Abstract
Understanding the origins and evolutionary trajectories of symbiotic partnerships remains a major challenge. Why are some symbioses lost over evolutionary time whereas others become crucial for survival? Here, we use a quantitative trait reconstruction method to characterize different evolutionary stages in the ancient symbiosis between legumes (Fabaceae) and nitrogen-fixing bacteria, asking how labile is symbiosis across different host clades. We find that more than half of the 1,195 extant nodulating legumes analyzed have a high likelihood (>95%) of being in a state of high symbiotic persistence, meaning that they show a continued capacity to form the symbiosis over evolutionary time, even though the partnership has remained facultative and is not obligate. To explore patterns associated with the likelihood of loss and retention of the N2-fixing symbiosis, we tested for correlations between symbiotic persistence and legume distribution, climate, soil and trait data. We found a strong latitudinal effect and demonstrated that low mean annual temperatures are associated with high symbiotic persistence in legumes. Although no significant correlations between soil variables and symbiotic persistence were found, nitrogen and phosphorus leaf contents were positively correlated with legumes in a state of high symbiotic persistence. This pattern suggests that highly demanding nutrient lifestyles are associated with more stable partnerships, potentially because they “lock” the hosts into symbiotic dependency. Quantitative reconstruction methods are emerging as a powerful comparative tool to study broad patterns of symbiont loss and retention across diverse partnerships.
Keywords: symbiosis, cooperation, deep history, reconstruction, persistence
Symbiotic partnerships have transformed the Earth’s nutrient cycles (1), driven the diversification of organisms (2, 3), and facilitated rapid adaptation of species to divergent new niches (4). Almost all organisms rely on symbiotic associations for some form of metabolism, protection, or energy (5, 6). These relationships can be stabilized if the interests of the partners are tightly aligned or partners have evolved mechanisms to control and coordinate interactions (7–9). However, symbioses can also be unstable over evolutionary time (10–12), and conflicts among partners can lead to breakdown of potentially beneficial relationships, and even drive the emergence of parasitism (13, 14). Although our theoretical understanding of the factors that promote cooperative relationships has increased (e.g., refs. 15–18), we have lacked comparative approaches that can be used across a diversity of symbiotic systems to ask why some partnerships persist over millions of years and others are more labile. What distinguishes host species that acquire highly stable symbioses from those species that do not? Do different hosts follow similar trajectories of symbiotic evolution? Can different stages of symbiotic evolution be correlated to specific ecological conditions? One tool to study broad patterns of symbiont evolution is quantitative trait reconstruction (19–21), a comparative method that allows us to (i) identify when symbioses become more or less persistent and (ii) correlate these levels of persistence to ecological factors and partner traits.
Persistent Symbioses
We define symbiotic persistence as the continued capacity of an organism to form a particular symbiosis over evolutionary time (e.g., millions to tens of millions of years). High symbiotic persistence means that the capacity to form the symbiosis is more likely to be retained in a species or lineage whereas highly labile symbioses are characterized by low persistence and frequent loss of symbiotic capacity. As defined here, symbiotic persistence is an inherently phylogenetic concept (i.e., increased evolutionary stability of symbioses), meaning that species with an increased symbiotic persistence may be phenotypically indistinguishable from more labile symbioses. Symbiotic persistence is not the same as a symbiosis being obligate or genetically integrated. Although we expect intracellularly integrated partnerships to typically have high symbiotic persistence, the reverse is not necessarily true. For instance, some symbioses successfully persist for millions of years, without ever integrating intracellularly into the host (9, 22–25). When a symbiosis has become fully persistent, the loss rate falls to zero, or nearly zero, because even some usually presumed permanent symbioses (e.g., mitochondria or chloroplasts in multicellular organisms) can be lost in extremely rare cases (26, 27).
The level of symbiotic persistence can be quantified using modern comparative methods, allowing us to estimate the likelihood that a symbiosis will be retained, rather than lost (20, 28). Loss rates across phylogenetic trees can be compared to identify species or clades associated with more persistent symbiotic partnerships. We can then attempt to correlate highly persistent symbioses with global distribution maps, climate and soil databases, or organismal trait databases to elucidate the environmental conditions and species characteristics associated with evolutionary persistence.
Testing for Persistence: The Case of the N2-Fixing Symbiosis
Our aim is to quantify symbiotic persistence in the mutualism between nodulating plants and their nitrogen-fixing bacteria. We focus specifically on plants in the Fabaceae (legumes) associating with a class of nitrogen-fixing bacteria known broadly as rhizobia (29). In exchange for atmospheric nitrogen fixed by bacterial partners, plants provide rhizobia with photosynthate for energy and highly specialized root structures, called nodules, to house bacteria (30). Although rhizobia can survive and reproduce in the soil, colonization of a legume and establishment of a successful symbiosis can lead to a 108-fold increase in a single rhizobial cell’s abundance (31), thereby creating a strong selection pressure for symbiotic engagement. Host plants can also survive in the absence of bacterial partners but do, in many ecological situations (32), benefit greatly from the nitrogen the bacteria provide (33, 34).
The legume–rhizobial symbiosis is a good case study to quantify symbiotic persistence for a number of reasons. First, the phylogenetic relationship among potential hosts is well-characterized (35), and a newly compiled database of which hosts engage in the rhizobial symbiosis is available (36; data available from Dryad, dx.doi.org/doi:10.5061/dryad.05k14). Second, the symbiosis is found across a range of plant hosts, spanning highly diverse habitats and ecosystems (37, 38), allowing for potential variation in symbiotic persistence and in the selection pressures driving it. Third, different morphological types of the symbiosis have evolved across host plants (30, 38, 39), suggesting that there is potential for variation in the evolutionary trajectory of the symbiosis. Lastly, because the symbiosis is facultative for both partners, it represents an interesting case in which high symbiotic persistence is not necessarily linked with strict mutual dependence or host–symbiont integration, as is the case with eukaryotic organelles or some insect endosymbionts (40, 41).
We previously used a phylogenetic reconstruction approach called “Hidden Rate Models” (HRMs) (20, 28) to study the evolutionary history of the N2-fixing symbiosis across the angiosperms (36). These models allow for heterogeneity in the loss rate of a trait, like a symbiotic partnership, and thus can identify the phylogenetic signal where host clades shift in and out of different evolutionary states (20, 28), such as when labile symbiotic associations become more stable. In our previous work, we applied such a model to 3,467 angiosperm species, 1,709 of which were in the four symbiotic nitrogen-fixation orders (Fabales, Fagales, Rosales, and Cucurbitales), known as the nitrogen-fixing clade (37, 42). We found that, across the nitrogen-fixing clade, symbiotic host plants can be in two different evolutionary states: a “regular fixing state” where the ability to form the symbiosis remains highly labile (1.17 losses per 100 million years per lineage) or a second class termed “stable fixing state” in which host plants are characterized by extremely low loss rates (0.02 losses per 100 million years per lineage): i.e., almost 60 times lower than that of hosts in the regular fixing state (36). The analysis suggested that, for plants in a state of stable fixing, it is almost impossible to lose the capacity to form the symbiosis (36).
What plant hosts have entered into this stable symbiotic state and why? Can specific ecological factors and host traits be correlated with the stable fixing state? To answer these questions, we further studied the 1,366 legume species in our analyzed dataset, of which 1,195 are capable of forming the symbiosis (data available from Dryad, dx.doi.org/doi:10.5061/dryad.05k14). Legumes are the best characterized order in the nitrogen-fixing clade and have a wide geographic range (37, 38), ensuring that we could explore a range of potential environmental conditions and host traits, and there is considerable variation in their ability to successfully form a symbiosis with rhizobia (30). We first used an HRM approach to test whether the best fit model of symbiosis evolution in angiosperms was similar to that in legume species. To establish if this model still had the best fit, we repeated our HRM analysis using the legume dataset and the same phylogeny, this time pruned to the legumes (35; data available from Dryad, dx.doi.org/10.5061/dryad.63q27.2). We found that, in agreement with the angiosperm analysis (36), our best model included both a regular fixing state and a stable fixing state with increased symbiotic persistence [89.6% Akaike information criterion (AIC) weight]. Additionally, we found a highly significant correlation between the stable fixing state likelihoods of extant nodulating legume species from our previous analysis (36) and our analysis of legumes only (r = 0.92, P << 0.01). These observations confirm that our model of symbiosis evolution across the angiosperms also provides an accurate representation of the evolution of stable fixation within the legumes.
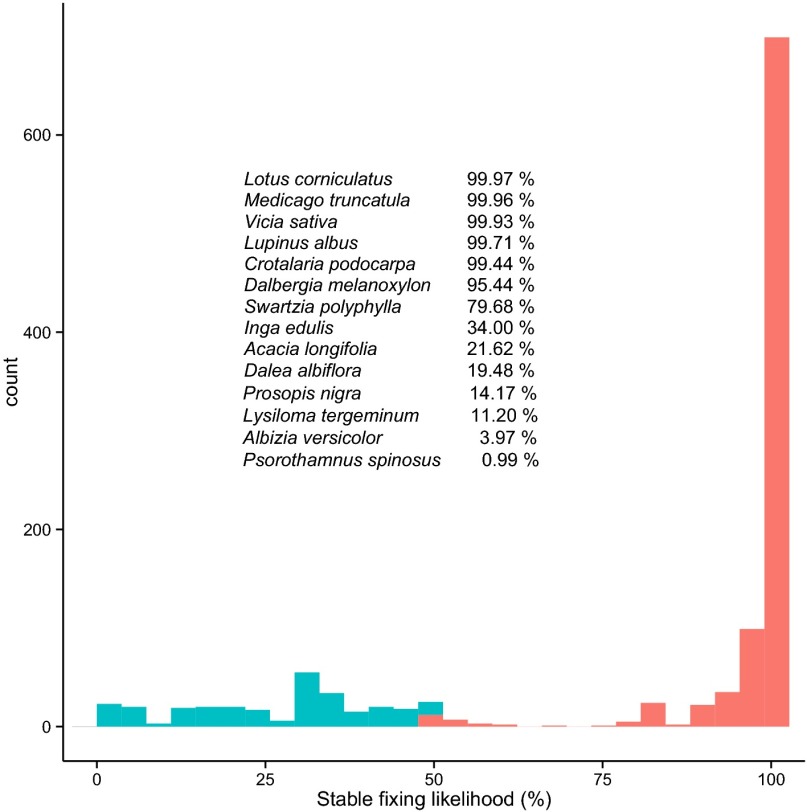
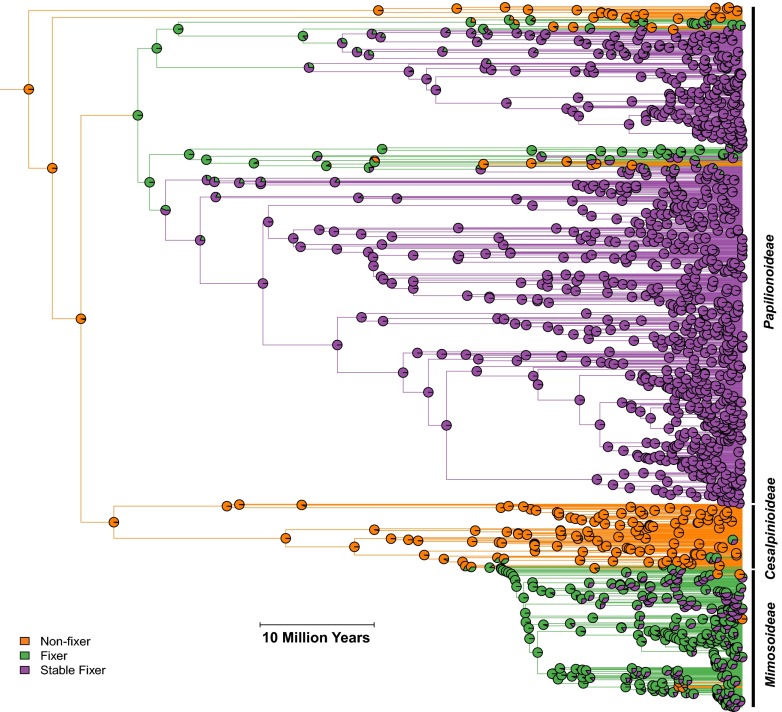
We then mapped our calculated stable fixing likelihoods (data available from Dryad, dx.doi.org/doi:10.5061/dryad.05k14) (Fig. 1) onto the legume phylogeny (Fig. 2) to analyze the evolutionary trajectory of the stable fixing state and to explore its correlation with ecological factors and host traits. The first observation is that there is considerable variation in symbiotic persistence. High stable fixing likelihoods (>95%) were found in 803 of the symbiotic legume species studied whereas 95 symbiotic species had very low likelihoods (<20%) of stable fixing (Fig. 1). The second observation is that the evolution to the stable fixing state is likely to have multiple origins across the legumes (Fig. 2). This finding suggests that there was no single event or innovation that allowed hosts to transition into a stable state, but rather that there were several parallel routes toward persistent capacity for symbiosis (36). Third, we observed that, within the Papilionoidea, there are repeated likely origins of stable fixing (Fig. 2). These origins range from the oldest over 50 million years ago, to a more recent origin a little over a million years ago, in the genus Amorpha (false indigo). This observation illustrates how, even within single clades, there is large variation in the timing of symbiotic stabilization. Fourth, we see that not all legumes have reached or are close to reaching the stable state (Fig. 1), which emphasizes that, in many legume clades like in the Mimosoideae (Fig. 2), the symbiosis is easier to lose.
Fig. 1.
Histogram of the stable fixing likelihood for all nodulating legume species in our reconstruction. To illustrate this variation, we list a range of species and their associated likelihoods of being stable fixers. Species with a stable fixing likelihood of over 50% are labeled red; the other species are labeled blue.
Fig. 2.
Pie charts indicate the likelihood of a host node being in either a nonsymbiotic state (orange), a “regular” N2-fixing state (green), or in a stable fixing state (purple), where the symbiosis has a high persistence. Represented are the legumes from their ancestral node about 62 million years ago. After the evolution of symbiotic nitrogen fixation (transition from orange to green), repeated evolution of stable fixing (transition from green to purple) is found across the legumes. Particularly within the Papilionoideae, legume species are very likely to currently be stable fixers, meaning that they show a continued capacity to form the symbiosis over evolutionary time. (Scale bar: 10 million years.)
Correlations Between Symbiosis Persistence and Ecological Factors or Host Traits
Quantitative reconstruction methods, such as the hidden rate models (20, 28), allow researchers to quantify the likelihood across the host phylogeny of evolutionary states that cannot necessarily be identified through phenotypic differences, such as the stable fixing state, and then ask whether there are ecological or trait commonalities among hosts in the stable fixing state. Specifically our aim was to ask how occurrence, climate, environment, and host trait data can be related to particular evolutionary states. We characterized symbiotic persistence in nodulating legumes by testing potential explanatory factors at four levels: (i) global geographical distribution, (ii) climate variables, (iii) local soil variables, and (iv) plant traits. For all variables considered, we explored the correlation of the potential explanatory variable (distribution, climate, etc.) with the underlying continuous likelihood of being a stable fixer as the response variable (36; data available from Dryad, dx.doi.org/doi:10.5061/dryad.05k14).
Latitudinal Pattern in Symbiotic Persistence Is Driven by Temperature.
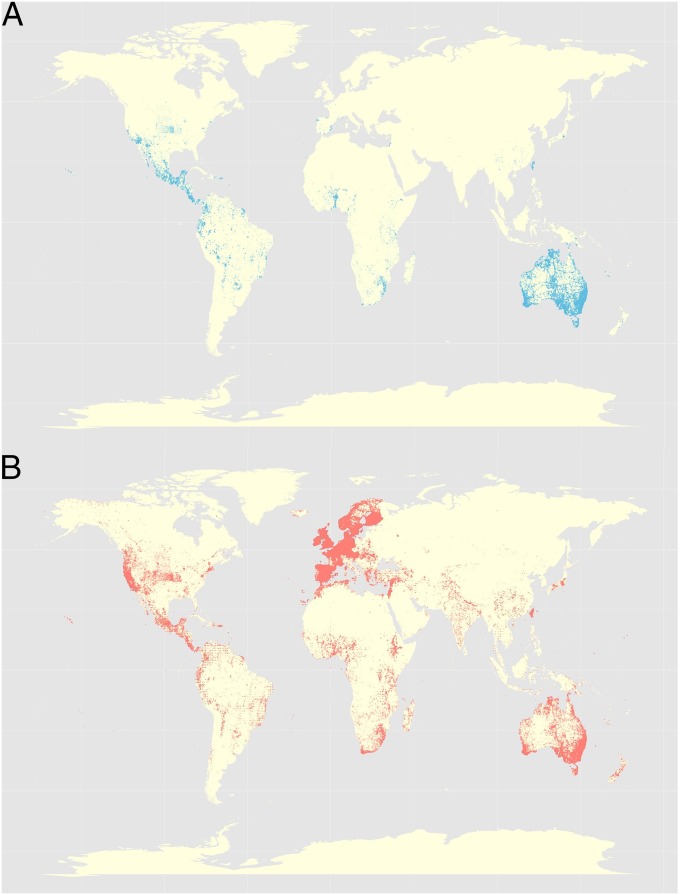
We first asked whether there were differences in global distribution between stable fixing legume species (>50% stable fixing likelihood) and legumes where the symbiosis is more easily lost (<50% stable fixing likelihood) (Fig. 1). We used the Global Biodiversity Information Facility (GBIF) to obtain (after quality control steps) a total of 3.2 million legume occurrences for 1,182 nodulating legume species in our analysis (43) (Methods). We plotted the occurrence of both regular (Fig. 3A) and stable fixers (Fig. 3B) onto the map of the world. We found that, in tropical regions, both regular and stable fixers are common. In contrast, in temperate, and particularly in polar regions, the legumes are almost exclusively characterized by highly persistent symbioses. When we considered the absolute median latitude of fixing legume species as a potential explanatory variable for stable fixing likelihood, we found a significant positive correlation using phylogenetic regression to correct for phylogenetic nonindependence (coefficient, 0.04% increase in stable fixing likelihood per degree absolute latitude, P = 0.02) (44, 45).
Fig. 3.
Occurrences of regular (A, blue) and stable fixing (B, red) legumes across the globe extracted from the Global Biodiversity Information Facility. Over 3.2 million occurrence records were plotted for the nodulating legumes analyzed in our dataset (Methods).
We then asked whether the correlation between latitude and symbiotic persistence could be explained by mean annual temperature (TMPMean). For each legume occurrence, we extracted the annual mean temperature (Fig. 3) from a global interpolated climate grid (46), obtaining a total of over 1.4 million temperature records, and calculated the median species-level TMPMean over all of the locations of a species for the 1,095 species for which sufficient data were available (Methods). We then used AIC criteria to test models that included temperature, latitude, and their interaction. We found that the only significant term was a negative correlation between the median annual mean temperature and stable fixing (P = 0.04), and that the previously significant latitude term was no longer significant in a full model containing both terms (phylogenetic regression, P = 0.41). The negative correlation between temperature and stable fixing was confirmed when we simplified the model to include only the significant model term temperature (coefficient, −0.1% change in stable fixing likelihood per degree TMPMean, P = 0.01). This finding suggests that the observed latitudinal pattern of stable symbiosis distribution is driven by temperature.
Soil Properties Are Not Correlated with Symbiotic Persistence in Legumes.
We then considered local environmental variables, specifically focusing on soil characteristics. We were interested in whether stable fixing hosts were positively correlated with low resource soils, particularly N availability. We extracted soil nitrogen, soil pH, and clay content based on our legume occurrence data (Fig. 2) from the Harmonized World Soil Database (47) and the International Soil Reference and Information Centre (ISRIC)-World Inventory of Soil Emission Potentials (WISE) database (48, 49), obtained a total of over 1.2 million records per soil variable and calculated species-level median values for 1,095 species (Methods). Using AIC criteria, we tested various models, which included the soil factors and their interactions. We found that none of the three soil variables improved the fit of our phylogenetic regression model compared with a model that fits only the intercept. We therefore rejected soil N, clay content, and pH as factors correlated with symbiotic persistence in the N2-fixing symbiosis in legumes.
High Host Phosphorus and Nitrogen Content Correlates with Symbiotic Persistence.
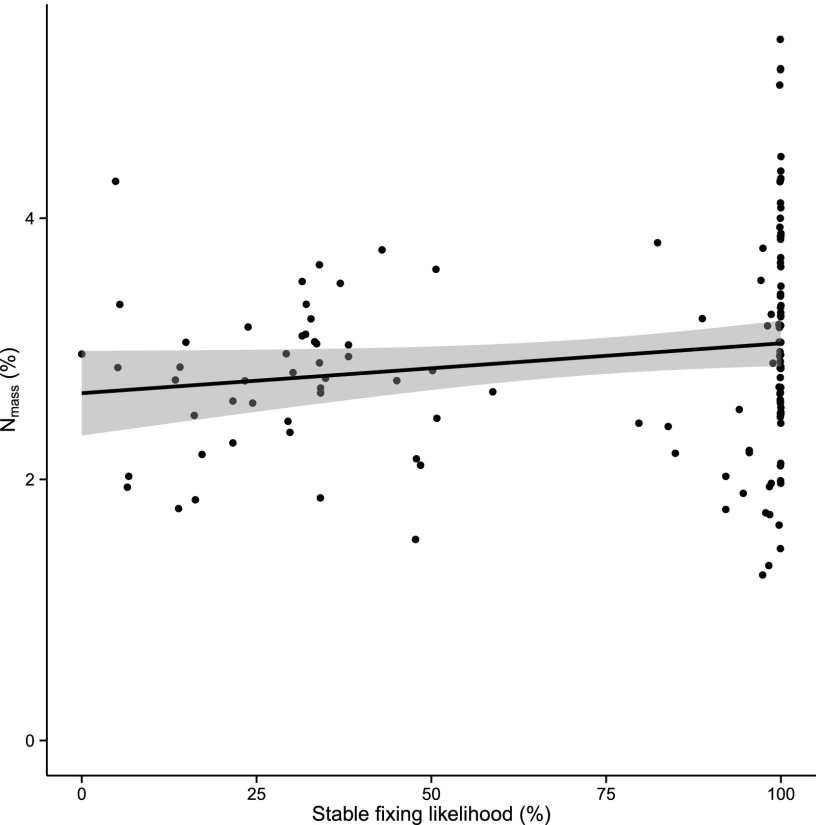
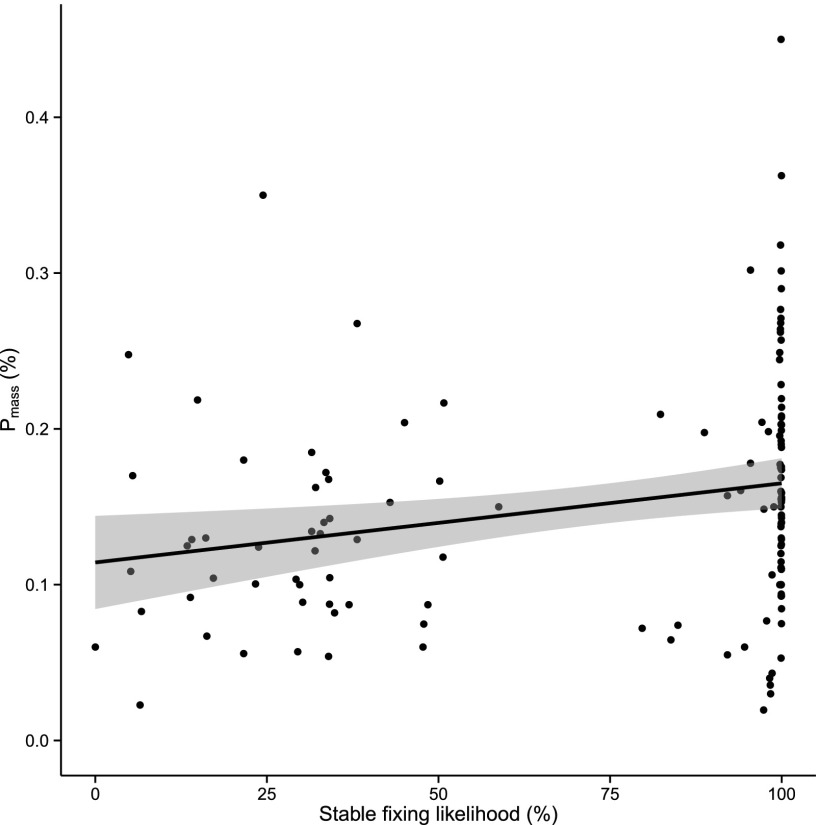
Lastly, we tested for correlations between stable fixing state likelihoods and host plant traits, focusing on leaf nitrogen and phosphorus content, asking whether the stable fixing state was associated with greater accumulation of nitrogen and phosphorus (32, 50). We obtained plant leaf nitrogen content (Nmass %; 1,114 records) and phosphorus content (Pmass %; 612 records) and calculated averages for 131 fixing legumes from our phylogenetic analysis for which data were available (Methods). We tested the association between stable fixing and host plant nutrient content in a phylogenetic regression model and found positive and significant relationships between Nmass % (coefficient 0.03, P = 0.03) (Fig. 4) and Pmass % (coefficient 0.05, P = 0.03) (Fig. 5) and the likelihood that a species is currently a stable fixer, as well as a negative interaction between Nmass % and Pmass % (coefficient, −0.01, P = 0.04). These positive correlations suggest that a higher symbiosis persistence is associated with a higher average leaf nitrogen and phosphorus content in legume hosts whereas the negative interaction term suggests that the positive relationship of either nutrient with symbiosis persistence diminishes with increases in the other nutrient.
Fig. 4.
Positive correlation between the calculated likelihood of extant nodulating legumes being in a stable fixing state and the average leaf nitrogen content of these same species (phylogenetic regression, P = 0.03).
Fig. 5.
Positive correlation between the calculated likelihood of extant nodulating legumes being in a stable fixing state and the average leaf phosphorus content of these same species (phylogenetic regression, P = 0.03).
Discussion
Symbiotic associations can be highly labile, with partnerships gained and lost over relatively short evolutionary time periods. Other symbioses show remarkable persistence over hundreds of millions of years. Although a suite of genomic tools has emerged to study symbiotic persistence as the physical process of host–symbiont integration (51–53), we lacked an approach that allowed researchers to ask broader scale questions about patterns and processes across diverse symbiotic systems. Quantitative phylogenetic reconstruction methods can be used to first identify clades with an increased symbiotic persistence, and then to analyze factors that can be correlated with persistence likelihoods. Studying symbiotic persistence in the legume–rhizobia mutualism is interesting because (i) there are legume clades in which the symbiosis is relatively labile over evolutionary times and others where it is quite stable (Figs. 1 and 2) and (ii) the symbiosis is not linked to mutual dependence or to intracellular integration (51, 52, 54), despite host benefits of N2 fixation in many ecological conditions (33, 34). These findings stress that not all highly persistent symbioses are on a route to intracellular lifestyles (55, 56). What factors determine whether a symbiosis is maintained or easily lost on an evolutionary time scale?
We performed a phylogenetic analysis of symbiotic persistence in legume hosts to explore possible geographic, climate, and soil variables and host traits that might correlate with symbiotic state likelihoods in legumes. Although research has demonstrated global variability in symbiosis investment (for example, physical intensity of host root colonization by symbiotic mycorrhizal fungi, ref. 57), there has been no attempt to map global patterns of evolutionary states of a symbiosis. First, we analyzed the geographical distribution of stable fixers across the globe and found a clear pattern showing that, at higher latitudes, legumes are generally stable fixers (Fig. 3A). In contrast, closer to the equator, the symbiosis can be highly persistent in some clades, as well as labile in others (Fig. 3B). These observations have an interesting parallel with a recent model that showed a latitudinal shift in N2-fixing tree strategies, with a constant rate of nitrogen fixation being favored at high latitudes and a plastic, flexible rate closer to the tropics (58). We found evidence that the latitudinal pattern of stable fixing is driven by temperature differences among species occurrences, with low annual temperatures associated with high symbiotic persistence. Although we cannot exclude the possibility that this pattern is driven by an unmeasured variable associated with global temperatures, this result is further confirmed by the relatively high occurrences of stable fixers in other low-temperature habitats at lower latitudes, such as the Himalayas and the Andes (Fig. 3B). An interesting hypothesis to consider is that the negative relationship between symbiosis persistence and temperature is linked with higher rates of molecular evolution in the tropics versus higher latitudes (59, 60). Experimental work has revealed that mutations in single genes can be sufficient to disrupt a legume’s capacity for symbiosis (61, 62). Although speculative, higher rates of molecular evolution driven by temperature could disrupt key gene networks involved in nodulation, thereby driving loss of the capacity for symbiosis. A similar positive effect of temperature on symbiosis loss has been reported experimentally in rhizobial bacteria (63). Whether this relationship between low temperature and high persistence holds for other symbiotic systems requires further exploration, but illustrates how patterns emerging from quantitative reconstructions can be used to answer broad questions about symbiosis evolution and stability. Another potential explanation for the observed latitudinal pattern relates to relative phosphorus and nitrogen limitations to plant hosts. Colder biomes are thought to be relatively nitrogen-limited, especially in formerly glaciated regions where time for nitrogen to accumulate from biotic N2 fixation has been relatively short (64–68). In contrast, the (sub)tropics tend to have very old, weathered soils depleted in phosphorus (68–70), where special adaptations to acquire phosphorus are presumably more important than N2 fixation. Such differences in soil type could favor the evolution of stable N2-fixing symbioses in polar regions.
Linking persistence of the N2-fixing symbiosis to N/P ratios remains an open question. However, we were able to extract three soil properties for our global legume occurrences to test the idea that stable fixing is associated with lower quality soils. We expected to find increased symbiotic persistence when environmental nitrogen availability, a function of soil pH, clay content, and nitrogen content (71), was lower because hosts theoretically derive more benefit from symbiosis in low-quality environments (72–74). Surprisingly, we found no significant relationship between soil variables and symbiotic persistence. This result could have a few potential explanations. The first is that most legumes are capable of down-regulating, and temporally controlling, nodulation in high soil nitrogen conditions (75, 76). Such control means that legumes can limit the potential negative effects of engaging in symbiosis in high nitrogen conditions and thus might not experience strong selection to lose the capacity for nodulation. However, this scenario assumes that there are few costs associated with hosts maintaining the symbiosis, which is unlikely (77). A second explanation is that we have analyzed only median total soil nitrogen whereas symbiosis stability in legumes might correlate with plant-available nitrogen and particularly with its lowest values, which fluctuate through the seasons (78–80). Third, a potential limitation of this analysis is that the scale of resolution of the soil databases we analyzed (1/2 a degree) might not be sufficient to detect variation in soil properties at a fine spatial scale. Although this smaller-scale variation is unlikely to affect global-scale patterns in soil properties, it might obscure relationships between soil variables at a finer scale and symbiotic persistence. A fourth explanation is that the soil environment data are a reflection, rather than a driver, of symbiotic persistence. The lack of correlation with low-quality soil and high symbiotic persistence could be an indication of the historic colonization of the environment by N2-fixing legumes, resulting in an influx of nitrogen to the soil (81, 82). This relationship illustrates a limitation of correlating particular habitat variables with evolutionary states: it can be difficult to determine cause and effect. This temporal problem can potentially be addressed by modeling evolution of the nitrogen fixation and soil variables simultaneously in more advanced phylogenetic frameworks (e.g., refs. 19, 21, 83, and 84), although evolutionary mapping of extrinsic traits like soil properties has attracted criticism (85).
Lastly, we found that leaf nitrogen and phosphorus contents were both positively correlated with highly persistent symbioses (Figs. 4 and 5). High N-demanding lifestyle and the capacity to engage in N2 fixation are thought to be linked (32, 50); our results are consistent with this idea and suggest that it also extends to symbiosis stability. Symbiotic N2 fixation allows for a more consistent source of nitrogen, and this link potentially drove the evolution of a high nitrogen lifestyle in some legumes, in turn favoring the symbiosis to persist. The positive correlation between leaf phosphorus content and persistent symbiosis is likewise expected because N2 fixation is linked with high phosphorus demand because N2-fixing efficiency is decreased by low phosphorus availability (32, 86). Another idea is that symbiotic N2 fixation may help alleviate phosphorus limitation because it allows for the secretion of more nitrogen-rich phosphatases, favoring plant phosphorus harvesting (87). Consequentially, high host nutrient demands may increase the benefits of being in a symbiotic relationship (88, 89), selecting for persistence of symbiotic capacity. It is still an open question as to whether a high nutrient lifestyle predated symbiosis persistence or the reverse. More detailed research is needed to determine the evolutionary order and causality of the correlation between persistence of nodulation capacity and high nutrient demands. Because a high-nutrient lifestyle is linked with fast growth and more investment in reproduction, that suite of traits is highly beneficial within a particular postdisturbance regeneration niche (sensu ref. 90). Legume clades experiencing such a selective regime may have effectively “locked” themselves into a symbiotic lifestyle.
A major question emerging from this research is why facultative symbioses, such as the N2-fixing symbiosis, are lost at all? In obligate symbioses, the absence of the symbiotic partner, or a single mutation disrupting the interaction, could immediately lead to extinction of the obligate partner. But why would a facultative species lose the capacity for symbiosis? In general, we would expect that three conditions need to be met: (i) an extended period in which the host plant does not need the symbiosis (for instance because the same benefits are provided by the environment); (ii) one or more mutations occur that cause the loss, which may be more or less likely in different clades; and (iii) the species needs to survive potential competition with other legumes that do retain the capacity for symbiosis, and then even perform better if conditions return in which being symbiotic is favorable. Thus, the capacity for symbiosis is lost if there is a long-term selective environment for loss (conditions i and iii) and genetic variation driving symbiosis loss (condition ii). If there is a cost, small or large, associated with maintaining the underlying genetic mechanism of N2-fixation capacity, this cost can contribute to a selective environment for loss of symbiotic capacity. Although there is some theoretical support for this idea (77, 91), it would be interesting to explicitly test for costs of maintaining facultative symbioses across diverse systems. Similarly, other factors we have considered, like high host nutrient demands, also affect the selective environment whereas factors like high molecular rates of evolution (59, 60) and genome duplication (92) impact upon the likelihood that mutations disrupt symbiotic capacity. By duplicating genes or whole genomes, redundant copies of key genes regulating the symbiosis can be created (92, 93). Because mutations are less likely to disrupt key symbiosis genes, duplication could result in a lower rate of symbiosis loss and higher retention of symbiotic capacity (36, 94). The potential link between symbiosis persistence and gene duplication is an interesting research avenue.
Although our aim is to illustrate how quantitative phylogenetic methods can be used to explore patterns of symbiosis evolution, we also recognize limitations to this approach. For example, our dataset of leaf nutrient contents was relatively small compared with our legume occurrence, temperature, and soil datasets (Methods). Additionally, these quantitative analyses are only correlative, and thus can only suggest patterns. Experiments are needed to compare the physiology of legumes and other hosts showing different likelihoods of symbiotic persistence (Fig. 1), allowing us to test for actual mechanisms that influence symbiosis stability (e.g., ref. 95). For instance, whereas there have been short-term evolutionary studies analyzing the mechanisms of symbiosis loss in rhizobia (96, 97), similar work studying the molecular mechanisms underlying nodulation loss in legumes is lacking. Lastly, our work is not a comprehensive analysis of all of the factors that could be involved in symbiotic persistence in N2 fixation. Rather, it illustrates the types of questions and variables that can be addressed. Other factors, such as historic atmospheric CO2 concentrations (98), have been thought to affect the evolution and stability of other plant root mutualisms (99, 100) and could likewise be considered in this framework.
The real power of a quantitative comparative approach is the ability to study evolutionary dynamics across a diversity of symbiosis types, from terrestrial (54, 101) to marine (102) habitats in various stages of persistence. For example, some squid species have a very intimate association with bioluminescent bacteria (25). Similar to the legume–rhizobial symbiosis, the bacterial symbionts retain a free-living stage and do not become physically integrated with the host. However, unlike legume hosts that are often found without rhizobial partners (30), adult squid hosts show a more consistent symbiotic engagement strategy with their bioluminescent symbionts (103). Why do these patterns differ and how is this trend reflected in the phylogenetic reconstruction of the partnerships? For instance, do we see fewer instances of loss for symbiotic capacity in squid lineages than legumes and why? Similarly, many insects have bacterial endosymbionts, some of which have been estimated to be associated with the same host clades for millions of years, and have become physically, genetically, and metabolically integrated with hosts (52, 54). One idea is that such integration is favored when symbionts access and deliver new forms of energy for their hosts, rather than simply increasing the efficiency of resource acquisition (56), as happens in the rhizobia–legume symbiosis. This idea could be tested in a comparative framework, asking what host or symbiont traits are associated with symbiont integration versus facultative associations? The first step to integration may be a switch from a more labile symbiotic state to a more persistent state, which can be phenotypically indistinguishable, but may be detected using phylogenetic comparative methods. Understanding these signals, and the general features that promote symbiosis evolution, will require quantitative reconstructions across a diversity of partnerships. Such comparative studies will shed light on how symbioses have solved the problem of aligning and controlling the interests of two species throughout the history of life (55).
Methods
We previously reconstructed the evolutionary history of N2 fixation using an HRM approach and a database of the nodulation status of 3,467 angiosperm species (36). Our subsequent analyses focused on the 1,366 legumes (Fabaceae) from our database to determine their stable fixing likelihoods and to test possible correlations between stable fixing likelihood and environmental and trait factors. We first confirmed our previous conclusion that nodulating legumes could be subdivided into stable fixers, with an increased symbiotic persistence, and in regular fixers, and that this finding also holds when analyzing only the legumes. We then tested for correlations between stable fixing likelihood in legumes capable of forming nodules (1,195 species; this subset is smaller because not all 1,366 legumes species can nodulate) (36) and (i) geographic, (ii) climate, and (iii) environmental variables and (iv) host traits. We obtained (i) location records for these species from the Global Biodiversity Information Facility (GBIF), (ii) temperature records for these locations from the Climate Research Unit (CRU) (46), (iii) soil clay content and pH values for all legume occurrences from the Harmonized World Soil Database (47) and soil nitrogen from the ISRIC-WISE database (48, 49), and (iv) plant leaf nitrogen and phosphorus concentrations from the TRY Plant Trait Database (104) and from the Yasuni Forest Dynamics database (105).
We used the R-package rgbif (43) to obtain location records for the nodulating legume species recorded in GBIF and our database, resulting in coverage of 1,182 species. We limited the maximum total number of locations per species to 100,000 (43). This procedure resulted in an analysis of almost 3.7 million locations. Subsequently, we scrubbed our location data to exclude all data points with spatial error codes (cdiv, cdout, cdrepf, cdreps, cucdmis, gdativ, preneglat, preneglon, preswcd, zerocd) and obtained a dataset of over 3.2 million legume locations across the globe (median 289 locations per species). We characterized species as stable fixers if they had a corrected stable fixing likelihood of more than 50% (Figs. 1 and 2), and otherwise as regular fixers for visualization purposes. We plotted these locations to the world map to identify general geographic patterns (Fig. 3) and found a strong latitudinal pattern. We calculated the median latitude per species across all their locations in our dataset and tested whether there was a latitudinal pattern in stable fixing likelihood by generating a phylogenetic linear regression model of stable fixing likelihood on absolute median latitude in the R-package phylolm, using Pagel’s lambda (44, 45). To correct for phylogenetic nonindependence, we pruned the full angiosperm phylogeny (35; data available from Dryad, dx.doi.org/10.5061/dryad.63q27.2) to include only those legumes for which we had both median latitude and stable fixing likelihoods.
We then obtained mean long-term annual temperature (TMPMean) for all locations in our cleaned location dataset, using a bilinear interpolation (106) to sample for the location from the climate grid (46). We calculated the median species-level TMPMean across all locations for each species, including only species for which we had temperature records that covered at least 10 different locations, resulting in a coverage of 1,095 legumes. We then explored the extent to which the observed latitudinal pattern was driven by temperature. As previously, we generated phylogenetic linear regression models of stable fixing likelihood as a response variable and its association with absolute median latitude and median TMPMean in phylolm using Pagel’s lambda (44, 45). We used AIC criteria to establish that the best model contained only temperature as an explanatory variable.
To analyze the correlation between stable fixing likelihoods and soil properties, we used a similar approach and used a bilinear interpolation to sample for the legume occurrence locations, which were then used to obtain soil clay content, soil pH, and total soil N levels from the Harmonized World Soil Database and the ISRIC-WISE database (both 1/2 a degree resolution) (47–49). These values were used to calculate species-level median values. We then generated phylogenetic linear regression models to explain the response variable stable fixing likelihoods in terms of these three soil variables. Using AIC criteria, we found that inclusion of none of these variables or their combinations provided a better fit than a model that included only an intercept.
We then studied the relationship between symbiotic persistence and host plant traits in nodulating legumes, and we obtained a database of plant leaf nitrogen and phosphorus contents from the TRY Plant Trait Database (104, 107–135) and from the Yasuni Forest Dynamics database (105). We calculated the mean nutrient content per species for both nitrogen (Nmass %) and phosphorus (Pmass %) and compiled a dataset of 131 symbiotic legumes for which we have both nitrogen and phosphorus data. Here, we found using AIC that the best phylogenetic linear model explaining stable fixing likelihood among legumes was one that included plant leaf phosphorus and nitrogen content as explanatory variables, as well as their interaction. We normalized Nmass and Pmass by scaling them by their respective SDs so that the regression coefficient could be compared between variables.
A potential limitation to the phylogenetic linear model analyses presented here is that we are taking the output of one phylogenetic analysis (i.e., stable fixing likelihoods) (36) and using it as an input for subsequent analysis. As defined here, symbiotic persistence can be discovered only in a phylogenetic reconstruction, not directly measured or observed in an organism, necessitating this approach. However, we can verify our results by testing for the phylogenetic correlation between the variables of interest for our best fit models and the underlying binary data on the presence or absence of nitrogen fixation capacity in legumes (data available from Dryad, dx.doi.org/doi:10.5061/dryad.05k14). We therefore generated phylogenetic generalized linear models in phylolm (44, 136), with host legume nodulation capacity as a dependent binary variable, and either mean annual temperature (TMPMean) or leaf nitrogen (Nmass %) and phosphorus (Pmass %) content and their interactions as explanatory variables. As previously, we scaled Nmass and Pmass by their respective SDs so that the regression coefficient could be compared between variables. We expect that, if there are relationships between these variables and stable fixing, they should also be recovered when analyzing the correlation between them and fixation per se. As expected, we recovered the same relationship of leaf nitrogen (coefficient, 1.22, P < 0.01) and phosphorus content (coefficient, 1.11, P < 0.01) and their interaction (coefficient, −0.37, P < 0.01) with symbiotic N2 fixation. Additionally, as expected, we recover that legumes at lower TMPMean are more likely to be able to form the symbiosis (coefficient, −0.10, P = 0.24). However, this association is not significant, indicating that this result is less robust than the link between symbiotic nitrogen fixation and host nutrient requirements.
Acknowledgments
We thank W. F. Doolittle, P. Keeling, J. McCutcheon, the Sackler Colloquium, L. M. Henry, L. A. A. Werner, J. I. Sprent, and two anonymous reviewers for providing feedback on our manuscript. This research was supported by Netherlands Organisation for Scientific Research Grants 836.10.001 and 864.10.005 (to E.T.K.) and European Research Council ERC Grant Agreement 335542 (to E.T.K.). The study has been supported by the TRY initiative on plant traits (https://www.try-db.org). The TRY initiative and database are hosted, developed, and maintained by J. Kattge and G. Bönisch (Max Planck Institute for Biogeochemistry, Jena, Germany). TRY is currently supported by DIVERSITAS/Future Earth and the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Symbioses Becoming Permanent: The Origins and Evolutionary Trajectories of Organelles,” held October 15–17, 2014, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Symbioses.
This article is a PNAS Direct Submission. J.P.M. is a guest editor invited by the Editorial Board.
References
- 1.Vitousek PM, Menge DNL, Reed SC, Cleveland CC. Biological nitrogen fixation: rates, patterns and ecological controls in terrestrial ecosystems. Philos Trans R Soc Lond B Biol Sci. 2013;368(1621):20130119. doi: 10.1098/rstb.2013.0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joy JB. Symbiosis catalyses niche expansion and diversification. Proc Biol Sci. 2013;280(1756):20122820. doi: 10.1098/rspb.2012.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber MG, Agrawal AA. Defense mutualisms enhance plant diversification. Proc Natl Acad Sci USA. 2014;111(46):16442–16447. doi: 10.1073/pnas.1413253111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moran NA. Symbiosis as an adaptive process and source of phenotypic complexity. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8627–8633. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bronstein JL. Our current understanding of mutualism. Q Rev Biol. 1994;69(1):31–51. [Google Scholar]
- 6.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110(9):3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiers ET, et al. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science. 2011;333(6044):880–882. doi: 10.1126/science.1208473. [DOI] [PubMed] [Google Scholar]
- 8.Aanen DK, et al. High symbiont relatedness stabilizes mutualistic cooperation in fungus-growing termites. Science. 2009;326(5956):1103–1106. doi: 10.1126/science.1173462. [DOI] [PubMed] [Google Scholar]
- 9.McFall-Ngai M. Divining the essence of symbiosis: Insights from the squid-vibrio model. PLoS Biol. 2014;12(2):e1001783. doi: 10.1371/journal.pbio.1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sachs JL, Skophammer RG, Regus JU. Evolutionary transitions in bacterial symbiosis. Proc Natl Acad Sci USA. 2011;108(Suppl 2):10800–10807. doi: 10.1073/pnas.1100304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiers ET, Palmer TM, Ives AR, Bruno JF, Bronstein JL. Mutualisms in a changing world: An evolutionary perspective. Ecol Lett. 2010;13(12):1459–1474. doi: 10.1111/j.1461-0248.2010.01538.x. [DOI] [PubMed] [Google Scholar]
- 12.Ghoul M, Griffin AS, West SA. Toward an evolutionary definition of cheating. Evolution. 2014;68(2):318–331. doi: 10.1111/evo.12266. [DOI] [PubMed] [Google Scholar]
- 13.Sachs JL, Simms EL. Pathways to mutualism breakdown. Trends Ecol Evol. 2006;21(10):585–592. doi: 10.1016/j.tree.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 14.Merckx V, Bidartondo MI. Breakdown and delayed cospeciation in the arbuscular mycorrhizal mutualism. Proc Biol Sci. 2008;275(1638):1029–1035. doi: 10.1098/rspb.2007.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leigh EG., Jr The evolution of mutualism. J Evol Biol. 2010;23(12):2507–2528. doi: 10.1111/j.1420-9101.2010.02114.x. [DOI] [PubMed] [Google Scholar]
- 16.Foster KR, Wenseleers T. A general model for the evolution of mutualisms. J Evol Biol. 2006;19(4):1283–1293. doi: 10.1111/j.1420-9101.2005.01073.x. [DOI] [PubMed] [Google Scholar]
- 17.Palmer TM, et al. Synergy of multiple partners, including freeloaders, increases host fitness in a multispecies mutualism. Proc Natl Acad Sci USA. 2010;107(40):17234–17239. doi: 10.1073/pnas.1006872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golubski AJ, Klausmeier CA. Control in mutualisms: Combined implications of partner choice and bargaining roles. J Theor Biol. 2010;267(4):535–545. doi: 10.1016/j.jtbi.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 19.Revell LJ. Ancestral character estimation under the threshold model from quantitative genetics. Evolution. 2014;68(3):743–759. doi: 10.1111/evo.12300. [DOI] [PubMed] [Google Scholar]
- 20.Beaulieu JM, O’Meara BC, Donoghue MJ. Identifying hidden rate changes in the evolution of a binary morphological character: The evolution of plant habit in campanulid angiosperms. Syst Biol. 2013;62(5):725–737. doi: 10.1093/sysbio/syt034. [DOI] [PubMed] [Google Scholar]
- 21.Felsenstein J. A comparative method for both discrete and continuous characters using the threshold model. Am Nat. 2012;179(2):145–156. doi: 10.1086/663681. [DOI] [PubMed] [Google Scholar]
- 22.Currie CR, Poulsen M, Mendenhall J, Boomsma JJ, Billen J. Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science. 2006;311(5757):81–83. doi: 10.1126/science.1119744. [DOI] [PubMed] [Google Scholar]
- 23.Remy W, Taylor TN, Hass H, Kerp H. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA. 1994;91(25):11841–11843. doi: 10.1073/pnas.91.25.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaltenpoth M, et al. Partner choice and fidelity stabilize coevolution in a Cretaceous-age defensive symbiosis. Proc Natl Acad Sci USA. 2014;111(17):6359–6364. doi: 10.1073/pnas.1400457111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kremer N, et al. The dual nature of haemocyanin in the establishment and persistence of the squid-vibrio symbiosis. Proc Biol Sci. 2014;281(1785):20140504. doi: 10.1098/rspb.2014.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hjort K, Goldberg AV, Tsaousis AD, Hirt RP, Embley TM. Diversity and reductive evolution of mitochondria among microbial eukaryotes. Philos Trans R Soc Lond B Biol Sci. 2010;365(1541):713–727. doi: 10.1098/rstb.2009.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merckx V, Freudenstein JV. Evolution of mycoheterotrophy in plants: A phylogenetic perspective. New Phytol. 2010;185(3):605–609. doi: 10.1111/j.1469-8137.2009.03155.x. [DOI] [PubMed] [Google Scholar]
- 28.O’Meara BC, Beaulieu JM. In: Modern Phylogenetic Comparative Methods and their Application in Evolutionary Biology. Garamszegi LZ, editor. Springer; Heidelberg: 2014. pp. 395–408. [Google Scholar]
- 29.Young JPW, Haukka KE. Diversity and phylogeny of rhizobia. New Phytol. 1996;133(1):87–94. [Google Scholar]
- 30.Sprent JI. Legume Nodulation: A Global Perspective. Wiley-Blackwell; Chichester, UK: 2009. [Google Scholar]
- 31.Denison RF, Kiers ET. Life histories of symbiotic rhizobia and mycorrhizal fungi. Curr Biol. 2011;21(18):R775–R785. doi: 10.1016/j.cub.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 32.Vitousek PM, et al. Towards an ecological understanding of biological nitrogen fixation. Biogeochemistry. 2002;77(58):1–45. [Google Scholar]
- 33.Burdon JJ, Gibson H, Searle SD, Woods MJ, Brockwell J. Variation in the effectiveness of symbiotic associations between native rhizobia and temperate Australian Acacia: Within-species interactions. J Appl Ecol. 1999;36(3):398–408. [Google Scholar]
- 34.Lee TD, Tjoelker MG, Reich PB, Russelle MP. Contrasting growth response of an N2-fixing and non-fixing forb to elevated CO2: Dependence on soil N supply. Plant Soil. 2003;255:475–486. [Google Scholar]
- 35.Zanne AE, et al. Three keys to the radiation of angiosperms into freezing environments. Nature. 2014;506(7486):89–92. doi: 10.1038/nature12872. [DOI] [PubMed] [Google Scholar]
- 36.Werner GDA, Cornwell WK, Sprent JI, Kattge J, Kiers ET. A single evolutionary innovation drives the deep evolution of symbiotic N2-fixation in angiosperms. Nat Commun. 2014;5:4087. doi: 10.1038/ncomms5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doyle JJ. 2011. Phylogenetic perspectives on the origins of nodulation. Mol Plant Microbe Interact 24(11):1289–1295.
- 38.Sprent JI, Ardley JK, James EK. From North to South: A latitudinal look at legume nodulation processes. S Afr J Bot. 2013;89:31–41. [Google Scholar]
- 39.Masson-Boivin C, Giraud E, Perret X, Batut J. Establishing nitrogen-fixing symbiosis with legumes: How many rhizobium recipes? Trends Microbiol. 2009;17(10):458–466. doi: 10.1016/j.tim.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Van Leuven JT, Meister RC, Simon C, McCutcheon JP. Sympatric speciation in a bacterial endosymbiont results in two genomes with the functionality of one. Cell. 2014;158(6):1270–1280. doi: 10.1016/j.cell.2014.07.047. [DOI] [PubMed] [Google Scholar]
- 41.Bennett GM, Moran NA. 2015. Heritable symbiosis: The advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci USA 112:10169–10176.
- 42.Soltis DE, et al. Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proc Natl Acad Sci USA. 1995;92(7):2647–2651. doi: 10.1073/pnas.92.7.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chamberlain S, Ram K, Barve V, McGlinn D. 2014. rgbif: Interface to the Global Biodiversity Information Facility API. R package version 0.7.7. Available at CRAN.R-project.org/package=rgbif.
- 44.Ho Ls, Ané C. A linear-time algorithm for Gaussian and non-Gaussian trait evolution models. Syst Biol. 2014;63(3):397–408. doi: 10.1093/sysbio/syu005. [DOI] [PubMed] [Google Scholar]
- 45.Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401(6756):877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- 46.New M, Lister D, Hulme M, Makin I. A high-resolution data set of surface climate over global land areas. Clim Res. 2002;21(1):1–25. [Google Scholar]
- 47.Nachtergaele F, van Velthuizen H, Verelst L, Wiberg D. 2012. Harmonized World Soil Database (FAO, Rome), Version 1.2.
- 48.Batjes NH. 2012. ISRIC-WISE derived soil properties on a 5 by 5 arc-minutes global grid (ver. 1.2.) (ISRIC-World Soil Information, Wageningen, The Netherlands), 52 p.
- 49.Batjes NH. A Homogenized Soil Data File for Global Environmental Research: A Subset of FAO, ISRCIC and NRCS Profiles. Wageningen; The Netherlands: 1995. [Google Scholar]
- 50.McKey D. 1994. The Nitrogen Factor, Advances in Legume Systematics, eds Sprent JI, McKey D (Royal Botanic Gardens, Kew, England), Part 5, pp 211–228.
- 51.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10(1):13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 52.Husnik F, et al. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell. 2013;153(7):1567–1578. doi: 10.1016/j.cell.2013.05.040. [DOI] [PubMed] [Google Scholar]
- 53.de Duve C. The origin of eukaryotes: A reappraisal. Nat Rev Genet. 2007;8(5):395–403. doi: 10.1038/nrg2071. [DOI] [PubMed] [Google Scholar]
- 54.Bennett GM, Moran NA. Small, smaller, smallest: The origins and evolution of ancient dual symbioses in a Phloem-feeding insect. Genome Biol Evol. 2013;5(9):1675–1688. doi: 10.1093/gbe/evt118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.West SA, Fisher RM, Gardner A, Kiers ET. 2015. Major evolutionary transitions in individuality. Proc Natl Acad Sci USA 112:10112–10119.
- 56.Kiers ET, West SA. Evolutionary biology: Evolving new organisms via symbiosis. Science. 2015;348(6233):392–394. doi: 10.1126/science.aaa9605. [DOI] [PubMed] [Google Scholar]
- 57. Soudzilovskaia NA, et al. (2015) Global patterns of plant root colonization intensity by mycorrhizal fungi explained by climate and soil chemistry. Glob Ecol Biogeogr 24:371–382.
- 58.Menge DNL, Lichstein JW, Angeles-Pérez G. Nitrogen fixation strategies can explain the latitudinal shift in nitrogen-fixing tree abundance. Ecology. 2014;95(8):2236–2245. doi: 10.1890/13-2124.1. [DOI] [PubMed] [Google Scholar]
- 59.Gillman LN, Keeling DJ, Ross HA, Wright SD. Latitude, elevation and the tempo of molecular evolution in mammals. Proc Biol Sci. 2009;276(1671):3353–3359. doi: 10.1098/rspb.2009.0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wright S, Keeling J, Gillman L. The road from Santa Rosalia: A faster tempo of evolution in tropical climates. Proc Natl Acad Sci USA. 2006;103(20):7718–7722. doi: 10.1073/pnas.0510383103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heckmann AB, et al. Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 2006;142(4):1739–1750. doi: 10.1104/pp.106.089508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soyano T, Kouchi H, Hirota A, Hayashi M. Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 2013;9(3):e1003352. doi: 10.1371/journal.pgen.1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zurkowski W. Molecular mechanism for loss of nodulation properties of Rhizobium trifolii. J Bacteriol. 1982;150(3):999–1007. doi: 10.1128/jb.150.3.999-1007.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pérez CA, et al. Ecosystem development in short-term postglacial chronosequences: N and P limitation in glacier forelands from Santa Inés Island, Magellan Strait. Austral Ecol. 2014;39(3):288–303. [Google Scholar]
- 65.Aerts R, Cornelissen JHC, Dorrepaal E. Plant performance in a warmer world: General responses of plants from cold, northern biomes and the importance of winter and spring events. Plant Ecol. 2006;182(1-2):65–77. [Google Scholar]
- 66.Shaver GR, et al. Species composition interacts with fertilizer to control long-term change in tundra productivity. Ecology. 2001;82(11):3163–3181. [Google Scholar]
- 67.Shaver GR, Chapin FS, III. Long-term responses to factorial, NPK fertilizer treatment by Alaskan wet and moist tundra sedge species. Ecography. 1995;18(3):259–275. [Google Scholar]
- 68.Lambers H, Raven JA, Shaver GR, Smith SE. Plant nutrient-acquisition strategies change with soil age. Trends Ecol Evol. 2008;23(2):95–103. doi: 10.1016/j.tree.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 69.Turner BL, et al. Soil microbial biomass and the fate of phosphorus during long-term ecosystem development. Plant Soil. 2013;367(1-2):225–234. [Google Scholar]
- 70.Lambers H, Mougel C, Jaillard B, Hinsinger P. Plant-microbe-soil interactions in the rhizosphere: An evolutionary perspective. Plant Soil. 2009;321(1-2):83–115. [Google Scholar]
- 71.Shaffer MJ, Ma L, Hansen S. Modeling Carbon and Nitrogen Dynamics for Soil Management. CRC; Boca Raton, FL: 2001. [Google Scholar]
- 72.Hochberg ME, Gomulkiewicz R, Holtà RD, Thompson JN. Weak sinks could cradle mutualistic symbioses: Strong sources should harbour parasitic symbioses. J Evol Biol. 2000;13(2):213–222. [Google Scholar]
- 73.Thrall PH, Hochberg ME, Burdon JJ, Bever JD. Coevolution of symbiotic mutualists and parasites in a community context. Trends Ecol Evol. 2007;22(3):120–126. doi: 10.1016/j.tree.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 74.Johnson NC. Resource stoichiometry elucidates the structure and function of arbuscular mycorrhizas across scales. New Phytol. 2010;185(3):631–647. doi: 10.1111/j.1469-8137.2009.03110.x. [DOI] [PubMed] [Google Scholar]
- 75.Batterman SA, Wurzburger N, Hedin LO. Nitrogen and phosphorus interact to control tropical symbiotic N2 fixation: A test in inga punctata. J Ecol. 2013;101(6):1400–1408. [Google Scholar]
- 76.Müller S, Pereira PAA, Martin P. Effect of different levels of mineral nitrogen on nodulation and N2 fixation of two cultivars of common bean (Phaseolus vulgaris L.) Plant Soil. 1993;152(1):139–143. [Google Scholar]
- 77.Menge DNL, Levin SA, Hedin LO. Evolutionary tradeoffs can select against nitrogen fixation and thereby maintain nitrogen limitation. Proc Natl Acad Sci USA. 2008;105(5):1573–1578. doi: 10.1073/pnas.0711411105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scogings PF, et al. Seasonal variations in nutrients and secondary metabolites in semi-arid savannas depend on year and species. J Arid Environ. 2015;114:54–61. [Google Scholar]
- 79.Lipson DA, Schmidt SK, Monson RK. Links between microbial population dynamics and nitrogen availability in an alpine ecosystem. Ecology. 1999;80(5):1623–1631. [Google Scholar]
- 80.Cain ML, Subler S, Evans JP, Fortin M-J. Sampling spatial and temporal variation in soil nitrogen availability. Oecologia. 1999;118(4):397–404. doi: 10.1007/s004420050741. [DOI] [PubMed] [Google Scholar]
- 81.Ledgard SF, Steele KW. Biological nitrogen fixation in mixed legume/grass pastures. Plant Soil. 1992;141(1-2):137–153. [Google Scholar]
- 82.Hiltbrunner E, et al. Ecological consequences of the expansion of N2-fixing plants in cold biomes. Oecologia. 2014;176(1):11–24. doi: 10.1007/s00442-014-2991-x. [DOI] [PubMed] [Google Scholar]
- 83.Kutsukake N, Innan H. In: Modern Phylogenetic Comparative Methods and their Application in Evolutionary Biology. Garamszegi LZ, editor. Springer; Heidelberg: 2014. pp. 409–424. [Google Scholar]
- 84.Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst Biol. 2004;53(5):673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- 85.Grandcolas P, Nattier R, Legendre F, Pellens R. Mapping extrinsic traits such as extinction risks or modelled bioclimatic niches on phylogenies: Does it make sense at all? Cladistics. 2011;27(2):181–185. doi: 10.1111/j.1096-0031.2010.00324.x. [DOI] [PubMed] [Google Scholar]
- 86.Vitousek PM, Howarth RW. Nitrogen limitation on land and in the sea: How can it occur? Biogeochemistry. 1991;13(2):87–115. [Google Scholar]
- 87.Houlton BZ, Wang Y-P, Vitousek PM, Field CB. A unifying framework for dinitrogen fixation in the terrestrial biosphere. Nature. 2008;454(7202):327–330. doi: 10.1038/nature07028. [DOI] [PubMed] [Google Scholar]
- 88.Werner GDA, et al. Evolution of microbial markets. Proc Natl Acad Sci USA. 2014;111(4):1237–1244. doi: 10.1073/pnas.1315980111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wyatt GAK, Kiers ET, Gardner A, West SA. A biological market analysis of the plant-mycorrhizal symbiosis. Evolution. 2014;68(9):2603–2618. doi: 10.1111/evo.12466. [DOI] [PubMed] [Google Scholar]
- 90.Grubb PJ. The maintenance of species-richness in plant communities: The importance of the regeneration niche. Biol Rev Camb Philos Soc. 1977;52:107–145. [Google Scholar]
- 91.Lahti DC, et al. Relaxed selection in the wild. Trends Ecol Evol. 2009;24(9):487–496. doi: 10.1016/j.tree.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 92.Li Q-G, Zhang L, Li C, Dunwell JM, Zhang Y-M. Comparative genomics suggests that an ancestral polyploidy event leads to enhanced root nodule symbiosis in the Papilionoideae. Mol Biol Evol. 2013;30(12):2602–2611. doi: 10.1093/molbev/mst152. [DOI] [PubMed] [Google Scholar]
- 93.Young ND, et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480(7378):520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cannon SB, et al. Multiple polyploidy events in the early radiation of nodulating and nonnodulating legumes. Mol Biol Evol. 2015;32(1):193–210. doi: 10.1093/molbev/msu296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Behm JE, Geurts R, Kiers ET. Parasponia: A novel system for studying mutualism stability. Trends Plant Sci. 2014;19(12):757–763. doi: 10.1016/j.tplants.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 96.Sachs JL, Ehinger MO, Simms EL. Origins of cheating and loss of symbiosis in wild Bradyrhizobium. J Evol Biol. 2010;23(5):1075–1089. doi: 10.1111/j.1420-9101.2010.01980.x. [DOI] [PubMed] [Google Scholar]
- 97.Sachs JL, Russell JE, Hollowell AC. Evolutionary instability of symbiotic function in Bradyrhizobium japonicum. PLoS ONE. 2011;6(11):e26370. doi: 10.1371/journal.pone.0026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rothman DH. Atmospheric carbon dioxide levels for the last 500 million years. Proc Natl Acad Sci USA. 2002;99(7):4167–4171. doi: 10.1073/pnas.022055499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Field KJ, et al. Contrasting arbuscular mycorrhizal responses of vascular and non-vascular plants to a simulated Palaeozoic CO2 decline. Nat Commun. 2012;3:835. doi: 10.1038/ncomms1831. [DOI] [PubMed] [Google Scholar]
- 100.Field KJ, et al. First evidence of mutualism between ancient plant lineages (Haplomitriopsida liverworts) and Mucoromycotina fungi and its response to simulated Palaeozoic changes in atmospheric CO2. New Phytol. 2015;205(2):743–756. doi: 10.1111/nph.13024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Koga R, Moran NA. Swapping symbionts in spittlebugs: Evolutionary replacement of a reduced genome symbiont. ISME J. 2014;8(6):1237–1246. doi: 10.1038/ismej.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Petersen JM, Wentrup C, Verna C, Knittel K, Dubilier N. Origins and evolutionary flexibility of chemosynthetic symbionts from deep-sea animals. Biol Bull. 2012;223(1):123–137. doi: 10.1086/BBLv223n1p123. [DOI] [PubMed] [Google Scholar]
- 103.Koch EJ, Miyashiro T, McFall-Ngai MJ, Ruby EG. Features governing symbiont persistence in the squid-vibrio association. Mol Ecol. 2014;23(6):1624–1634. doi: 10.1111/mec.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kattge J, et al. TRY: A global database of plant traits. Glob Change Biol. 2011;17(9):2905–2935. [Google Scholar]
- 105.Kraft NJB, Ackerly DD. Functional trait and phylogenetic tests of community assembly across spatial scales in an Amazonian forest. Ecol Monogr. 2010;80(3):401–422. [Google Scholar]
- 106.Hijmans RJ. 2014. raster: Geographic Data Analysis and Modeling. R package version 2.3-12.
- 107.Cornelissen JHC, et al. Leaf digestibility and litter decomposability are related in a wide range of subarctic plant species and types. Funct Ecol. 2004;18(6):779–786. [Google Scholar]
- 108.Milla R, Reich PB. Multi-trait interactions, not phylogeny, fine-tune leaf size reduction with increasing altitude. Ann Bot (Lond) 2011;107(3):455–465. doi: 10.1093/aob/mcq261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Han W, Fang J, Guo D, Zhang Y. Leaf nitrogen and phosphorus stoichiometry across 753 terrestrial plant species in China. New Phytol. 2005;168(2):377–385. doi: 10.1111/j.1469-8137.2005.01530.x. [DOI] [PubMed] [Google Scholar]
- 110.Prentice IC, et al. Evidence of a universal scaling relationship for leaf CO2 drawdown along an aridity gradient. New Phytol. 2011;190(1):169–180. doi: 10.1111/j.1469-8137.2010.03579.x. [DOI] [PubMed] [Google Scholar]
- 111.Fitter A, Peat H. The Ecological Flora Database. J Ecol. 1994;82:415–425. [Google Scholar]
- 112.Louault F, Pillar VD, Aufrère J, Garnier E, Soussana JF. Plant traits and functional types in response to reduced disturbance in a semi-natural grassland. J Veg Sci. 2005;16(2):151–160. [Google Scholar]
- 113.Reich PB, Oleksyn J, Wright IJ. Leaf phosphorus influences the photosynthesis-nitrogen relation: A cross-biome analysis of 314 species. Oecologia. 2009;160(2):207–212. doi: 10.1007/s00442-009-1291-3. [DOI] [PubMed] [Google Scholar]
- 114.Niinemets Ü. Global-scale climatic controls of leaf dry mass per area, density, and thickness in trees and shrubs. Ecology. 2001;82(2):453–469. [Google Scholar]
- 115.Reich PB, et al. Scaling of respiration to nitrogen in leaves, stems and roots of higher land plants. Ecol Lett. 2008;11(8):793–801. doi: 10.1111/j.1461-0248.2008.01185.x. [DOI] [PubMed] [Google Scholar]
- 116.Wright IJ, et al. The worldwide leaf economics spectrum. Nature. 2004;428(6985):821–827. doi: 10.1038/nature02403. [DOI] [PubMed] [Google Scholar]
- 117.Penuelas J, et al. Faster returns on “leaf economics” and different biogeochemical niche in invasive compared with native plant species. Glob Change Biol. 2010;16(8):2171–2185. [Google Scholar]
- 118.Preston KA, Cornwell WK, Denoyer JL. Wood density and vessel traits as distinct correlates of ecological strategy in 51 California coast range angiosperms. New Phytol. 2006;170(4):807–818. doi: 10.1111/j.1469-8137.2006.01712.x. [DOI] [PubMed] [Google Scholar]
- 119.Domingues TF, Martinelli LA, Ehleringer JR. Ecophysiological traits of plant functional groups in forest and pasture ecosystems from eastern Amazônia, Brazil. Plant Ecol. 2007;193(1):101–112. [Google Scholar]
- 120.Han W, et al. Floral, climatic and soil pH controls on leaf ash content in China’s terrestrial plants. Glob Ecol Biogeogr. 2012;21(3):376–382. [Google Scholar]
- 121.Chen Y, Han W, Tang L, Tang Z, Fang J. Leaf nitrogen and phosphorus concentrations of woody plants differ in responses to climate, soil and plant growth form. Ecography (Cop) 2013;36(2):178–184. [Google Scholar]
- 122.Kattge J, Knorr W, Raddatz T, Wirth C. Quantifying photosynthetic capacity and its relationship to leaf nitrogen content for global-scale terrestrial biosphere models. Glob Change Biol. 2009;15(4):976–991. [Google Scholar]
- 123.Blonder B, Violle C, Bentley LP, Enquist BJ. Venation networks and the origin of the leaf economics spectrum. Ecol Lett. 2011;14(2):91–100. doi: 10.1111/j.1461-0248.2010.01554.x. [DOI] [PubMed] [Google Scholar]
- 124.Campetella G, et al. Patterns of plant trait-environment relationships along a forest succession chronosequence. Agric Ecosyst Environ. 2011;145(1):38–48. [Google Scholar]
- 125.Spasojevic MJ, Suding KN. Inferring community assembly mechanisms from functional diversity patterns: The importance of multiple assembly processes. J Ecol. 2012;100(3):652–661. [Google Scholar]
- 126.Vergutz L, Manzoni S, Porporato A, Novais R, Jackson R. 2012. A Global Database of Carbon and Nutrient Concentrations of Green and Senesced Leaves (Oak Ridge National Laboratory Distributed Active Archive Center, Oak Ridge, TN), data set.
- 127.Craven D, et al. 2007. Between and within-site comparisons of structural and physiological characteristics and foliar nutrient content of 14 tree species at a wet, fertile site and a dry, infertile site in Panama. For Ecol Manage 238(1-3):335–346.
- 128.Cornelissen J, et al. Functional traits of woody plants: Correspondence of species rankings between field adults and laboratory-grown seedlings? J Veg Sci. 2003;14:311–322. [Google Scholar]
- 129.Kerkhoff AJ, Fagan WF, Elser JJ, Enquist BJ. Phylogenetic and growth form variation in the scaling of nitrogen and phosphorus in the seed plants. Am Nat. 2006;168(4):E103–E122. doi: 10.1086/507879. [DOI] [PubMed] [Google Scholar]
- 130.Baraloto C, et al. Decoupled leaf and stem economics in rain forest trees. Ecol Lett. 2010;13(11):1338–1347. doi: 10.1111/j.1461-0248.2010.01517.x. [DOI] [PubMed] [Google Scholar]
- 131.Ordoñez JC, et al. Plant strategies in relation to resource supply in mesic to wet environments: Does theory mirror nature? Am Nat. 2010;175(2):225–239. doi: 10.1086/649582. [DOI] [PubMed] [Google Scholar]
- 132.Fyllas N, et al. Basin-wide variations in foliar properties of Amazonian forest: Phylogeny, soils and climate. Biogeosciences. 2009;6:2677–2708. [Google Scholar]
- 133.Garnier E, et al. Assessing the effects of land-use change on plant traits, communities and ecosystem functioning in grasslands: A standardized methodology and lessons from an application to 11 European sites. Ann Bot (Lond) 2007;99(5):967–985. doi: 10.1093/aob/mcl215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Butterfield BJ, Briggs JM. Regeneration niche differentiates functional strategies of desert woody plant species. Oecologia. 2011;165(2):477–487. doi: 10.1007/s00442-010-1741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Domingues TF, et al. Co-limitation of photosynthetic capacity by nitrogen and phosphorus in West Africa woodlands. Plant Cell Environ. 2010;33(6):959–980. doi: 10.1111/j.1365-3040.2010.02119.x. [DOI] [PubMed] [Google Scholar]
- 136.Ives AR, Garland T., Jr Phylogenetic logistic regression for binary dependent variables. Syst Biol. 2010;59(1):9–26. doi: 10.1093/sysbio/syp074. [DOI] [PubMed] [Google Scholar]