Abstract
Pelvic organ prolapse (POP) is a common highly disabling disorder with a large hereditary component. It is characterized by a loss of pelvic floor support that leads to the herniation of the uterus in or outside the vagina. Genome-wide linkage studies have shown an evidence of POP association with the region 9q21 and six other loci in European pedigrees. The aim of our study was to test the above associations in a case-control study in Russian population. Twelve SNPs including SNPs cited in the above studies and those selected using the RegulomeDB annotations for the region 9q21 were genotyped in 210 patients with POP (stages III-IV) and 292 controls with no even minimal POP. Genotyping was performed using the polymerase chain reaction with confronting two-pair primers (PCR–CTPP). Association analyses were conducted for individual SNPs, 9q21 haplotypes, and SNP-SNP interactions. SNP rs12237222 with the highest RegulomeDB score 1a appeared to be the key SNP in haplotypes associated with POP. Other RegulomeDB Category 1 SNPs, rs12551710 and rs2236479 (scores 1d and 1f, resp.), exhibited epistatic effects. In this study, we verified the region 9q21 association with POP in Russians, using RegulomeDB annotations.
1. Introduction
Pelvic organ prolapse (POP) is the dropping of the pelvic organs caused by weakness or damage to the normal support of the pelvic floor. Prevalence of a disease state (stage II prolapse or greater) varies by data source from 3% of parous women [1] to 19% of women with advanced disease requiring surgery [2]. As many as 50% of women may have some degree of POP. Clinical manifestations related to POP often become evident after menopause [3], though it is becoming a serious health problem for women of all ages with first symptoms that might be experienced before age 30 [4]. POP rarely occurs as a separate condition and often correlates with urinary and faecal incontinence, sexual dysfunction, psychological, and social disadaptation [5]. Risk factors, which predispose to POP, include pelvic floor injury (vaginal parity and pelvic floor trauma during childbirth), lifestyle and health conditions (mainly, menopause, body mass index (BMI), chronic cough, constipation, and heavy lifting), genetic-related conditions (ethnicity, connective tissue disorders, and familial character of prolapse) [6, 7]. Forty-three percent of the variation in susceptibility for POP may be explained by genetic effects [8].
The majority of multifactorial disorders are characterized by a large spectrum of genetic variations in disease modifying genes, whereas information about causative polymorphic genes is scarce. In contrast, the genetic studies of POP have been mainly focused on a limited number of causative genes, among them are the genes controlling the collagen and elastin synthesis and remodeling [9–12], extracellular matrix metabolism [9, 13–16], and hormone receptors [17–19]. There are also three genome-wide linkage studies of the same group of researchers that have determined chromosome 9q21 [20], six other loci [21], and chromosomes 10q24–26 and 17q25 [22] as the regions associated with a predisposition for advanced POP in European pedigrees. It is known that family-based genetic studies may be unsuccessful for complex traits in general population [23] and the results should be validated in independent studies. No candidate gene studies have been performed yet from these genetic regions.
Functional SNPs in a specified chromosome region can be chosen with a powerful tool, RegulomeDB, a database which provides functional annotations of SNPs in the human genome using data sets from ENCODE and other sources [24]. These annotations include data on chromatin structure, methylation, protein motifs, and binding. RegulomeDB presents a scoring system, with categories ranging from 1 to 7, where category 7 variants lack evidence of regulatory function, while category 1 variants are those “likely to affect binding and linked to expression of a gene target” [24]. Category 1 is further divided into 1a–1f subcategories. A variant scored as 1a has the highest confidence on functionality. To date, functional annotation data have been mostly used for a post hoc analysis of GWAS data [25–28] (Hong et al., 2014; Rajkumar et al., 2014; Rosenthal et al., 2014; Zia et al., 2015) while selection of putative functional SNPs may also be useful in future studies.
To verify GWAS findings on POP association with the 9q21 chromosome region [20] and six other loci (rs1455311 (4q21), rs1036819 (8q24), rs430794 (9q22), rs8027714 (15q11), rs1810636 (20p13), and rs2236479 (21q22)) [21], we conducted an association study, in a Russian population, between POP risk and twelve SNPs that have been identified in the above studies or selected with RegulomeDB annotations for the region 9q21.
2. Materials and Methods
2.1. Subjects
The study was approved by the Ethics Committee of the Pirogov Russian National Research Medical University (university review board approval number 117 (April 16, 2012)) and was conducted according to the principles of the Declaration of Helsinki; all patients provided written informed consent.
Based on the POP-Q examination, POP patients were diagnosed with stages III-IV prolapse and controls without prolapse and prior history of prolapse surgery (stage 0). Subjects were recruited from the clinical bases of the Department of Obstetrics and Gynecology of the Pirogov Russian National Research Medical University (RNRMU), Moscow, Russia (from December 2011 to September 2013). Exclusion criteria for the group under study included family history of pelvic organ prolapse, lack of informed consent, pregnancy, gynecologic malignancy, and hereditary (Marfan or Ehlers-Danlos syndromes) or acquired connective tissue disorders (rheumatoid arthritis, systemic lupus erythematosus, scleroderma, polymyositis, and dermatomyositis). We also carefully collected data on perineal trauma and episiotomy (surgical incision, which is made to prevent more extensive vaginal traumas during childbirth). These data were collected based on the copies of the hospital medical records and the results of physical examination (the scars). Women with perineal trauma and episiotomy were combined in the group “perineal trauma in childbirth,” since both conditions lead to pelvic muscle weakness, higher risk of third- and fourth-degree traumas, and pelvic floor disorders [29].
To compare the studied genotypes distribution in Russian population and HapMap populations, we included the following populations from the project [30]: Caucasian populations, Utah residents with Northern and Western European ancestry (СEU), and Toscans in Italy (TSI); Asian populations, Chinese in Metropolitan Denver, Colorado (CHD), and Japanese in Tokyo (JPT); African populations, Yoruba in Ibadan, Nigeria (YRI), and Luhya in Webuye, Kenya (LWK); and two other populations, Gujarati Indians in Houston, Texas (GIH), and Mexican ancestry in Los Angeles, California (MEX). SNP genotype data were downloaded from the HapMap project site (The International HapMap Consortium, 2003), all HapMap samples were divided by gender, and only female samples (CEU, n = 89; TSI, n = 44; CHD, n = 44; JPT, n = 45; YRI, n = 80; LWK, n = 45; GIH, n = 43; MEX, n = 43) were included in a principal component analysis.
2.2. SNPs Selection and Genotyping
The purpose of this study was to replicate the results of genome-wide linkage and genome-wide association studies for pelvic organ prolapse. In the linkage study of high-risk POP pedigrees, 9q21 region (between rs4077632 and rs10868525 (Assembly GRCh37.p13, 9: 81163473–89618028 bp)) has shown an evidence for a predisposition to POP [20]. The top effect was found for rs11139451. Some other intergenic markers (e.g., rs4077632) and possible genes candidates in the region (e.g., TLE1 and TLE4) have been mentioned. Additionally to the SNPs cited by the authors (rs4077632 and rs11139451), we chose four markers utilizing RegulomeDB functional annotations for the region 9q21. There are SNPs rs2777781 in the gene TLE1 (RegulomeDB score 3a), rs2807303 in the gene TLE4 (RegulomeDB score 2a), and rs12237222 (RegulomeDB score 1a) and rs12551710 in the gene FRMD3 (RegulomeDB score 1d). Six other SNPs have been associated with POP in the more recent genome-wide association study of Allen-Brady et al. [21], that is, rs1455311 (4q21), rs1036819 (8q24), rs430794 (9q22), rs8027714 (15q11), rs1810636 (20p13), and rs2236479 (21q22). These SNPs were also included in our investigation.
DNA was isolated from 200 μL of blood using gDNA purification kit Diatom DNA Prep 200 (Isogene laboratory, Moscow, Russia). The genotyping was performed with a PCR-CTPP (polymerase chain reaction with confronting two-pair primers) [31]. Amplification was carried out using an ABI thermal cycler with two external and two internal sequence-specific primers (Supplementary Table 1, in Supplementary Material available online at http://dx.doi.org/10.1155/2015/837904) and tubes PCR MasterMix (Isogene laboratory, Moscow, Russia). The PCR products were analyzed in ethidium bromide-stained 2% agarose gel. Blinded duplicates (10%) of randomly taken DNA samples for each SNP were genotyped once more with 100% concordance in genotype calling.
2.3. Statistical Analysis
Categorical demographic and clinical variables were assessed using Fisher exact test (two-tailed). Since continuous demographic and clinical variables did not assume a normal distribution, the Mann-Whitney U nonparametric test was used to compare such variables. Deviation from Hardy-Weinberg equilibrium was assessed by χ 2 analysis. Logistic regression analysis implemented in SNPStats package [32] was performed to assess associations between studied SNPs and POP. In multivariate analysis, we adjusted adjusted for main covariates: age, BMI, vaginal parity, and vaginal trauma in childbirth. Genetic model was selected using the Akaike Information Criterion (AIC) value with the lowest AIC value considered corresponding to the best-fitting model for the fitted variant. For genotypes with minor allele frequencies <5% only dominant and additive genetic models were evaluated. A two-tailed P value < 0.05 was considered statistically significant.
Pairwise linkage disequilibrium (LD) values were measured as Lewontin's D′-values and estimated from genotype data using the expectation-maximization algorithm implemented in SNPStats software. The SNPStats software was also used to evaluate haplotype association with POP by individual haplotype P values.
Gene-gene interactions were estimated using “SNPassoc” package [33] within R statistical software [34] performing log-likelihood ratio tests (LRTs).
WINPEPI test power calculator [35] was used to assess sample size and test power. Assuming that odds ratio (OR) ≥ 2.0 is clinically meaningful [36], we established test power in the range of 70.30% to 96.06% (at two-sided significance level) for this effect size, sample size n = 500 (300 controls and 200 cases), and risk genotype frequencies in a range of 10%–50%.
Successful replication study presents a statistically significant association of the same genetic variant in the same direction [37]. Our study cannot be considered a classic replication study since prior results have been obtained in a genome-wide linkage analysis for a gene region. Nevertheless, our results provide verification of the association of pelvic organ prolapse with the 9q21 haplotype in Caucasians. It is accepted that replication studies do not require genome-wide multiple testing correction [38]. In our study, we also did not adjust for multiple comparisons since we tested robust statistical associations [39, 40]. The Bonferroni correction was applied when testing for SNP interaction.
To detect admixture, we investigated Russian and HapMap populations structure via the discriminant analysis of principal components (DAPC) [41]. It was evaluated using the adegenet package in R statistical software [42].
Interpopulation differences in allele frequencies were assessed with pairwise Weir and Cockerham's Fst estimator (Fst), using package diversity [43].
3. Results
3.1. SNPs Characteristics
SNPs considered in the study are described in Table 1. Of the 12 studied SNPs, five SNPs are intergenic, while seven SNPs are located in the genes. Among these seven SNPs, five SNPs are related to the protein-coding genes, TLE1 (rs2777781), TLE4 (rs2807303), COL18A1 (rs2236479), ZFAT (rs1036819), and FRMD3 (rs12551710), and two SNPs are mapped to the RNA gene LINC01088 (rs1455311) and uncharacterized gene LOC102723989 (rs12237222). With few exceptions (rs2807303 and rs1036819), the majority of the SNPs assigned to the genes lie within intron regions. The choice of the genetic markers rs12237222 (score 1a) and rs12551710 (score 1d) in the region 9q21 was based on the RegulomeDB annotations. We present here the comparative qualitative and quantitative characteristics of the RegulomeDB SNPs in the categories 1a–1f in the whole human genome and in the region 9q21 (Table 2). Among the relatively small number of the SNPs scored 1a in the whole human genome, only one SNP with a score 1a was found in the region 9q21. This is the SNP rs12237222.
Table 1.
SNPs considered in this study.
| rs | Chromosome region | Chromosome position assembly GRCh37.p13 |
Gene | Function | RegulomeDB score | Alleles | MAF (Allele) |
|---|---|---|---|---|---|---|---|
| rs1455311 | 4q21 | 79964587 | LINC01088 | Intron | 5 | C:T | 0.217 (C) |
| rs1036819 | 8q24 | 135611945 | ZFAT-AS1, ZFAT | NearGene-3 | 4 | A:C | 0.128 (C) |
| rs4077632 | 9q21 | 81163473 | — | — | No data | A:G | 0.310 (G) |
| rs2807303 | 9q21 | 82187095 | TLE4 | NearGene-5 | 2a | C:T | 0.336 (T) |
| rs2777781 | 9q21 | 84215034 | TLE1 | Intron | 3a | A:T | 0.309 (T) |
| rs11139451 | 9q21 | 84495608 | — | — | 5 | C:T | 0.221 (C) |
| rs12237222 | 9q21 | 85834743 | LOC102723989 | Intron | 1a | G:T | 0.425 (G) |
| rs12551710 | 9q21 | 86088295 | FRMD3 | Intron | 1d | C:T | 0.042 (T) |
| rs430794 | 9q22 | 93852815 | — | — | 5 | A:C | 0.272 (A) |
| rs8027714 | 15q11 | 24964597 | — | — | 6 | A:G | 0.040 (A) |
| rs1810636 | 20p13 | 2654925 | — | — | 4 | G:T | 0.332 (T) |
| rs2236479 | 21q22 | 46919132 | COL18A1 | Intron | 1f | A:G | 0.339 (A) |
Table 2.
The distribution of RegulomeDB category 1a–1f SNPs in the whole human genome and in the region 9q21.
| RegulomeDB category | Category description | Number of SNPs with RegulomeDB score 1a–1f | |
|---|---|---|---|
| In the whole human genome | In the region 9q21 | ||
| 1a | eQTL + TF binding + matched TF motif + matched DNase footprint + DNase peak | 352 | 1 |
| 1b | eQTL + TF binding + any motif + DNase footprint + DNase peak | 2568 | 17 |
| 1c | eQTL + TF binding + matched TF motif + DNase peak | 85 | 0 |
| 1d | eQTL + TF binding + any motif + DNase peak | 1668 | 7 |
| 1e | eQTL + TF binding + matched TF motif | 54 | 0 |
| 1f | eQTL + TF binding/DNase peak | 34706 | 127 |
3.2. Characteristics of the Study Population
In total, 210 patients with pelvic organ prolapse (stages III-IV) and 292 control subjects with no even minimal prolapse were included in the study. The description of the study population is presented in Supplementary Table 2. Briefly, our cases and controls were similar in ethnicity (Caucasians from the European region of Russian Federation) and age (57.65 ± 10.80 and 57.25 ± 12.70, resp.). Higher BMI (29.17 ± 5.85 and 27.46 ± 6.56, P = 5.6 × 10−4), vaginal parity (1.69 ± 0.65 and 1.46 ± 0.54, P = 0.014), and perineal trauma in childbirth (52.45% versus 32.75%, P = 1.7 × 10−5) were found in POP patients than in matched controls.
3.3. Single-Locus Analysis
All SNPs were in Hardy-Weinberg equilibrium in the cases and in the controls. In the crude analysis, the minor rs12237222-G allele frequency was found to be higher in the cases than in the controls under the dominant model (Table 3). This effect disappeared in the multivariate analysis. Other SNPs were not associated with POP neither in the crude nor in the multivariate analysis (Supplementary Table 3).
Table 3.
The distribution of rs12237222 genotypes among cases and controls.
| Genotypes | Control | POP |
Crude P value, OR (95% CI) |
Adjusted P valueaOR, (95% CI) | ||
|---|---|---|---|---|---|---|
| Number (%) n = 291 |
HWP | Number (%) n = 210 |
HWP | |||
| rs12237222 | ||||||
| T/T | 104 (35.7) | 0.63 | 55 (26.2) | 0.21 | 0.023 (dom) 1.57 (1.06–2.31) |
0.12 (dom) 1.41 (0.91–2.18) |
| T/G | 136 (46.7) | 114 (54.3) | ||||
| G/G | 51 (17.5) | 41 (19.5) | ||||
HWP: Hardy-Weinberg probability; OR: odds ratio; CI: confidence interval; dom: dominant model.
aAdjusted by age, body mass index (BMI), perineal trauma in childbirth, and vaginal parity. In multivariate analysis, there were 271 controls and 198 cases.
3.4. Haplotype Analysis
For the haplotype analysis, we included all studied SNPs located in the region 9q21 (Table 4). Haplotype number four comprising rs12237222-G allele was significantly associated with POP in the analysis adjusted for age, BMI, vaginal parity, and peritoneal trauma in childbirth. When we excluded this SNP from the haplotype analysis, we found that haplotype number one that included risk alleles from the remaining five SNPs became the reference haplotype (the most common). The pronounced effect was also found for the two-SNP-haplotype rs2777781-A/rs12237222-G with total haplotype frequency 0.3087 (Table 4). No significant association was observed between other haplotypes and POP development.
Table 4.
The distribution of 9q21 haplotypes among cases and controls.
| Number | rs4077632 | rs2807303 | rs2777781 | rs11139451 | rs12237222 | rs12551710 | Frequencies | P value | OR (95% CI) |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Controls | POP | |||||||||
| Six SNP-haplotype associations with response | ||||||||||
| 1 | A | T | A | T | T | C | 0.1334 | 0.0985 | — | 1.00 |
| 2 | A | C | A | T | T | C | 0.1185 | 0.0943 | 0.38 | 0.59 (0.19–1.90) |
| 3 | G | C | A | T | G | C | 0.0436 | 0.0937 | 0.24 | 1.92 (0.65–5.71) |
| 4 | A | C | A | T | G | C | 0.059 | 0.1019 | 0.029 | 3.36 (1.13–9.95) |
|
| ||||||||||
| Five SNP-haplotype associations with response | ||||||||||
| 1 | A | C | A | T | — | C | 0.1807 | 0.205 | — | 1.00 |
| 2 | A | T | A | T | — | C | 0.1662 | 0.1324 | 0.96 | 1.02 (0.48–2.16) |
| 3 | G | C | A | T | — | C | 0.1146 | 0.1171 | 0.68 | 0.85 (0.39–1.86) |
| 4 | A | C | T | T | — | C | 0.0949 | 0.0763 | 0.63 | 0.81 (0.34–1.92) |
|
| ||||||||||
| Two SNP-haplotype associations with response | ||||||||||
| 1 | — | — | A | — | T | — | 0.4581 | 0.3663 | — | 1.00 |
| 2 | — | — | A | — | G | — | 0.2782 | 0.3575 | 0.0091 | 1.72 (1.15–2.59) |
| 3 | — | — | T | — | T | — | 0.1328 | 0.167 | 0.064 | 1.62 (0.97–2.70) |
| 4 | — | — | T | — | G | — | 0.1309 | 0.1092 | 0.63 | 0.88 (0.53–1.47) |
Only haplotypes with total frequencies ≥10% are considered. Analysis is adjusted by age, body mass index (BMI), peritoneal trauma in childbirth, and vaginal parity. Significant results are in bold.
It is interesting to note that rs12237222 is in weak but highly significant linkage disequilibrium with rs11139451 (D′ = 0.257, P = 1.45 × 10−6) that has been found to show the maximum HLOD score in the study of Allen-Brady et al. [20] (Supplementary Table 4).
3.5. Analysis of SNP Interaction
Figure 1 presents a heat plot color map for an analysis of SNP interaction. Cells are colored according to interaction P value (less significant effects, light yellow; more significant effects, green). The most significant interactions (dark green cells) were observed for two SNP pairs, rs12551710 (RegulomeDB score 1d) and rs1455311 (RegulomeDB score 5) (P = 7.3 × 10−5), from first interaction pair and rs2236479 (RegulomeDB score 1f) and rs2777781 (RegulomeDB score 3a) (P = 1.7 × 10−4) from second interaction pair. It should be highlighted that these highly significant results did not reflect statistical interaction, since the above SNPs were not associated with POP in our single-locus analysis. Given that interaction results were novel, we used correction for multiplicity considering all possible combinations (12 × 12 = 144). The results remained significant after Bonferroni correction for twelve SNP pairs (P = 0.011 and P = 0.024, resp.).
Figure 1.
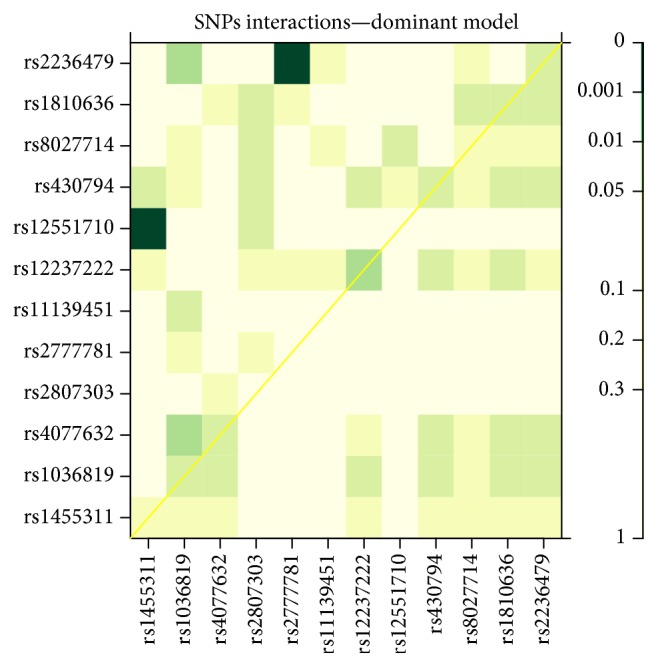
Gene-gene interaction plot. The plot shows the significance levels of gene-gene interactions and pelvic organ prolapse (POP) in the dominant model. Each plot indicated the P values preceded by different likelihood ratio tests. Different colors show different levels of statistical significance. The diagonal line contains the P values from likelihood ratio test for the crude effect of each SNP. The upper triangle in the matrix presents the P values for the interaction (epistasis) log-likelihood ratio test. The lower triangle shows the P values from likelihood ratio test comparing the two-SNP additive likelihood to the best of the single-SNP models.
3.6. Analysis of Population Structure
The discriminant analysis of principal components showed that Russian population has no internal stratification (Figure 2). Russian population was located close to other Caucasian populations, CEU and TSI. African YRI and LWK populations and Asian CHD and JPT populations were far from Caucasian populations, while GIH and MEX populations took an intermediate position (Figure 2).
Figure 2.
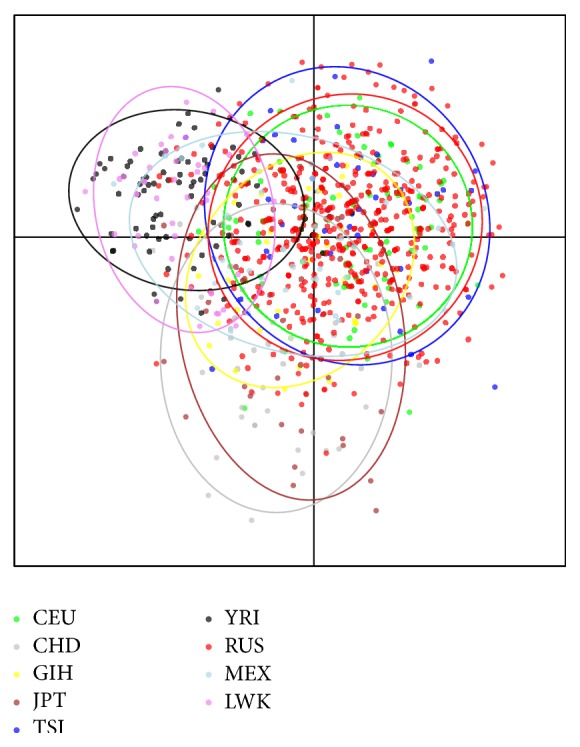
Structure analysis of 12 SNPs in this data (RUS) and in other Caucasian populations (CEU, Utah residents from Northern and Western European and TSI, Toscans from Italy), Asian populations (CHD, Chinese from Metropolitan Denver, Colorado, and JPT, Japanese from Tokyo), African populations (YRI, Yoruba from Ibadan, Nigeria, and LWK, Luhya from Webuye, Kenya), and two other populations (GIH, Gujarati Indians from Houston, Texas, and MEX, Mexican ancestry in Los Angeles, California). Only female samples were included in a principal component analysis. HapMap data were absent in TSI, MEX, GIH, CHD, and LWK populations for rs1223722 and rs12551710 and in CHD population for rs8027714.
Fst statistics are presented in Supplementary Table 5. The pairwise Fst values between the examined populations were in line with the data of DAPC. The lowest pairwise Fst values (Fst = 0) were revealed for CEU-RUS and CEU-TSI populations.
4. Discussion
In this gene candidate study, we tested the association of the region 9q21 and six other loci [20, 21] with advanced POP in Caucasians. We used RegulomeDB resource to select regulatory SNPs in the region 9q21. There was only one SNP rs12237222 in the region with the best RegulomeDB score 1a. This SNP appeared to be the key SNP for the 9q21 haplotype associated with POP. Two other category 1 variants, which are also characterized as eQTL, rs2236479 (21q22), and rs12551710 (9q21) exhibited epistatic (nonlinear interaction) effects with rs2777781 and rs1455311, respectively. To control for the confounding effect of known clinical covariates (age, BMI, vaginal parity, and pelvic floor trauma in childbirth), we adjusted for these factors. To assess hidden population stratification, we compared the genotypic frequencies of 12 studied SNPs to those of HapMap ethnic groups and found that our sample showed little structure.
In terms of ancestry, Russians are Caucasians. The study of Russian population was representative, from an ethnic standpoint of the other Caucasian populations (CEU and TSI). Other HapMap populations in this study, YRI, LWK, JPT, and CHD populations differed significantly from all three Caucasian populations, Russian, CEU, and TSI. It is worth noting that RUS, CEI, and TSI populations and relatively close to them GIH and MEX populations are characterized by high prevalence of POP [4, 44, 45]. In contrast, African and Asian women presented in our study by YRI, LWK, JPT, and CHD populations are less prone to POP development [4, 44, 46, 47]. This may be explained mainly by anatomical features [48, 49]. Nevertheless, it is an interesting challenge for future research directions to determine whether ethnic-specific genotype frequencies of the studied SNPs make any contribution to higher prevalence of POP in Caucasian, GIH, and MEX females compared with Asian and African females.
All our patients had an advanced prolapse (stages III-IV on the POP-Q examination), while our controls had not even minimal prolapse. Consideration of more extreme phenotypes is recommended for the genetic association studies [50]. Utilizing extreme phenotypes increases the association effects, with improved statistical power of relatively modest sample sizes [51]. In relation to POP, this direction could be taken into account not only for patients but for controls as well. In a recent study of Wu et al. [15], their control group included controls with no or minimal prolapse, who were older than patients. We applied another approach for recruitment of an appropriate comparison group. Should POP arise, we would expect that subclinical manifestation progresses to more severe symptoms. The rate of this progress depends on many variables including the history of vaginal childbirth and other clinical conditions (e.g., hysterectomy), age, frequent heavy lifting, and intense physical activity leading to the repetitive increase of pelvic pressure [52]. For this reason, we purposely included women without any subclinical POP in our control group.
It is known that over 98% of the human genome comprises noncoding DNA. However, the Encyclopedia of DNA elements (ENCODE) project has recently demonstrated that a biochemical function could be assigned to 80% of the human genome [53]. Regulatory activity of noncoding DNA explains the fact that a very high amount (90%) of GWAS-identified SNPs associated with multifactorial diseases and complex traits are located in noncoding regions [54]. RegulomeDB includes data sets from ENCODE and other sources to predict regulatory function of a SNP in a score based system. Lower scores mean increasing evidence that a variant is located in a functional region: category 1 indicates that a variant has the highest likelihood of being linked to the expression of a target gene and category 1a is characterized by a full set of features (eQTL + TF binding + matched TF motif + matched DNase Footprint + DNase peak). RegulomeDB project provides an important tool for both tasks, planning an association study and post hoc analysis; its data are useful for generating hypotheses and explaining possible epistasis interactions.
SNP interaction might be common among eQTL SNPs that influence gene expression. Epistatic interactions involve local or distant factors acting in cis or trans. Cis-interaction usually occurs within 500 kb region and may sometimes reflect an underlying haplotype effect, while transinteraction indicates interaction between SNPs from distant regions [55]. Loci involved in gene-gene interactions may not show associations on their own. It is also known that in replication studies some loci associations could not be verified, but the corresponding regions might show clear interaction signals [56]. We could not replicate all associations from the studies of Allen-Brady et al. [20, 21], but the interaction effects revealed in our study for several SNPs are in line with the associations found in the above studies for the relevant SNPs.
It is proposed that epistasis among regulatory variants can have strong phenotypic effects, since gene regulation has an important role in adaptation to different harmful stimuli. Regulatory SNPs often lie in noncoding sequences and affect transcription of coding and noncoding RNAs or RNA functions and processing [57]. In our study, among SNPs associated with POP in the haplotype and/or interaction analysis, three SNPs (TLE1 rs2777781, FRMD3 rs12551710, and COL18A1 rs2236479) were located within the genes with possible impact upon development and organization of muscle and connective tissue of the pelvic floor [20, 21]. Their effects may be realized based on regulation of the expression of these genes or via transregulatory activity. Two other SNPs, rs1455311 and rs12237222 (the top SNP in this assay), within the nonprotein coding genes were also associated with POP in the haplotype or interaction analysis. Based on data of SCAN SNP and CNV Annotation Database [58] assayed in lymphoblastoid cells, these SNPs appeared to exhibit transregulatory activity in CEU population: rs1455311 was associated with the expression of the NAPRT1 (nicotinate phosphoribosyl transferase domain containing 1) gene and rs12237222 influenced the expression of the CAT (catalase) and GYPE (glycophorin E) genes. NAPRT1 and CAT genes encode antioxidant enzymes and control response to oxidative stress, which is known to play a role in the development of many chronic or late-onset diseases including POP [59]. Regulatory SNPs may exert a tissue-dependent effect on gene expression but in general, “allele specific expression differences between individuals dominate over tissue-specific effects” [60]. Basing on these data, we can speculate that the above SNPs may be associated with POP in haplotype or interaction dependent mode, in particular through influence on efficacy of oxidative stress response.
A limitation of our study is that, firstly, our sample size was modest and the study is powered to detect only relatively large effect sizes (minimum detectable OR~1.7). Secondly, the study is limited to the population of Russian Federation. The strength of the study is in the sampling of extreme phenotypes, the inclusion of all main risk factors for POP in multivariate regression analysis, little population structure, and using of RegulomeDB resource to select functional SNPs.
5. Conclusions
The findings of the present study verified the results of the genome-wide linkage analysis in European pedigrees with POP. We found that the key SNP in the haplotype 9q21 associated with POP in our Russian population had the best RegulomeDB score 1a and two other SNPs category 1 were involved in SNP-SNP interaction associations. To the best of our knowledge, this is the first successful research which was performed utilizing RegulomeDB annotations a priori, for planning investigations. RegulomeDB annotations can be useful for designing an association study of the chromosome region.
Supplementary Material
Table S1. Summary on primer sequences, amplicons, and PCR cycling conditions for individual SNPs analyzed in the study.
Table S2. Characteristics of the POP and control groups involved in the study.
Table S3. The distribution of genotypes among cases and controls.
Table S4. Linkage disequilibrium analysis of six SNPs in the region 9q21.
Table S5. Fst pairwise values between populations.
Acknowledgment
This work was supported by the Russian Foundation for Basic Research, Project 15-04-02378.
Conflict of Interests
The authors declare no potential conflict of interests.
References
- 1.Nygaard I., Barber M. D., Burgio K. L., et al. Prevalence of symptomatic pelvic floor disorders in US women. The Journal of the American Medical Association. 2008;300(11):1311–1316. doi: 10.1001/jama.300.11.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith F. J., Holman C. D. J., Moorin R. E., Tsokos N. Lifetime risk of undergoing surgery for pelvic organ prolapse. Obstetrics & Gynecology. 2010;116(5):1096–1100. doi: 10.1097/aog.0b013e3181f73729. [DOI] [PubMed] [Google Scholar]
- 3.Mannella P., Palla G., Bellini M., Simoncini T. The female pelvic floor through midlife and aging. Maturitas. 2013;76(3):230–234. doi: 10.1016/j.maturitas.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Kumari S., Walia I., Singh A. Self-reported uterine prolapse in a resettlement colony of North India. The Journal of Midwifery & Women's Health. 2000;45(4):343–350. doi: 10.1016/s1526-9523(00)00033-7. [DOI] [PubMed] [Google Scholar]
- 5.Jelovsek J. E., Barber M. D. Women seeking treatment for advanced pelvic organ prolapse have decreased body image and quality of life. The American Journal of Obstetrics & Gynecology. 2006;194(5):1455–1461. doi: 10.1016/j.ajog.2006.01.060. [DOI] [PubMed] [Google Scholar]
- 6.Lukacz E. S., Lawrence J. M., Contreras R., Nager C. W., Luber K. M. Parity, mode of delivery, and pelvic floor disorders. Obstetrics & Gynecology. 2006;107(6):1253–1260. doi: 10.1097/01.aog.0000218096.54169.34. [DOI] [PubMed] [Google Scholar]
- 7.Miedel A., Tegerstedt G., Mæhle-Schmidt M., Nyrén O., Hammarström M. Nonobstetric risk factors for symptomatic pelvic organ prolapse. Obstetrics and Gynecology. 2009;113(5):1089–1097. doi: 10.1097/aog.0b013e3181a11a85. [DOI] [PubMed] [Google Scholar]
- 8.Altman D., Forsman M., Falconer C., Lichtenstein P. Genetic influence on stress urinary incontinence and pelvic organ prolapse. European Urology. 2008;54(4):918–923. doi: 10.1016/j.eururo.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Ferrari M. M., Rossi G., Biondi M. L., Viganò P., Dell'Utri C., Meschia M. Type I collagen and matrix metalloproteinase 1, 3 and 9 gene polymorphisms in the predisposition to pelvic organ prolapse. Archives of Gynecology and Obstetrics. 2012;285(6):1581–1586. doi: 10.1007/s00404-011-2199-9. [DOI] [PubMed] [Google Scholar]
- 10.Ferrell G., Lu M., Stoddard P., et al. A single nucleotide polymorphism in the promoter of the loxl1 gene and its relationship to pelvic organ prolapse and preterm premature rupture of membranes. Reproductive Sciences. 2009;16(5):438–446. doi: 10.1177/1933719108330567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khadzhieva M. B., Kamoeva S. V., Chumachenko A. G., et al. Fibulin-5 (FBLN5) gene polymorphism is associated with pelvic organ prolapse. Maturitas. 2014;78(4):287–292. doi: 10.1016/j.maturitas.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Kluivers K. B., Dijkstra J. R., Hendriks J. C. M., Lince S. L., Vierhout M. E., van Kempen L. C. L. COL3A1 2209G>A is a predictor of pelvic organ prolapse. The International Urogynecology Journal. 2009;20(9):1113–1118. doi: 10.1007/s00192-009-0913-y. [DOI] [PubMed] [Google Scholar]
- 13.Nikolova G., Lee H., Berkovitz S., et al. Sequence variant in the laminin γ1 (LAMC1) gene associated with familial pelvic organ prolapse. Human Genetics. 2007;120(6):847–856. doi: 10.1007/s00439-006-0267-1. [DOI] [PubMed] [Google Scholar]
- 14.Skorupski P., Jankiewicz K., Miotła P., Marczak M., Kulik-Rechberger B., Rechberger T. The polymorphisms of the MMP-1 and the MMP-3 genes and the risk of pelvic organ prolapse. The International Urogynecology Journal. 2013;24(6):1033–1038. doi: 10.1007/s00192-012-1970-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J. M., Visco A. G., Grass E. A., et al. Matrix metalloproteinase-9 genetic polymorphisms and the risk for advanced pelvic organ prolapse. Obstetrics & Gynecology. 2012;120(3):587–593. doi: 10.1097/aog.0b013e318262234b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J. M., Visco A. G., Grass E. A., et al. Comprehensive analysis of LAMC1 genetic variants in advanced pelvic organ prolapse. The American Journal of Obstetrics & Gynecology. 2012;206(5):447.e1–447.e6. doi: 10.1016/j.ajog.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H.-Y., Chung Y.-W., Lin W.-Y., Chen W.-C., Tsai F.-J., Tsai C.-H. Estrogen receptor alpha polymorphism is associated with pelvic organ prolapse risk. The International Urogynecology Journal. 2008;19(8):1159–1163. doi: 10.1007/s00192-008-0603-1. [DOI] [PubMed] [Google Scholar]
- 18.Chen H.-Y., Chung Y.-W., Lin W.-Y., Wang J.-C., Tsai F.-J., Tsai C.-H. Collagen type 3 alpha 1 polymorphism and risk of pelvic organ prolapse. The International Journal of Gynecology and Obstetrics. 2008;103(1):55–58. doi: 10.1016/j.ijgo.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 19.Chen H.-Y., Chung Y.-W., Lin W.-Y., Chen W.-C., Tsai F.-J., Tsai C.-H. Progesterone receptor polymorphism is associated with pelvic organ prolapse risk. Acta Obstetricia et Gynecologica Scandinavica. 2009;88(7):835–838. doi: 10.1080/00016340902822073. [DOI] [PubMed] [Google Scholar]
- 20.Allen-Brady K., Norton P. A., Farnham J. M., Teerlink C., Cannon-Albright L. A. Significant linkage evidence for a predisposition gene for pelvic floor disorders on chromosome 9q21. The American Journal of Human Genetics. 2009;84(5):678–682. doi: 10.1016/j.ajhg.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen-Brady K., Cannon-Albright L., Farnham J. M., et al. Identification of six loci associated with pelvic organ prolapse using genome-wide association analysis. Obstetrics & Gynecology. 2011;118(6):1345–1353. doi: 10.1097/aog.0b013e318236f4b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen-Brady K., Cannon-Albright L. A., Farnham J. M., Norton P. A. Evidence for pelvic organ prolapse predisposition genes on chromosomes 10 and 17. The American Journal of Obstetrics & Gynecology. 2015;212(6):771.e1–771.e7. doi: 10.1016/j.ajog.2014.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bush W. S., Moore J. H. Chapter 11: genome-wide association studies. PLoS Computational Biology. 2012;8(12) doi: 10.1371/journal.pcbi.1002822.e1002822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyle A. P., Hong E. L., Hariharan M., et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Research. 2012;22(9):1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong K.-W., Jeong S. W., Chung M., Cho S. B. Association between expression quantitative trait loci and metabolic traits in two korean populations. PLoS ONE. 2014;9(12) doi: 10.1371/journal.pone.0114128.e114128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajkumar A. P., Christensen J. H., Mattheisen M., et al. Analysis of t(9;17)(q33.2;q25.3) chromosomal breakpoint regions and genetic association reveals novel candidate genes for bipolar disorder. Bipolar Disorders. 2015;17(2):205–211. doi: 10.1111/bdi.12239. [DOI] [PubMed] [Google Scholar]
- 27.Rosenthal S. L., Barmada M. M., Wang X., Demirci F. Y., Kamboh M. I. Connecting the dots: potential of data integration to identify regulatory snps in late-onset Alzheimer's disease GWAS findings. PLoS ONE. 2014;9(4) doi: 10.1371/journal.pone.0095152.e95152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zia A., Bhatti A., John P., Kiani A. K. Data interpretation: deciphering the biological function of Type 2 diabetes associated risk loci. Acta Diabetologica. 2015;52(4):789–800. doi: 10.1007/s00592-014-0700-1. [DOI] [PubMed] [Google Scholar]
- 29.Memon H. U., Handa V. L. Vaginal childbirth and pelvic floor disorders. Women's Health. 2013;9(3):265–277. doi: 10.2217/whe.13.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 31.Hamajima N. PCR–CTPP: a new genotyping technique in the era of genetic epidemiology. Expert Review of Molecular Diagnostics. 2001;1(1):119–123. doi: 10.1586/14737159.1.1.119. [DOI] [PubMed] [Google Scholar]
- 32.Solé X., Guinó E., Valls J., Iniesta R., Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 33.González J. R., Armengol L., Solé X., et al. SNPassoc: an R package to perform whole genome association studies. Bioinformatics. 2007;23(5):644–645. doi: 10.1093/bioinformatics/btm025. [DOI] [PubMed] [Google Scholar]
- 34.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. http://www.R-project.org/ [Google Scholar]
- 35.Abramson J. H. WINPEPI updated: computer programs for epidemiologists, and their teaching potential. Epidemiologic Perspectives and Innovations. 2011;8, article 1 doi: 10.1186/1742-5573-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shapiro S. Looking to the 21st century: have we learned from our mistakes, or are we doomed to compound them? Pharmacoepidemiology and Drug Safety. 2004;13(4):257–265. doi: 10.1002/pds.903. [DOI] [PubMed] [Google Scholar]
- 37.Chanock S. J., Manolio T., Boehnke M., et al. Replicating genotype-phenotype associations. Nature. 2007;447(7145):655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 38.Thomas D. C., Casey G., Conti D. V., Haile R. W., Lewinger J. P., Stram D. O. Methodological issues in multistage genome-wide association studies. Statistical Science. 2009;24(4):414–429. doi: 10.1214/09-STS288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gladman D. D., Farewell V. T. HLA studies in psoriatic arthritis: current situation and future needs. The Journal of Rheumatology. 2003;30(1):4–6. [PubMed] [Google Scholar]
- 40.Westfall P., Young S. S. Contradictions in highly cited clinical research. The Journal of the American Medical Association. 2005;294(21):2695–2696. doi: 10.1001/jama.294.21.2695-c. [DOI] [PubMed] [Google Scholar]
- 41.Jombart T., Devillard S., Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics. 2010;11, article 94 doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jombart T., Ahmed I. Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics. 2011;27(21):3070–3071. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keenan K., Mcginnity P., Cross T. F., Crozier W. W., Prodöhl P. A. DiveRsity: an R package for the estimation and exploration of population genetics parameters and their associated errors. Methods in Ecology and Evolution. 2013;4(8):782–788. doi: 10.1111/2041-210x.12067. [DOI] [Google Scholar]
- 44.Whitcomb E. L., Rortveit G., Brown J. S., et al. Racial differences in pelvic organ prolapse. Obstetrics & Gynecology. 2009;114(6):1271–1277. doi: 10.1097/aog.0b013e3181bf9cc8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu J. M., Vaughan C. P., Goode P. S., et al. Prevalence and trends of symptomatic pelvic floor disorders in U.S. women. Obstetrics and Gynecology. 2014;123(1):141–148. doi: 10.1097/AOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brizzolara S., Grandinetti A., Mor J. Percentage of hysterectomy of pelvic organ prolapse in five ethnic groups. International Urogynecology Journal. 2002;13(6):372–376. doi: 10.1007/s001920200082. [DOI] [PubMed] [Google Scholar]
- 47.Zacharin R. F. ‘A Chinese anatomy’: the pelvic supporting tissues of the Chinese and occidental female compared and contrasted. The Australian and New Zealand Journal of Obstetrics and Gynaecology. 1977;17(1):1–11. doi: 10.1111/j.1479-828x.1977.tb02661.x. [DOI] [Google Scholar]
- 48.Yang J.-M., Yang S.-H., Huang W.-C. Biometry of the pubovisceral muscle and levator hiatus in nulliparous Chinese women. Ultrasound in Obstetrics & Gynecology. 2006;28(5):710–716. doi: 10.1002/uog.3825. [DOI] [PubMed] [Google Scholar]
- 49.Handa V. L., Lockhart M. E., Fielding J. R., et al. Racial differences in pelvic anatomy by magnetic resonance imaging. Obstetrics & Gynecology. 2008;111(4):914–920. doi: 10.1097/aog.0b013e318169ce03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marian A. J. Molecular genetic studies of complex phenotypes. Translational Research. 2012;159(2):64–79. doi: 10.1016/j.trsl.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang K., Li W.-D., Zhang C. K., et al. A genome-wide association study on obesity and obesity-related traits. PLoS ONE. 2011;6(4) doi: 10.1371/journal.pone.0018939.e18939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Dell K. K., Morse A. N. It's not all about birth: biomechanics applied to pelvic organ prolapse prevention. The Journal of Midwifery and Women's Health. 2008;53(1):28–36. doi: 10.1016/j.jmwh.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 53.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hindorff L. A., Sethupathy P., Junkins H. A., et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turner S. D., Bush W. S. Multivariate analysis of regulatory SNPS: empowering personal genomics by considering cis-epistasis and heterogeneity. Proceedings of the 16th Pacific Symposium on Biocomputing (PSB '11); January 2011; pp. 276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y., Xu H., Chen S., et al. Genome-wide interaction-based association analysis identified multiple new susceptibility loci for common diseases. PLoS Genetics. 2011;7(3) doi: 10.1371/journal.pgen.1001338.e1001338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sadee W., Hartmann K., Seweryn M., Pietrzak M., Handelman S. K., Rempala G. A. Missing heritability of common diseases and treatments outside the protein-coding exome. Human genetics. 2014;133(10):1199–1215. doi: 10.1007/s00439-014-1476-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gamazon E. R., Zhang W., Konkashbaev A., et al. SCAN: SNP and copy number annotation. Bioinformatics. 2010;26(2):259–262. doi: 10.1093/bioinformatics/btp644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim E. J., Chung N., Park S. H., et al. Involvement of oxidative stress and mitochondrial apoptosis in the pathogenesis of pelvic organ prolapse. The Journal of Urology. 2013;189(2):588–594. doi: 10.1016/j.juro.2012.09.041. [DOI] [PubMed] [Google Scholar]
- 60.Gutierrez-Arcelus M., Ongen H., Lappalainen T., et al. Tissue-specific effects of genetic and epigenetic variation on gene regulation and splicing. PLOS Genetics. 2015;11(1) doi: 10.1371/journal.pgen.1004958.e1004958 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary on primer sequences, amplicons, and PCR cycling conditions for individual SNPs analyzed in the study.
Table S2. Characteristics of the POP and control groups involved in the study.
Table S3. The distribution of genotypes among cases and controls.
Table S4. Linkage disequilibrium analysis of six SNPs in the region 9q21.
Table S5. Fst pairwise values between populations.


