Abstract
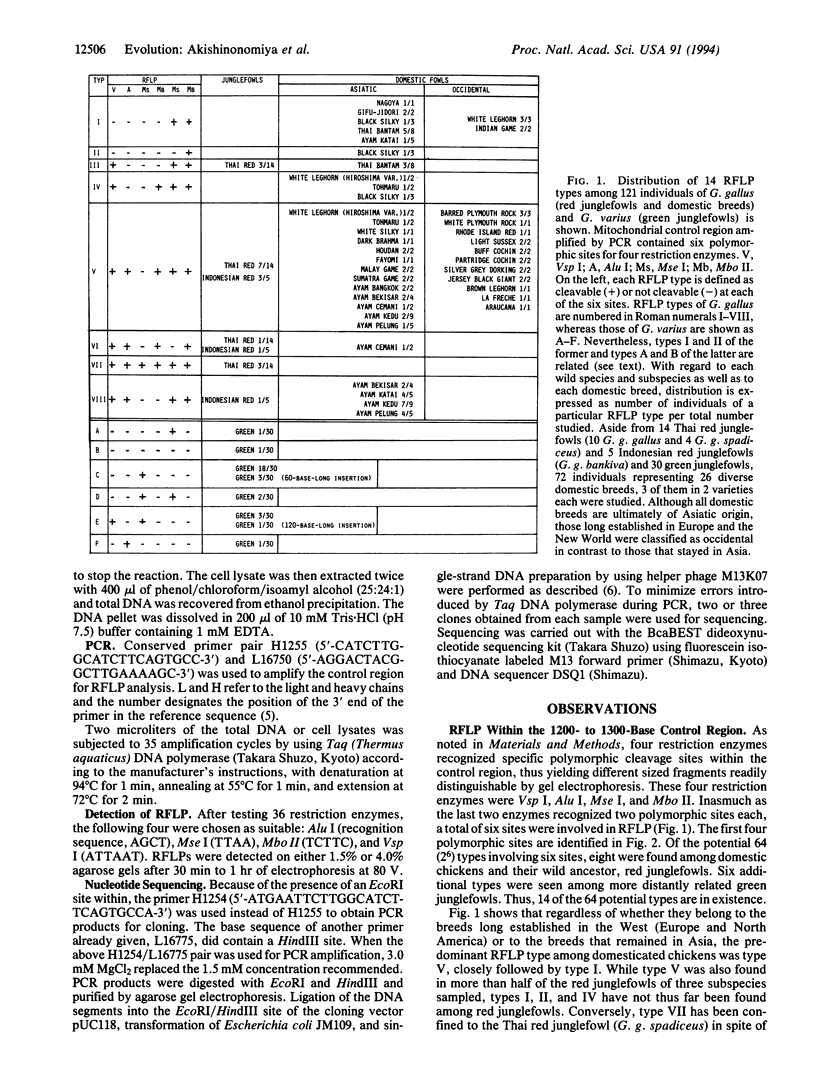
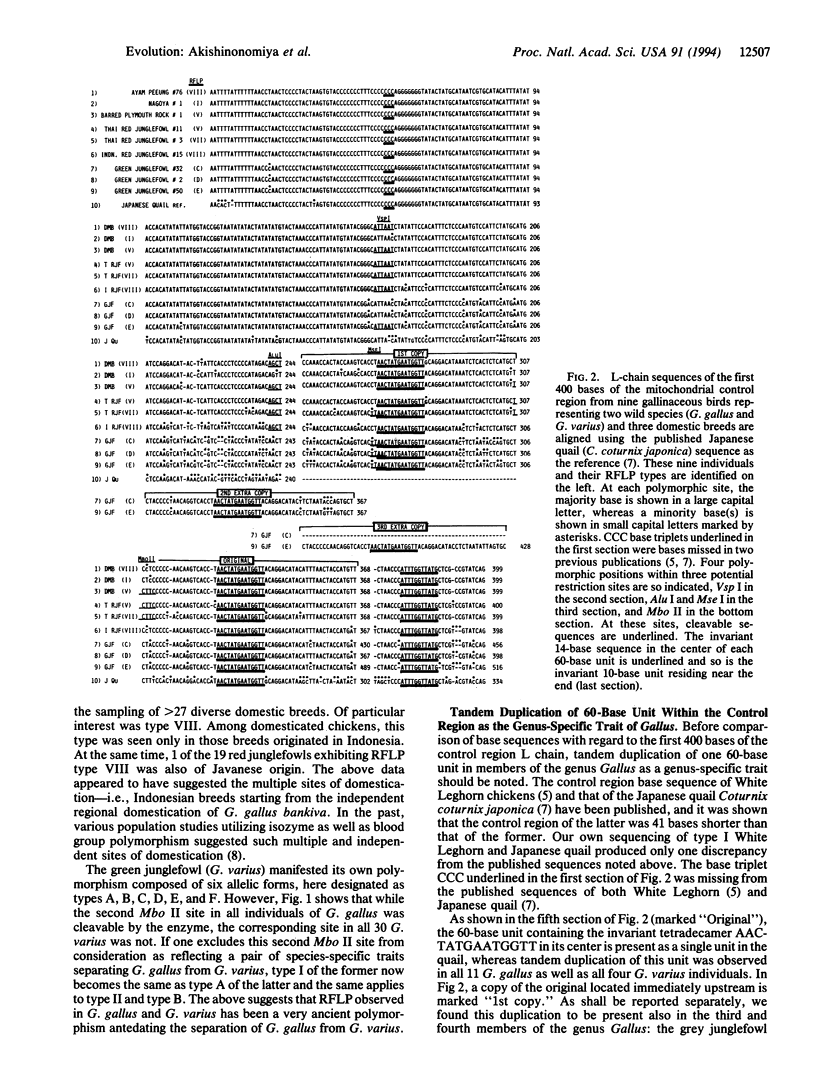
The noncoding control region of the mitochondrial DNA of various gallinaceous birds was studied with regard to its restriction fragment length polymorphism (RFLP) and sequences of the first 400 bases. Tandem duplication of the 60-base unit was established as a trait unique to the genus Gallus, which is shared neither by pheasants nor by quails. Unlike its close ally Gallus varius (green junglefowl), the red junglefowl Gallus gallus is a genetically very diverse species; the 7.0% sequence divergence was seen between those from Thailand (G. g. gallus and G. g. spadiceus) and the other from the Indonesian island of Java (G. g. Bankiva). Furthermore, the divergence increased to 27.83% if each transversion is regarded as an equivalent of 10 transitions. On the other hand, a mere 0.5-3.0% difference (all transitions) separated various domestic breeds of the chicken from two G. g. gallus of Thailand, thus indicating a single domestication event in the area inhabited by this subspecies of the red junglefowl as the origin of all domestic breeds. Only transitions separated six diverse domesticated breeds. Nevertheless, a 2.75% difference was seen between RFLP type I breeds (White Leghorn and Nagoya) and a RFLP type VIII breed (Ayam Pelung). The above data suggested that although the mitochondrion of RFLP type V was the main contributor to domestication, hens of other RFLP types also contributed to this event.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cann R. L., Stoneking M., Wilson A. C. Mitochondrial DNA and human evolution. Nature. 1987 Jan 1;325(6099):31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- Desjardins P., Morais R. Nucleotide sequence and evolution of coding and noncoding regions of a quail mitochondrial genome. J Mol Evol. 1991 Feb;32(2):153–161. doi: 10.1007/BF02515387. [DOI] [PubMed] [Google Scholar]
- Desjardins P., Morais R. Sequence and gene organization of the chicken mitochondrial genome. A novel gene order in higher vertebrates. J Mol Biol. 1990 Apr 20;212(4):599–634. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- Di Rienzo A., Wilson A. C. Branching pattern in the evolutionary tree for human mitochondrial DNA. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1597–1601. doi: 10.1073/pnas.88.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HALEY L. E. SERUM ALBUMIN POLYMORPHISM IN QUAIL AND CHICKEN-QUAIL HYBRIDS. Genetics. 1965 Jun;51:983–986. doi: 10.1093/genetics/51.6.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus R. T., MacHugh D. E., Bradley D. G., Sharp P. M., Cunningham P. Evidence for two independent domestications of cattle. Proc Natl Acad Sci U S A. 1994 Mar 29;91(7):2757–2761. doi: 10.1073/pnas.91.7.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart D. T., Baker A. J. Patterns of sequence variation in the mitochondrial D-loop region of shrews. Mol Biol Evol. 1994 Jan;11(1):9–21. doi: 10.1093/oxfordjournals.molbev.a040096. [DOI] [PubMed] [Google Scholar]
- Vigilant L., Pennington R., Harpending H., Kocher T. D., Wilson A. C. Mitochondrial DNA sequences in single hairs from a southern African population. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9350–9354. doi: 10.1073/pnas.86.23.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]


