Abstract
Apicomplexans are a major lineage of parasites, including causative agents of malaria and toxoplasmosis. How such highly adapted parasites evolved from free-living ancestors is poorly understood, particularly because they contain nonphotosynthetic plastids with which they have a complex metabolic dependency. Here, we examine the origin of apicomplexan parasitism by resolving the evolutionary distribution of several key characteristics in their closest free-living relatives, photosynthetic chromerids and predatory colpodellids. Using environmental sequence data, we describe the diversity of these apicomplexan-related lineages and select five species that represent this diversity for transcriptome sequencing. Phylogenomic analysis recovered a monophyletic lineage of chromerids and colpodellids as the sister group to apicomplexans, and a complex distribution of retention versus loss for photosynthesis, plastid genomes, and plastid organelles. Reconstructing the evolution of all plastid and cytosolic metabolic pathways related to apicomplexan plastid function revealed an ancient dependency on plastid isoprenoid biosynthesis, predating the divergence of apicomplexan and dinoflagellates. Similarly, plastid genome retention is strongly linked to the retention of two genes in the plastid genome, sufB and clpC, altogether suggesting a relatively simple model for plastid retention and loss. Lastly, we examine the broader distribution of a suite of molecular characteristics previously linked to the origins of apicomplexan parasitism and find that virtually all are present in their free-living relatives. The emergence of parasitism may not be driven by acquisition of novel components, but rather by loss and modification of the existing, conserved traits.
Keywords: Apicomplexa, Chromera, Colpodella, plastid organelle, parasitism origin
Apicomplexans are globally important parasites of humans and animals that include Plasmodium (malaria), Toxoplasma (toxoplasmosis), and Cryptosporidium (cryptosporidiosis). Their success as parasites rests on several highly specialized structures and systems that enable them to gain entry to and divide within cells or tissues of their hosts. These structures include the multimembrane pellicle, a relict nonphotosynthetic plastid (absent in Cryptosporidium), and the apical complex, which is made up of cytoskeletal and secretory elements (e.g., the conoid and rhoptries, respectively). Many specific characteristics of apicomplexans make attractive drug targets, and others may have played a key role in the origin of parasitism. Indeed, the question of apicomplexan origins has been of interest in general but is challenged by a paucity of comparable information from free-living relatives. Several apicomplexan relatives are known, some photosynthetic and others predatory (1, 2), but we lack a comprehensive understanding of their biology because they have either been discovered only recently, or are difficult to establish and maintain in culture. In general, photosynthetic apicomplexan relatives are referred to as chromerids (including Chromera and Vitrella) (1, 3) whereas predators are referred to as colpodellids (including Colpodella, Alphamonas, and Voromonas) (2, 4). Predatory and photosynthetic varieties are often treated as monophyletic (3), but exactly how chromerids, colpodellids, and apicomplexans are related to one another remains unresolved.
The absence of a robust phylogenetic framework is evident in our inability to reconstruct the evolution of even the most obvious of characters, plastid organelles. Nonphotosynthetic plastids were discovered in apicomplexans over a decade ago (5, 6), and plastids were present in the common ancestor of not only apicomplexans, chromerids, and colpodellids, but also dinoflagellates (7), a lineage collectively called the myzozoans (4). Photosynthesis has been lost multiple times in the group (7), but how many times and whether plastids were retained in nonphotosynthetic species are unknown. Myzozoans are a potentially useful system for characterizing the still mysterious forces that control plastid retention and loss in general (8). There is now evidence for the retention of plastid-derived genes in some nonphotosynthetic myzozoans (9, 10), but plastid distribution alone is of a limited benefit. Reconstruction of dependencies on plastid metabolic pathways in light of their cytosolic analogs and compound uptake would allow us to go beyond patterns to identifying processes that explain why plastids have been retained or lost in myzozoans as well as other eukaryotes.
The origin of the apicomplexan parasitic mechanism has been even more elusive. This elusiveness is partly because of their diverse strategies of interacting with the host cell and immunity: Many early-branching apicomplexans do not invade cells completely (e.g., some gregarines), many secreted proteins are species-specific (e.g., ROP proteins), and both structures and functions may be lost (e.g., Theileria lacks an apical complex). Other apicomplexan characteristics are ancestral, but their original function is unclear because structures involved in invasion have been identified in free-living relatives: an open conoid and rhoptry-like organelles are found in colpodellids, Chromera, and dinoflagellate relatives (11, 12) whereas the multimembrane alveolar pellicle is found across an even broader group (4, 13). Their homology is generally inferred from morphological similarity, but how deeply their functions are linked at the molecular level is unknown. Similarly, apparently unique characteristics in many apicomplexan proteins (14, 15) could be linked to the evolution of parasitism, but this idea cannot be tested without knowing their real distribution.
Here, we examine the origin and early evolution of apicomplexans based on analysis of molecular data from their free-living relatives. We first surveyed the global diversity of apicomplexan-related lineages from environmental sequence data, which reveal a number of uncharacterized groups in previously overlooked environments. Five species, two photosynthetic chromerids and three predatory colpodellids, spanning the majority of this diversity were then cultured, and their transcriptomes were sequenced. A concatenated phylogeny of 85 nuclear proteins unambiguously rejected the monophyly of both chromerids and colpodellids, resolved their relationship to apicomplexans, and showed multiple independent losses of photosynthesis in early apicomplexan evolution. The metabolic basis for plastid dependency was reconstructed, pointing to an ancient reliance on organelle isoprenoid synthesis and a mechanism of plastid and plastid genome retention or loss. Finally, the origin of parasitism itself was examined by determining the broader distribution of putatively apicomplexan-specific features. Many of them could be traced back to free-living relatives, or beyond, and few characteristics are unequivocally associated with the origin of apicomplexans, suggesting that the boundary between parasites and their free-living relatives is not likely to be represented by a readily identifiable suite of character-state changes.
Results and Discussion
Diversity and Distribution of Apicomplexan Relatives.
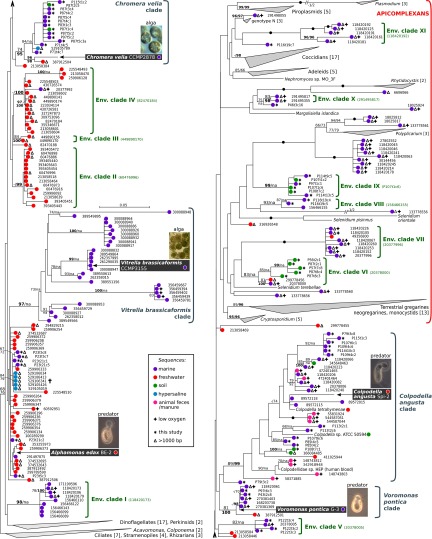
Understanding the diversity of the closest relatives of apicomplexans is critical to understanding their origin, but the only direct survey to date (9) used plastid markers, which may be misleading if plastid genomes are not universally retained. Accordingly, we identified apicomplexan-related lineages in nuclear 18S rDNA environmental sequences from GenBank, augmented with 78 sequences amplified from marine, soil, and freshwater samples using apicomplexan-specific primers (SI Appendix, Table S1 and Materials and Methods). Phylogenetic analysis of 226 environmental sequences added to a reference alignment including 106 eukaryotic 18S rDNAs of an established topology (SI Appendix, Fig. S1) were congruent with the reference tree and consistently recovered major apicomplexan and eukaryotic subgroups (Fig. 1 and SI Appendix, Figs. S1 and S2). In addition to many individual sequences, 11 strongly supported clades known only from environmental sequences were identified. Six of these clades branched within the apicomplexans (clades VI to XI) (Fig. 1). Most branched close to gregarines; however, a small group is related to coccidians and hematozoans (Fig. 1 and SI Appendix, Fig. S2). These sequences come from oxygen-depleted marine sediment in Greenland (clade XI), with the exception of a single sequence obtained in this study from a coral reef sample.
Fig. 1.
Unsuspected diversity and novel clades and habitats in apicomplexan relatives. Maximum likelihood phylogeny (RAxML) of 106 eukaryotic and 226 environmental 18S rDNA sequences, color-coded according to their environmental source (box). The first set of support values (300 nonparametric bootstraps) corresponds to the topology shown; the second set corresponds to the analysis of sequences longer than 1,000 nucleotides (“na” denotes support not applicable in the second set). Only values greater than 65% are shown for clarity; those over 95% are highlighted. Full black circles denote 100/100 support. New environmental clades (green) are followed by reference accessions. The cross indicates the position of ∼1,700 sequences from a hypersaline lake. Other large clades are indicated by triangles, with the number of sequences in square brackets. Note that Env. clade I was related to apicomplexans rather than dinoflagellates in the analysis of sequences >1,000 nucleotides.
The majority of environmental sequences (77%) branched basal to apicomplexans (Fig. 1), including five previously unidentified clades of apicomplexan relatives, dominated by either freshwater (clades II, III, and IV) or marine (clades I and V) samples. Most environmental sequences, however, fell into enlarged clades encompassing known genera, and some shared similar habitats: Vitrella and Voromonas are marine and are surrounded by marine sequences, and the relatives of Vitrella are specifically derived from CaCO3-dominated environments (SI Appendix, Table S1), consistent with previous evidence (9). In contrast, other groups are mixed: Chromera is marine and falls within a predominantly marine clade, but its closest relative is from fresh water, and other relatives comprise a mix of marine, hypersaline, and soil sequences (Fig. 1). The Colpodella clade (commonly described from fresh water and soil) contains marine sequences, as well as sequences from animal feces, calves with diarrhea, moa coprolites, oocysts in ostrich feces, and one previously reported from human blood (16). Several of these sequences were interpreted as Cryptosporidium parasites, and their consistent recovery from animal hosts, sometimes associated with disease, is suggestive of parasitism or opportunism. Additional sequences come from environments where apicomplexan relatives are known but little understood [hypersaline habitats in Fig. 1 (2) and coral reefs in clade V and SI Appendix, Fig. S2 (9)], or where none have yet been described (low oxygen environments; 26% of all environmental sequences) (SI Appendix, Table S1). Overall, much of the diversity of apicomplexan relatives remains uncharacterized, but the few chromerids and colpodellids that have been characterized in any detail are scattered broadly among this diversity, suggesting that they altogether represent a great potential for understanding deep apicomplexan evolution.
A Complex Distribution of Photosynthesis.
To reconstruct major transitions around the origin of apicomplexans, we carried out transcriptomic surveys from five species that cover much of the diversity of free-living apicomplexan relatives (Fig. 1): both photosynthetic chromerids (Chromera and Vitrella) and three nonphotosynthetic colpodellids (Voromonas, Alphamonas, and Colpodella). Both chromerids are available in culture, but colpodellids are generally not because they require eukaryotic prey that is itself heterotrophic. All three genera were successfully cultured on enrichments of Procryptobia, Spumella, and Parabodo, and transcriptomes of all five were sequenced by Illumina 100-bp paired-end technology and assembled. Transcriptomes were generated from two cultured strains of Colpodella angusta of different cell size (Spi-2 and BE-6), which were nearly identical at the nucleotide level and pooled. Chromera and Voromonas contigs were also pooled with those generated in recent independent studies (Materials and Methods) (10).
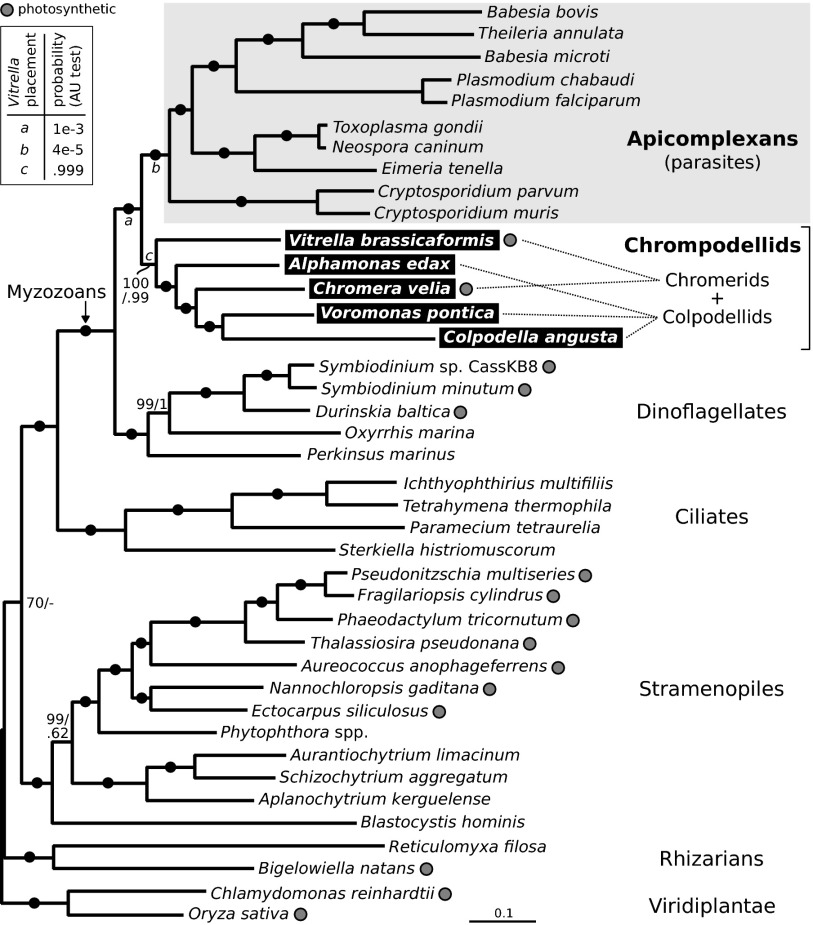
To determine the relationships among these five lineages and parasitic apicomplexans, a phylogeny was inferred from a concatenated alignment of 40 taxa and 85 nucleus-encoded proteins previously tested in eukaryote-wide phylogenies (17). Individual phylogenies of all 85 proteins had congruent topologies and were combined into a supermatrix minimizing missing information (Materials and Methods). The resulting multiprotein phylogeny is strongly and consistently resolved by both maximum likelihood (ML) (RAxML LG+GAMMA model) and Bayesian (Phylobayes; GTR+CAT+GAMMA model) analyses (Fig. 2). Contrary to expectations based on smaller datasets (7), all chromerids and colpodellids form a single monophyletic sister group to apicomplexans with strong support, which we will informally refer to as “chrompodellids.” Within this group, however, neither chromerids nor colpodellids are monophyletic: Chromera is closely related to Voromonas and Colpodella whereas Vitrella branches at the base of the group (alternative positions with apicomplexans or deeper are rejected by approximately unbiased tests) (Fig. 2). This phylogeny shows that photosynthesis was lost, not only in the ancestor of apicomplexans themselves but also in at least two additional lineages: in the branches leading to Alphamonas, and to Voromonas and Colpodella (Fig. 2). This number may increase further depending on the nature of the yet uncharacterized lineages (Fig. 1).
Fig. 2.
Chrompodellids represent a large sister group of apicomplexans with a complex distribution of photosynthesis. Maximum likelihood and Bayesian analyses inferred from the concatenation of 85 proteins (23,111 amino acids) resolve the relationships among apicomplexans and their relatives and demonstrate that the photosynthetic chromerids and predatory colpodellids are paraphyletic. Best RAxML tree (LG+GAMMA model) is shown with nonparametric bootstraps/Phylobayes posterior probabilities at branches. Full black circles indicate 100/1.0 support. Two alternative placements for Vitrella (at positions marked a and b) were rejected by approximately unbiased (AU) test at P = 0.005 (box).
The Principles of Dependency on Nonphotosynthetic Plastids.
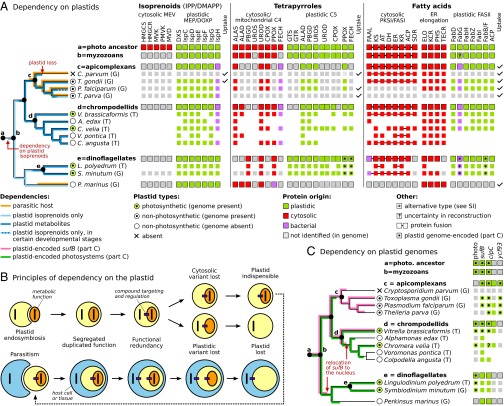
The functions of apicomplexan plastids are now well-established (18, 19), but why it was retained is not necessarily obvious given these functions. This uncertainty is because, in the early stages of its endosymbiotic integration, many core biosynthetic functions would have existed in both the host and symbiont. The evolution of efficient transport of metabolites would lead to functional redundancy, which in turn may result in the loss of metabolic capacity in one or the other, subfunctionalization, or, rarely, their merger. If cytoplasmic loss or subfunctionalization takes place, then the host becomes dependent on the plastid even if other plastid functions, including photosynthesis, are lost. Photosynthesis has therefore been lost many times, but plastid loss is very rare (20) because the compounds supplied by plastid biosynthetic pathways are rarely available directly from the environment. To identify metabolic functions that may have led to plastid dependency in apicomplexans and their relatives, we reconstructed the distribution of three key metabolic pathways: isoprenoid (IPP and DMAPP), tetrapyrrole, and fatty acid biosynthesis. The evolutionary history of the plastid versions of all three pathways has been well-characterized in apicomplexans (18, 19), but their history in other myzozoans and the fate of the cytosolic versions are less clear. We identified plastid and cytosolic candidate genes for all enzymes in all three pathways from apicomplexans and chrompodellids, as well as dinoflagellates and their sisters, perkinsinds (Materials and Methods and SI Appendix, Table S2). In the parasites, uptake of corresponding metabolites from hosts was also summarized from published predictions based on both direct and indirect evidence.
The results (Fig. 3 A and B) predict that plastid dependency was established early, before the split of apicomplexans and dinoflagellates, and that the key event was the loss of cytosolic isoprenoid biosynthesis in the common ancestor of apicomplexans and dinoflagellates. Plastid isoprenoid biosynthesis has been maintained in all myzozoans, with the exception of Cryptosporidium, and was previously proposed to be the only indispensable plastid pathway in the Plasmodium blood stage, and therefore a strong selective explanation for its retention (21). Altogether, this evidence suggests that dependency on plastid isoprenoids is not only ancestral, but also the most significant barrier to plastid loss over a much longer evolutionary time span. Biosynthesis of tetrapyrroles and fatty acids may also contribute to plastid dependency, but only in certain subgroups. Photosynthetic dinoflagellates seem to rely on plastid tetrapyrrole biosynthesis in the absence of its mitochondrial/cytosolic counterpart (22), and chrompodellids and apicomplexans rely on several plastid-localized steps (18, 23). Perkinsus seems to have retained the ability to synthesize tetrapyrroles in the cytosol (three enzymes were not identified, however). Fatty acid biosynthesis and elongation is the most complex and least understood of the three pathways, but the plastid variant seems to be dispensable in some nonphotosynthetic species or their life stages (24).
Fig. 3.
Mechanisms of dependency on plastids and plastid genomes in myzozoans. (A) A phylogeny of myzozoans with genomic (G) and/or transcriptomic (T) data are shown (Left), together with a reconstruction of plastid and nonplastid variants of core metabolic pathways (on Right with pathway above and individual enzymes below; see SI Appendix, Table S2 for abbreviations of enzyme names) color-coded as to presence/absences and source (Below). Proteins were assigned to compartment and pathways by phylogeny and similarity, and plastid copies have targeting extensions where available. Uptake of compounds was based on published reports where available. Perkinsus was based on an unpublished draft genome. Loss of cytosolic isoprenoid biosynthesis before the divergence of myzozoans established a unilateral metabolic dependency on the plastid, which was relieved by uptake of host compounds by parasites. (B) Diagram showing how unilateral dependency on the plastid may result from functional redundancy during endosymbiosis. Additional symbioses, including parasitism, may reestablish functional redundancy leading to plastid loss (or retention, which is not illustrated). Photosynthesis is ignored and may be lost at different stages of plastid endosymbiosis. (C) Dependency on plastid genomes in myzozoans. Photosynthetic species rely on plastid-encoded photosystem subunits (photo) whereas apicomplexans rely on plastid-encoded sufB, clpC, and, potentially, ycf93. The discovery of nuclear sufB and clpC provides a rationale for plastid genome loss in Perkinsus, nonphotosynthetic dinoflagellates, and, most likely, colpodellids. ClpC occurs in two forms in some species, but their relationships are not clear (SI Appendix, Fig. S4) (plastid-encoded clpC in Chromera is probably a pseudogene).
Plastid dependency can be lost if the compounds supplied by it can be acquired from the environment (Fig. 3B). In the case of parasites (and symbionts), independence from metabolic pathways typically means salvaging metabolites from their host and exploiting a niche that sufficiently covers demands throughout the lifecycle. For plastids in myzozoa, independence from plastid-synthesized fatty acids, heme, and especially isoprenoids would be required. Cryptosporidium is currently the only myzozoan where complete loss of these biosynthetic pathways is known to have taken place, and plastids have been lost outright (20). Interestingly, Cryptosporidium is predicted to have a very low requirement for heme (25) and has retained the fatty acid elongation pathway in the cytosol and endoplasmic reticulum (ER) (26). We hypothesize that another major factor in its plastid independence is the exploitation of a single host niche (gut epithelium) throughout its lifecycle. That implies that isoprenoid precursors may be more readily available in gut epithelium than other cell types and that other species with similar infection strategies (like gregarines) are more likely to lack plastids. For generalists like Toxoplasma (27, 28), or species that pass through blood like Perkinsus, Theileria, or Plasmodium (whose blood stage is dependent on de novo biosynthesis of isoprenoid precursors) (21), plastids are more likely to be retained because, in at least one life stage, plastid metabolism is essential.
All free-living phototrophs and heterotrophs retain the capability to synthesize isoprenoids, tetrapyrroles, and fatty acids (as is also apparent here) (Fig. 3A), suggesting that they are not readily available from food or the environment. The ancient establishment of plastid dependency in myzozoa therefore predicts that plastids have been maintained in free-living apicomplexan relatives. Indeed, all three colpodellid predators that we examined were found to contain abundant evidence for plastids with metabolism almost identical to that of apicomplexans (Fig. 3A). Plastid isoprenoid and heme synthesis is common to all (and chromerids) whereas fatty acid biosynthesis is differentially represented, much like in the apicomplexans. Alphamonas expresses primarily the plastid FASII (similar to Plasmodium), Voromonas and Colpodella express the cytosolic PKSI/FASI (similar to Cryptosporidium), and Chromera and Vitrella express both (similar to Toxoplasma). Altogether, chrompodellids retained all 34 proteins found in apicomplexans and six additional proteins relating to the same pathways) (Fig. 3A).
Because this metabolic reconstruction is based on transcriptome data, interpreting the absence of a single gene is not straightforward. In this case, however, the data do support the conclusion that cytosolic isoprenoid biosynthesis is absent because all five genes of the entire pathway are uniformly absent from all five chrompodellid species, as well as being absent from all complete apicomplexan genomes. At the same time, the conclusion that these genes indicate the presence of a cryptic plastid in colpodellids is based on the presence of plastid-derived proteins whose localization has been examined in model systems. The subcellular localization of individual proteins is not known and may differ in rare cases (particularly in the complex tetrapyrrole pathway). However, there is no evidence for a massive relocation of any of the three pathways (here or in any system), and plastid localizations in chrompodellids are supported by the presence of canonical bipartite N-terminal presequences (SI Appendix, Table S3). Lastly, additional pathways could localize to nonphotosynthetic plastids and contribute to their dependency. Certain amino acids are synthesized in the plastids of plants and algae; their localization in chrompodellids and dinoflagellates remains unknown although they are apparently not plastidic in apicomplexans (19). Fe-S cluster synthesis and the ferredoxin redox system represent a different case: they are indispensable for plastid function in apicomplexans (and are also found in chrompodellids) (SI Appendix, Table S3), but their function is not required in the rest of the cell (29), suggesting that they do not represent a barrier to plastid loss.
Plastid Genome Dependency.
If all essential plastid genes are relocated to the nucleus and their products are targeted back to the plastid, then the genome can be lost with no functional repercussions. Nevertheless, almost all plastids retain a genome, and its loss is considered unlikely (30); indeed, the first conclusive case has only recently been reported in a green alga (31). There are multiple metabolic and regulatory genes contributing to the dependency on most plastid genomes, but, in apicomplexan plastids, only two such genes have been retained (6): sufB (a subunit of the Fe-S cluster assembly) and clpC (a subunit of the ATP-dependent Clp protease). Both genes are nucleus-encoded in green algae and plants, indicating that their relocation to the nucleus is feasible, so we characterized their distribution in myzozoans.
Orthologs of sufB were found in dinoflagellates, Perkinsus, and all chrompodellids. All myzozoan sufBs cluster with plastid-encoded and cyanobacterial orthologs in protein ML phylogenies, and this topology is retained when fast-evolving sequences are excluded (SI Appendix, Fig. S3 and Materials and Methods). This tree is consistent with the expected cyanobacterial origin for all myzozoan sufBs, contrary to recent conclusions (10). With the exception of Vitrella and apicomplexans, however, all myzozoan sufB sequences are predicted to be nucleus-encoded: The genomic sequences in Perkinsus and Symbiodinium each contain multiple introns, and transcript sequences in Chromera, Voromonas, and Colpodella encode N-terminal extensions consistent with plastid-targeting sequences (SI Appendix, Fig. S3) (the N terminus was not sequenced from Alphamonas). This distribution suggests that sufB was transferred to the nucleus at least twice independently and represents no barrier to plastid genome loss in nonphotosynthetic myzozoans other than apicomplexans (Fig. 3C).
Nucleus-encoded clpC homologs were also identified in multiple species (Perkinsus, dinoflagellates, Chromera, Vitrella, Theileria, and Plasmodium, where it is plastid-targeted) (32) and once again grouped specifically with plastid-encoded and cyanobacterial orthologs in protein phylogenies (SI Appendix, Fig. S4). Some species retained both nucleus and plastid-encoded paralogues (Fig. 3C), and the origin of this paralogy is poorly resolved. However, the existence of a single nucleus-encoded clpC in Perkinsus and Symbiodinium genomes strongly suggests that this form is sufficient for function. Similarly, a single nuclear clpC was identified in Colpodella, and the plastid-encoded form in Chromera is likely a pseudogene, suggesting that clpC may also not represent an absolute barrier to plastid genome loss (although Theileria has both but depends on the plastid copy).
Other than sufB and clpC, myzozoan plastid-encoded genes of known function relate to transcription and translation (or photosynthesis in chromerids and dinoflagellates) (6, 7), which alone cannot account for genome retention. Plastid-encoded tRNA-fMet was hypothesized to function in apicomplexan mitochondria (30); however, the absence of methionyl-tRNA formyltransferase in piroplasmids such as Theileria (33) suggests that it is not essential. Plastid-encoded tRNA-Glu (30) is required for tetrapyrrole synthesis in some organisms but is not required for the C4 pathway used by apicomplexans, chrompodellids, or Perkinsus (Fig. 3A). We predict that dinoflagellates import tRNA-Glu from the nucleus. Plastid-encoded ORFs of unknown function could also have indispensable roles, but there is no evidence for this conclusion, and syntenic conservation suggests that several correspond to ribosomal proteins (7). The recently identified ycf93 represents the only exception in this respect because it is expressed, syntenic among apicomplexans, and plastid membrane-localized in Plasmodium falciparum (34). However, ycf93 is fast-evolving and has not been unambiguously identified outside apicomplexans: Vitrella contains a short ORF at the same position, but it lacks the transmembrane region. Although the function of the ycf93 remains unknown, it could contribute to the retention of the apicomplexan plastid genomes (particularly in the piroplasmids such as Theileria where clpC is the only other plausible candidate), but there is no evidence that it necessitates plastid genome dependence in chrompodellids.
Altogether, the discovery of nuclear-encoded sufB and clpC creates a simple framework to explain plastid genome loss in myzozoans: Once these genes are moved to the nucleus, the plastid genome is dispensable if photosynthesis is lost (Fig. 3C). The Perkinsus draft genome contains no plastid-encoded genes or nucleus-encoded genes for plastid genome expression but does have nucleus-encoded sufB and clpC. In chrompodellids, sufB was relocated to the nucleus just after the divergence of Vitrella, and clpC is either nuclear or unknown. No colpodellid genes relating to plastid genome expression were found, and genomic DNA sequence surveys of Alphamonas and Voromonas (Materials and Methods) identified mitochondria-encoded genes, but no traces of plastid genomes. About half of known dinoflagellates are nonphotosynthetic, and the ancestral relocation of sufB and clpC to the nucleus predicts that many or all of these species lack plastid genomes as well. Ironically, the apicomplexan plastid genome has informed our thinking about why organellar genomes persist, but it now seems that it is the exception among its closest relatives and simply a coincidence of the retention of sufB and clpC.
Early Origin of Apicomplexan-Specific Characteristics in Their Free-Living Relatives.
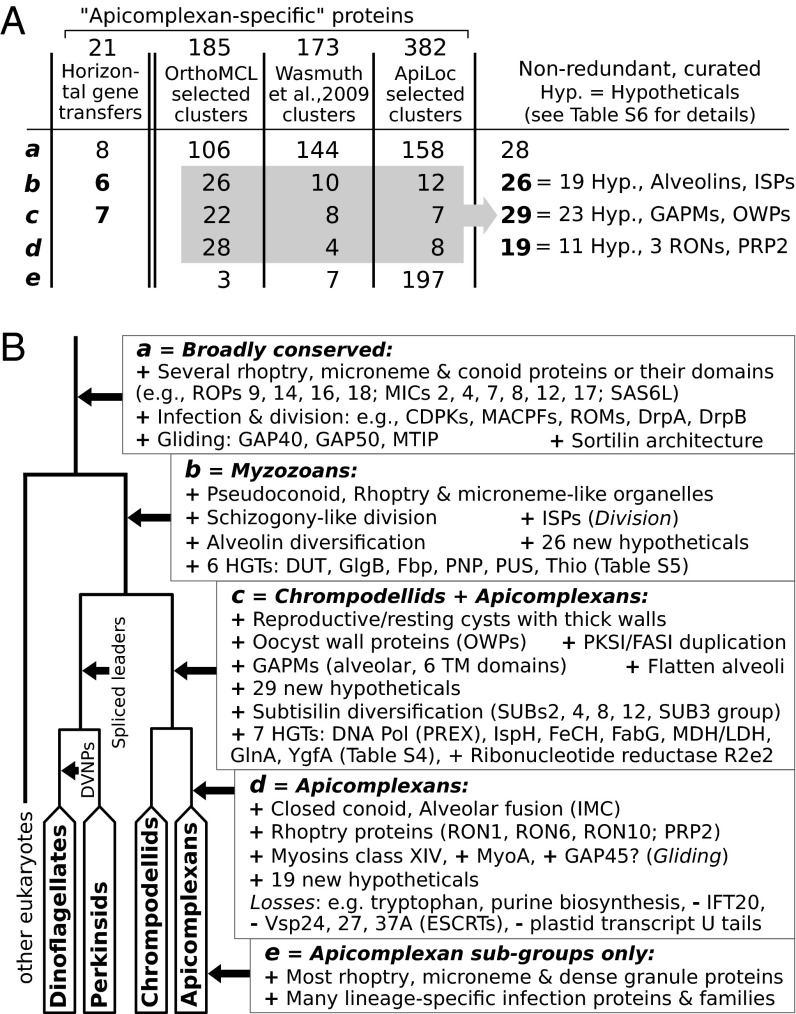
The apicomplexan infection machinery is distinctive, of obvious interest due to their medical significance, and potentially linked to the emergence of parasitism. However, once again, we have no information on the distribution of proteins related to infection in free-living relatives. Accordingly, we carried out a detailed search in the chrompodellid transcriptomes for proteins related to key apicomplexan functions, especially those inferred to be unique to the group and those linked to parasitism, based on published studies (14, 15), OrthoMCL (35), and Apiloc (Materials and Methods). This dataset included representative proteins that are involved in cellular invasion, gliding motility, or division, or that have original structural or functional properties (apicomplexan-specific or horizontally acquired). Homologs were then sought in all chrompodellids and other eukaryotes using similarity searches and were sorted into five categories based on their distribution (categories a–e in Fig. 4 A and B).
Fig. 4.
Summary of “apicomplexan-specific” characteristics in their free-living relatives. (A) Classification of four groups of apicomplexan proteins based on distribution: horizontal gene transfers (HGTs), unique protein clusters [OrthoMCL and Wasmuth et al. (14)], and proteins associated with infection (Apiloc). Categories a–e correspond to the phylogenetic distribution of these proteins when free-living relatives are considered, which is broken down by specific processes and proteins in B. Note that putative GAP45 in Cryptosporidium is highly divergent (indicated by “?” in category d).
The first category (category a) includes proteins that may have an apicomplexan-specific function but contain regions found in a wide range of eukaryotes (including chrompodellids). The presence of these proteins alone is therefore not linked to the origin of parasitism, and their roles in parasites and free-living relatives cannot be assessed without direct functional evidence. Several proteins associated with the apical complex itself belong to this category, including rhoptry and microneme proteins (SI Appendix, Table S4) and cytoskeletal SAS6L (36). If chrompodellid homologs of these proteins are associated with structures for invasion or feeding, it would support the homology of these structures and the apical complex (37). Many apicomplexan proteins involved in infection also belong to this category, including MAC perforins (MACPFs), rhomboid proteases (ROMs), calcium-dependent protein kinases (CDPKs), dynamins (DrpA, DrpB), and conserved components of the gliding machinery, including myosin anchor proteins GAP40 and GAP50 (Fig. 4B and SI Appendix, Table S4). Unexpectedly, many proteins predicted to be unique to apicomplexans by OrthoMCL or Wasmuth et al. (14) also contain regions with significant similarity to other eukaryotes (Fig. 4A). Similarly, out of 21 bacterial genes predicted to have been acquired in the apicomplexan ancestor (SI Appendix, Table S4) (15, 38), none are in fact apicomplexan-specific: 13 are found in apicomplexan relatives (next paragraph) and 8 have even broader distribution (Fig. 4A).
The second and third category of proteins (categories b and c) are those found only in apicomplexans and other myzozoans. Thirteen apicomplexan genes of bacterial origin are found in apicomplexans and chrompodellids, and 6 of them also in other myzozoans (SI Appendix, Table S5). These genes cover a range of functions, including the plastid isoprenoid (IspH), tetrapyrrole (FECH), and fatty acid (FabG) biosynthesis, glutamine (GlnA) and folate metabolism (YgfA), nucleotide interconversion (PNP, PUS, DUT), and malate and lactate dehydrogenases (Fig. 4B and SI Appendix, Fig. S5). Chromerids also contain the multisubunit DNA polymerase (PREX) involved in apicomplexan plastid genome replication (38); it is absent in colpodellids, consistent with their lacking a plastid genome (SI Appendix, Fig. S5). A unique clade of ribonucleotide reductase (R2e2) known only from apicomplexans (39) is also found in chrompodellids, and the phylogeny reveals it in fact originated by duplication from the canonical eukaryotic type (SI Appendix, Fig. S6). Chrompodellids also share an expansion in subtilisin proteases (SI Appendix, Fig. S7), which are secreted from micronemes and important for adhesion to host cell during infection (40). Lastly, two groups of proteins are uniquely shared between apicomplexans and their closest relatives and potentially linked to shared morphologies (SI Appendix, Table S6). The first are inner membrane complex (alveolar) proteins, which are important in cell division, establishing cell shape and polarity, and motility. GAPMs (with six transmembrane regions) and ALP1 are found in apicomplexans and chrompodellids (41, 42) whereas ISPs (43) and alveolins are common to all myzozoans (the latter are also present in ciliates but diversified in myzozoans). Interestingly, chromerids, colpodellids, Perkinsus, and certain dinoflagellates (3, 44) are known to multiply by a process similar to apicomplexan schizogony. The second group are oocyst wall proteins (OWPs), which are exclusively found in species that form reproductive/resting cysts: Cryptosporidium, coccidians, and all chrompodellids (3, 12, 44, 45). Cell-wall maintenance has also been linked to the presence of multidomain polyketide synthases (PKSI/FASI) (26, 46), which are found in all myzozoans (Fig. 3A and SI Appendix, Table S7) but specifically duplicated in the ancestor of apicomplexans and chrompodellids based on phylogeny of their single-copy domains (FAAL, SDR) (Fig. 4B). This finding suggests that apicomplexan and chrompodellid schizogony and cyst production rely on common morphological and molecular grounds and predate the emergence of parasitism.
The last two categories of proteins (categories d and e) are those unique to apicomplexans and absent in all other eukaryotes. The large majority of them are specific to apicomplexan subgroups including the most rhoptry, microneme, and dense body proteins (SI Appendix, Table S4). Only a handful of these proteins are shared by all apicomplexans: four rhoptry and three secreted proteins (SI Appendix, Table S6), the apicomplexan myosins class XIV including the gliding motor (MyoA) and, perhaps, GAP45 (Fig. 4B). For most apicomplexan-specific proteins, however, there is little or no functional information (Fig. 4A), and further investigation of chrompodellid genomes could erode this already small list.
In contrast to acquisitions of complete proteins, apicomplexan origin may be more strongly correlated with protein loss, family diversification, and modification of protein structure. Well-known examples of loss include de novo purine and tryptophan biosynthesis, polyU tails in plastid transcripts (47), and conserved eukaryotic components of flagellar transport (IFT20) (48) and endosomal sorting complexes (ESCRTs; Vsp24, Vsp27, and Vsp37A) (49). All of them are present in chrompodellids, confirming that their loss in the ancestor of apicomplexans coincides with the origin of parasitism (Fig. 4B). Whether the same is true for diversification of protein families and domain architectures remains to be tested by using chrompodellid genomes. Few such characteristics are known at the moment, however, and others may be misleading. Four specific insertions in the apicomplexan sortilin were suggested to account for its functional specificity—an essential role in biogenesis of rhoptries and micronemes, gliding, and host cell invasion (50)—but we find that three are present in other eukaryotes and the last is specific to coccidians.
Overall, correlations observed in the absence of data from the closest known free-living relatives of apicomplexans were more a reflection of the distant relationship between apicomplexans and available comparators, rather than a transition to parasitism, where few changes can now be identified. Many “apicomplexan-specific” genes are found in their free-living relatives, and the functions of these proteins should be examined directly for a better understanding of their potential roles in the origin of parasitism.
Conclusions
Chromerids and colpodellids are a diverse but monophyletic clade that is the closest known relative of the apicomplexan parasites. Phylogenomics shows that photosynthesis has been lost multiple times independently in these organisms, and reconstruction of plastid and cytosolic metabolism points to an ancient, obligatory plastid dependency primarily related to biosynthesis of isoprenoids. We propose that plastid loss is rare and dependent upon multiple factors: low requirements for metabolites made by plastids, low levels of inherent plastid metabolic dependency, and exploitation environments rich in these metabolites throughout the lifecycle. Plastid genome loss, in contrast, is not related to function but can be traced to the relocation of two genes: sufB and clpC. This simple model suggests that independent loss of plastid genomes may turn out to be common in myzozoans. Lastly, multiple apicomplexan-specific genes (related, e.g., to division or oocyst walls) are shared with relatives, and few genes can be linked with the origin of parasitism. This observation suggests that the evolution of parasitism is not primarily linked to the acquisition of novel structures or components, but rather to loss and modification of those already present. Once established, the persistence of parasitism is likely governed by very different factors: Adapting to the host and immunity, in particular, would involve significant changes not primarily related to loss of function. Further illumination of this phase of apicomplexan evolution will require a similar strategy of establishing a phylogenetic framework and more comprehensive characterization of the earliest diverging apicomplexans, such as gregarines.
Materials and Methods
Seventy-eight environmental 18S rDNA sequences were generated from samples collected at several locations around Vancouver, BC, Canada, and 174 sequences were obtained from GenBank, after a removal of unrelated, short, chimeric, fast-evolving, and nonoverlapping sequences in both sets (see SI Appendix for details). The final reference dataset containing 106 eukaryotes (SI Appendix, Fig. S1) was analyzed with all deep-branching environmental sequences, those longer than 1,000 nucleotides (Fig. 1), or those branching within apicomplexan clades (SI Appendix, Fig. S2). Culturing of chrompodellids, sequencing, and assembling their transcriptomic and genomic surveys are described in SI Appendix, SI Materials and Methods. The multiprotein phylogenetic dataset (Fig. 2) was build by identifying conserved phylogenetic markers (17) in predicted proteins from the five chrompodellids, a broad selection of apicomplexans, and other eukaryotes. The final 85 proteins were selected to minimize the missing information in chrompodellids: No complete missing sequence was allowed in any of the five species, and fewer than 10 in other taxa could be absent (23,111 total positions; 9% missing). Individual trees of all 85 protein markers were analyzed to confirm their congruence and identify possible contaminants. Sequences were aligned in MAFFT v. 7.127b (l-INS-i algorithm), and variable sites were excluded in BMGE 1.1 using −h 0.4 −g 0.6 parameters. Phylogenies were calculated in RAxML 7.04 (GTR+GAMMA+4 model, 20 standard replicates, and 300 nonparametric bootstraps). Phylobayes MPI v. 1.4f was run using the GTR+CAT model as two chains for 30 000 cycles (burn-in of 5,000). Distribution of plastid proteins (Fig. 3) was determined using custom eukaryotic and prokaryotic databases (SI Appendix, SI Materials and Methods) and GenBank, BLASTp searches, and RAxML phylogenies (100 rapid bootstraps). Domains in PKSI/FASI polypeptides were identified using National Center for Biotechnology Information Conserved Domain searches and analyzed separately. Apicomplexan FabD fell at an unresolved position between bacteria and eukaryotes but is known to be plastid-localized. Several dinoflagellate proteins had a distinct evolutionary origin from apicomplexans [asterisks in Fig. 3: PPOX and FabF were related to viridiplantae, FeCH to Bdellovibrio and red algae (SI Appendix, Fig. S5), and FabG to proteobacteria]. PhyML 3.0 (LG+GAMMA+4) phylogenies (aLRT branch supports, 10 random starts) were used in datasets where many species or few positions (SI Appendix, Figs. S4 and S5) were available. Genomic DNA surveys were queried for mitochondrion- and plastid-encoded genes: mitochondrial cox1, cox3, and cob were detected in both samples, but no hits to plastid genes were identified. Four strategies were used to identify proteins or protein clusters that have apicomplexan-specific distribution or relate to apicomplexan functions associated with parasitism (Fig. 4)—those predicted to have been acquired by the apicomplexan ancestor from bacteria, those predicted to be unique and ancestral to apicomplexans by OrthoMCL and Wasmuth et al. (14), and those belonging to one or more of the following Apiloc categories based on subcellular localization: Apical, Exported, Membrane, IMC, Oocyst wall, and Golgi (a control for proteins of eukaryotic origin). Proteins were sorted by eye based on distribution and 1e−5 evalue cutoff and were refined by a second round of in-depth searches (SI Appendix, SI Materials and Methods).
Supplementary Material
Acknowledgments
J.J. thanks Ales Horak for help with phylogenies and Forest L. Rohwer (San Diego State University) for providing conditions to finish this research. This work was supported by Canadian Institutes for Health Research Grant MOP-42517 (to P.J.K.), by Russian Science Foundation Grant 14-14-00515 (to D.V.T.), by Russian Foundation for Basic Research Grants 14-04-000500 and 14-04-000554 (to A.P.M.), and by the Gordon and Betty Moore Foundation through Grant 2637 to the National Center for Genome Resources (NCGR). Samples MMETSP0288 and MMETSP0290 were sequenced, assembled, and annotated at the NCGR. J.J. is a Global Scholar, and P.J.K. is a Senior Fellow of the Canadian Institute for Advanced Research. F.B. and M.K. were supported by a grant from the Tula Foundation to the Center for Microbial Biodiversity and Evolution.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Symbioses Becoming Permanent: The Origins and Evolutionary Trajectories of Organelles,” held October 15–17, 2014, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Symbioses.
This article is a PNAS Direct Submission. J.P.M. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KP213181–KP213258).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423790112/-/DCSupplemental.
References
- 1.Moore RB, et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;451(7181):959–963. doi: 10.1038/nature06635. [DOI] [PubMed] [Google Scholar]
- 2.Simpson AGB, Patterson DJ. Ultrastructure and identification of the predatory flagellate Colpodella pugnax Cienkowski (Apicomplexa) with a description of Colpodella turpis n. sp. and a review of the genus. Syst Parasitol. 1996;33(3):187–198. [Google Scholar]
- 3.Oborník M, et al. Morphology, ultrastructure and life cycle of Vitrella brassicaformis n. sp., n. gen., a novel chromerid from the Great Barrier Reef. Protist. 2012;163(2):306–323. doi: 10.1016/j.protis.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Cavalier-Smith T, Chao EE. Protalveolate phylogeny and systematics and the origins of Sporozoa and dinoflagellates (phylum Myzozoa nom. nov.) Eur J Protistol. 2004;40(3):185–212. [Google Scholar]
- 5.McFadden GI, Reith ME, Munholland J, Lang-Unnasch N. Plastid in human parasites. Nature. 1996;381(6582):482. doi: 10.1038/381482a0. [DOI] [PubMed] [Google Scholar]
- 6.Wilson RJ, et al. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J Mol Biol. 1996;261(2):155–172. doi: 10.1006/jmbi.1996.0449. [DOI] [PubMed] [Google Scholar]
- 7.Janouškovec J, Horák A, Oborník M, Lukeš J, Keeling PJ. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci USA. 2010;107(24):10949–10954. doi: 10.1073/pnas.1003335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodył A, Stiller JW, Mackiewicz P. Chromalveolate plastids: Direct descent or multiple endosymbioses? Trends Ecol Evol. 2009;24(3):119–121, author reply 121–122. doi: 10.1016/j.tree.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Janouškovec J, Horák A, Barott KL, Rohwer FL, Keeling PJ. Global analysis of plastid diversity reveals apicomplexan-related lineages in coral reefs. Curr Biol. 2012;22(13):R518–R519. doi: 10.1016/j.cub.2012.04.047. [DOI] [PubMed] [Google Scholar]
- 10.Gile GH, Slamovits CH. Transcriptomic analysis reveals evidence for a cryptic plastid in the colpodellid Voromonas pontica, a close relative of chromerids and apicomplexan parasites. PLoS ONE. 2014;9(5):e96258. doi: 10.1371/journal.pone.0096258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mylnikov A, Mylnikova Z. Feeding spectra and pseudoconoid structure in predatory alveolate flagellates. Inland Water Biol. 2008;1(3):210–216. [Google Scholar]
- 12.Oborník M, et al. Morphology and ultrastructure of multiple life cycle stages of the photosynthetic relative of apicomplexa, Chromera velia. Protist. 2011;162(1):115–130. doi: 10.1016/j.protis.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Tikhonenkov DV, et al. Description of Colponema vietnamica sp.n. and Acavomonas peruviana n. gen. n. sp., two new alveolate phyla (Colponemidia nom. nov. and Acavomonidia nom. nov.) and their contributions to reconstructing the ancestral state of alveolates and eukaryotes. PLoS ONE. 2014;9(4):e95467. doi: 10.1371/journal.pone.0095467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wasmuth J, Daub J, Peregrín-Alvarez JM, Finney CAM, Parkinson J. The origins of apicomplexan sequence innovation. Genome Res. 2009;19(7):1202–1213. doi: 10.1101/gr.083386.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J, et al. Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum. Genome Biol. 2004;5(11):R88. doi: 10.1186/gb-2004-5-11-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan CL, et al. Colpodella spp.-like parasite infection in woman, China. Emerg Infect Dis. 2012;18(1):125–127. doi: 10.3201/eid1801.110716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burki F, Okamoto N, Pombert J-F, Keeling PJ. The evolutionary history of haptophytes and cryptophytes: Phylogenomic evidence for separate origins. Proc Biol Sci. 2012;279(1736):2246–2254. doi: 10.1098/rspb.2011.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ralph SA, et al. Tropical infectious diseases: Metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat Rev Microbiol. 2004;2(3):203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- 19.Seeber F, Soldati-Favre D. 2010. Metabolic pathways in the apicoplast of apicomplexa. International Review of Cell and Molecular Biology (Elsevier), 161–228. Available at linkinghub.elsevier.com/retrieve/articleSelectSinglePerm?Redirect=www.sciencedirect.com/science/article/pii/S1937644810810056?via%3Dihub&key=ab8c9b9be510bed9d51196fc9df26ca6a3ef88cd. Accessed September 1, 2014.
- 20.Abrahamsen MS, et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304(5669):441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 21.Yeh E, DeRisi JL. Chemical rescue of malaria parasites lacking an apicoplast defines organelle function in blood-stage Plasmodium falciparum. PLoS Biol. 2011;9(8):e1001138. doi: 10.1371/journal.pbio.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butterfield ER, Howe CJ, Nisbet RER. An analysis of dinoflagellate metabolism using EST data. Protist. 2013;164(2):218–236. doi: 10.1016/j.protis.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Kořený L, Sobotka R, Janouškovec J, Keeling PJ, Oborník M. Tetrapyrrole synthesis of photosynthetic chromerids is likely homologous to the unusual pathway of apicomplexan parasites. Plant Cell. 2011;23(9):3454–3462. doi: 10.1105/tpc.111.089102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaughan AM, et al. Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell Microbiol. 2009;11(3):506–520. doi: 10.1111/j.1462-5822.2008.01270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Dooren GG, Kennedy AT, McFadden GI. The use and abuse of heme in apicomplexan parasites. Antioxid Redox Signal. 2012;17(4):634–656. doi: 10.1089/ars.2012.4539. [DOI] [PubMed] [Google Scholar]
- 26.Zhu G, Marchewka MJ, Woods KM, Upton SJ, Keithly JS. Molecular analysis of a Type I fatty acid synthase in Cryptosporidium parvum. Mol Biochem Parasitol. 2000;105(2):253–260. doi: 10.1016/s0166-6851(99)00183-8. [DOI] [PubMed] [Google Scholar]
- 27.Mazumdar J, Striepen B. Make it or take it: Fatty acid metabolism of apicomplexan parasites. Eukaryot Cell. 2007;6(10):1727–1735. doi: 10.1128/EC.00255-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z-H, Ramakrishnan S, Striepen B, Moreno SNJ. Toxoplasma gondii relies on both host and parasite isoprenoids and can be rendered sensitive to atorvastatin. PLoS Pathog. 2013;9(10):e1003665. doi: 10.1371/journal.ppat.1003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dellibovi-Ragheb TA, Gisselberg JE, Prigge ST. Parasites FeS up: Iron-sulfur cluster biogenesis in eukaryotic pathogens. PLoS Pathog. 2013;9(4):e1003227. doi: 10.1371/journal.ppat.1003227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barbrook AC, Howe CJ, Purton S. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 2006;11(2):101–108. doi: 10.1016/j.tplants.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Smith DR, Lee RW. A plastid without a genome: Evidence from the nonphotosynthetic green algal genus Polytomella. Plant Physiol. 2014;164(4):1812–1819. doi: 10.1104/pp.113.233718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El Bakkouri M, et al. The Clp chaperones and proteases of the human malaria parasite Plasmodium falciparum. J Mol Biol. 2010;404(3):456–477. doi: 10.1016/j.jmb.2010.09.051. [DOI] [PubMed] [Google Scholar]
- 33.Pino P, et al. Mitochondrial translation in absence of local tRNA aminoacylation and methionyl tRNA Met formylation in Apicomplexa. Mol Microbiol. 2010;76(3):706–718. doi: 10.1111/j.1365-2958.2010.07128.x. [DOI] [PubMed] [Google Scholar]
- 34.Goodman CD, McFadden GI. Ycf93 (Orf105), a small apicoplast-encoded membrane protein in the relict plastid of the malaria parasite Plasmodium falciparum that is conserved in Apicomplexa. PLoS ONE. 2014;9(4):e91178. doi: 10.1371/journal.pone.0091178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen F, Mackey AJ, Stoeckert CJ, Jr, Roos DS. OrthoMCL-DB: Querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34(Database issue):D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Leon JC, et al. A SAS-6-like protein suggests that the Toxoplasma conoid complex evolved from flagellar components. Eukaryot Cell. 2013;12(7):1009–1019. doi: 10.1128/EC.00096-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okamoto N, Keeling PJ. The 3D structure of the apical complex and association with the flagellar apparatus revealed by serial TEM tomography in Psammosa pacifica, a distant relative of the Apicomplexa. PLoS ONE. 2014;9(1):e84653. doi: 10.1371/journal.pone.0084653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seow F, et al. The plastidic DNA replication enzyme complex of Plasmodium falciparum. Mol Biochem Parasitol. 2005;141(2):145–153. doi: 10.1016/j.molbiopara.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 39.Munro JB, Jacob CG, Silva JC. A novel clade of unique eukaryotic ribonucleotide reductase R2 subunits is exclusive to apicomplexan parasites. J Mol Evol. 2013;77(3):92–106. doi: 10.1007/s00239-013-9583-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lagal V, et al. Toxoplasma gondii protease TgSUB1 is required for cell surface processing of micronemal adhesive complexes and efficient adhesion of tachyzoites. Cell Microbiol. 2010;12(12):1792–1808. doi: 10.1111/j.1462-5822.2010.01509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bullen HE, et al. A novel family of Apicomplexan glideosome-associated proteins with an inner membrane-anchoring role. J Biol Chem. 2009;284(37):25353–25363. doi: 10.1074/jbc.M109.036772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordon JL, Buguliskis JS, Buske PJ, Sibley LD. Actin-like protein 1 (ALP1) is a component of dynamic, high molecular weight complexes in Toxoplasma gondii. Cytoskeleton (Hoboken) 2010;67(1):23–31. doi: 10.1002/cm.20414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beck JR, et al. A novel family of Toxoplasma IMC proteins displays a hierarchical organization and functions in coordinating parasite division. PLoS Pathog. 2010;6(9):e1001094. doi: 10.1371/journal.ppat.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mylnikov AP. Ultrastructure and phylogeny of colpodellids (Colpodellida, Alveolata) Biol Bull. 2009;36(6):582–590. [PubMed] [Google Scholar]
- 45.Templeton TJ, et al. The Cryptosporidium oocyst wall protein is a member of a multigene family and has a homolog in Toxoplasma. Infect Immun. 2004;72(2):980–987. doi: 10.1128/IAI.72.2.980-987.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bushkin GG, et al. Evidence for a structural role for acid-fast lipids in oocyst walls of Cryptosporidium, Toxoplasma, and Eimeria. MBio. 2013;4(5):e00387–13. doi: 10.1128/mBio.00387-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dorrell RG, Drew J, Nisbet RER, Howe CJ. Evolution of chloroplast transcript processing in Plasmodium and its chromerid algal relatives. PLoS Genet. 2014;10(1):e1004008. doi: 10.1371/journal.pgen.1004008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Briggs LJ, Davidge JA, Wickstead B, Ginger ML, Gull K. More than one way to build a flagellum: Comparative genomics of parasitic protozoa. Curr Biol. 2004;14(15):R611–R612. doi: 10.1016/j.cub.2004.07.041. [DOI] [PubMed] [Google Scholar]
- 49.Tomavo S, Slomianny C, Meissner M, Carruthers VB. Protein trafficking through the endosomal system prepares intracellular parasites for a home invasion. PLoS Pathog. 2013;9(10):e1003629. doi: 10.1371/journal.ppat.1003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sloves P-J, et al. Toxoplasma sortilin-like receptor regulates protein transport and is essential for apical secretory organelle biogenesis and host infection. Cell Host Microbe. 2012;11(5):515–527. doi: 10.1016/j.chom.2012.03.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.