Abstract
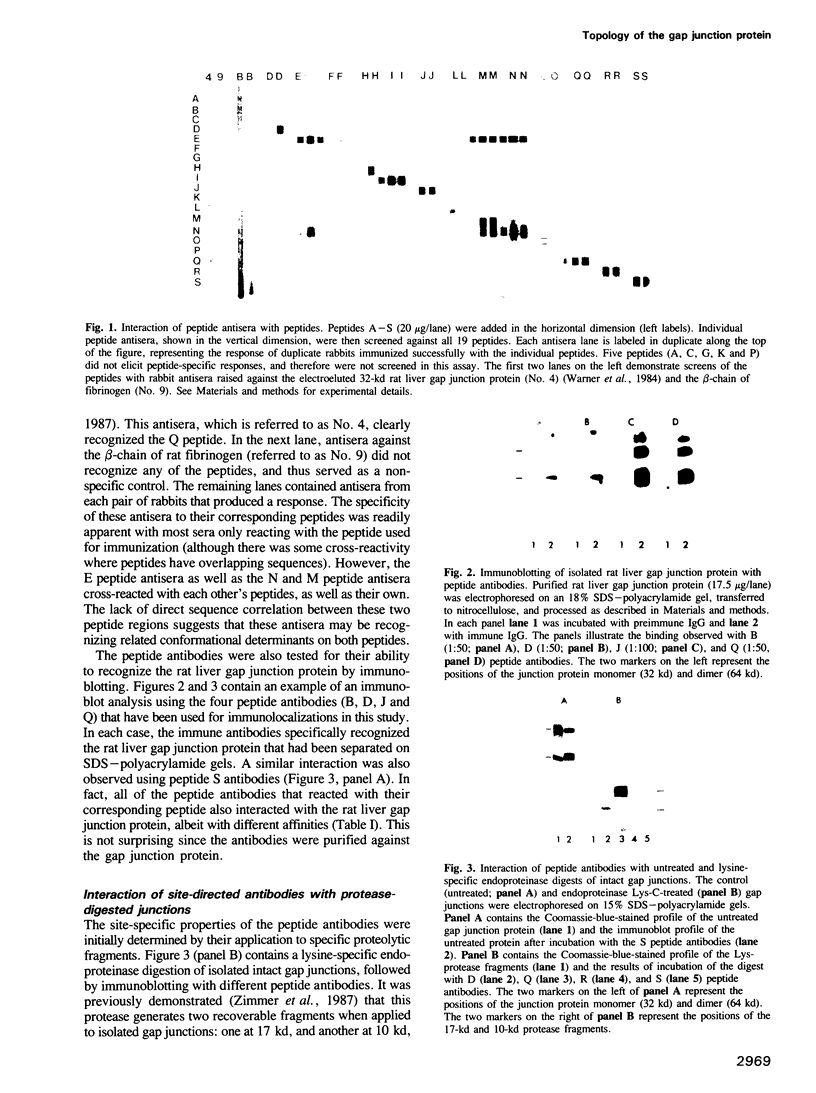
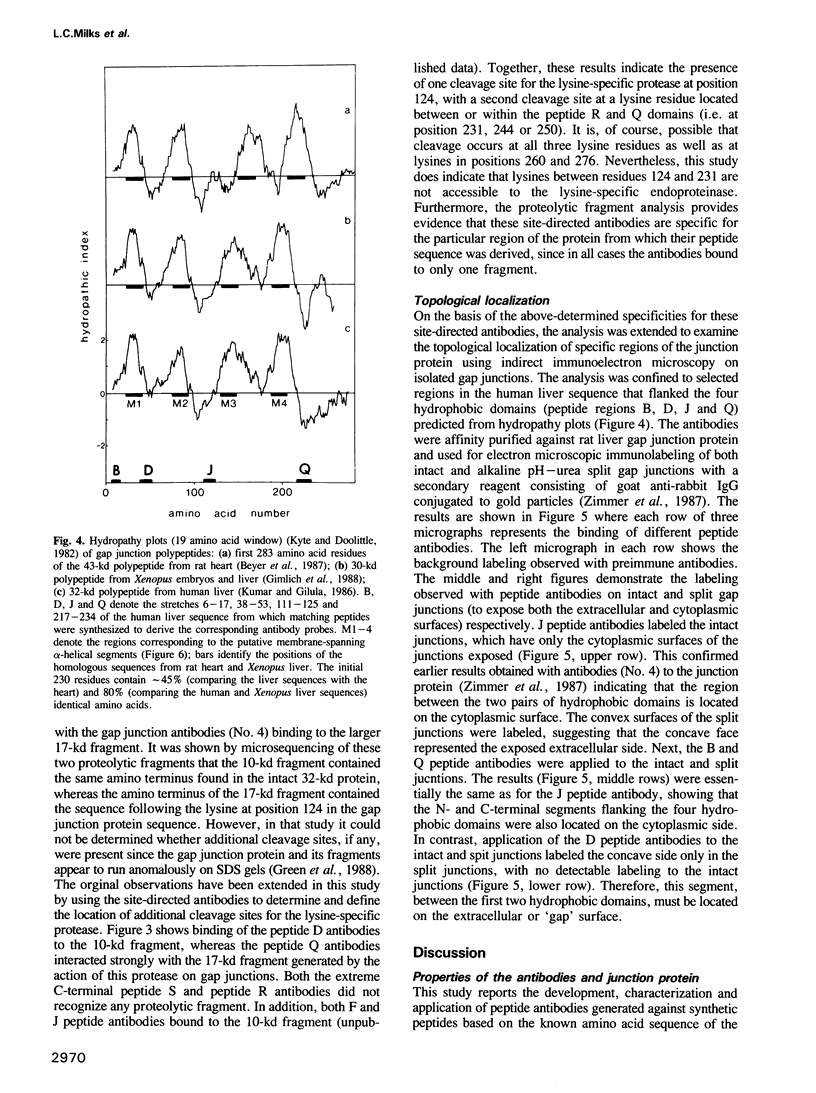
Synthetic peptides corresponding to sequences in the human liver gap junction protein were chemically synthesized and used for generation of peptide antisera to defined sequences in the protein. The antibodies were affinity purified and characterized by demonstrating that they specifically recognized both their corresponding synthetic peptide (as indicated by dot blot analysis) and the native 32-kd gap junction protein (by immunoblotting). The specificity of a subset of the different site-specific antibodies was subsequently confirmed by demonstration of their binding to specific gap junction fragments produced by treatment with a lysine-specific endoproteinase. Immunoelectron microscopy was used to localize the specific peptide antibody epitopes to either the cytoplasmic or extracellular surfaces of the gap junction. Results indicate a transmembrane orientation for the protein with the amino and carboxyl termini located on the cytoplasmic side of the membrane. Based on these data, a model is proposed for the transmembrane folding of the gap junction protein.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avrameas S., Ternynck T. The cross-linking of proteins with glutaraldehyde and its use for the preparation of immunoadsorbents. Immunochemistry. 1969 Jan;6(1):53–66. doi: 10.1016/0019-2791(69)90178-5. [DOI] [PubMed] [Google Scholar]
- Beyer E. C., Paul D. L., Goodenough D. A. Connexin43: a protein from rat heart homologous to a gap junction protein from liver. J Cell Biol. 1987 Dec;105(6 Pt 1):2621–2629. doi: 10.1083/jcb.105.6.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulinski J. C. Peptide antibodies: new tools for cell biology. Int Rev Cytol. 1986;103:281–302. doi: 10.1016/s0074-7696(08)60838-4. [DOI] [PubMed] [Google Scholar]
- Dermietzel R., Leibstein A., Frixen U., Janssen-Timmen U., Traub O., Willecke K. Gap junctions in several tissues share antigenic determinants with liver gap junctions. EMBO J. 1984 Oct;3(10):2261–2270. doi: 10.1002/j.1460-2075.1984.tb02124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermietzel R., Yancey B., Janssen-Timmen U., Traub O., Willecke K., Revel J. P. Simultaneous light and electron microscopic observation of immunolabeled liver 27 KD gap junction protein on ultra-thin cryosections. J Histochem Cytochem. 1987 Mar;35(3):387–392. doi: 10.1177/35.3.3029214. [DOI] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A. The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell. 1981 Feb;23(2):411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- Fraser S. E., Green C. R., Bode H. R., Gilula N. B. Selective disruption of gap junctional communication interferes with a patterning process in hydra. Science. 1987 Jul 3;237(4810):49–55. doi: 10.1126/science.3037697. [DOI] [PubMed] [Google Scholar]
- Gilula N. B., Reeves O. R., Steinbach A. Metabolic coupling, ionic coupling and cell contacts. Nature. 1972 Feb 4;235(5336):262–265. doi: 10.1038/235262a0. [DOI] [PubMed] [Google Scholar]
- Green C. R., Harfst E., Gourdie R. G., Severs N. J. Analysis of the rat liver gap junction protein: clarification of anomalies in its molecular size. Proc R Soc Lond B Biol Sci. 1988 Mar 22;233(1271):165–174. doi: 10.1098/rspb.1988.0016. [DOI] [PubMed] [Google Scholar]
- Green N., Alexander H., Olson A., Alexander S., Shinnick T. M., Sutcliffe J. G., Lerner R. A. Immunogenic structure of the influenza virus hemagglutinin. Cell. 1982 Mar;28(3):477–487. doi: 10.1016/0092-8674(82)90202-1. [DOI] [PubMed] [Google Scholar]
- Henderson D., Eibl H., Weber K. Structure and biochemistry of mouse hepatic gap junctions. J Mol Biol. 1979 Aug 5;132(2):193–218. doi: 10.1016/0022-2836(79)90391-7. [DOI] [PubMed] [Google Scholar]
- Henderson R., Unwin P. N. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975 Sep 4;257(5521):28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- Hendrickson W. A., Ward K. B. Pseudosymmetry in the structure of myohemerythrin. J Biol Chem. 1977 May 10;252(9):3012–3018. [PubMed] [Google Scholar]
- Hertzberg E. L. A detergent-independent procedure for the isolation of gap junctions from rat liver. J Biol Chem. 1984 Aug 10;259(15):9936–9943. [PubMed] [Google Scholar]
- Hertzberg E. L., Gilula N. B. Isolation and characterization of gap junctions from rat liver. J Biol Chem. 1979 Mar 25;254(6):2138–2147. [PubMed] [Google Scholar]
- Houghten R. A. General method for the rapid solid-phase synthesis of large numbers of peptides: specificity of antigen-antibody interaction at the level of individual amino acids. Proc Natl Acad Sci U S A. 1985 Aug;82(15):5131–5135. doi: 10.1073/pnas.82.15.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler J., Christie D., Bullivant S. Homologies between gap junction proteins in lens, heart and liver. Nature. 1988 Feb 25;331(6158):721–723. doi: 10.1038/331721a0. [DOI] [PubMed] [Google Scholar]
- Kreil G. Transfer of proteins across membranes. Annu Rev Biochem. 1981;50:317–348. doi: 10.1146/annurev.bi.50.070181.001533. [DOI] [PubMed] [Google Scholar]
- Kumar N. M., Gilula N. B. Cloning and characterization of human and rat liver cDNAs coding for a gap junction protein. J Cell Biol. 1986 Sep;103(3):767–776. doi: 10.1083/jcb.103.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lee S., Gilula N. B., Warner A. E. Gap junctional communication and compaction during preimplantation stages of mouse development. Cell. 1987 Dec 4;51(5):851–860. doi: 10.1016/0092-8674(87)90108-5. [DOI] [PubMed] [Google Scholar]
- Liu F. T., Zinnecker M., Hamaoka T., Katz D. H. New procedures for preparation and isolation of conjugates of proteins and a synthetic copolymer of D-amino acids and immunochemical characterization of such conjugates. Biochemistry. 1979 Feb 20;18(4):690–693. doi: 10.1021/bi00571a022. [DOI] [PubMed] [Google Scholar]
- Loewenstein W. R. Junctional intercellular communication: the cell-to-cell membrane channel. Physiol Rev. 1981 Oct;61(4):829–913. doi: 10.1152/physrev.1981.61.4.829. [DOI] [PubMed] [Google Scholar]
- Makowski L., Caspar D. L., Phillips W. C., Goodenough D. A. Gap junction structures. II. Analysis of the x-ray diffraction data. J Cell Biol. 1977 Aug;74(2):629–645. doi: 10.1083/jcb.74.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan A. D., Bloomer A. C., Butler P. J. Structural repeats and evolution of tobacco mosaic virus coat protein and RNA. J Mol Biol. 1980 Jan 25;136(3):203–224. doi: 10.1016/0022-2836(80)90372-1. [DOI] [PubMed] [Google Scholar]
- Michel H., Weyer K. A., Gruenberg H., Dunger I., Oesterhelt D., Lottspeich F. The 'light' and 'medium' subunits of the photosynthetic reaction centre from Rhodopseudomonas viridis: isolation of the genes, nucleotide and amino acid sequence. EMBO J. 1986 Jun;5(6):1149–1158. doi: 10.1002/j.1460-2075.1986.tb04340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson B., Dermietzel R., Teplow D., Traub O., Willecke K., Revel J. P. Two homologous protein components of hepatic gap junctions. Nature. 1987 Oct 22;329(6141):732–734. doi: 10.1038/329732a0. [DOI] [PubMed] [Google Scholar]
- Paul D. L. Molecular cloning of cDNA for rat liver gap junction protein. J Cell Biol. 1986 Jul;103(1):123–134. doi: 10.1083/jcb.103.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B. R., Siliciano J. D., Mooseker M. S., Goodenough D. A. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol. 1986 Sep;103(3):755–766. doi: 10.1083/jcb.103.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin P. N., Ennis P. D. Two configurations of a channel-forming membrane protein. Nature. 1984 Feb 16;307(5952):609–613. doi: 10.1038/307609a0. [DOI] [PubMed] [Google Scholar]
- Unwin P. N., Zampighi G. Structure of the junction between communicating cells. Nature. 1980 Feb 7;283(5747):545–549. doi: 10.1038/283545a0. [DOI] [PubMed] [Google Scholar]
- Walter G. Production and use of antibodies against synthetic peptides. J Immunol Methods. 1986 Apr 17;88(2):149–161. doi: 10.1016/0022-1759(86)90001-3. [DOI] [PubMed] [Google Scholar]
- Warner A. E., Guthrie S. C., Gilula N. B. Antibodies to gap-junctional protein selectively disrupt junctional communication in the early amphibian embryo. Nature. 1984 Sep 13;311(5982):127–131. doi: 10.1038/311127a0. [DOI] [PubMed] [Google Scholar]
- Weber P. C., Salemme F. R. Structural and functional diversity in 4-alpha-helical proteins. Nature. 1980 Sep 4;287(5777):82–84. doi: 10.1038/287082a0. [DOI] [PubMed] [Google Scholar]
- Young J. D., Cohn Z. A., Gilula N. B. Functional assembly of gap junction conductance in lipid bilayers: demonstration that the major 27 kd protein forms the junctional channel. Cell. 1987 Mar 13;48(5):733–743. doi: 10.1016/0092-8674(87)90071-7. [DOI] [PubMed] [Google Scholar]
- Zimmer D. B., Green C. R., Evans W. H., Gilula N. B. Topological analysis of the major protein in isolated intact rat liver gap junctions and gap junction-derived single membrane structures. J Biol Chem. 1987 Jun 5;262(16):7751–7763. [PubMed] [Google Scholar]