contents during pyroptotic cell death. Remarkably, these extracellular ASC specks, which contain both NLRP3 and ASC, were shown to induce speck formation in neighboring cells, thus propagating inflammation from single cells to broader tissue environment. This novel extracellular function of the ASC specks is consistent with the presence of the extracellular specks in patients with chronic inflammatory diseases such as CAPS (cryopyrin-associated periodic syndromes), as well as the presence of autoantibodies against the ASC specks in patients with autoimmune disorders (77, 79). Importantly, NLRP3 harboring mutations identified in CAPS patients spontaneously forms specks even in the absence of the adapter ASC, demonstrating a correlation between the gain-of-function mutations in NLRP3, its ability to form specks, and clinical autoinflammatory phenotype (77, 79). The above studies not only revealed a completely new functionality of the inflammasome in the extracellular space but also opened the door for therapeutic targeting the ASC speck formation as a means to modulate inflammation for various clinical conditions (80, 81).
Regulation of AIM2 and IFI16 by their respective PYD domains
Because immune responses to nucleic acids are highly proinflammatory, it is perhaps not surprising that multiple mechanisms have evolved to regulate the activation of such immune receptors. In the absence of DNA, the AIM2 PYD was shown to associate with the AIM2 HIN domain through electrostatic and hydrophobic interactions (82). This intramolecular interaction may prevent spurious activation by retaining the receptor in an autoinhibited resting state, which can be released by dsDNA engagement. Such mechanism of autoinhibition has been observed for the NLRC4 inflammasome (83), as well as other nucleic acid-sensing receptors such as RIG-I (84). In contrast, the IFI16 PYD does not participate in autoinhibition but instead enhances DNA binding by the full-length receptor (78). The comparatively weak DNA-binding affinities of the individual IFI16 HIN domains are much enhanced in the context of the full-length IFI16 protein. Mechanistically, the PYD–PYD interactions may facilitate the oligomerization of IFI16 at long dsDNA fragments, as well as present well-defined oligomeric PYD–PYD interfaces for association with downstream signaling partners. Conceptually, the two apparently contradictory roles of the PYD domain are not necessarily mutually exclusive. In the absence of the dsDNA ligand, intramolecular domain–domain interaction may elevate the threshold of inflammasome activation to prevent spurious inflammatory responses; the presence of long dsDNA fragments may facilitate the release of the autoinhibition as well as the formation of receptor oligomers stabilized by both PYD–PYD domain interactions and multivalent DNA binding, resulting in highly cooperative signaling complex assembly.
Regulation of the AIM2 and IFI16 inflammasomes by HIN domain- and PYD domain-containing proteins
It has been reported that both IFI16 and a mouse protein p202, which encodes homologous tandem HIN domains as IFI16 but lacks the PYD domain, regulate the activation of the AIM2 inflammasome (7, 85). The p202 HINa domain has a higher affinity for dsDNA than AIM2 and IFI16, and it employs a DNA-binding surface that is located on the opposite side of the HIN domain compared with those from AIM2 and IFI16 (26, 27). In contrast, the p202 HINb domain has minimal affinity for DNA but instead forms a tetramer that directly binds the AIM2 HIN domain to prevent productive dsDNA association, which explains its specific inhibition of AIM2 (26). IFI16 was also shown to associate with AIM2 and suppress its activation of caspase-1 in THP-1 cells (85). It is not clear whether this suppressive effect was due to the modulation of the AIM2 receptor, direct competition for ligand or adapter binding, or other indirect mechanisms.
Both the AIM2 and IFI16 inflammasomes are regulated by host and microbial proteins that contain PYD domains (POPs). For example, the POP3 protein was shown to directly bind the AIM2 and IFI16 PYD domains, thus inhibiting their association with the adapter ASC and suppressing inflammasome activation induced by DNA viruses (86). By contrast, the POP1 protein directly associates with the adapter ASC to inhibit inflammasome activation (87). Similar to POP1, poxvirus-encoded PYD domain-containing proteins M013 (88) and gp013L (89) were shown to directly bind the adapter ASC to suppress the processing of IL-1 β and IL-18 as an immune evasion mechanism.
Future directions
Higher order signaling filaments are emerging as one of the general mechanisms for the assembly of signal transduction platforms such as the inflammasomes and the RLR receptor complexes (74, 90). The benefit of such highly polymerized filaments is efficient activation and propagation of signals following initial triggering event. Nonetheless, a potential issue with such higher order signaling machineries is whether they could be disassembled to revert inflammatory conditions to homeostatic states. The AIM2 and NLRP3 inflammasomes have been shown to undergo ubiquitination and be recruited to autophagosomes, thus demonstrating a mechanism to eliminate the activated inflammasomes (91). It remains to be determined if other inflammasomes can also be recycled through the autophagy pathway.
The molecular characterization of the ASC PYD filament has provided novel insights on the highly polymerized signaling platform. Future studies of additional PYD and CARD filaments will undoubtedly shed lights on how other inflammasomes are assembled. It would be of particular interest to examine whether the formation of the non-canonical inflammasomes also involves similar filament formation, and how the caspase-4/5/11 CARD may contribute to both LPS binding and oligomerization/activation of the catalytic domain. In general, more extensive structural and biophysical dissection of the inflammasome components and complexes are needed for comprehensive understanding of the molecular mechanisms of inflammasome activation and assembly.
Despite progress on the inflammasome filament formation mediated by PYD–PYD and CARD–CARD interactions, receptor oligomerization mediated by the ATP-binding NOD domains from the NLR inflammasomes remain poorly characterized. ATP was shown to moderately enhance the full-length NLRP3-mediated ASC filament formation, but ATP binding to NLRP3 alone was not sufficient to trigger the filament formation (73). The NLRP1 inflammasome was reconstituted in the presence of ATP and muramyl dipeptide (68), whereas the NAIP5–NLRC4 inflammasome was purified directly from HEK293 cells expressing NAIP5, NLRC4, and flagellin with over molar ratio of NAIP5 compared with NLRC4 and flagellin (69). The assembly of the NAIP5–NLRC4 inflammasome did not require ATP binding to NAIP5. Whether this is true for NLRC4 remains to be determined, even though the inflammasome appears to be intact in the presence of EDTA that chelates magnesium ions important for ATP binding. Future studies of the NLR inflammasomes in the presence or absence of their ligands and ATP will be essential to dissect their roles in receptor conformational change, the inflammasome complex assembly, and the activation of caspases.
Emerging evidence suggests that the activation of inflammasomes is cell type specific and often species specific. The distinct roles of IFI16 in macrophages and CD4+ T cells during immune responses to HIV-1 infection highlight the importance of investigating cell type-specific factors that modulate innate immune responses. Understanding the mechanisms of the pyroptotic T-cell death mediated by IFI16 activation will be a critical area of research for acquired immunodeficiency syndrome therapy. Furthermore, species-specific inflammasome responses to nucleic acids as exemplified by the NLRP3 inflammasome (33) cautions the use of appropriate models for studying inflammasome functions applicable to clinical pathologies.
Acknowledgements
The author would like to acknowledge support from the Case Research Institute and University Hospitals Case Medical Center.
Footnotes
The author has no conflicts of interest to declare.
References
- 1.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 2.Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park HH, Lo Y-C, Lin S-C, Wang L, Yang JK, Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandes-Alnemri T, Yu J-W, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bürckstümmer T, et al. An orthogonal proteomicgenomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 7.Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 8.Kerur N, et al. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20:319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 11.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 12.Franchi L, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1β in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 13.Miao EA, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 14.Amer A, et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem. 2006;281:35217–35223. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- 15.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wlodarska M, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045–1059. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khare S, et al. An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity. 2012;36:464–476. doi: 10.1016/j.immuni.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vladimer GI, et al. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity. 2012;37:96–107. doi: 10.1016/j.immuni.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi J, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 20.Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013;341:1250–1253. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kayagaki N, et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341:1246–1249. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- 22.Shenoy AR, et al. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 2012;336:481–485. doi: 10.1126/science.1217141. [DOI] [PubMed] [Google Scholar]
- 23.Meunier E, et al. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509:366–370. doi: 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- 24.Pilla DM, et al. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc Natl Acad Sci USA. 2014;111:6046–6051. doi: 10.1073/pnas.1321700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin T, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin Q, et al. Molecular mechanism for p202-mediated specific inhibition of AIM2 inflammasome activation. Cell Rep. 2013;4:327–339. doi: 10.1016/j.celrep.2013.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ru H, et al. Structural basis for termination of AIM2-mediated signaling by p202. Cell Res. 2013;23:855–858. doi: 10.1038/cr.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Wang J, Wang J, Cao LS, Wang ZX, Wu JW. Structural mechanism of DNA recognition by the p202 HINa domain: insights into the inhibition of Aim2-mediated inflammatory signalling. Acta Crystallogr F Struct Biol Commun. 2014;70:21–29. doi: 10.1107/S2053230X1303135X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanneganti T-D, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nat Cell Biol. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 30.Allen IC, et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimada K, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kailasan Vanaja S, et al. Bacterial RNA:DNA hybrids are activators of the NLRP3 inflammasome. Proc Natl Acad Sci USA. 2014;111:7765–7770. doi: 10.1073/pnas.1400075111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sha W, et al. Human NLRP3 inflammasome senses multiple types of bacterial RNAs. Proc Natl Acad Sci USA. 2014;111:16509–16054. doi: 10.1073/pnas.1412487111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta R, et al. RNA and b-hemolysin of group B Streptococcus induce interleukin-1β (IL-1β) by activating NLRP3 inflammasomes in mouse macrophages. J Biol Chem. 2014;289:13701–13705. doi: 10.1074/jbc.C114.548982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franchi L, et al. Cytosolic double-stranded RNA activates the NLRP3 inflammasome via MAVS-induced membrane permeabilization and K+ efflux. J Immunol. 2014;193:4114–4222. doi: 10.4049/jimmunol.1400582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitoma H, et al. The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity. 2013;39:123–135. doi: 10.1016/j.immuni.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weng L, et al. The E3 ubiquitin ligase tripartite motif 33 is essential for cytosolic RNA-induced NLRP3 inflammasome activation. J Immunol. 2014;193:3676–3682. doi: 10.4049/jimmunol.1401448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choubey D. Interferon-inducible Ifi200-family genes as modifiers of lupus susceptibility. Immunol Lett. 2012;147:10–17. doi: 10.1016/j.imlet.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uchida K, et al. Identification of specific autoantigens in Sjögren's syndrome by SEREX. Immunology. 2005;116:53–63. doi: 10.1111/j.1365-2567.2005.02197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mondini M, et al. Role of the interferon-inducible gene IFI16 in the etiopathogenesis of systemic autoimmune disorders. Ann N Y Acad Sci USA. 2007;1110:47–56. doi: 10.1196/annals.1423.006. [DOI] [PubMed] [Google Scholar]
- 41.Dombrowski Y, et al. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci Transl Med. 2011;3:82ra38. doi: 10.1126/scitranslmed.3002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lande R, et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 43.Morizane S, et al. Cathelicidin antimicrobial peptide LL-37 in psoriasis enables keratinocyte reactivity against TLR9 ligands. J Invest Dermatol. 2011;132:135–143. doi: 10.1038/jid.2011.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeYoung KL, et al. Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene. 1997;15:453–457. doi: 10.1038/sj.onc.1201206. [DOI] [PubMed] [Google Scholar]
- 45.Cresswell KS, Clarke CJP, Jackson JT, Darcy PK, Trapani JA, Johnstone RW. Biochemical and growth regulatory activities of the HIN-200 family member and putative tumor suppressor protein, AIM2. Biochem Biophys Res Commun. 2005;326:417–424. doi: 10.1016/j.bbrc.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 46.Rathinam VAK, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandes-Alnemri T, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones JW, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci USA. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sauer J-D, Witte CE, Zemansky J, Hanson B, Lauer P, Portnoy DA. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe. 2010;7:412–419. doi: 10.1016/j.chom.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim S, et al. Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur J Immunol. 2010;40:1545–1551. doi: 10.1002/eji.201040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warren SE, et al. Cutting edge: cytosolic bacterial DNA activates the inflammasome via Aim2. J Immunol. 2010;185:818–821. doi: 10.4049/jimmunol.1000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reinholz M, et al. HPV16 activates the AIM2 inflammasome in keratinocytes. Arch Dermatol Res. 2013;305:723–732. doi: 10.1007/s00403-013-1375-0. [DOI] [PubMed] [Google Scholar]
- 53.Albrecht M, Choubey D, Lengauer T. The HIN domain of IFI-200 proteins consists of two OB folds. Biochem Biophys Res Commun. 2005;327:679–687. doi: 10.1016/j.bbrc.2004.12.056. [DOI] [PubMed] [Google Scholar]
- 54.Seelig HP, Ehrfeld H, Renz M. Interferon-γ–inducible protein p16. a new target of antinuclear antibodies in patients with systemic lupus erythematosus. Arthritis Rheum. 1994;37:1672–1683. doi: 10.1002/art.1780371117. [DOI] [PubMed] [Google Scholar]
- 55.Jakobsen MR, et al. PNAS plus: from the cover: IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc Natl Acad Sci USA. 2013;110:E4571–E4580. doi: 10.1073/pnas.1311669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doitsh G, et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2013;505:509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Monroe KM, et al. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2013;343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berg RK, et al. T cells detect intracellular DNA but fail to induce type I IFN responses: implications for restriction of HIV replication. PLoS ONE. 2014;9:e84513. doi: 10.1371/journal.pone.0084513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liao JCC, et al. Interferon-inducible protein 16: insight into the interaction with tumor suppressor p53. Structure. 2011;19:418–429. doi: 10.1016/j.str.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xin H, Curry J, Johnstone RW, Nickoloff BJ, Choubey D. Role of IFI 16, a member of the interferon-inducible p200-protein family, in prostate epithelial cellular senescence. Oncogene. 2003;22:4831–4840. doi: 10.1038/sj.onc.1206754. [DOI] [PubMed] [Google Scholar]
- 61.Trapani JA, et al. A novel gene constitutively expressed in human lymphoid cells is inducible with interferon-gamma in myeloid cells. Immunogenetics. 1992;36:369–376. doi: 10.1007/BF00218044. [DOI] [PubMed] [Google Scholar]
- 62.Fujiuchi N, et al. Requirement of IFI16 for the maximal activation of p53 induced by ionizing radiation. J Biol Chem. 2004;279:20339–20344. doi: 10.1074/jbc.M400344200. [DOI] [PubMed] [Google Scholar]
- 63.Ting JPY, Willingham SB, Bergstralh DT. NLRs at the intersection of cell death and immunity. Nat Rev Immunol. 2008;8:372–379. doi: 10.1038/nri2296. [DOI] [PubMed] [Google Scholar]
- 64.Gross O, Thomas CJ, Guarda G, Tschopp J. The inflammasome: an integrated view. Immunol Rev. 2011;243:136–151. doi: 10.1111/j.1600-065X.2011.01046.x. [DOI] [PubMed] [Google Scholar]
- 65.Qi S, et al. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell. 2010;141:446–457. doi: 10.1016/j.cell.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 66.Yuan S, Yu X, Topf M, Ludtke SJ, Wang X, Akey CW. Structure of an apoptosome-procaspase-9 CARD complex. Structure. 2010;18:571–583. doi: 10.1016/j.str.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yuan S, et al. Structure of the drosophila apoptosome at 6.9 å resolution. Structure. 2011;19:128–140. doi: 10.1016/j.str.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Faustin B, et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 69.Halff EF, Diebolder CA, Versteeg M, Schouten A, Brondijk THC, Huizinga EG. Formation and structure of a NAIP5-NLRC4 inflammasome induced by direct interactions with conserved N-and C-terminal regions of flagellin. J Biol Chem. 2012;287:38460–38472. doi: 10.1074/jbc.M112.393512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fernandes-Alnemri T, et al. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Masumoto J, et al. ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J Biol Chem. 1999;274:33835–33838. doi: 10.1074/jbc.274.48.33835. [DOI] [PubMed] [Google Scholar]
- 72.Vajjhala PR, et al. Identification of multifaceted binding modes for pyrin and ASC pyrin domains gives insights into pyrin inflammasome assembly. J Biol Chem. 2014;289:23504–23519. doi: 10.1074/jbc.M114.553305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu A, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cai X, et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin S-C, Lo Y-C, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Park HH, et al. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128:533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baroja-Mazo A, et al. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nat Immunol. 2014;15:738–748. doi: 10.1038/ni.2919. [DOI] [PubMed] [Google Scholar]
- 78.Morrone SR, Wang T, Constantoulakis LM, Hooy RM, Delannoy MJ, Sohn J. Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proc Natl Acad Sci USA. 2013;111:E62–E71. doi: 10.1073/pnas.1313577111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Franklin BS, et al. The adaptor ASC has extracellular and “prionoid” activities that propagate inflammation. Nat Immunol. 2014;15:727–737. doi: 10.1038/ni.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de Rivero Vaccari JP, Lotocki G, Marcillo AE, Dietrich WD, Keane RW. A molecular platform in neurons regulates inflammation after spinal cord injury. J Neurosci. 2008;28:3404–3414. doi: 10.1523/JNEUROSCI.0157-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ibrahim E, et al. Neutralization of ASC improves sperm motility in men with spinal cord injury. Hum Reprod. 2014;29:2368–2373. doi: 10.1093/humrep/deu230. [DOI] [PubMed] [Google Scholar]
- 82.Jin T, Perry A, Smith P, Jiang J, Xiao TS. Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J Biol Chem. 2013;288:13225–13235. doi: 10.1074/jbc.M113.468033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hu Z, et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 2013;341:172–175. doi: 10.1126/science.1236381. [DOI] [PubMed] [Google Scholar]
- 84.Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 85.Veeranki S, Duan X, Panchanathan R, Liu H, Choubey D. IFI16 protein mediates the anti-inflammatory actions of the type-I interferons through suppression of activation of caspase-1 by inflammasomes. PLoS ONE. 2011;6:e27040. doi: 10.1371/journal.pone.0027040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khare S, et al. The PYRIN domain-only protein POP3 inhibits ALR inflammasomes and regulates responses to infection with DNA viruses. Nat Immunol. 2014;15:343–353. doi: 10.1038/ni.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stehlik C, Krajewska M, Welsh K, Krajewski S, Godzik A, Reed JC. The PAAD/PYRIN-only protein POP1/ASC2 is a modulator of ASC-mediated nuclear-factor-kappa B and pro-caspase-1 regulation. Biochem J. 2003;373:101–113. doi: 10.1042/BJ20030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Johnston JB, et al. A poxvirus-encoded pyrin domain protein interacts with ASC-1 to inhibit host inflammatory and apoptotic responses to infection. Immunity. 2005;23:587–598. doi: 10.1016/j.immuni.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 89.Dorfleutner A, et al. A Shope Fibroma virus PYRIN-only protein modulates the host immune response. Virus Genes. 2007;35:685–694. doi: 10.1007/s11262-007-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peisley A, Bin W, Yao H, Walz T, Hur S. RIG-I forms signaling-competent filaments in an ATP-dependent. Ubiquitin-independent manner. Mol Cell. 2013;51:1–11. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 91.Shi C-S, et al. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]