Abstract
Background
Acinetobacter baumannii is a common opportunistic pathogen that causes major nosocomial infections in hospitals. In this study, we hypothesized a high prevalence of A. baumanni ESBL (extended-spectrum beta-lactamase) among all collected isolates.
Methods
A. baumannii isolates (n = 107) from ICU (Intensive care unit) of local hospitals in Makkah were phenotypically and genotypically characterized. The identity and antibiotic susceptibility of A. baumannii strains were determined using the Vitek-2 system. The identified ESBL producers were further analyzed by PCR and sequencing followed by MLST typing. blaTEM, blaSHV, and the blaCTX-M-group genes 1, 2, 8, 9, and 25 were investigated. Furthermore, blaOXA51-like and blaOXA23-like genes were also examined in the carbapenem-resistant A. baumannii isolates.
Results
Our data indicated a high prevalence of A. baumannii ESBL producers among the collected strains. Of the 107 A. baumannii isolates, 94 % were found to be resistant to cefepime and ceftazidime, and aztreonam using the Vitek 2 system. The genes detected encoded TEM, OXA-51-like and OXA-23-like enzymes, and CTX-M-group proteins 1, 2, 8, 9, and 25. MLST typing identified eight sequence type (ST) groups. The most dominant STs were ST195 and ST557 and all of them belong to worldwide clonal complex (CC) 2.
Conclusions
This study has shown that there is a high prevalence of antimicrobial resistance in A. baumannii. The diversity of STs may suggest that new ESBL strains are constantly emerging. The molecular diversity of the ESBL genes in A. baumannii may have contributed to the increased antimicrobial resistance among all isolates.
Keywords: Acinetobacter baumannii, Phenotyping, Genotyping, Saudi Arabia
Background
Acinetobacter baumannii is an opportunistic and rapidly emerging pathogen. It is an important agent of nosocomial infections worldwide, such as urinary tract infections, septicemia, pneumonia, burns, meningitis, and wound infections in hospitals, due to its remarkable propensity to rapidly acquire resistance determinants to a wide range of antibacterial agents [1–4]. Many studies have documented high rates of multidrug-resistance (MDR) in A. baumannii [4–6]. The development of resistance to the third-generation cephalosporins was a major breakthrough in the fight against MDR strains. However, due to the frequent use of these agents, new plasmids encoding β-lactamase capable of hydrolyzing extended-spectrum cephalosporins were first reported in 1983 [7, 8]. These extended-spectrum β-lactamases (ESBLs) are mutant, plasmid-mediated, and produced by gram negative bacilli that mediate resistance to penicillin, cephalosporins, and monobactams [9]. These ESBLs are commonly recognized in Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter baumannii and are found worldwide [10]. The majority of ESBLs are members of either the TEM, SHV, or CTX-M (class A) families based on the Ambler molecular classification of β-lactamase genes [11, 12]. One of the major genes of ESBL family is the CTX-M, which is divided into five phylogenetic groups based on amino acid sequence identity: the CTX-M-1 group, the CTX-M-2 group, the CTX-M-8 group, the CTX-M-9 group, and the CTX-M-25 group. The presence and prevalence of these different groups are variable depending on the geographical locale [13, 14]. In Saudi Arabia, the high prevalence of ESBL A. baumannii was reported in several studies [15–17]. The PCR technology is widely used technique to screen for ESBL in modern hospitals. A specific multiplex PCR assay has been optimized to screen for multiple ESBL genes to facilitate and monitoring the spread and emergence of ESBL-producing bacteria [18]. The epidemiologic characterization of A. baumannii by multilocus sequence typing (MLST) is a highly used method and has been applied successfully [19]. With reports on the high prevalence of ESBL production in members of A. baumannii globally and a paucity of information specifically regarding the emergence of ESBL A. baumannii in major Saudi general hospitals in Makkah, this study reports the analysis of the antibiotic susceptibility profiles and molecular characterization of 107 A. baumannii ESBL producers isolated from ICU ward based on the phenotypic and genotypic approach. Understanding the molecular nature of the spread of A. baumannii in local hospitals is important, especially in hospitals that admit thousands of local and foreign people during their holy journey to Makkah. This work may enhance our understanding of the extent of the epidemiologic re-emergence of this bacterium. The genes that were investigated from A. baumannii isolates by PCR were blaTEM, blaSHV, and the blaCTX-M-group genes 1, 2, 8, 9, and 25. Furthermore, blaOXA51-like and blaOXA23-like enzymes were examined in carbapenem-resistant A. baumannii. This work may partially contribute to the global effort to map the molecular signature of A. baumannii.
Methods
Study design
A total of 107 bacterial isolates were collected from different ICU patients from clinical labs at local general hospitals in Makkah during 2 years from 2012 to 2014. Samples were subjected to a conventional microbiology analysis, phenotyping, and genotyping characterizations at the national center for biotechnology, KACST. The nature of the samples were blood, and skin wound infections predominantly.
Species identification and antimicrobials susceptibilities
Bacterial identities were confirmed using the Vitek 2 system (GN ID Card, bioMérieux, Craponne, France) and PCR. Antibiotic susceptibility testing was conducted according to the manufacturer’s recommendations (gram negative antimicrobial susceptibility testing (AST) cards, bioMérieux, Craponne, France). The extraction of genomic DNA was performed using QIAGEN kits (QIAamp DNA Mini Kit, cat# 69506, QIAGen, Valencia, CA, USA) according to the manufacturer’s recommendations and or the MagNA Pure LC DNA Isolation Kit III Bacteria, Fungi (Roche, Basel, Switzerland). The results of Vitek ESBL susceptibility test were reported according to the CLSI criteria. Quality-control bacterial strains (E. coli ATCC 35218 and Pseudomonas aeruginosa ATCC 27853) were used in all tests.
Detection of ESBL and carbapenem genes by PCR
All of the positive ESBL isolates according to phenotypic assays (n = 100) were further confirmed by PCR and sequencing. The genes investigated in this study were the blaTEM, blaSHV, and blaCTX-M-group genes 1, 2, 8, 9, and 25. Furthermore, blaOXA51-like and blaOXA23-like enzymes were tested for carbapenem-resistant A. baumannii. The gDNA was extracted using a QIAamp Genomic DNA kit (QIAGEN, Venlo, Netherlands) and used for PCR directly, or overnight cultures were boiled at 95 °C for 10 min to produce a bacterial gDNA/plasmid lysate that was diluted 1:10 with ddH2O before it was used for PCR. PCR amplification was performed with either 1 µl of pure gDNA or 10 µl of gDNA/plasmid lysate as a template. Final reactions of 25 μl of illustra PuReTaq Ready-To-Go PCR beads (GE Health Biosciences, USA) were used in the PCR reaction according to the manufacturer’s recommendations. The reactions were set up as follows: 10–22 μl of nuclease-free water (Promega) depending on the DNA templates being used; 2 µl of 10 pmol of each blaTEM, blaSHV, and blaCTX-M-group genes 1, 2, 8, 9, and 25; blaOXA51-like and blaOXA23-like forward and reverse primers (Eurofins MWG Operon, Germany); and 1–10 µl of DNA template or bacterial lysate were used (Table 1) [18]. The cycling conditions of the PCR are illustrated in Table 1. All of the amplicons were size fractionated using 1 % agarose gel electrophoresis and visualized under ultraviolet illumination using the Gel Doc EZ system (Bio-Rad, Hercules, CA, USA).
Table 1.
Primers for the rapid characterization of A. baumannii by multiplex PCR
| No. | blaOXA-like enzymes of A. baumannii | Amplification conditions | ||
|---|---|---|---|---|
| 1 | blaOXA-51 F | 5′-TAA TGC TTT GAT CGG CCT TG | 353 bp | Initial denaturation at 94 °C for 5 min, followed by 30 cycles of 94 °C for 25 s, 52 °C for 40 s and 72 °C for 50 s, and a final elongation at 72 °C for 6 min |
| 2 | blaOXA-51R | 5′-TGG ATT GCA CTT CAT CTT GG | ||
| 3 | blaOXA-23-F | 5′-GAT CGG ATT GGA GAA CCA GA | 501 bp | |
| 4 | blaOXA-23-R | 5′-ATT TCT GAC CGC ATT TCC AT | ||
| blaCTX-M genes | ||||
| 7 | Group 1-F | 5′-AAA AAT CAC TGC GCC AGT TC | 415 bp | Initial denaturation at 94 °C for 5 min, followed by 30 cycles of 94 °C for 25 s, 52 °C for 40 s and 72 °C for 50 s, and a final elongation at 72 °C for 6 min |
| 8 | Group 1-R | 5′-AGC TTA TTC ATC GCC ACG TT | ||
| 9 | Group 2-F | 5′- CGA CGC TAC CCC TGC TAT T | 552 bp | |
| 10 | Group 2-R | 5′-CCA GCG TCA GAT TTT TCA GG | ||
| 11 | Group 9-F | 5′-CAA AGA GAG TGC AAC GGATG | 205 bp | |
| 12 | Group 9-R | 5′-ATT GGA AAG CGT TCA TCA CC | ||
| 13 | Group 8F | 5′-TCG CGT TAA GCG GAT GAT GC | 666 bp | |
| 14 | Group 8R | 5′-AAC CCA CGA TGT GGG TAG C | ||
| 15 | Group 25F | 5′-GCA CGA TGA CAT TCG GG | 327 bp | |
| 16 | Group 25R | 5′-AAC CCA CGA TGT GGG TAG C | ||
| 1 | TEM-F | 5′-CATTTCCGTGTCGCCCTTATTC | 800 bp | Initial denaturation at 94 °C for 10 min, followed by 30 cycles at 94 °C for 40 s, 60 °C for 40 s, and 72 °C for 1 min, and a final elongation step at 72 °C for 7 min |
| 2 | TEM-R | 5′-CGTTCATCCATAGTTGCCTGAC | ||
| 3 | SHV-F | 5′-AGCCGCTTGAGCAAATTAAAC | 713 bp | |
| 4 | SHV-R | 5′-ATCCCGCAGATAAATCACCAC | ||
| 1 | 16S rRNA 8F | 5′-GCG GAT CCG CGG CCG CTG CAG AGT TTG ATC CTG GCT CAG | 797 bp | Initial denaturation at 94 °C for 5 min, followed by 35 cycles at 94 °C for 60 s, 55 °C for 30 s, and 72 °C for 60 s, and a final elongation step at 72 °C for 7 min |
| 2 | 16S rRNA 805R | 5′-GCG GAT CCG CGG CCG CGG ACT ACC AGG GTA TCT AAT | ||
Amplification and sequencing of the 16S rRNA gene
Amplification and sequencing of 16S rRNA were performed to confirm the identity of A. baumannii used in this study [20]. In the PCR amplification, each reaction contained 25 μl of illustra PuReTaq Ready-To-Go PCR beads (GE Health Biosciences, USA). The reaction was set up as follows: 22 μl of nuclease-free water (Promega), 2 µl of 10 pmol of each forward and reverse primer (Eurofins MWG Operon, Germany) were used (Table 1) [18]. Exactly 1 μl of 100 ng/µl DNA template was added to the beads. The amplification conditions are highlighted in Table 1. The amplification products were subjected to gel electrophoresis in 1 % agarose followed by ethidium bromide staining and were visualized under ultraviolet illumination using the Gel Doc EZ system (Bio-Rad, Hercules, CA, USA). Sense and anti-sense strands of PCR amplicons were purified and sequenced in an ABI 3130 Genetic Analyzer (Life Technologies, Carlsbad, CA, USA) using ABI BigDye terminator cycle sequencing ready reaction kit chemistry according to the manufacturer’s recommendations. Following sequencing, the data were identified using a basic local alignment search tool BLAST-n (http://www.ncbi.nlm.nih.gov/BLAST) or RDP database [21]. The identification of A. baumannii using the 16S rRNA was unequivocal. Therefore, there was no need to use additional confirmatory targets such as rpoB and gyrB genes [22].
Multilocus sequence typing (MLST)
The Acinetobacter baumannii complex MLST typing was performed by utilizing seven house-keeping genes: Citrate synthase (gltA), DNA gyrase subunit B (gyrB), Glucose dehydrogenase B (gdhB), Homologous recombination factor (recA), 60-kDa chaperonin (cpn60), Glucose-6-phosphate isomerase (gpi), RNA polymerase sigma factor (rpoD). The primers used for amplification and sequencing are illustrated in Table 2 [19]. The PCR amplifications were completed in a MasterCycler nexus (Eppendorf, Hamburg, Germany) with the following conditions: 35 cycles of initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, and extension at 72 °C for 2 min and 4 min final extension at 72 °C. The PCR products were directly verified by 1 % agarose gel electrophoresis before they were purified from the reaction mixture for sequencing. Bidirectional sequencing was performed for each isolate. Different allele sequences were assigned for each locus with an arbitrary allele number for identification. Each bacterial isolate was characterized by a pattern of numbers defining its sequence type (ST). The sequences of the seven housekeeping genes were analyzed by using an A. baumannii database (http://pubmlst.org/abaumannii/) [23]. The allelic profile similarities were produced by BioNumerics version (7) created by Applied Maths NV. Available from (http://www.applied-maths.com).
Table 2.
Primers used in PCR to amplify the seven housekeeping genes in A. baumannii isolates
| No. | Locus | Primer | Sequences | Amplicon size (bp) | Usage |
|---|---|---|---|---|---|
| 1 | gltA | Citrato F1 | AAT TTA CAG TGG CAC ATT AGG TCC C | 722 | Amp/seq |
| Citrato R12 | GCA GAG ATA CCA GCA GAG ATA CAC G | Amp/seq | |||
| 2 | gyrB | gyrB_F | TGA AGG CGG CTT ATC TGA GT | 594 | Amp/seq |
| gyrB_R | GCT GGG TCT TTT TCC TGA CA | Amp/seq | |||
| 3 | gdhB | GDHB 1F | GCT ACT TTT ATG CAA CAG AGC C | 774 | Amp |
| GDH SEC F | ACC ACA TGC TTT GTT ATG | Seq | |||
| GDHB 775R | GTT GAG TTG GCG TAT GTT GTG C | Amp | |||
| GDH SEC R | GTT GGC GTA TGT TGT GC | Seq | |||
| 4 | recA | RA1 | CCT GAA TCT TCY GGT AAA AC | 425 | Amp/seq |
| RA2 | GTT TCT GGG CTG CCA AAC ATT AC | Amp/seq | |||
| 5 | cpn60 | cpn60_F | GGT GCT CAA CTT GTT CGT GA | 640 | Amp/seq |
| cpn60_R | CAC CGA AAC CAG GAG CTT TA | Amp/seq | |||
| 6 | gpi | gpi_F | GAA ATT TCC GGA GCT CAC AA | 456 | Amp/seq |
| gpi_R | TCA GGA GCA ATA CCC CAC TC | Amp/seq | |||
| 7 | rpoD | rpoD-F | ACC CGT GAA GGT GAA ATC AG | 672 | Amp/seq |
| rpoD-R | TTC AGC TGG AGC TTT AGC AAT | Amp/seq |
Ethics statement
Ethical approval and consent were not required for this project because no human nor animal subjects were used.
Results
Antimicrobial susceptibility testing and screening for ESBL
In this study, 94 % (100/107) of A. baumannii were MDR. Among the 107 isolates of A. baumannii tested, one hundred isolates were confirmed as ESBL producers by phenotypic and genotypic assays, four isolates were susceptible to the third generation cephalosporins (Figs. 1, 2) and three isolates were not confirmed as A. baumannii by 16S rRNA PCR. The ESBL A. baumannii were recovered from different clinical specimens, blood, and skin wound infections predominantly. The susceptibility data of the ESBL-producing A. baumannii showed that 94 % of the 107 isolates resistant to the panel of the VITEK 2 gram negative Susceptibility Card, whereas 4 % were sensitive isolates based on CLSI criteria.
Fig. 1.
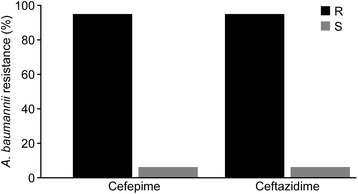
Cephalosporin susceptibility pattern by A. baumannii isolates. Among the 107 isolates of A. baumannii tested, 100 isolates (94 %) were confirmed as ESBL producers by phenotypic assay
Fig. 2.
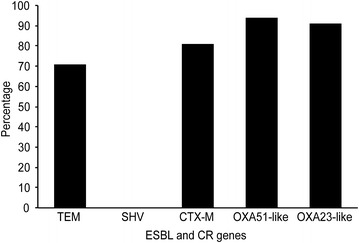
The overall distribution of ESBL and carbapenemase genes detected in A. baumannii isolates
16S rRNA identification and the detection of ESBL and carbapenemase genes
The 16S rRNA sequencing of all isolates (n = 107) generated a high score (≥97 % in total) of A. baumannii identity using the BLAST and Ribosomal Database Project (http://rdp.cme.msu.edu/) [21, 24]. To determine the extent of genotypic diversity among the MDR A. baumannii, PCR and sequencing of blaTEM, blaSHV, and the blaCTX-M-group genes 1, 2, 8, 9, and 25 and the blaOXA51-like, and blaOXA23-like genes were employed. All of the PCR-based ESBL-positive A. baumannii isolates (n = 100) were concordant with the phenotyping data. Of these isolates, seventy-one (71 %) harbored the blaTEM gene. None of them contained the blaSHV gene and eighty-one isolates (81 %) encoded blaCTX-M-group genes 1, 2, 8, 9, and 25. Finally, ninety-four (94 %) isolates carried the carbapenemase gene OXA51-like, and ninety-one isolates (91 %) contained OXA23-like (Table 3; Fig. 2). The sequencing analysis of all of the genes showed approximately 90 % sequence similarity to the submitted sequences that are related to the genes deposited in GenBank.
Table 3.
Detection and ESBL genotyping of 107 Acinetobacter baumannii clinical isolates
| PCR size | 501 bp | 353 bp | 713 bp | 800 bp | 327 bp | 205 bp | 666 bp | 552 bp | 415 bp |
|---|---|---|---|---|---|---|---|---|---|
| Gene | CTX-M1 | CTX-M2 | CTX-M8 | CTX-M9 | CTX-M25 | TEM | SHV | OXA- 51-like | OXA-23-like |
| Positive isolates of 107 isolates | 9 | 73 | 72 | 10 | 61 | 73 | 0 | 100 | 97 |
| Percentage (%) | 81 | 71 | 0 | 94 | 91 | ||||
Multilocus sequence typing analysis
MLST and sequence-based typing of ESBL and carbapenemase isolates were performed to analyze the genetic relationship of all of the isolates. The MLST anaylsis contains 97 isolates of 102. Five isolates were not typable due to low quality traces files and were not assigned STs but were included in the dendrogram. The MLST analysis allowed us to group the A. baumannii isolates into eight STs (Figs. 3, 4). MLST typing showed that the most dominant sequence type was ST195 (n = 69), followed by ST557 (n = 6), ST 208 (n = 4), ST499 (n = 2), ST218 (n = 2), ST231 (n = 1), ST222 (n = 1), and ST286 (n = 2). All of STs except ST 231 belonge to clonal complexity 2 (CC2) and lineage clone 2. The tree (Fig. 3) is based on the nucleotide sequence of at least 6 or 7 housekeeping genes. The analysis was based on data sets that include all STs in the Pasteur MLST databases of A. baumannii (http://pubmlst.org/abaumannii/).
Fig. 3.
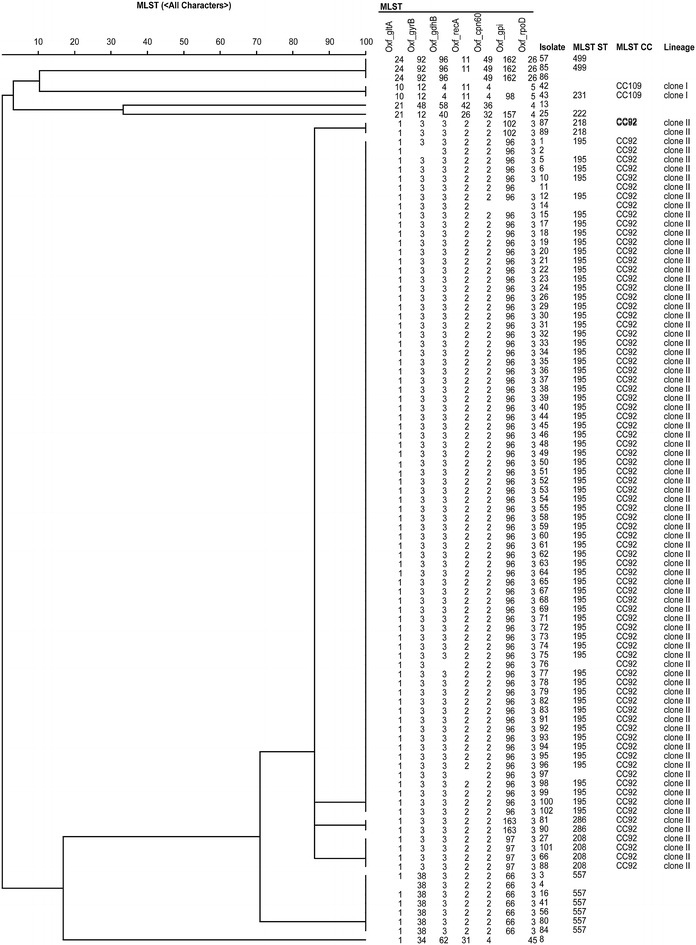
UPGMA (unweighted pair group method with arithmetic mean) dendrogram based on the catagorical coefficient applied to the allele IDs. All isolates with at least six loci amplified were included. The dendrogram was generated by BioNumerics 7 software. The ST numbers assigned for each isolate were generated by the Pasteur MLST scheme (http://pubmlst.org/abaumannii/). The tree is a rooted based on the nucleotide sequence of the six and seven housekeeping genes. The analysis was based on data sets that include all STs in the Pasteur MLST databases. The first clade consists of ST195, 208, 218 and 286; the second clade of ST231; the third clade of ST499; the fourth clade of ST 557; the fifth clade of ST222. The sixth and seventh clades have two nontypeable isolates due to low quality sequencing trace files
Fig. 4.
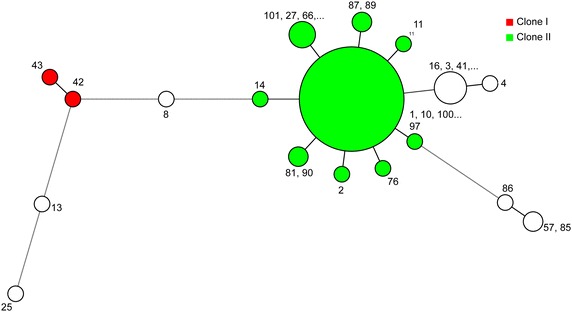
Minimum spanning tree constructed based on the allele IDs
Discussion
In this study, we detected and characterized the phenotypic and genotypic nature of ESBL producers in A. baumannii, which were isolated from general hospitals in Makkah, Saudi Arabia. At least 107 A. baumannii isolates were characterized by the Vitek-2 system and PCR-sequencing followed by MLST typing. Our data indicated a high prevalence of A. baumannii ESBL producers among the collected isolates. A remarkable outcome of this study was the large number of antibiotic resistance genes found in these isolates. Ninety-four percent of A. baumannii isolates were found to have three major resistant determinants. We speculate that if more drug-resistant genes were screened, we would have found pan-resistant A. baumannii isolates.
CTX-M β-lactamases produced by A.baumannii strains is plasmid-mediated hence the wide spread and long time survival in hospitals. The CTX-M gene activity conferring resistance to cefotaxime and ceftazidime. We detected CTX-M group 1, 2, 8, 9, 25 in our current study (81 %). The high rate of prevalence of CTX-M resistance in gram negative bacteria may be influenced by mobile genetic elements around these genes which include transposon, insertion sequences (IS) and integrons [25]. Consistent with our study, all gram negative CTX-M producing bacteria are often associated with other families of other β-lactamases resistance causing multi-drug resistance phenomena. The high prevalence rate around the world of CTX-M makes it a predominant drug resistant gene in gram negative bacteria [26].
We studied the dynamic spread of A.baumannii in our population by MLST. The discriminatory power of the MLST system is comparable to other techniques such as pulsed field gel electrophoresis (PFGE). Yet, MLST provides a quick and easy method to study the epidemiology of ESBL-producing bacteria and to monitor the international emergence of multidrug resistant bacteria. Consistent with other studies that used MLST in the epidemiologic characterization of clinically important bacterial pathogens such as A.baumannii, Streptococcus pneumoniae, Streptococcus pyogenes, Neisseria meningitidis, Campylobacter jejuni, Staphylococcus aureus, Enterococcus faecium, Haemophilus influenza, and Vibriocholera, our study has detected different allelic diversity (STs) which belongs to clonal complex (CC)2 which is globally distributed in Europe, Asia, Africa, Australia, USA, South America [19, 27–29].
The drug of choice to treat nosocomial infection caused by A.baumannii is the carbapenems. However, there is an increasing rate of carbapenem-resistant A.baumannii around the world caused by OXA23-like enzume or OXA51-like enzyme acitivies [30]. The first OXA23-like enzyme with carbapenem-activity to A. baumannii was isolated and characterized in Scotland in 1985. This drug-resistant determinant is encoded by the plasmid therefore it is transferable [31]. This may explain the high prevalence of carbapenemase-producing A. baumannii in hospitals around the world. The other gene cluster in the OXA family is the blaOXA-51-like gene which is chromosomally encoded and naturally occurs in A. baumannii. The functional product of this gene delivers carbapenemase resistance to meropenem and imipenem; its role in carbapenem resistance may be influenced by the presence of ISAba1. PCR mapping studies have found that the absence of this sequence upstream of blaOXA-51-like gene may contribute to a minimal effect on carbapenem susceptibility [32–36].
A recent study in the Gulf Countries Council (GCC) [37], namely, Saudi Arabia, the United Arab Emirates, Oman, Qatar, Bahrain, and Kuwait, suggested a high prevalence of carbapenemase resistance in A. baumannii, Escherichia coli and Klebsiella pneumonia.A. baumannii (n = 117) was studied as clusters in seven different sequence types: ST195, ST208, ST229, ST436, ST450, ST452 and ST499. Three of these sequences were identified in our study, including ST195, ST499, and ST208, which may suggest the circulation of these three STs in GCC countries [17, 37]. The circulation of the STs within GCC may be due to the closeness of these countries to each other. Recent reports have been accumulating from Saudi Arabia due to the wide and rapid spread of carbapenem-resistant gram negative bacteria isolated from local hospitals specially during high season [38–40].
The high level of detection of ESBL and carbapenemase resistance among local isolates may suggest an increasing incidence rate of infection with ESBL-producing A. baumannii. Such high rates of ESBL-producing bacteria may impose a burden on routine clinical practice, especially for patients with chronic diseases and immunocompromised patients. Although national surveillance data are lacking, outbreaks of infection due to ESBL-producing A. baumannii have been reported by many hospitals within the Kingdom of Saudi Arabia. The true prevalence of ESBL producers is not known and is likely underestimated because of the difficulties encountered in their detection by most local hospitals. However, it is clear that ESBL-producing bacteria are distributed worldwide and their prevalence is increasing [2, 6, 41, 42]. Therefore, periodic screening of ESBL-producing A. baumannii during the high hospital visitation season is recommended in all local hospitals to establish national surveillance data archives of the level of spread of ESBL producers.
Conclusions
In this study, we randomly surveyed and characterized ESBL-producing A. baumannii from ICU of local hospitals in Makkah city, Saudi Arabia. Our data indicated a high prevalence of A. baumannii ESBL producers among the collected isolates. Based on MLST typing, we have evidence of eight STs groups in our isolates. The epidemiologic diversity of these isolates may suggest that new ESBL strains are constantly emerging. The molecular diversity of the ESBL genes in A. baumannii may have contributed to the increased antimicrobial resistance among all isolates. Therefore, periodic screening of ESBL-producing A. baumannii during the high hospital visitation season is recommended in all local hospitals.
Authors’ contributions
EA and MK contributed to study design. RB, FB, BA, MA collected the samples and all contributed equally to the lab experiments. All authors contributed to data interpretation. EA and MK drafted and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This study was supported by internal grant from the National Center for Biotechnology at King Abdulaziz City for Science and Technology in Riyadh.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Contributor Information
Essam J. Alyamani, Phone: +966 1 481 3806, Email: eyamani@kacst.edu.sa
Mohamed A. Khiyami, Email: mkhiyami@kacst.edu.sa
Rayan Y. Booq, Email: rbooq@kacst.edu.sa
Basel M. Alnafjan, Email: bnafjan@kacst.edu.sa
Musaad A. Altammami, Email: mtammami@kacst.edu.sa
Fayez S. Bahwerth, Email: Fayez-Bahwerth@hotmail.com
References
- 1.Eliopoulos GM, Maragakis LL, Perl TM. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin Infect Dis. 2008;46(8):1254–1263. doi: 10.1086/529198. [DOI] [PubMed] [Google Scholar]
- 2.Antunes L, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis. 2014;71(3):292–301. doi: 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- 3.Srinivasan VB, Rajamohan G, Pancholi P, Stevenson K, Tadesse D, Patchanee P, Marcon M, Gebreyes WA. Genetic relatedness and molecular characterization of multidrug resistant Acinetobacter baumannii isolated in central Ohio, USA. Ann Clin Microbiol Antimicrob. 2009;8(1):21. doi: 10.1186/1476-0711-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergogne-Berezin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9(2):148–165. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21(3):538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51(10):3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knothe H, Shah PDP, Krcmery V, Antal M, Mitsuhashi S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection. 1983;11(6):315–317. doi: 10.1007/BF01641355. [DOI] [PubMed] [Google Scholar]
- 8.Philippon A, Labia R, Jacoby G. Extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1989;33(8):1131. doi: 10.1128/AAC.33.8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rawat D, Nair D. Extended-spectrum β-lactamases in Gram negative bacteria. J Glob Infect Dis. 2010;2(3):263. doi: 10.4103/0974-777X.68531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fluit AC, Visser MR, Schmitz FJ. Molecular detection of antimicrobial resistance. Clin Microb Rev. 2001;14(4):836–871. doi: 10.1128/CMR.14.4.836-871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrob Agents Chemother. 2010;54(3):969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naas T, Oxacelay C, Nordmann P. Identification of CTX-M-type extended-spectrum-β-lactamase genes using real-tme PCR and pyrosequencing. Antimicrob Agents Chemother. 2007;51(1):223–230. doi: 10.1128/AAC.00611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naas T, Cuzon G, Bogaerts P, Glupczynski Y, Nordmann P. Evaluation of a DNA microarray (Check-MDR CT102) for rapid detection of TEM, SHV, and CTX-M extended-spectrum beta-lactamases and of KPC, OXA-48, VIM, IMP, and NDM-1 carbapenemases. J Clin Microbiol. 2011;49(4):1608–1613. doi: 10.1128/JCM.02607-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al Johani SM, Akhter J, Balkhy H, El-Saed A, Younan M, Memish Z. Prevalence of antimicrobial resistance among gram-negative isolates in an adult intensive care unit at a tertiary care center in Saudi Arabia. Ann Saudi Med. 2010;30(5):364. doi: 10.4103/0256-4947.67073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Agamy MH, Shibl AM, Ali MS, Khubnani H, Radwan HH, Livermore DM. Distribution of β-lactamases in carbapenem-non-susceptible Acinetobacter baumannii in Riyadh, Saudi Arabia. J Glob Antimicrob Resist. 2014;2(1):17–21. doi: 10.1016/j.jgar.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Zowawi HM, Sartor AL, Sidjabat HE, Balkhy HH, Walsh TR, Al Johani SM, AlJindan RY, Alfaresi M, Ibrahim E, Al-Jardani A, et al. Molecular epidemiology of carbapenem resistant Acinetobacter baumannii in the Gulf Cooperation Council States. Dominance of OXA-23-type producers. J Clin Microb. 2015;53:896–903. doi: 10.1128/JCM.02784-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(3):490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 19.Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodriguez-Valera F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005;43(9):4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosshard PP, Abels S, Altwegg M, Böttger EC, Zbinden R. Comparison of conventional and molecular methods for identification of aerobic catalase-negative gram-positive cocci in the clinical laboratory. J Clin Microbiol. 2004;42(5):2065–2073. doi: 10.1128/JCM.42.5.2065-2073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maidak BL, Olsen GJ, Larsen N, Overbeek R, McCaughey MJ, Woese CR. The Ribosomal database project (RDP) Nucleic Acids Res. 1996;24(1):82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee MJ, Jang SJ, Li XM, Park G, Kook J-K, Kim MJ, Chang Y-H, Shin JH, Kim SH, Kim D-M, et al. Comparison of rpoB gene sequencing, 16S rRNA gene sequencing, gyrB multiplex PCR, and the VITEK2 system for identification of Acinetobacter clinical isolates. Diagn Microbiol Infect Dis. 2014;78(1):29–34. doi: 10.1016/j.diagmicrobio.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Jolley K, Maiden M. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010;11(1):595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsen N, Olsen GJ, Maidak BL, McCaughey MJ, Overbeek R, Macke TJ, Marsh TL, Woese CR. The ribosomal database project. Nucleic Acids Res. 1993;21(13):3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lahlaoui H, Ben Haj Khalifa A, Ben Moussa M. Epidemiology of Enterobacteriaceae producing CTX-M type extended spectrum beta-lactamase (ESBL) Med Mal Infect. 2014;44(9):400–404. doi: 10.1016/j.medmal.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Canton R, Novais A, Valverde A, Machado E, Peixe L, Baquero F, Coque TM. Prevalence and spread of extended-spectrum beta-lactamase-producing Enterobacteriaceae in Europe. Clin Microbiol Infect. 2008;14(Suppl 1):144–153. doi: 10.1111/j.1469-0691.2007.01850.x. [DOI] [PubMed] [Google Scholar]
- 27.Huber CA, Sartor AL, McOdimba F, Shah R, Shivachi P, Sidjabat HE, Revathi G, Paterson DL. Outbreaks of multidrug-resistant Acinetobacter baumannii strains in a Kenyan teaching hospital. J Glob Antimicrob Resist. 2014;2(3):190–193. doi: 10.1016/j.jgar.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 28.Jeannot K, Diancourt L, Vaux S, Thouverez M, Ribeiro A, Coignard B, Courvalin P, Brisse S. Molecular epidemiology of carbapenem non-susceptible Acinetobacter baumannii in France. PLoS One. 2014;9(12):e115452. doi: 10.1371/journal.pone.0115452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pournaras S, Gogou V, Giannouli M, Dimitroulia E, Dafopoulou K, Tsakris A, Zarrilli R. Single-locus-sequence-based typing of blaOXA-51-like genes for rapid assignment of acinetobacter baumannii clinical isolates to international clonal lineages. J Clin Microbiol. 2014;52(5):1653–1657. doi: 10.1128/JCM.03565-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans BA, Hamouda A, Amyes SG. The rise of carbapenem-resistant Acinetobacter baumannii. Curr Pharm Des. 2013;19(2):223–238. doi: 10.2174/138161213804070285. [DOI] [PubMed] [Google Scholar]
- 31.Paton R, Miles RS, Hood J, Amyes SG, Miles RS, Amyes SG. ARI 1: beta-lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int J Antimicrob Agents. 1993;2(2):81–87. doi: 10.1016/0924-8579(93)90045-7. [DOI] [PubMed] [Google Scholar]
- 32.Brown S, Young HK, Amyes SG. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2005;11(1):15–23. doi: 10.1111/j.1469-0691.2004.01016.x. [DOI] [PubMed] [Google Scholar]
- 33.Coelho JM, Turton JF, Kaufmann ME, Glover J, Woodford N, Warner M, Palepou M-F, Pike R, Pitt TL, Patel BC. Occurrence of carbapenem-resistant Acinetobacter baumannii clones at multiple hospitals in London and Southeast England. J Clin Microbiol. 2006;44(10):3623–3627. doi: 10.1128/JCM.00699-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walther-Rasmussen J, Høiby N. OXA-type carbapenemases. J Antimicrob Chemother. 2006;57(3):373–383. doi: 10.1093/jac/dki482. [DOI] [PubMed] [Google Scholar]
- 35.Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, Pitt TL. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett. 2006;258(1):72–77. doi: 10.1111/j.1574-6968.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- 36.Karah N, Sundsfjord A, Towner K, Samuelsen O. Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist Updates Rev Comment Antimicrob Anticancer Chemother. 2012;15(4):237–247. doi: 10.1016/j.drup.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 37.Zowawi HM, Sartor AL, Balkhy HH, Walsh TR, Al Johani SM, AlJindan RY, Alfaresi M, Ibrahim E, Al-Jardani A, Al-Abri S, et al. Molecular characterization of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in the countries of the Gulf cooperation council: dominance of OXA-48 and NDM producers. Antimicrob Agents Chemother. 2014;58(6):3085–3090. doi: 10.1128/AAC.02050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Memish ZA, Assiri A, Almasri M, Roshdy H, Hathout H, Kaase M, Gatermann SG, Yezli S. Molecular characterization of carbapenemase production among gram-negative bacteria in Saudi Arabia. Microb Drug Resist. 2015;21(3):307–314. doi: 10.1089/mdr.2014.0121. [DOI] [PubMed] [Google Scholar]
- 39.Memish ZA. Infection control in Saudi Arabia: meeting the challenge. Am J Infect Control. 2002;30(1):57–65. doi: 10.1067/mic.2002.120905. [DOI] [PubMed] [Google Scholar]
- 40.Aly M, Tayeb HT, Al Johani SM, Alyamani EJ, Aldughaishem F, Alabdulkarim I, Balkhy HH. Genetic diversity of OXA-51-like genes among multidrug-resistant Acinetobacter baumannii in Riyadh, Saudi Arabia. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol. 2014;33(7):1223–1228. doi: 10.1007/s10096-014-2068-0. [DOI] [PubMed] [Google Scholar]
- 41.Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol. 2007;5(12):939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 42.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One. 2010;5(4):e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]


