Abstract
In virtually all multicellular eukaryotes, mitochondria are transmitted exclusively through one parent, usually the mother. In this short review, we discuss some of the major consequences of uniparental transmission of mitochondria, including deleterious effects in males and selection for increased transmission through females. Many of these consequences, particularly sex ratio distortion, have well-studied parallels in other maternally transmitted genetic elements, such as bacterial endosymbionts of arthropods. We also discuss the consequences of linkage between mitochondria and other maternally transmitted genetic elements, including the role of cytonuclear incompatibilities in maintaining polymorphism. Finally, as a case study, we discuss a recently discovered maternally transmitted sex ratio distortion in an insect that is associated with extraordinarily divergent mitochondria.
Keywords: symbiosis, cytoplasmic male sterility, Wolbachia, reproductive parasitism, genetic conflict
By virtue of their symbiotic origin, mitochondria are special (1). They have retained their own genome (with a few interesting exceptions), despite the fact that the vast majority of mitochondrial proteins are encoded in the much larger nuclear genome. A functioning organelle thus requires the tight regulation and coordination of two genomes with very different properties, histories, and locations. In addition, mitochondrial genomes reproduce asexually and are cytoplasmically inherited, typically through one sex, usually the female. This mode of transmission differs from most nuclear genomes, and has important consequences on an organism’s fitness. There have been many excellent reviews on the different evolutionary trajectories of mitochondrial and nuclear genomes, including how these can result in genetic conflicts and incompatibility (e.g., refs. 2–10). In this short review we focus on the relationship between mitochondria and sex ratio distortion. We discuss how maternal transmission can drive the evolution of mitochondria (and other symbionts) that increase the frequency of females. We also consider how linkage between mitochondria and other maternally transmitted genetic elements, such as sex ratio distorters, can result in cytonuclear incompatibilities that may ultimately affect the persistence of the distorter. Finally, as a case study, we discuss a recently discovered maternally transmitted sex ratio distortion in a booklouse that is associated with extraordinarily divergent mitochondria.
In almost all multicellular eukaryotes, as well as many unicellular ones (i.e., microbial eukaryotes), transmission of mitochondria is strictly uniparental (4, 11). This is not just a consequence of eggs being much larger than sperm, as mitochondria are transmitted exclusively via males in some lineages, such as many conifer species (12). Organisms have independently evolved diverse, sophisticated strategies to target and destroy mitochondria from the opposite sex, even in species with sperm containing very few mitochondria (11, 13, 14). This is best explained as a mechanism of control by the host, to reduce conflict and to prevent the spread of selfish mitochondria (5, 15). Uniparental inheritance prevents mixing of different cytoplasmic lineages, and is thus expected to reduce competitive interactions between mitochondrial variants. Even with strict uniparental transmission, many generations of asexual reproduction within a host may allow mitochondrial genomes that have a replication advantage but that are ultimately deleterious to increase in frequency. In general, the frequency and fitness consequences of such selfish mitochondria have been little studied, although these have been documented in diverse organisms, including nematodes and yeast (16, 17). In humans, there are many documented cases of mitochondrial genomes with deleterious mutations reaching high frequencies within an individual, with negative health consequences (18).
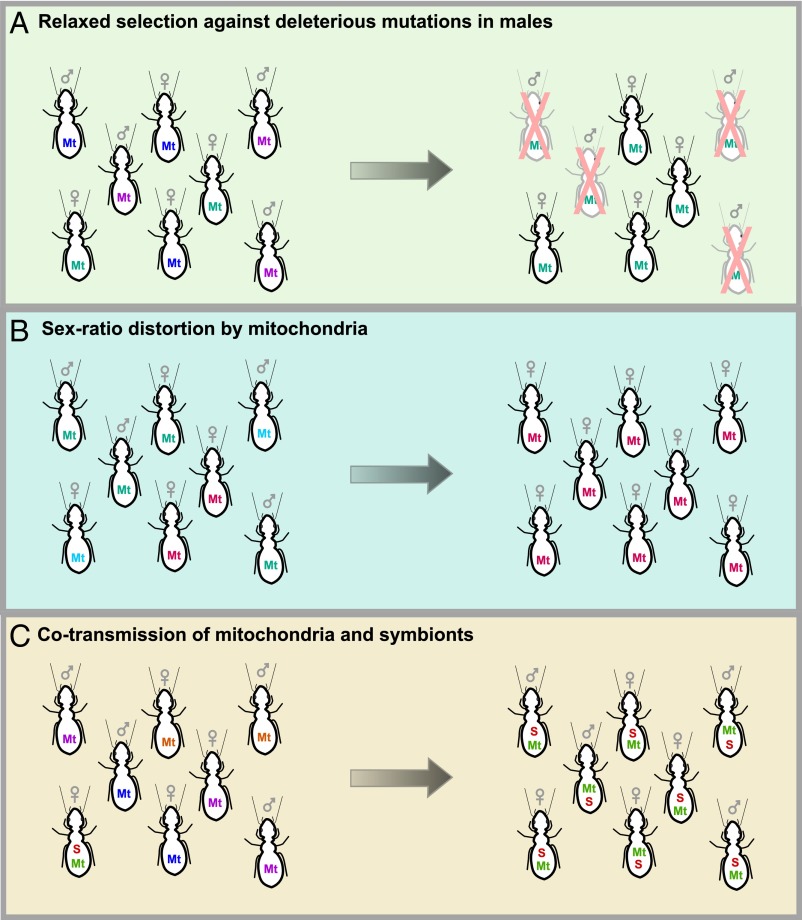
Uniparental transmission means that one sex is an evolutionary dead end, and this plays a major role in shaping the evolutionary trajectory of mitochondria (19, 20). For most multicellular organisms, transmission of mitochondria is maternal, and we focus the rest of our discussion on this mode of inheritance. First, the combination of asexual reproduction and small, serially bottlenecked populations has resulted in the persistent accumulation of slightly deleterious mutations in mitochondria (21). This pattern has been observed across a wide range of organisms, and in addition to mitochondria, we also see it in maternally transmitted microbial endosymbionts (22). This phenomenon is exacerbated in the nontransmitting sex: a major consequence of maternal transmission of mitochondria is that mutations that are deleterious in males can reach high frequencies if they are neutral (or advantageous, or even slightly deleterious) in females (20, 23) (Fig. 1A). This finding helps explain why male infertility in humans is commonly a result of mitochondrial mutations (24, 25). A recent beautiful study in Drosophila melanogaster fruit flies demonstrated the pervasive effects of this hidden mitochondrial variation on male fitness (26). The authors established fly lines with different mitochondrial genomes but the same nuclear genetic background. Despite the fact that these flies differed only with respect to their tiny mitochondrial genomes, there were large fitness consequences, but only in males. One mitonuclear combination resulted in male sterility, and in all combinations gene expression in males (but not females) was radically altered, with over 1,000 nuclear genes affected, especially in male-specific tissues.
Fig. 1.
Three possible consequences of maternal transmission of mitochondria (and other maternally transmitted organelles and symbionts). (A) Mutations that are deleterious in males can become common if they do not decrease female fitness. (B) Maternally transmitted lineages that increase the frequency of females will be favored by selection. (C) Mitochondria and maternally transmitted symbionts are linked, such that symbionts that spread in a population will bring their associated mitochondrial haplotype along with them. Different colors represent different haplotypes. Mt, mitochondria; S, symbiont.
Another striking consequence of maternal transmission is sex ratio distortion (Fig. 1B). Maternally transmitted lineages that increase the frequency of females will be favored by selection; this may result in conflicts between cytoplasmic and nuclear genes over optimal offspring sex ratios (27, 28). Female-biased sex distortion is best known in maternally transmitted microbial symbionts of arthropods (19, 29, 30), in which at least five different lineages of intracellular bacteria, such as Wolbachia, Rickettsia, and Spiroplasma (31–33), and one lineage of intracellular microbial eukaryote, Microsporidia (34), have independently evolved the ability to manipulate reproduction in a wide range of hosts. These symbionts distort sex ratios in three sophisticated ways: by causing infected females to reproduce asexually (parthenogenesis-induction), by transforming infected males into females (feminization), or by killing the sons of infected females early in development (male-killing). These strategies have different predicted equilibrium frequencies and evolutionary outcomes. For example, symbionts that induce parthenogenesis are more likely to become fixed in a population, because males are no longer required for reproduction. Although there has been much recent work on reproductive manipulators, we are probably only at the tip of the iceberg in describing the diversity of manipulators, because only terrestrial arthropods have been surveyed in any detail. Interestingly, many strategies that have been predicted to occur (19, 35), such as symbionts that distort sex ratios by preventing fertilization of Y chromosome-bearing sperm, have not yet been discovered.
What about sex ratio distortion by organelles? As far as we are aware, there are no known cases of distortion by plastids (and there has been relatively little work on consequences of uniparental transmission and sex-specific effects of plastids). On the other hand, mitochondria that distort sex ratios are well known; this is extremely common in plants in a phenomenon called cytoplasmic male sterility (5, 36). Cytoplasmic male sterility has evolved independently hundreds of times in hermaphroditic plant species, and in many different ways, from causing sterile or inviable pollen to preventing the proper development of male reproductive organs. This process results in an individual that is female. Cytoplasmic male sterility has been studied in great detail, in part because of its agricultural importance as an effective tool to prevent selfing. Two additional features of cytoplasmic male sterility stand out. First, its genetic basis is striking, as it typically involves the evolution of novel mitochondrial genes (5, 36, 37), as opposed to accumulation of sex-specific deleterious mutations in genes that are already present, although the novel genes have often incorporated truncations and fusions of other mitochondrial genes. Second, cytoplasmic male sterility has repeatedly been followed by the evolution of nuclear genes that suppress male sterility. In many cases, both sterility and suppressor genes become fixed in a population, such that sterility is only uncovered through genetic crosses between different populations. There is also evidence for evolutionary arms races between sterilizing and suppressing genes (38), highlighting the importance of conflict in shaping the evolution of sex ratio distortion. Although the genetic basis of a number of cytoplasmic male sterility and nuclear suppressor systems is now known, the mechanisms involved are still poorly understood.
Why are there no known cases of mitochondrial sex ratio distortion in animals, or for that matter, in any organisms other than plants? Another way of asking this question is whether there is something special about plants and their mitochondria. Plant mitochondrial genomes are incredibly dynamic (39–41) and have a great propensity for horizontal transfer and acquisition of novel genes, as seen in the many different ways cytoplasmic male sterility has evolved. Indeed, the first documented case of lateral gene transfer in eukaryotes that did not involve mobile genetic elements was in plant mitochondria (42). This was recently shown to be taken to an extreme degree in Amborella trichopoda, whose enormous ∼4-Mb mitochondrial genome has acquired the equivalent of six foreign mitochondrial genomes from algae, mosses, and other flowering plants (43). Another recent study showed that some mitochondrial tRNAs in liverworts were likely acquired from Chlamydia (44). However, horizontal transfer in mitochondrial genomes is not unique to plants, and has been reported in diverse lineages, including fungi, sponges, and corals, often associated with mobile introns (45–50). We would not be surprised if many more cases of mitochondrial horizontal transfer will be reported, including transfers from intracellular microbial endosymbionts, which include many known sex ratio distorters, occur in high numbers within cells and in close proximity to mitochondria, and are common sources of transfer to nuclear genomes (51). One lineage of endosymbionts in ticks, Candidatus Midichloria mitochondrii, is even known to reside within mitochondria (52). In sum, we do not see any clear reason why we should not find mitochondrial distortion in lineages other than plants. Because cytoplasmic male sterility in plants is always found in association with hermaphroditism, perhaps a useful place to start to look would be in lineages that contain hermaphrodites.
One especially promising lineage to study mitochondrial involvement in sex distortion is that of some bivalves. These are the only animals that are known to deviate from uniparental transmission of mitochondria (53, 54). Some bivalves have two distinct types of mitochondria. One type is transmitted from mothers to all their offspring (sons and daughters), whereas the other is transmitted exclusively from fathers to sons. This unusual mode of mitochondrial transmission is called doubly uniparental inheritance and it has been speculated that it evolved from paternal mitochondria escaping targeted destruction by the host (55). One fascinating consequence of doubly uniparental inheritance is that the two types of mitochondria are very different: they have different dimensions, tissue distributions, and are highly divergent at the sequence level. Strikingly, both mitochondrial types have acquired new genes (55–57), confirming that animal mitochondrial genomes are capable of evolving novelty. Little is known about the function of these novel genes, although recent studies have shown that they are transcribed and translated into proteins (58). Although they have no clear homologs, it has been suggested that at least one of the novel genes may have a viral origin (55, 57, 58). Understanding the function of these novel genes will not only gain insight into the mechanism of doubly uniparental inheritance, but it may also shed light on how sex itself is determined in bivalves, as this is not yet known. Interestingly, it has been speculated that mitochondria themselves might determine sex, as only males contain male-specific mitochondria and their unique genes (56). Finally, unusual sex ratio distortion has been documented in Mytilus mussels and Ruditapes clams (57, 59, 60), with individuals from the same population producing female-biased, male-biased, or 50:50 sex ratios. In Mytilus, female bias appears to be under maternal nuclear control. Sex ratio distortion in bivalves is an intriguing system to look for antagonistic interactions between distorting mitochondria and nuclear suppressors, similar to cytoplasmic male sterility in plants.
Another consequence of maternal transmission is that all genetic entities that are exclusively maternally transmitted, such as organelles, endosymbionts, and female-limited (W) sex chromosomes, are in perfect linkage (8) (Fig. 1C). As a result, their evolutionary fates are bound together. This has been studied in great detail in symbiont–mitochondria associations (61, 62), particularly with respect to the population genetic consequences of cotransmission; in contrast, there have been few studies on W chromosome–mitochondria associations (63), probably because until recently there have been few available W chromosome markers. Mitochondrial markers are often used to track and to age maternally transmitted microbial symbiont infections, including sex ratio distorters. Many studies have shown that symbionts decrease mitochondrial genetic variation as they spread through the population, replacing uninfected individuals with infected ones, along with their associated mitochondrial genome (61, 64–66). At the same time, the effective population size of mitochondria in the remaining uninfected individuals will be greatly reduced, further affecting variation. This phenomenon is especially common in symbionts that cause cytoplasmic incompatibility, in which uninfected females produce few or no offspring when they mate with infected males. As a result, infected females are at a reproductive advantage over their uninfected counterparts and rapidly replace them (67), purging mitochondrial variation.
On the other hand, it has also been shown that mitochondrial polymorphisms can persist in a population because of linkage with inherited symbionts (62, 68). For example, the ladybird beetle Adalia bipunctata is polymorphic both for mitochondrial haplotypes and at least two strains of male-killing Rickettsia (62). The deeply divergent mitochondria suggest that these male-killer infections are old, but it is not known how or why both the male-killers (and their mitochondrial partners) have persisted. In some cases, inherited symbionts and their associated mitochondrial partner have been introduced into a new host via hybridization. For example, the fly Drosophila quinaria harbors two extremely divergent mitochondria (69). One is perfectly linked with infection with a strain of the symbiont Wolbachia (whose effect on its host is not known, but it does not appear to cause cytoplasmic incompatibility or sex ratio distortion), and it is suggested that this mitochondrial haplotype actually came from a now extinct species that was the original host for this Wolbachia.
What are the functional consequences of mitochondrial polymorphism and linkage? A number of studies have begun to examine functional differences between mitochondrial variants, with consequences on host fitness. For example, a recent study in the neotropical pseudoscorpion Cordylochernes scorpioides found that trade-offs explained the persistence of two divergent mitochondrial genomes; although males carrying one of these genomes had higher sperm competitive ability, females with this mitochondrial genome had reduced sexual receptivity (70). In warblers, hybridization has resulted in the introgression of a mitochondrial variant that is associated with differences in flight efficiency and migratory potential (71). Little work has been done on functional consequences of linkage between mitochondria and symbionts, and how a symbiont’s persistence and spread depend on its mitochondrial partner (and vice versa) is generally not known. Perhaps the most detailed work has been in Drosophila simulans, which segregates numerous mitochondrial haplotypes with different respiration efficiencies, and that are linked to different strains of Wolbachia (72, 73). Some of the mitochondria associated with symbionts appear to be so deeply divergent that one might wonder how and whether they are coadapted with the nuclear genome. Studies in a wide range of organisms, including copepods (74) and wasps (75), have shown that rapid coevolution between mitochondria and nuclear genomes can result in hybrid mitonuclear incompatibilities, and we might expect symbionts and other sex ratio distortions to be constrained by similar incompatibilities.
Case Study: Extraordinary Sex Ratio Distortion and Mitochondrial Polymorphism in an Insect
We recently found an unusual case of extreme sex ratio distortion in a booklouse. Booklice are the closest free-living relatives of parasitic lice; both are members of the insect order Psocodea (76). The distortion occurs in a recently discovered sexual booklouse that is closely related to Liposcelis bostrychophila (Psocodea: Liposcelidae), a worldwide pest of stored grains and domestic kitchens that reproduces via apomictic parthenogenesis and is universally infected with a Rickettsia endosymbiont (77–79). The sexual form is not a pest; we collect it under dead yucca leaves and leaf litter in the Chiricahua Mountains of southeastern Arizona. Morphologically, the sexual form is virtually indistinguishable from L. bostrychophila. Because they are genetically distinct and reproductively isolated by virtue of their mode of reproduction, we refer to the sexual form as L. nr. bostrychophila.
When we confirmed that the sexual form is obligately sexual (i.e., virgin females will never produce offspring), we were surprised to find that our laboratory cultures of L. nr. bostrychophila were polymorphic for two types of females. One type of female never produces sons, whereas the other produces a mixed sex ratio (Fig. S1). The inheritance of this extreme sex ratio distortion is strictly maternal (i.e., females whose mothers produced only daughters will do the same, whereas females whose mothers produced sons and daughters will produce a mixed sex ratio). Although, in our experience, this polymorphism can be stably maintained in mixed laboratory cultures, we now culture the two types of females separately, adding males from the “normal” line to mate with “distorter” females every generation. We have ruled out the possibility that distorter females are gynogenetic sperm parasites (i.e., parthenogenetic lineages that require male sperm from a close relative to trigger development) (80), by recovering paternal alleles in the daughters of distorter females (Fig. S2).
Because the distortion is maternally inherited, we searched for maternally transmitted microbial symbionts that might be causing it, as these are of course well known in insects. Despite extensive searches, using targeted and untargeted molecular screens, as well as microscopy, we have found no evidence of a microbial symbiont. (This is in contrast to the asexual L. bostrychophila, which harbors Rickettsia.) We also find no evidence for male-killing in the distorter line, as there is no difference in the number of eggs that hatch and ultimately develop into adults compared with normal females (generalized linear model: df = 18, P = 0.113). Instead, we were surprised to find that mitochondrial genes in the distorter and normal lines are highly divergent. We focused our efforts on sequencing the mitochondrial genomes of these two different lines, to explore the possibility that the distortion might be caused by mitochondria.
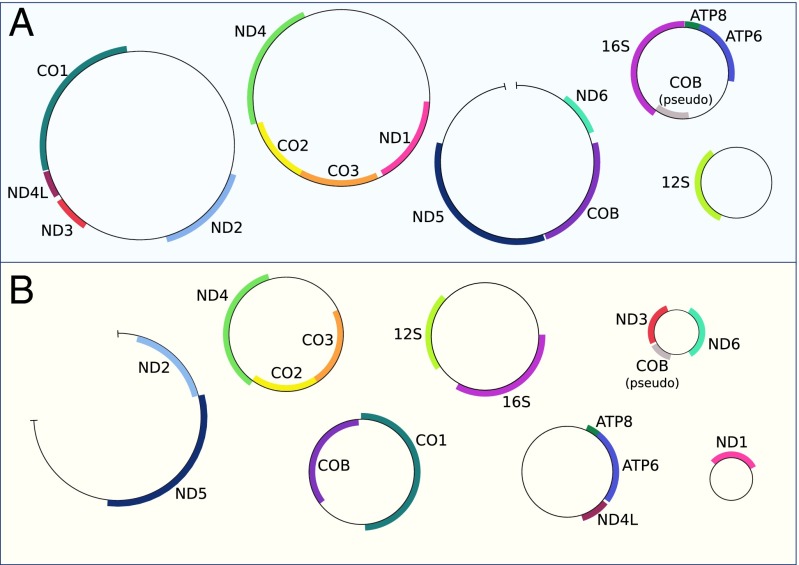
Sequencing the mitochondrial genomes of distorter and normal individuals proved to be quite a surprise (and also quite a challenge) (Fig. 2). Not only were they incredibly divergent (ranging from ∼53–77% similarity at protein-coding genes), they also had radically different gene order and genome structure. Both distorter and normal individuals had multipartite mitochondrial genomes, consisting of at least five and seven minicircles, respectively. Minicircle mitochondrial genomes have been documented in a number of Psocodea (81), ranging from two in L. bostrychophila and two other Liposcelis species (82, 83) [although another species, Liposcelis decolor, has a single “canonical” chromosome (84)], all the way to the extreme case in the human louse, Pediculus humanus, whose mitochondrial genome consists of 18 minicircles (85). Although many other mitochondria (and plastids) have evolved minicircle genome architecture (86, 87), in arthropods this organization is apparently restricted to Psocodea. Mitochondrial genes in Psocodea also appear to evolve extremely rapidly, in both parasitic and free-living species (76, 88). Much work remains to be done to understand how sex is determined in L. nr. bostrychophila and to identify the genetic basis of the extreme sex distortion, such as whether it is caused by mitochondria or by another female-limited portion of the genome, perhaps a B chromosome or a distorting X chromosome. Little is known about sex determination in Liposcelis and other Psocodea; this lineage typically exhibits XO male sex determination (89).
Fig. 2.
Distorter (A) and normal (B) L. nr. bostrychophila have radically different mitochondrial genome order and organization. Protein-coding and ribosomal genes are labeled; genes on the forward/complementary strand are on the outside/inside of the circles. Similarity between genes ranges from 53–80%: ATP6 (75.4%), ATP8 (65.4%), CO1 (76.6%), CO2 (73.9%), CO3 (70.6%), COB (76.8%), ND1 (76.1%), ND2 (73.8%), ND3 (76.8%), ND4 (72.4%), ND4L (75.9%), ND5 (70.9%), ND6 (53.1%), 12S (80.1%), 16S (80.1%). Although all circles have been closed using PCR, a few have not been completely sequenced, and these are indicated by the small gaps. Normal minicircle sizes (minicircles are named for their largest gene): 16S (3,265 bp), ND4 (3,426 bp), CO1 (3,147 bp), ATP6 (2,714 bp), ND6 (1,354 bp), ND1 (1,275 bp), ND5 (>3,717 bp). Distorter minicircle sizes: 16S (2,746 bp), ND4 (5,312 bp), CO1 (5,626 bp), 12S (2,131 bp), ND5 (∼4,600 bp).
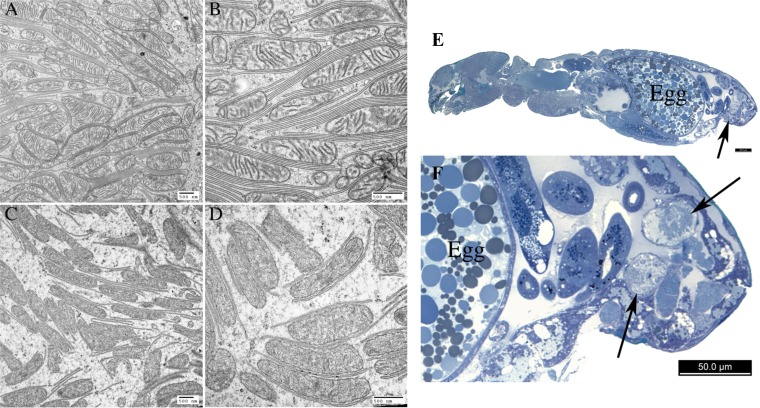
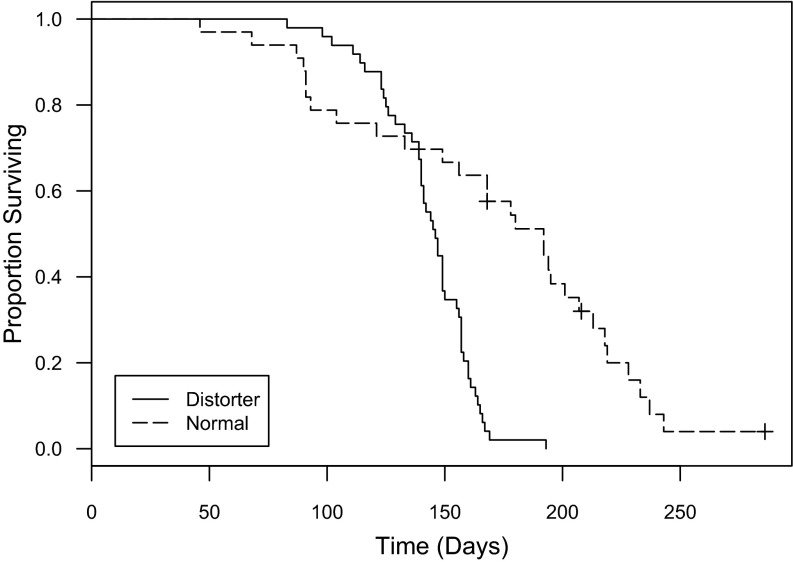
The degree of divergence between the normal and distorter mitochondrial genomes is striking, and we hypothesize that it has functional consequences. Interestingly, we have found a consistent morphological difference between distorter and normal mitochondria. We first uncovered this difference while performing electron microscopy to search for microbes that might be causing the sex ratio distortion. We did not find any microbes, but instead found that mitochondria in the paired rectal glands (90) of distorter females look unusual (Fig. 3). In normal females, these glands are packed with mitochondria that are tightly associated with membrane, forming mitochondrial–scalariform junction complexes. These complexes are only known in arthropods and are commonly found in the rectum, where they play an important role in ion transport and osmoregulation (91, 92). In the rectal glands of distorter females of the same age, on the other hand, there are few or no scalariform junctions, and mitochondria have few or no cristae and irregular shapes; this morphology is reminiscent of aged or damaged mitochondria (93, 94). Mitochondria in other tissues in distorter females do not appear different from normal females. We speculate that the unusual mitochondria in distorter rectal glands may be a result of cytonuclear incompatibilities that are exposed in these tissues because they are so metabolically active (and packed with mitochondria). Further support for cytonuclear incompatibilities in distorter females comes from the observation that they have a reduced lifespan relative to normal females (coxph: df = 1, P < 0.001) (Fig. 4); this is also intriguing, given the mitochondrion’s well-known role in longevity (72). Thus, even if mitochondria are not the cause of the sex ratio distortion, incompatibilities between distorter mitochondria and the nuclear genome may play a major role in shaping how the distortion persists in the wild (and in our mixed laboratory cultures), as we might otherwise expect distorter females to overtake their normal counterparts because they only produce females, which would then lead to extinction.
Fig. 3.
Distinctive mitochondrial morphology in rectal glands of distorter females. (A) TEM of 4-wk-old normal female rectal gland, showing mitochondria with intact cristae, many intact scalariform junctions (i.e., parallel plasma membranes), and even ground substance (i.e., cytosol) between the two. (B) Close-up of previous image. (C) TEM of 4-wk-old distorter female rectal gland, showing abnormal mitochondria with fragmented, electron dense material within and few cristae, few intact scalariform junctions, and condensed ground substance between the two. (D) Close-up of previous image. (E and F) Light micrograph of sagittal section of a normal female with arrows to indicate the location of the glands. (Scale bars: 500 nm in A–D and 50 μm in E and F.)
Fig. 4.
Distorter L. nr. bostrychophila females have a significantly shorter lifespan than normal females (coxph: df = 1, P < 0.001; n = 51 and 32 for distorter and normal females, respectively). Crosses indicate censored data (two individuals were lost during the experiment and one individual survived past the end of the experiment).
Conclusion
We predict that the coming years will see the discovery of many novel cases of sex ratio distortion, such as the extreme case in booklice described here, as well as the discovery of nonplant mitochondrial distorters. This will be spurred in part by the growing realization of the importance of microbial symbionts in shaping the ecology and evolution of multicellular organisms. It will also be facilitated by the ease of DNA sequencing, which will make it much easier to develop markers for sex chromosomes and selfish genetic elements. Of course, the easiest place to start looking for interesting systems is in cases of deeply divergent mitochondrial polymorphisms, and this will be facilitated by the (fortuitous) choice of the mitochondrial cytochrome c oxidase gene as the marker of choice in animal DNA barcoding studies that are currently cataloguing the planet’s biodiversity (95). Finally, we speculate that the persistence of many sex ratio distortion systems, as well as other interesting and unusual reproductive systems with maternal inheritance (96, 97), may be affected by mitonuclear incompatibilities.
Methods
Insect Rearing.
Distorter and normal females were kept in separate cultures in glass jars (125 mL). We used a 1:10 (by weight) mixture of Rice Krispies (Kellogg’s) and Cracked Wheat (Bob’s Red Mill) to rear insects. Colonies were maintained at 75% relative humidity and 27 °C. We added males to distorter female colonies weekly to ensure females had an opportunity to mate.
Mitochondrial Sequencing and Annotation.
We sequenced the mitochondrial genome of distorter and normal females with a combination of Illumina and Sanger sequencing. For Illumina sequencing, we extracted DNA from ethanol-preserved distorter and normal females (∼35 pooled individuals per line) using a Qiagen DNeasy Blood and Tissue kit. Libraries for each line were constructed and sequenced by Beckman Coulter Genomics, providing ∼4 × 107 100-bp PE reads per line. Draft assemblies for each line were generated with Ray v2.20 (k = 31) (98). We searched assemblies for mitochondrial genes using tblastx, with sequenced L. bostrychophila mitochondrial genomes as queries (82). Pieces of retrieved genes (400–800 bp) were then used as seeds in mitoBim (99) (proofreading mode) to corroborate and extend minicircles, before validation by PCR and Sanger sequencing (see Dataset S1 for primer sequence and PCR conditions). We sequenced most of the PCR products directly but in some cases products were cloned using StrataClone PCR cloning kits (Agilent Technologies). All Sanger sequencing was carried out with total DNA extractions from 16 females in 60 μL of PrepMan Ultra (Life Technologies). We annotated the mitochondrial protein coding regions by extracting ORFs longer than 120 bp from the minicircle assemblies using getorf (EMBOSS) and using blastp searches against the nonredundant protein (nr) database (National Center for Biotechnology Information). We manually identified rRNA coding regions using Geneious v6.1 by performing nucleotide alignments using the default parameters with the rRNA coding regions from L. bostrychophila. Mitochondrial genome sequences have been deposited in GenBank under the following accession nos. KP641133, KP657691–657699, and KP671844–671845.
We also completed a series of PCR reactions in individual booklice to explore mitochondrial variation within each female type. For eight individual females of each female type (i.e., distorter or normal), we amplified five different regions of the mitochondrial genome that were expected to range in size from 1,200 to 3,000 bp. Single female DNA extractions were carried out in 20 μL of PrepMan Ultra. For all five regions, all eight females produced a single band of the expected size, suggesting that there is little within-type variation.
Microscopy.
Adult insects were processed using standard transmission electron microscopy (TEM) methodology (100): double-fixation and embedding into Epon. For light microscopy, 0.5-μm sections were stained in Richardson’s Stain (Azure II and Methylene Blue in Borax solution). Next, 85-nm-thick TEM sections were stained in uranyl acetate and lead citrate and viewed in a Hitachi H7000 TEM at 75 kV. Images were captured using an AMT 2k × 2k CCD camera.
Longevity and Male-Killing.
We set up jars containing 150 late-instar females of each type (distorter or normal) along with 75 males. After 5 days the females were reproductively mature and mated. We then transferred the females into ∼5 g of cracked wheat to lay eggs. After 24 h the females were transferred to another jar containing 5 g of cracked wheat. After we removed the females from egg-laying jars, 10 eggs from the jar were transferred into a Petri dish (35-mm diameter) containing 0.7 g of Rice Krispies and cracked wheat. We prepared two Petri dishes containing 10 eggs every day for each female type. We did this for 5 d, resulting in 10 replicate containers for each female type.
Three weeks after the eggs were laid, we began checking for adults. We recorded when individuals completed development and transferred females into a new Petri dish containing 0.7 g of food. Females raised in the same Petri dish were kept together as adults. We recorded when males completed development but then discarded them. We checked females approximately three times a week and recorded female longevity (from the date eggs were laid until death) as well as the number and sex of individuals that developed from each container.
We analyzed data using Rstudio v3.1.0 (101). We performed a survival analysis for the data assessing longevity of females with the package survival (102) using a Cox proportional hazards (coxph) model. We assessed whether the different female types differed in longevity, clustering individuals by container. We also assessed whether there was any evidence of male killing by examining whether there was a difference in the number of individuals (males and females) that developed from a container depending on the type of individuals in the container.
Supplementary Material
Acknowledgments
We thank Ed Mockford, who has taught us so much about Liposcelis. This research was supported by a National Sciences and Engineering Research Council of Canada Discovery grant (to S.J.P.); the Canadian Institute for Advanced Research (S.J.P.); and a National Sciences and Engineering Research Council of Canada scholarship (to P.T.H.).
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Symbioses Becoming Permanent: The Origins and Evolutionary Trajectories of Organelles,” held October 15–17, 2014, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Symbioses.
This article is a PNAS Direct Submission. J.P.M. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KP641133, KP657691–KP657699, and KP671844–KP671845).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421391112/-/DCSupplemental.
References
- 1.Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 2012;4(9):a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballard J, Rand D. The population biology of mitochondrial DNA and its phylogenetic implications. Annu Rev Ecol Evol Syst. 2005;36:621–642. [Google Scholar]
- 3.Ballard JW, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13(4):729–744. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- 4.Birky CW., Jr The inheritance of genes in mitochondria and chloroplasts: Laws, mechanisms, and models. Annu Rev Genet. 2001;35:125–148. doi: 10.1146/annurev.genet.35.102401.090231. [DOI] [PubMed] [Google Scholar]
- 5.Burt A, Trivers R. Genes in Conflict: The Biology of Selfish Genetic Elements. Belknap; Cambridge, MA: 2006. Selfish mitochondrial DNA; pp. 142–184. [Google Scholar]
- 6.Burton RS, Pereira RJ, Barreto FS. Cytonuclear genomic interactions and hybrid breakdown. Annu Rev Ecol Evol Syst. 2013;44:281–302. [Google Scholar]
- 7.Dowling DK, Friberg U, Lindell J. Evolutionary implications of non-neutral mitochondrial genetic variation. Trends Ecol Evol. 2008;23(10):546–554. doi: 10.1016/j.tree.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Hurst GD, Jiggins FM. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: The effects of inherited symbionts. Proc Biol Sci. 2005;272(1572):1525–1534. doi: 10.1098/rspb.2005.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rand DM, Haney RA, Fry AJ. Cytonuclear coevolution: The genomics of cooperation. Trends Ecol Evol. 2004;19(12):645–653. doi: 10.1016/j.tree.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Wolff JN, Ladoukakis ED, Enríquez JA, Dowling DK. Mitonuclear interactions: Evolutionary consequences over multiple biological scales. Philos Trans R Soc Lond B Biol Sci. 2014;369(1646):20130443. doi: 10.1098/rstb.2013.0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birky CW., Jr Uniparental inheritance of mitochondrial and chloroplast genes: Mechanisms and evolution. Proc Natl Acad Sci USA. 1995;92(25):11331–11338. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neale DB, Marshall KA, Sederoff RR. Chloroplast and mitochondrial DNA are paternally inherited in Sequoia sempervirens D. Don Endl. Proc Natl Acad Sci USA. 1989;86(23):9347–9349. doi: 10.1073/pnas.86.23.9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al Rawi S, et al. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334(6059):1144–1147. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- 14.Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334(6059):1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- 15.Frank SA. Host-symbiont conflict over the mixing of symbiotic lineages. Proc Biol Sci. 1996;263(1368):339–344. doi: 10.1098/rspb.1996.0052. [DOI] [PubMed] [Google Scholar]
- 16.Taylor DR, Zeyl C, Cooke E. Conflicting levels of selection in the accumulation of mitochondrial defects in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2002;99(6):3690–3694. doi: 10.1073/pnas.072660299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark KA, et al. Selfish little circles: Transmission bias and evolution of large deletion-bearing mitochondrial DNA in Caenorhabditis briggsae nematodes. PLoS ONE. 2012;7(7):e41433. doi: 10.1371/journal.pone.0041433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6(5):389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Werren JH, O'Neill SL. The evolution of heritable symbionts. In: O’Neill SL, Hoffmann AA, Werren JH, editors. Influential Passengers: Inherited Microorganisms and Arthropod Reproduction. Oxford Univ Press; Oxford: 1997. pp. 1–42. [Google Scholar]
- 20.Frank SA, Hurst LD. Mitochondria and male disease. Nature. 1996;383(6597):224. doi: 10.1038/383224a0. [DOI] [PubMed] [Google Scholar]
- 21.Nachman MW, Brown WM, Stoneking M, Aquadro CF. Nonneutral mitochondrial DNA variation in humans and chimpanzees. Genetics. 1996;142(3):953–963. doi: 10.1093/genetics/142.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moran NA. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci USA. 1996;93(7):2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camus MF, Clancy DJ, Dowling DK. Mitochondria, maternal inheritance, and male aging. Curr Biol. 2012;22(18):1717–1721. doi: 10.1016/j.cub.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 24.Ruiz-Pesini E, et al. Human mtDNA haplogroups associated with high or reduced spermatozoa motility. Am J Hum Genet. 2000;67(3):682–696. doi: 10.1086/303040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kao SH, Chao HT, Wei YH. Multiple deletions of mitochondrial DNA are associated with the decline of motility and fertility of human spermatozoa. Mol Hum Reprod. 1998;4(7):657–666. doi: 10.1093/molehr/4.7.657. [DOI] [PubMed] [Google Scholar]
- 26.Innocenti P, Morrow EH, Dowling DK. Experimental evidence supports a sex-specific selective sieve in mitochondrial genome evolution. Science. 2011;332(6031):845–848. doi: 10.1126/science.1201157. [DOI] [PubMed] [Google Scholar]
- 27.Cosmides LM, Tooby J. Cytoplasmic inheritance and intragenomic conflict. J Theor Biol. 1981;89(1):83–129. doi: 10.1016/0022-5193(81)90181-8. [DOI] [PubMed] [Google Scholar]
- 28.Lewis D. Male sterility in natural populations of hermaphrodite plants: The equilibrium between females and hermaphrodites to be expected with different types of inheritance. New Phytol. 1941;40(1):56–63. [Google Scholar]
- 29.Stouthamer R, Breeuwer JA, Hurst GD. Wolbachia pipientis: Microbial manipulator of arthropod reproduction. Annu Rev Microbiol. 1999;53:71–102. doi: 10.1146/annurev.micro.53.1.71. [DOI] [PubMed] [Google Scholar]
- 30.Engelstaedter J, Hurst GDD. The ecology and evolution of microbes that manipulate host reproduction. Annu Rev Ecol Evol Syst. 2009;40:127–149. [Google Scholar]
- 31.Poulson DF, Sakaguchi B. Nature of “sex-ratio” agent in Drosophila. Science. 1961;133(3463):1489–1490. doi: 10.1126/science.133.3463.1489. [DOI] [PubMed] [Google Scholar]
- 32.Stouthamer R, Luck RF, Hamilton WD. Antibiotics cause parthenogenetic Trichogramma (Hymenoptera/Trichogrammatidae) to revert to sex. Proc Natl Acad Sci USA. 1990;87(7):2424–2427. doi: 10.1073/pnas.87.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Werren JH, et al. Rickettsial relative associated with male killing in the ladybird beetle (Adalia bipunctata) J Bacteriol. 1994;176(2):388–394. doi: 10.1128/jb.176.2.388-394.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bulnheim HP, Vávra J. Infection by the microsporidian Octosporea effeminans sp. n., and its sex determining influence in the amphipod Gammarus duebeni. J Parasitol. 1968;54(2):241–248. [PubMed] [Google Scholar]
- 35.Hurst LD. The incidences, mechanisms and evolution of cytoplasmic sex-ratio distorters in animals. Biol Rev Camb Philos Soc. 1993;68(1):121–194. [Google Scholar]
- 36.Chase CD. Cytoplasmic male sterility: A window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 2007;23(2):81–90. doi: 10.1016/j.tig.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 37.Luo D, et al. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat Genet. 2013;45(5):573–577. doi: 10.1038/ng.2570. [DOI] [PubMed] [Google Scholar]
- 38.Fujii S, Bond CS, Small ID. Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc Natl Acad Sci USA. 2011;108(4):1723–1728. doi: 10.1073/pnas.1007667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knoop V. The mitochondrial DNA of land plants: Peculiarities in phylogenetic perspective. Curr Genet. 2004;46(3):123–139. doi: 10.1007/s00294-004-0522-8. [DOI] [PubMed] [Google Scholar]
- 40.Sloan DB, et al. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012;10(1):e1001241. doi: 10.1371/journal.pbio.1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sloan DB, Barr CM, Olson MS, Keller SR, Taylor DR. Evolutionary rate variation at multiple levels of biological organization in plant mitochondrial DNA. Mol Biol Evol. 2008;25(2):243–246. doi: 10.1093/molbev/msm266. [DOI] [PubMed] [Google Scholar]
- 42.Bergthorsson U, Adams KL, Thomason B, Palmer JD. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature. 2003;424(6945):197–201. doi: 10.1038/nature01743. [DOI] [PubMed] [Google Scholar]
- 43.Rice DW, et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science. 2013;342(6165):1468–1473. doi: 10.1126/science.1246275. [DOI] [PubMed] [Google Scholar]
- 44.Knie N, Polsakiewicz M, Knoop V. Horizontal gene transfer of chlamydial-like tRNA genes into early vascular plant mitochondria. Mol Biol Evol. 2015;32(3):629–634. doi: 10.1093/molbev/msu324. [DOI] [PubMed] [Google Scholar]
- 45.Bilewitch JP, Degnan SM. A unique horizontal gene transfer event has provided the octocoral mitochondrial genome with an active mismatch repair gene that has potential for an unusual self-contained function. BMC Evol Biol. 2011;11:228. doi: 10.1186/1471-2148-11-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fukami H, Chen CA, Chiou CY, Knowlton N. Novel group I introns encoding a putative homing endonuclease in the mitochondrial cox1 gene of Scleractinian corals. J Mol Evol. 2007;64(5):591–600. doi: 10.1007/s00239-006-0279-4. [DOI] [PubMed] [Google Scholar]
- 47.Pont-Kingdon GA, et al. A coral mitochondrial mutS gene. Nature. 1995;375(6527):109–111. doi: 10.1038/375109b0. [DOI] [PubMed] [Google Scholar]
- 48.Rot C, Goldfarb I, Ilan M, Huchon D. Putative cross-kingdom horizontal gene transfer in sponge (Porifera) mitochondria. BMC Evol Biol. 2006;6:71. doi: 10.1186/1471-2148-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vallès Y, Halanych KM, Boore JL. Group II introns break new boundaries: Presence in a bilaterian’s genome. PLoS ONE. 2008;3(1):e1488. doi: 10.1371/journal.pone.0001488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu B, Hao W. Horizontal transfer and gene conversion as an important driving force in shaping the landscape of mitochondrial introns. G3. 2014;4(4):605–612. doi: 10.1534/g3.113.009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kondo N, Nikoh N, Ijichi N, Shimada M, Fukatsu T. Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect. Proc Natl Acad Sci USA. 2002;99(22):14280–14285. doi: 10.1073/pnas.222228199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sassera D, et al. ‘Candidatus Midichloria mitochondrii’, an endosymbiont of the tick Ixodes ricinus with a unique intramitochondrial lifestyle. Int J Syst Evol Microbiol. 2006;56(Pt 11):2535–2540. doi: 10.1099/ijs.0.64386-0. [DOI] [PubMed] [Google Scholar]
- 53.Zouros E, Oberhauser Ball A, Saavedra C, Freeman KR. An unusual type of mitochondrial DNA inheritance in the blue mussel Mytilus. Proc Natl Acad Sci USA. 1994;91(16):7463–7467. doi: 10.1073/pnas.91.16.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zouros E. Biparental inheritance through uniparental transmission: The doubly uniparental inheritance (DUI) of mitochondrial DNA. Evol Biol. 2013;40(1):1–31. [Google Scholar]
- 55.Milani L, Ghiselli F, Guerra D, Breton S, Passamonti M. A comparative analysis of mitochondrial ORFans: New clues on their origin and role in species with doubly uniparental inheritance of mitochondria. Genome Biol Evol. 2013;5(7):1408–1434. doi: 10.1093/gbe/evt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Breton S, et al. Novel protein genes in animal mtDNA: A new sex determination system in freshwater mussels (Bivalvia: Unionoida)? Mol Biol Evol. 2011;28(5):1645–1659. doi: 10.1093/molbev/msq345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Milani L, Ghiselli F, Nuzhdin SV, Passamonti M. Nuclear genes with sex bias in Ruditapes philippinarum (Bivalvia, veneridae): Mitochondrial inheritance and sex determination in DUI species. J Exp Zoolog B Mol Dev Evol. 2013;320:442–454. doi: 10.1002/jez.b.22520. [DOI] [PubMed] [Google Scholar]
- 58.Milani L, Ghiselli F, Maurizii MG, Nuzhdin SV, Passamonti M. Paternally transmitted mitochondria express a new gene of potential viral origin. Genome Biol Evol. 2014;6(2):391–405. doi: 10.1093/gbe/evu021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saavedra C, Reyero MI, Zouros E. Male-dependent doubly uniparental inheritance of mitochondrial DNA and female-dependent sex-ratio in the mussel Mytilus galloprovincialis. Genetics. 1997;145(4):1073–1082. doi: 10.1093/genetics/145.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kenchington E, MacDonald B, Cao L, Tsagkarakis D, Zouros E. Genetics of mother-dependent sex ratio in blue mussels (Mytilus spp.) and implications for doubly uniparental inheritance of mitochondrial DNA. Genetics. 2002;161(4):1579–1588. doi: 10.1093/genetics/161.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ballard JW, Hatzidakis J, Karr TL, Kreitman M. Reduced variation in Drosophila simulans mitochondrial DNA. Genetics. 1996;144(4):1519–1528. doi: 10.1093/genetics/144.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiggins FM, Tinsley MC. An ancient mitochondrial polymorphism in Adalis bipunctata linked to a sex-ratio-distorting bacterium. Genetics. 2005;171(3):1115–1124. doi: 10.1534/genetics.105.046342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berlin S, Tomaras D, Charlesworth B. Low mitochondrial variability in birds may indicate Hill-Robertson effects on the W chromosome. Heredity (Edinb) 2007;99(4):389–396. doi: 10.1038/sj.hdy.6801014. [DOI] [PubMed] [Google Scholar]
- 64.Turelli M, Hoffmann AA, McKechnie SW. Dynamics of cytoplasmic incompatibility and mtDNA variation in natural Drosophila simulans populations. Genetics. 1992;132(3):713–723. doi: 10.1093/genetics/132.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiggins FM. Male-killing Wolbachia and mitochondrial DNA: Selective sweeps, hybrid introgression and parasite population dynamics. Genetics. 2003;164(1):5–12. doi: 10.1093/genetics/164.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shoemaker DD, Dyer KA, Ahrens M, McAbee K, Jaenike J. Decreased diversity but increased substitution rate in host mtDNA as a consequence of Wolbachia endosymbiont infection. Genetics. 2004;168(4):2049–2058. doi: 10.1534/genetics.104.030890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turelli M, Hoffmann AA. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature. 1991;353(6343):440–442. doi: 10.1038/353440a0. [DOI] [PubMed] [Google Scholar]
- 68.Xiao JH, et al. Wolbachia infection and dramatic intraspecific mitochondrial DNA divergence in a fig wasp. Evolution. 2012;66(6):1907–1916. doi: 10.1111/j.1558-5646.2011.01561.x. [DOI] [PubMed] [Google Scholar]
- 69.Dyer KA, Burke C, Jaenike J. Wolbachia-mediated persistence of mtDNA from a potentially extinct species. Mol Ecol. 2011;20(13):2805–2817. doi: 10.1111/j.1365-294X.2011.05128.x. [DOI] [PubMed] [Google Scholar]
- 70.Padua MV, Zeh DW, Bonilla MM, Zeh JA. Sisters’ curse: Sexually antagonistic effects constrain the spread of a mitochondrial haplogroup superior in sperm competition. Proc Biol Sci. 2014;281(1797):20141686. doi: 10.1098/rspb.2014.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toews DPL, Mandic M, Richards JG, Irwin DE. Migration, mitochondria, and the yellow-rumped warbler. Evolution. 2014;68(1):241–255. doi: 10.1111/evo.12260. [DOI] [PubMed] [Google Scholar]
- 72.Melvin RG, Ballard JW. Intraspecific variation in survival and mitochondrial oxidative phosphorylation in wild-caught Drosophila simulans. Aging Cell. 2006;5(3):225–233. doi: 10.1111/j.1474-9726.2006.00211.x. [DOI] [PubMed] [Google Scholar]
- 73.Pichaud N, Ballard JWO, Tanguay RM, Blier PU. Naturally occurring mitochondrial DNA haplotypes exhibit metabolic differences: Insight into functional properties of mitochondria. Evolution. 2012;66(10):3189–3197. doi: 10.1111/j.1558-5646.2012.01683.x. [DOI] [PubMed] [Google Scholar]
- 74.Ellison CK, Burton RS. Disruption of mitochondrial function in interpopulation hybrids of Tigriopus californicus. Evolution. 2006;60(7):1382–1391. [PubMed] [Google Scholar]
- 75.Ellison CK, Niehuis O, Gadau J. Hybrid breakdown and mitochondrial dysfunction in hybrids of Nasonia parasitoid wasps. J Evol Biol. 2008;21(6):1844–1851. doi: 10.1111/j.1420-9101.2008.01608.x. [DOI] [PubMed] [Google Scholar]
- 76.Yoshizawa K, Johnson KP. Phylogenetic position of Phthiraptera (Insecta: Paraneoptera) and elevated rate of evolution in mitochondrial 12S and 16S rDNA. Mol Phylogenet Evol. 2003;29(1):102–114. doi: 10.1016/s1055-7903(03)00073-3. [DOI] [PubMed] [Google Scholar]
- 77.Yusuf M, Turner B. Characterisation of Wolbachia-like bacteria isolated from the parthenogenetic stored-product pest psocid Liposcelis bostrychophila (Badonnel) (Psocoptera) J Stored Prod Res. 2004;40(2):207–225. [Google Scholar]
- 78.Perlman SJ, Zchori-Fein E, Hunter MS. The emerging diversity of Rickettsia. Proc Biol Sci. 2006;273(1598):2097–2106. doi: 10.1098/rspb.2006.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Behar A, McCormick LJ, Perlman SJ. Rickettsia felis infection in a common household insect pest, Liposcelis bostrychophila (Psocoptera: Liposcelidae) Appl Environ Microbiol. 2010;76(7):2280–2285. doi: 10.1128/AEM.00026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Beukeboom L, Vrijenhoek R. Evolutionary genetics and ecology of sperm-dependent parthenogenesis. J Evol Biol. 1998;11(6):755–782. [Google Scholar]
- 81.Cameron SL, Yoshizawa K, Mizukoshi A, Whiting MF, Johnson KP. Mitochondrial genome deletions and minicircles are common in lice (Insecta: Phthiraptera) BMC Genomics. 2011;12:394. doi: 10.1186/1471-2164-12-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wei DD, et al. The multipartite mitochondrial genome of Liposcelis bostrychophila: Insights into the evolution of mitochondrial genomes in bilateral animals. PLoS ONE. 2012;7(3):e33973. doi: 10.1371/journal.pone.0033973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen SC, et al. Evolution of multipartite mitochondrial genomes in the booklice of the genus Liposcelis (Psocoptera) BMC Genomics. 2014;15:861. doi: 10.1186/1471-2164-15-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen SC, Wei DD, Shao R, Dou W, Wang JJ. The complete mitochondrial genome of the booklouse, Liposcelis decolor: Insights into gene arrangement and genome organization within the genus Liposcelis. PLoS ONE. 2014;9(3):e91902. doi: 10.1371/journal.pone.0091902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shao R, Kirkness EF, Barker SC. The single mitochondrial chromosome typical of animals has evolved into 18 minichromosomes in the human body louse, Pediculus humanus. Genome Res. 2009;19(5):904–912. doi: 10.1101/gr.083188.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burger G, Gray MW, Lang BF. Mitochondrial genomes: Anything goes. Trends Genet. 2003;19(12):709–716. doi: 10.1016/j.tig.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 87.Zhang Z, Green BR, Cavalier-Smith T. Single gene circles in dinoflagellate chloroplast genomes. Nature. 1999;400(6740):155–159. doi: 10.1038/22099. [DOI] [PubMed] [Google Scholar]
- 88.Yoshizawa K, Johnson KP. Changes in base composition bias of nuclear and mitochondrial genes in lice (Insecta: Psocodea) Genetica. 2013;141(10-12):491–499. doi: 10.1007/s10709-013-9748-z. [DOI] [PubMed] [Google Scholar]
- 89.Golub NV, Nokkala S. Chromosome numbers in eight species of Palaearctic Psocoptera (Insecta) Comp Cytogenet. 2009;3(1):33–41. [Google Scholar]
- 90.Noland RC. The anatomy of Troctes divinatorius, Mull. Trans Wis Acad Sci Arts Lett. 1924;21:195–211. [Google Scholar]
- 91.Noble-Nesbitt J. Hindgut with rectum. In: Harrison FW, Loche M, editors. Microscopic Anatomy of Invertebrates, Volume 11B: Insecta. Wiley-Liss; New York: 1998. pp. 759–808. [Google Scholar]
- 92.Noirot C, Noirot-Timothee C. Cell associations. In: Harrison FW, Loche M, editors. Microscopic Anatomy of Invertebrates, Volume 11A: Insecta. Wiley-Liss; New York: 1998. pp. 27–49. [Google Scholar]
- 93.Yasuda K, et al. Age-related changes of mitochondrial structure and function in Caenorhabditis elegans. Mech Ageing Dev. 2006;127(10):763–770. doi: 10.1016/j.mad.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 94.Oda Y, Yui R, Sakamoto K, Kita K, Matsuura ET. Age-related changes in the activities of respiratory chain complexes and mitochondrial morphology in Drosophila. Mitochondrion. 2012;12(2):345–351. doi: 10.1016/j.mito.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 95.Hebert P, Ratnasingham S, deWaard J. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci. 2003;270(Suppl 1):S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hoekstra HE. Unequal transmission of mitochondrial haplotypes in natural populations of field mice with XY females (genus Akodon) Am Nat. 2003;161(1):29–39. doi: 10.1086/344910. [DOI] [PubMed] [Google Scholar]
- 97.Normark BB, Ross L. Genetic conflict, kin and the origins of novel genetic systems. Philos Trans R Soc Lond B Biol Sci. 2014;369(1642):20130364. doi: 10.1098/rstb.2013.0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Boisvert S, Laviolette F, Corbeil J. Ray: Simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol. 2010;17(11):1519–1533. doi: 10.1089/cmb.2009.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hahn C, Bachmann L, Chevreux B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—A baiting and iterative mapping approach. Nucleic Acids Res. 2013;41(13):e129. doi: 10.1093/nar/gkt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hayat MA. 1989. Principles and Techniques of Electron Microscopy: Biological Applications (CRC Press, Boca Raton, FL), 3rd Ed, 469 pp.
- 101. R Core Team (2014) R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, Vienna, Austria) Available at www.R-project.org/
- 102.Therneau T. A Package for Survival Analysis in S. R package version 2.37-7. 2014 Available at cran.r-project.org/web/packages/survival/index.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.