Abstract
Haloperidol is an effective antipsychotic drug for treatment of schizophrenia, but prolonged use can lead to debilitating side effects. To better understand the effects of long-term administration, we measured global metabolic changes in mouse brain following 3 mg/kg/day haloperidol for 28 days. These conditions lead to movement-related side effects in mice akin to those observed in patients after prolonged use. Brain tissue was collected following microwave tissue fixation to arrest metabolism and extracted metabolites were assessed using both liquid and gas chromatography mass spectrometry (MS). Over 300 unique compounds were identified across MS platforms. Haloperidol was found to be present in all test samples and not in controls, indicating experimental validity. Twenty-one compounds differed significantly between test and control groups at the p < 0.05 level. Top compounds were robust to analytical method, also being identified via partial least squares discriminant analysis. Four compounds (sphinganine, N-acetylornithine, leucine and adenosine diphosphate) survived correction for multiple testing in a non-parametric analysis using false discovery rate threshold < 0.1. Pathway analysis of nominally significant compounds (p < 0.05) revealed significant findings for sphingolipid metabolism (p = 0.02) and protein biosynthesis (p = 0.03). Altered sphingolipid metabolism is suggestive of disruptions to myelin. This interpretation is supported by our observation of elevated N-acetylaspartylglutamate in the haloperidol-treated mice (p = 0.004), a marker previously associated with demyelination. This study further demonstrates the utility of murine neurochemical metabolomics as a method to advance understanding of CNS drug effects.
Keywords: metabolomics, antipsychotics, myelin, sphingolipids, extrapyramidal symptoms
Introduction
Antipsychotic drugs are central to the treatment of schizophrenia, a serious and debilitating mental disorder characterized by delusions, hallucinations, social withdrawal and cognitive deficits (van Os and Kapur, 2009). Haloperidol was among the first antipsychotics and was widely hailed as a breakthrough upon its introduction in the 1950s (Lopez-Munoz and Alamo, 2009). Since then, haloperidol has been used extensively in psychiatry and its effectiveness is well established (Adams et al., 2013). However, it is now realized that haloperidol can lead to severe, debilitating and sometimes irreversible side effects (Rosebush and Mazurek, 1999; Gao et al., 2008).
Among the side effects of haloperidol, the risk for extrapyramidal symptoms (EPS) is perhaps the most salient. In a meta-analysis of antipsychotic clinical trials, haloperidol had the highest risk of EPS among all 15 antipsychotic drugs studied (Leucht et al., 2013). EPS are a complex and potentially heterogeneous set of symptoms, characterized by involuntary movement and loss of motor control. These include parkinsonism, akathisia and tardive dyskinesia (TD), which can sometimes persist long after drug treatment has ended (Marsden and Jenner, 1980). While haloperidol has a high risk for EPS, more modern antipsychotic drugs are not without risk for these side effects (Peluso et al., 2012). EPS therefore represent an ongoing issue with antipsychotic drug treatment. Understanding the biological mechanisms that lead to haloperidol-induced EPS could conceivably guide future drug design. If we can identify the pathways leading to EPS, new candidate compounds could be evaluated for their toxicity relative to activation of these pathways.
Research into the biology of EPS has traditionally focused on dopaminergic neurotransmission (Meltzer and Stahl, 1976). Specifically, antipsychotics are hypothesized to produce EPS due to blockade of dopamine D2 receptors in the striatum, which acts as an interface between higher brain function in the cortex and motor control in the basal ganglia (Meltzer and Stahl, 1976; Glazer, 2000). While dopamine receptor blockade plays an established role in antipsychotic action (Meltzer, 2013), it cannot explain some critical features of EPS, such as the persistence of symptoms after drug treatment has ended. TD, for example, develops in approximately one third of patients undergoing long-term treatment with "typical" first generation antipsychotics, including haloperidol, and may be irreversible in up to half of such cases (Soares-Weiser and Fernandez, 2007). The biological basis of TD is essentially unknown (Crowley et al., 2012a), suggesting that new directions in EPS research may be warranted. In this respect, the use of massively-parallel, discovery-oriented "omics" technologies could be fruitful. One such potentially powerful tool for understanding the global biological consequences of drug administration is metabolomics (Clayton et al., 2006; Kaddurah-Daouk et al., 2008; Patti et al., 2012).
When using metabolomic approaches to understand the consequences of CNS drugs, researchers have generally either utilized accessible human body fluids such as blood, or have obtained tissue from laboratory model organisms, in particular rodents. To the best of our knowledge, only one previous study has used metabolomics methods to examine the biochemical effects of antipsychotics in the rodent brain. Specifically, McLoughlin et al. (2009) examined the effects of several different classes of psychotropic drugs, including haloperidol, in rat brain using NMR. However, drawbacks of their study included the relative insensitivity of NMR relative to MS analysis and the relatively small number of compounds reported (n=15).
We have previously employed methods to assay the neurochemical metabolic effects of CNS drugs in mice (McClay et al., 2013), measuring over 300 unique compounds using a mix of MS methods, and considered that application of these methods to haloperidol could shed light on biochemical mechanisms. A particular advantage of our strategy is the use of focused beam microwave irradiation (FBMI) to instantaneously and permanently denature all enzymes in the brain at time of sacrifice (de Graaf et al., 2009). In a high-intensity microwave pulse lasting only milliseconds, brain metabolism is arrested completely and metabolites are preserved in the pre-sacrifice state (Ikarashi et al., 1985; Login and Dvorak, 1994). Considering EPS specifically, an established rodent model of TD has been developed to study the phenotype (Turrone et al., 2002). In this model, chronic administration of antipsychotics, including haloperidol, leads to vacuous chewing movements similar to those observed in patients with TD (Crowley et al., 2012a). Therefore, in the present study, we report untargeted MS-based metabolomic analysis of murine brain tissue following exposure to long-term haloperidol administration at doses previously demonstrated to produce behavioral symptoms that model EPS in patients.
Methods
Subjects
Eight week old male C57BL/6J mice were obtained from the Jackson Laboratory (Bar Harbor, ME). Mice were allowed to acclimate to the vivarium (AAALAC-accredited) for 1 week prior to testing and were housed up to four per cage. Food (7012 Teklad LM-485 Sterilizable Diet, Harlan Laboratories, Indianapolis, Indiana) and water were available ad libitum under a 12-h/12-h light/dark cycle (lights on at 0700–1,900 h) with all testing occurring during the light phase. All procedures were approved by the Institutional Animal Care and Use Committee of Virginia Commonwealth University.
Drug administration
Our aim was to achieve steady-state concentrations of haloperidol that mimic clinical doses in patients (10–50 nM or 3.75–19 ng/ml) (Hsin-tung and Simpson, 2000). Previous work (Crowley et al., 2012a; Crowley et al., 2012b) indicated that 3.0 mg/kg/day haloperidol delivered via continuous 60-day slow release subcutaneous pellets (Innovative Research of America, Sarasota, FL) yielded plasma haloperidol concentrations in mice in the 10– 50 nM range with lower variation as compared to minipumps, repeated injections or oral administration. Furthermore, this dose and delivery method has been previously demonstrated to produce vacuous chewing movements in C57BL/6J mice that peaked around four weeks and persisted for over one year, long after drug administration ended (Crowley et al., 2012a). Therefore, for our metabolomics experiments, eleven animals received haloperidol pellets delivering 3.0 mg/kg/day, while ten control animals received placebo pellets containing the same matrix materials but no drug. Pellets were implanted subcutaneously, centrally above the scapulae, under isoflurane anesthesia and the incision sealed with VetBond (3M, St. Paul, MN). After recovery, animals were returned to the standard housing conditions outlined above.
Sample preparation
After 28 days, animals were sacrificed via focused beam microwave irradiation (FBMI), using a 10 kW microwave tissue fixation system (Muromachi TMW-4012C, Tokyo, Japan). Brain tissue was collected by dissection immediately after fixation, followed by snap freezing in liquid nitrogen. Samples were stored at -80 °C prior to overnight shipment on dry ice to the metabolomics facility (Metabolon, Research Triangle Park, NC). Brain tissue was homogenized using a GenoGrinder (OPS Diagnostics, Lebanon, NJ) and extracted into two fractions using proprietary solvents (Metabolon, Research Triangle Park, NC). The first fraction was for analysis via liquid chromatography mass spectrometry (LC/MS), while the second was for analysis via gas chromatography mass spectrometry (GC/MS). A TurboVap (Zymark, Hopkinton, MA) was used to quickly remove organic solvent before samples were frozen and vacuum dried in preparation for loading. Aliquots from each experimental sample were pooled and these “matrix” samples were injected throughout the platform day run and served as technical replicates. In this manner, variability in quantitation of the experimental samples was monitored.
Liquid chromatography mass spectometry (LC/MS, LC/MS2): The LC/MS component of our metabolomics approach was previously described in detail by (Evans et al., 2009). Briefly, the LC/MS platform used a Waters (Milford, MA) Acquity ultra performance liquid chromatography (UPLC) and a Thermo-Finnigan (Thermo Fisher, Waltham, MA) LTQ mass spectrometer, consisting of an electrospray ionization (ESI) source and linear ion-trap (LIT) mass analyzer. The sample extract was split into two aliquots, dried, then reconstituted in acidic or basic LC-compatible solvents, each of which contain eleven injection standards at fixed concentrations. One aliquot was analyzed using acidic positive ion optimized conditions and the other using basic negative ion optimized conditions in two independent injections using separate dedicated columns. Extracts reconstituted in acidic conditions were gradient eluted using water and methanol, both containing 0.1% formic acid, while the basic extracts, which also use water/methanol, contained 6.5 mM ammonium bicarbonate. The MS analysis was alternated between MS and data-dependent MS2 scans using dynamic exclusion (McClay et al., 2013).
The LC/MS mass accurate portion of the platform used a Surveyor high performance liquid chromatography (HPLC) and a Thermo-Finnigan LTQ-FT mass spectrometer, with a linear ion-trap front end and a Fourier transform ion cyclotron resonance mass spectrometer backend. For ion counts more than 2 million, an accurate mass measurement was performed. These were made on the parent ion as well as fragments and the typical mass error was under 5 ppm. Ions with counts under two million required more effort to characterize. Fragmentation spectra (MS/MS) were typically generated in data dependent manner, but where necessary, targeted MS/MS was employed, such as in the case of lower level signals (McClay et al., 2013).
Gas chromatography / Mass Spectroscopy (GC/MS)
A previous implementation of the GC/MS platform was described by (Ohta et al., 2009). Samples undergoing GC/MS analysis were redried under vacuum for at least 24 hours prior to being derivatized under dried nitrogen using bistrimethyl-silyl-triflouroacetamide (BSTFA). The GC column was 5% phenyl and the temperature ramp was 40° to 300° C in 16 minutes. Samples were analyzed on a Thermo-Finnigan Trace DSQ fast-scanning single-quadrupole mass spectrometer using electron impact ionization.
Data pre-processing
Proprietary in-house software (Metabolon, Research Triangle Park, NC) was used to perform detection and integration of MS peaks as described previously (Evans et al., 2009). Briefly, extracted ion chromatograms were binned by mass, baseline noise was determined, peaks areas were calculated, and peak thresholds including minimum height, signal-to-noise, width, symmetry, and area were applied. MS peaks passing the above threshold criteria were grouped based on peak apex retention times. Deconvolution and identification of correlated ion features was carried out as described previously (Dehaven et al., 2010), with correlated ion features from multiple platforms belonging to a given biochemical organized into a single ion group. This ion group is then used to represent that metabolite in statistical analyses. Specific biochemicals were identified by comparison to library entries of purified standards (approximately 1500 for the GC and LC platforms combined). Haloperidol was added to the compound library so that it could be identified for validation purposes.
Following deconvolution and library matching, each identified compound was represented by a single, uncorrected quantitative variable. The following steps were taken to process the data prior to statistical analysis: 1) metabolites with >50% missing values were dropped, 2) for the remaining metabolites, missing values were imputed with half of the minimum positive value of each variable in the original data under the assumption that they were below the limits of detection, 3) metabolites with low variance, specifically the 10% with the lowest standard deviations, were dropped from further analysis, and 4) data normalization was carried out via autoscaling, whereby each variable was mean-centered and divided by the standard deviation.
Statistical Analysis
All analysis was carried out using the MetaboAnalyst package, version 2.0 (Xia and Wishart, 2011; Xia et al., 2012). After data quality control and normalization as described above, we first tested for significant differences in the levels of each compound between the haloperidol and control groups using Student's t-test. We then used Statistical Analysis of Microarrays (SAM) (Tusher et al., 2001), a non-parametric method implemented for metabolomics in MetaboAnalyst, to calculate significance and correct for multiple testing using a false discovery rate (FDR) threshold of 0.1 (van den Oord and Sullivan, 2003). We also implemented multivariate analysis in the form of partial least squares discriminant analysis (PLS-DA), a supervised discriminatory method, to inspect the differences between the two experimental groups. Validity of the PLS-DA model was assessed using cross-validation methods, as recommended by (Bijlsma et al., 2006). Finally, overrepresentation analysis (ORA) was used to test the set of nominally significant metabolites (p < 0.05) from our univariate analysis for enrichment within known biological pathways. A hypergeometric test was used to evaluate if our metabolite set was represented more than expected by chance within canonical biological pathways.
Results
Animals receiving haloperidol tolerated the drug well and all implants remained intact at 28 days. Metabolomic analysis of brain tissue detected 302 unique compounds across both MS platforms. Of these, 201 were identified with confidence while the rest were consistently detected entities that could not be identified because no matching entries were present in our compound library. However, these unidentified compounds were included in our analyses under the assumption that any showing significant associations with haloperidol exposure could be followed-up in subsequent experiments, if necessary. As a validation measure, haloperidol was added to our library and investigated in our metabolomic analysis. We observed detectable levels of haloperidol in all test samples and in no control samples. This confirmed experimental validity as it shows the drug was present in the tissue of interest (brain) at the time of sampling. Previous work (Crowley et al., 2012a) showed that the dose and delivery method used (3 mg/kg/day) led to blood concentrations comparable to clinical doses in humans.
Our initial analysis aimed to detect the compounds that differed the most between the haloperidol-treated animals and controls. After data quality control and normalization, we performed t-tests for all compounds and identified 21 significant findings at p < 0.05 (Table 1). Of these 12 were up-regulated and 9 were down-regulated in the haloperidol-treated animals relative to controls. The most significant up-regulated compounds were N-acetyl-aspartyl-glutamate (NAAG), a common peptide neurotransmitter compound in the mammalian brain, and 2-stearoyl glycerophosphocholine, a lysophosphatidylcholine and component of biological membranes. The most significant down-regulated compounds were leucine, a common amino acid, and sphinganine, an important sphingolipid metabolite and intermediate in the synthesis of ceramide. Several energy-related compounds, such as ADP and glucose, were also altered in the haloperidol-treated animals.
Table 1.
All nominally significant (p < 0.05) metabolite findings in a comparison of brain tissue from animals undergoing chronic haloperidol treatment versus controls. Compounds names "Y-…" indicate recurrent, unique compounds that could not be assigned an identity because no corresponding entry existed in our library. ChEBI is the compound ID from the Chemical Entities of Biological Interest database from the European Bioinformatics Institute (Hastings et al., 2013). P-values are from t-tests conducted using the "MetaboAnalyst" software package. Effect direction is given for the haloperidol-treated animals versus controls.
| Compound | ChEBI ID | p-value | Haloperidol effect direction |
|---|---|---|---|
| leucine | 15603 | 0.00281 | − |
| N-acetyl-aspartyl-glutamate (NAAG) | 73688 | 0.00375 | + |
| sphinganine | 16566 | 0.00378 | − |
| adenosine 5'-diphosphate (ADP) | 16761 | 0.00388 | − |
| N-acetylornithine | 16543 | 0.00466 | − |
| 2-stearoyl glycerophosphocholine | 76076 | 0.00558 | + |
| glutaroyl carnitine | 82952 | 0.01214 | − |
| Y-12422 (recurrent unidentified compound) | n/a | 0.01476 | + |
| Glucose | 17234 | 0.01684 | + |
| guanosine 5'- diphosphate (GDP) | 17552 | 0.01868 | − |
| Homoserine | 15699 | 0.01948 | + |
| phosphocholine | 18132 | 0.02618 | − |
| homocarnosine | 28050 | 0.03059 | + |
| sphingosine | 16393 | 0.03410 | − |
| 5-methylthioadenosine (MTA) | 17509 | 0.03452 | + |
| Y-12421(recurrent unidentified compound) | n/a | 0.03499 | + |
| tyrosine | 17895 | 0.04282 | + |
| adenosine 5'-monophosphate (AMP) | 16027 | 0.04310 | − |
| 24(S)-hydroxycholesterol | 34310 | 0.04607 | + |
| oleate (18:1n9) | 30823 | 0.04737 | + |
| fructose 1,6-diphosphate | 78682 | 0.04756 | + |
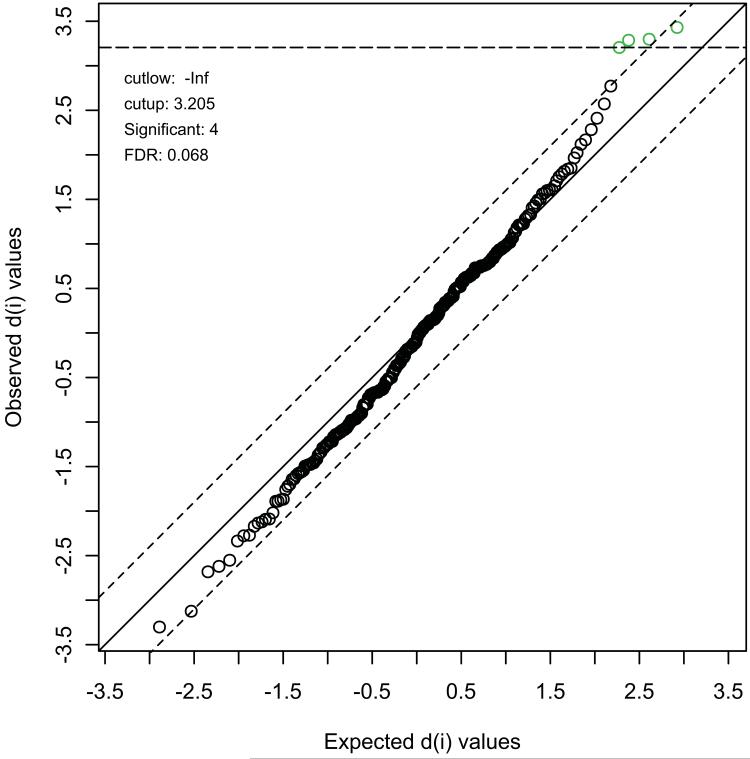
Considering we used a significance threshold of p < 0.05, with the expectation of one false positive finding (type I error) for every 20 tests, and that we tested 302 unique compounds, the expected number of type I errors was 15.1. We exceeded this by detecting 21 significant effects. However, to more formally address the issue of multiple testing, we used SAM (statistical analysis of microarrays). This non-parametric test was designed for use in high-dimensional biological experiments with limited numbers of samples and uses permutations to obtain empirical levels of significance. The results from SAM are shown in Figure 1. We observed four compounds that were significant at a false discovery rate (FDR) threshold of 0.068. We typically use an FDR threshold of 0.1 to declare findings significant as this provides an optimum balance between detecting effects and controlling type I errors (van den Oord and Sullivan, 2003). The four significant compounds, starting with the most significant, were leucine, sphinganaine, ADP and N-acetylornithine, all of which were down-regulated in the haloperidol-treated animals. NAAG, the most up-regulated compound, just failed to reach significance in this analysis.
Figure 1.
Statistical Analysis of Microarrays (SAM), as implemented in MetaboAnalyst and applied to our metabolomics data. The plot shows observed (y-axis) compared to expected (x-axis) values of the d(i) statistic. This represents the ratio of difference in metabolite levels between groups to the variation observed in the data for that metabolite. Thresholds (delta = 0.6) were adjusted to control the FDR < 0.1. Four compounds, all down-regulated in haloperidol-treated animals, passed FDR control at threshold = 0.068, as indicated in the plot.
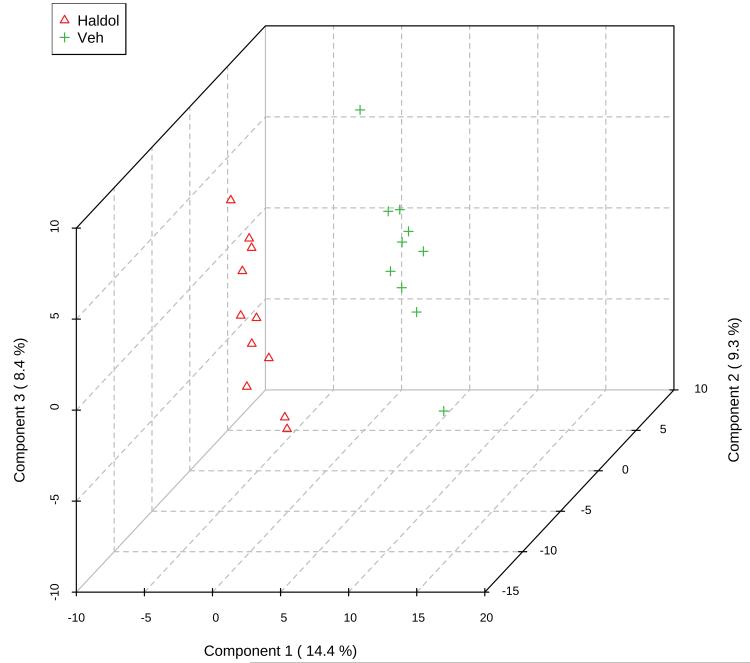
In addition to analysis of individual compounds, a common multivariate analysis technique employed in metabolomic studies is partial least squares discriminant analysis (PLS-DA). PLS-DA projects the data into a low-dimensional space that maximizes the separation between experimental groups using a small number of latent variables. Figure 2A shows a clear separation of the two treatment groups using this method. Both 10-fold cross-validation and leave-one-out cross-validation (LOOCV) methods indicated a four dimension model was optimal in explaining the maximum amount of variation using the cross-validated sum of squares. The most important compounds in the PLS-DA projection, i.e. those that matter most in discriminating between haloperidol-treated animals and controls, are shown in Figure 2B. There was very substantial agreement between the compounds indicated by PLS-DA and those observed in the univariate analysis. Specifically, the top six compounds were leucine, NAAG, sphinganine, ADP, N-acetylornithine and 2-stearoyl glycerophosphocholine. These compounds, as a group, were noticeably distinct from the next most relevant compounds (Figure 2B).
Figure 2A.
Three dimensional plot of the top 3 components in PLS-DA of our metabolomics data, showing clear separation of the groups by metabolite profiles. "Haldol" (triangles) indicates the haloperidol-treated animals, while "Veh" (crosses) are the controls.
To test for functional themes in our results, we performed a pathway analysis on significant (p < 0.05) compounds in our univariate analysis. Starting with the list of 21 compounds, we removed recurrent unidentified compounds as these cannot be assigned to pathways. We also removed basic energy metabolites, such as glucose and ADP, because of their ubiquitous nature. This left 14 compounds, of which 11 could be mapped to the compound database. These were entered into pathway analysis in the MetaboAnalyst package and the results are shown in Table 2. The most significant pathway was sphingolipid metabolism, reiterating the significant finding with sphinganine in all previous analyses, but also including sphingosine that was nominally significant in the univariate analysis.
Table 2.
Pathway analysis of nominally significant compounds. "Total" gives the number of compounds in the reference pathway, "expected" is the number of hits expected by chance, while "hits" is the observed number in our data.
| Pathway | total | expected | Hits | p-value |
|---|---|---|---|---|
| SPHINGOLIPID METABOLISM | 15 | 0.2 | 2 | 0.0154 |
| PROTEIN BIOSYNTHESIS | 19 | 0.253 | 2 | 0.0244 |
| METHIONINE METABOLISM | 24 | 0.32 | 2 | 0.038 |
Discussion
Our study involved the untargeted analysis of 302 unique metabolites to compare global neurochemistry of mice treated with chronic haloperidol for 28 days versus controls. Advantages of our study included 1) a high degree of experimental control, with genetically homogenous mice housed in constant conditions, 2) a proven drug delivery method that produces blood haloperidol concentrations in mice similar to clinical doses in humans, 3) a dose and duration of haloperidol treatment previously shown to produce movement-related side effects in C57BL/6 mice that serves as a model of human EPS, 4) FBMI to denature brain enzymes and instantly arrest metabolism at time of sacrifice, and 5) a multi-platform metabolomics strategy that yielded data on over 300 compounds.
Several analytical methods converged on key compounds that were significantly altered in haloperidol-treated animals versus controls. Our overall top finding was leucine. While an important amino acid, it is somewhat ubiquitous in metabolism and therefore does not give any substantial, specific insights into the action of haloperidol. Several of our other top findings were similarly ubiquitous, particularly energy metabolites such as ADP. These compounds are recognized to be highly variable from sample to sample, and therefore it can be difficult to obtain reliable findings. Nevertheless, recent studies have shown that molecular pathways associated with brain energy metabolism are altered by antipsychotic treatments in human patients (Ma et al., 2009). Thus, our energy-related findings are broadly consistent with human studies of antipsychotic drugs.
Other top findings in our study included sphinganine and N-acetylornithine, both of which passed FDR control in our SAM analysis. N-acetylornithine is a minor component of deproteinized mammalian blood (Armstrong, 1979) and is an intermediate in arginine metabolism. It has been linked to chronic kidney disease and suggested as a possible biomarker for kidney function in humans (Suhre et al., 2011). However, no previous study has linked N-acetylornithine to antipsychotics, nor is there a characterized role for this compound in the CNS. Therefore, a causal relationship between decreased N-acetylornithine and haloperidol exposure, as observed here, is difficult to speculate upon. On the other hand, our finding with sphinganine suggests very specific CNS-related mechanisms affected by haloperidol.
Decreased sphinganine was observed in the haloperidol-treated animals relative to controls. Furthermore, our pathway analysis revealed sphingolipid metabolism as the top result. The compounds contributing to this pathway analysis finding were sphinganine and sphingosine. These are both sphingolipid metabolites that may function as second messengers or regulators of signal transduction that affect events ranging from apoptosis to regulation of the cell cycle (Hoekstra et al., 2003). They form part of the broader sphingolipid class of compounds, which are one of the three major classes of membrane lipids in eukaryotic cells (Levy and Futerman, 2010). Sphingolipids have important roles in cell recognition, neurotransmission and nervous system cell signaling (Hoekstra et al., 2003). Specifically, sphingosine is an 18-carbon aliphatic amino alcohol that is phosphorylated to shingosine-1-phosphate, a potent signaling lipid with activity in the CNS (Okada et al., 2009; Martin and Sospedra, 2014). Furthermore, sphingosine is converted to ceramide via ceramide synthetase, in addition to being a breakdown product of ceramide via alkaline ceramidase, while sphinganine is an intermediate in ceramide synthesis (Figure 3). Another compound among our nominally significant findings was phosphocholine. The addition of a phosphocholine residue from CDP-choline to ceramide via ceramide cholinephosphotransferase produces sphingomyelin, the major component of the myelin sheath that surrounds and insulates nerve cell axons (Slotte, 2013) (Figure 3). Taken together, our findings appear to converge on sphingomyelin.
Figure 3.
Functional relationships between selected top metabolite findings. Compounds measured in our study are color-coded to illustrate their significance levels in the haloperidol (haldol) versus control analysis. Effect directions in the haloperidol-treated animals are illustrated by arrows to the right of measured compounds. Ceramide synthesis takes place in the endoplasmic reticulum, while synthesis of sphingomyelin occurs in the golgi body (see Small molecular Pathway Database SMPDB00034 sphingolipid metabolism (Frolkis et al., 2010)). Sphingomyelin is the major component of the insulating myelin sheath that surrounds axons. Demyelination has been previously associated with elevated N-acetyl-aspartyl-glutamate (NAAG) levels, also observed in our study.
In our data, sphingosine, sphinganine and phosphocholine were all depleted in the haloperidol treated mice. Considering previous rodent studies of antipsychotic effects on CNS metabolites, the most notable study to date was published by McLoughlin et al. (2009). In this study, the authors investigated several drugs, including haloperidol. Differences in their approach from that used in our current study included 1) the use of rats rather mice, 2) 1.0 mg/kg/day haloperidol for 21 days administered orally rather than 3.0 mg/kg/day for 28 days via subcutaneous implant, and 3) the use of NMR to study 15 unique compounds, rather than MS to study over 300. Despite these differences, phosphocholine was among the 15 compounds they measured and they also observed diminished levels of this metabolite in haloperidol-treated animals. Thus, our phosphocholine finding is essentially a replication of that by McLoughlin et al. (2009). While the other sphingolipids identified our study, sphinganine and sphingosine, were not measured by McLoughlin et al., the replication of phosphocholine adds further validity to our findings.
As outlined, our findings with sphingolipids and phosphocholine appear to converge on sphingomyelin. This could suggest a disruption to myelin following long-term haloperidol treatment. In support of this interpretation, our most significant up-regulated compound in the haloperidol-treated animals was N-acetyl-aspartyl-glutamate (NAAG). NAAG is a peptide neurotransmitter that is synthesized in neurons and present at high concentrations in the CNS. It modulates glutamatergic neurotransmission and may have roles in synaptic plasticity and neuroprotection (Benarroch, 2008). Impaired NAAG signaling has been implicated in schizophrenia and some studies have suggested NAAG plays a role in antipsychotic drug mechanisms of action (Ghose et al., 2009; Olszewski et al., 2012; Jessen et al., 2013). Of particular relevance to our current study, elevated NAAG has been associated with hypomyelination disorders such as Canavan's disease and Pelizaeus-Merzbacher disease (PMD) (Kolodziejczyk et al., 2009). Symptoms of these disorders include a diminished motor control. Elevated NAAG has also been observed in a mouse model of PMD (Takanashi et al., 2012). The model involves a myelin synthesis-deficient mouse, which has a loss of motor control as a characteristic phenotype. In other human studies, demyelination has been directly linked to extrapyramidal movement abnormalities in cases not involving antipsychotics (van der Knaap et al., 2007).
Our group previously conducted a genome-wide association study of EPS in 738 patients with schizophrenia, most of whom were treated with atypical antipsychotics (Aberg et al., 2010). One of our genome-wide significant findings was with genetic variants in ZNF202, a gene encoding a transcriptional repressor. ZNF202 controls expression of PLP1, which encodes a major protein in myelin. Mutations in PLP1 cause PMD and indeed this gene is deleted in the myelin synthesis-deficient mouse model of PMD mentioned above. Together, these observations could indicate that haloperidol-induced EPS may result, at least in part, from demyelination. There is a lack of focus on this mechanism in the literature with respect to antipsychotic-induced EPS. If targeted studies can confirm this hypothesis, future drug studies could examine the demyelination potential of new candidate antipsychotic compounds to limit EPS in patients. It should be emphasized that while haloperidol has one of the highest incidences of EPS of any antipsychotic (Leucht et al., 2013), more recent antipsychotics are not free from EPS (Peluso et al., 2012). Any improvement in our ability to limit these side effects could have substantial clinical benefit.
Our study provides new data on the CNS effects of chronic haloperidol administration, that suggests a role for demyelination in the pathophysiology of EPS. Expanding these methods to investigate other antipsychotics could be a fruitful avenue to identify novel biological mechanisms of efficacy and unwanted side-effects.
Figure 2B.
Variables of importance in projection (VIP) scores for the most relevant metabolites in PLS-DA. The panel to the right indicates relative effect direction in the two groups.
Acknowledgements
We gratefully acknowledge the assistance of Danny Alexander, Edward Karoly and John Luster at Metabolon, Inc, Research Triangle Park, North Carolina. We also acknowledge the assistance of Lindsey King and Krista Scoggins at Virginia Commonwealth University in expediting the experiments. This work was supported by grants from the US National Institutes of Health to EJvdO (R21DA021411) and a NARSAD (National Alliance for Research on Schizophrenia and Depression, now the Brain Research Foundation) Young Investigator Award to JLM. Both JLM and EJvdO are also partly supported by grant 1R01MH097283 from the US National Institute of Mental Health.
Footnotes
Conflict of Interest Statement
After working on this project, REV left Virginia Commonwealth University and is now an employee of Eli Lilly. All other authors declare no potential conflicts of interest.
References
- Adams CE, Bergman H, Irving CB, Lawrie S. Haloperidol versus placebo for schizophrenia. Cochrane Database Syst Rev. 2013;11:CD003082. doi: 10.1002/14651858.CD003082.pub3. [DOI] [PubMed] [Google Scholar]
- Armstrong MD. N-delta-acetylornithine and S-methylcysteine in blood plasma. Biochim Biophys Acta. 1979;587:638–642. doi: 10.1016/0304-4165(79)90015-1. [DOI] [PubMed] [Google Scholar]
- Benarroch EE. N-acetylaspartate and N-acetylaspartylglutamate: neurobiology and clinical significance. Neurology. 2008;70:1353–1357. doi: 10.1212/01.wnl.0000311267.63292.6c. [DOI] [PubMed] [Google Scholar]
- Bijlsma S, Bobeldijk I, Verheij ER, Ramaker R, Kochhar S, Macdonald IA, van Ommen B, Smilde AK. Large-scale human metabolomics studies: a strategy for data (pre-) processing and validation. Anal Chem. 2006;78:567–574. doi: 10.1021/ac051495j. [DOI] [PubMed] [Google Scholar]
- Clayton TA, Lindon JC, Cloarec O, Antti H, Charuel C, Hanton G, Provost JP, Le Net JL, Baker D, Walley RJ, Everett JR, Nicholson JK. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature. 2006;440:1073–1077. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- Crowley JJ, Adkins DE, Pratt AL, Quackenbush CR, van den Oord EJ, Moy SS, Wilhelmsen KC, Cooper TB, Bogue MA, McLeod HL, Sullivan PF. Antipsychotic-induced vacuous chewing movements and extrapyramidal side effects are highly heritable in mice. Pharmacogenomics J. 2012a;12:147–155. doi: 10.1038/tpj.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley JJ, Kim Y, Szatkiewicz JP, Pratt AL, Quackenbush CR, Adkins DE, van den Oord E, Bogue MA, Yang H, Wang W, Threadgill DW, de Villena FP, McLeod HL, Sullivan PF. Genome-wide association mapping of loci for antipsychotic-induced extrapyramidal symptoms in mice. Mamm Genome. 2012b;23:322–335. doi: 10.1007/s00335-011-9385-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf RA, Chowdhury GM, Brown PB, Rothman DL, Behar KL. In situ 3D magnetic resonance metabolic imaging of microwave-irradiated rodent brain: a new tool for metabolomics research. J Neurochem. 2009;109:494–501. doi: 10.1111/j.1471-4159.2009.05967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehaven CD, Evans AM, Dai H, Lawton KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform. 2010;2:9. doi: 10.1186/1758-2946-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- Frolkis A, Knox C, Lim E, Jewison T, Law V, Hau DD, Liu P, Gautam B, Ly S, Guo AC, Xia J, Liang Y, Shrivastava S, Wishart DS. SMPDB: The Small Molecule Pathway Database. Nucleic Acids Res. 2010;38:D480–487. doi: 10.1093/nar/gkp1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K, Kemp DE, Ganocy SJ, Gajwani P, Xia G, Calabrese JR. Antipsychotic-induced extrapyramidal side effects in bipolar disorder and schizophrenia: a systematic review. J Clin Psychopharmacol. 2008;28:203–209. doi: 10.1097/JCP.0b013e318166c4d5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose S, Gleason KA, Potts BW, Lewis-Amezcua K, Tamminga CA. Differential expression of metabotropic glutamate receptors 2 and 3 in schizophrenia: a mechanism for antipsychotic drug action? Am J Psychiatry. 2009;166:812–820. doi: 10.1176/appi.ajp.2009.08091445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer WM. Extrapyramidal side effects, tardive dyskinesia, and the concept of atypicality. J Clin Psychiatry 61 Suppl. 2000;3:16–21. [PubMed] [Google Scholar]
- Hastings J, de Matos P, Dekker A, Ennis M, Harsha B, Kale N, Muthukrishnan V, Owen G, Turner S, Williams M, Steinbeck C. The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic Acids Res. 2013;41:D456–463. doi: 10.1093/nar/gks1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra D, Maier O, van der Wouden JM, Slimane TA, van ISC. Membrane dynamics and cell polarity: the role of sphingolipids. J Lipid Res. 2003;44:869–877. doi: 10.1194/jlr.R300003-JLR200. [DOI] [PubMed] [Google Scholar]
- Hsin-tung E, Simpson G. Medication-induced movement disorders. In: Kaplan H, Sadock BJ, editors. Comprehensive Textbook of Psychiatry. Lippincott, Williams & Wilkins; Philadelphia: 2000. [Google Scholar]
- Ikarashi Y, Sasahara T, Maruyama Y. Postmortem changes in catecholamines, indoleamines, and their metabolites in rat brain regions: prevention with 10-kW microwave irradiation. J Neurochem. 1985;45:935–939. doi: 10.1111/j.1471-4159.1985.tb04083.x. [DOI] [PubMed] [Google Scholar]
- Jessen F, Fingerhut N, Sprinkart AM, Kuhn KU, Petrovsky N, Maier W, Schild HH, Block W, Wagner M, Traber F. N-acetylaspartylglutamate (NAAG) and N-acetylaspartate (NAA) in patients with schizophrenia. Schizophr Bull. 2013;39:197–205. doi: 10.1093/schbul/sbr127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- Kolodziejczyk K, Hamilton NB, Wade A, Karadottir R, Attwell D. The effect of N-acetyl-aspartyl-glutamate and N-acetyl-aspartate on white matter oligodendrocytes. Brain. 2009;132:1496–1508. doi: 10.1093/brain/awp087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucht S, Cipriani A, Spineli L, Mavridis D, Orey D, Richter F, Samara M, Barbui C, Engel RR, Geddes JR, Kissling W, Stapf MP, Lassig B, Salanti G, Davis JM. Comparative efficacy and tolerability of 15 antipsychotic drugs in schizophrenia: a multiple-treatments meta-analysis. Lancet. 2013;382:951–962. doi: 10.1016/S0140-6736(13)60733-3. [DOI] [PubMed] [Google Scholar]
- Levy M, Futerman AH. Mammalian ceramide synthases. IUBMB Life. 2010;62:347–356. doi: 10.1002/iub.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Login GR, Dvorak AM. Application of microwave fixation techniques in pathology to neuroscience studies: a review. J Neurosci Methods. 1994;55:173–182. doi: 10.1016/0165-0270(94)90209-7. [DOI] [PubMed] [Google Scholar]
- Lopez-Munoz F, Alamo C. The consolidation of neuroleptic therapy: Janssen, the discovery of haloperidol and its introduction into clinical practice. Brain Res Bull. 2009;79:130–141. doi: 10.1016/j.brainresbull.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Ma D, Guest PC, Bahn S. Metabonomic studies of schizophrenia and psychotropic medications: focus on alterations in CNS energy homeostasis. Bioanalysis. 2009;1:1615–1626. doi: 10.4155/bio.09.144. [DOI] [PubMed] [Google Scholar]
- Marsden CD, Jenner P. The pathophysiology of extrapyramidal side-effects of neuroleptic drugs. Psychol Med. 1980;10:55–72. doi: 10.1017/s003329170003960x. [DOI] [PubMed] [Google Scholar]
- Martin R, Sospedra M. Sphingosine-1 phosphate and central nervous system. Curr Top Microbiol Immunol. 2014;378:149–170. doi: 10.1007/978-3-319-05879-5_7. [DOI] [PubMed] [Google Scholar]
- McClay JL, Adkins DE, Vunck SA, Batman AM, Vann RE, Clark SL, Beardsley PM, van den Oord EJ. Large-scale neurochemical metabolomics analysis identifies multiple compounds associated with methamphetamine exposure. Metabolomics. 2013;9:392–402. doi: 10.1007/s11306-012-0456-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoughlin GA, Ma D, Tsang TM, Jones DN, Cilia J, Hill MD, Robbins MJ, Benzel IM, Maycox PR, Holmes E, Bahn S. Analyzing the effects of psychotropic drugs on metabolite profiles in rat brain using 1H NMR spectroscopy. J Proteome Res. 2009;8:1943–1952. doi: 10.1021/pr800892u. [DOI] [PubMed] [Google Scholar]
- Meltzer HY. Update on typical and atypical antipsychotic drugs. Annu Rev Med. 2013;64:393–406. doi: 10.1146/annurev-med-050911-161504. [DOI] [PubMed] [Google Scholar]
- Meltzer HY, Stahl SM. The dopamine hypothesis of schizophrenia: a review. Schizophr Bull. 1976;2:19–76. doi: 10.1093/schbul/2.1.19. [DOI] [PubMed] [Google Scholar]
- Ohta T, Masutomi N, Tsutsui N, Sakairi T, Mitchell M, Milburn MV, Ryals JA, Beebe KD, Guo L. Untargeted metabolomic profiling as an evaluative tool of fenofibrate-induced toxicology in Fischer 344 male rats. Toxicol Pathol. 2009;37:521–535. doi: 10.1177/0192623309336152. [DOI] [PubMed] [Google Scholar]
- Okada T, Kajimoto T, Jahangeer S, Nakamura S. Sphingosine kinase/sphingosine 1-phosphate signalling in central nervous system. Cell Signal. 2009;21:7–13. doi: 10.1016/j.cellsig.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Olszewski RT, Bzdega T, Neale JH. mGluR3 and not mGluR2 receptors mediate the efficacy of NAAG peptidase inhibitor in validated model of schizophrenia. Schizophr Res. 2012;136:160–161. doi: 10.1016/j.schres.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso MJ, Lewis SW, Barnes TR, Jones PB. Extrapyramidal motor side-effects of first- and second-generation antipsychotic drugs. Br J Psychiatry. 2012;200:387–392. doi: 10.1192/bjp.bp.111.101485. [DOI] [PubMed] [Google Scholar]
- Rosebush PI, Mazurek MF. Neurologic side effects in neuroleptic-naive patients treated with haloperidol or risperidone. Neurology. 1999;52:782–785. doi: 10.1212/wnl.52.4.782. [DOI] [PubMed] [Google Scholar]
- Slotte JP. Biological functions of sphingomyelins. Prog Lipid Res. 2013;52:424–437. doi: 10.1016/j.plipres.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Soares-Weiser K, Fernandez HH. Tardive dyskinesia. Semin Neurol. 2007;27:159–169. doi: 10.1055/s-2007-971169. [DOI] [PubMed] [Google Scholar]
- Suhre K, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanashi J, Saito S, Aoki I, Barkovich AJ, Ito Y, Inoue K. Increased N-acetylaspartate in model mouse of Pelizaeus-Merzbacher disease. J Magn Reson Imaging. 2012;35:418–425. doi: 10.1002/jmri.22817. [DOI] [PubMed] [Google Scholar]
- Turrone P, Remington G, Nobrega JN. The vacuous chewing movement (VCM) model of tardive dyskinesia revisited: is there a relationship to dopamine D(2) receptor occupancy? Neurosci Biobehav Rev. 2002;26:361–380. doi: 10.1016/s0149-7634(02)00008-8. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Oord EJ, Sullivan PF. False discoveries and models for gene discovery. Trends Genet. 2003;19:537–542. doi: 10.1016/j.tig.2003.08.003. [DOI] [PubMed] [Google Scholar]
- van der Knaap MS, Linnankivi T, Paetau A, Feigenbaum A, Wakusawa K, Haginoya K, Kohler W, Henneke M, Dinopoulos A, Grattan-Smith P, Brockmann K, Schiffmann R, Blaser S. Hypomyelination with atrophy of the basal ganglia and cerebellum: follow-up and pathology. Neurology. 2007;69:166–171. doi: 10.1212/01.wnl.0000265592.74483.a6. [DOI] [PubMed] [Google Scholar]
- van Os J, Kapur S. Schizophrenia. Lancet. 2009;374:635–645. doi: 10.1016/S0140-6736(09)60995-8. [DOI] [PubMed] [Google Scholar]
- Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. MetaboAnalyst 2.0--a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012;40:W127–133. doi: 10.1093/nar/gks374. [DOI] [PMC free article] [PubMed] [Google Scholar]