Abstract
Transcriptional regulation and posttranscriptional processing underlie many cellular and organismal phenotypes. We used RNA sequence data generated by Genotype-Tissue Expression (GTEx) project to investigate the patterns of transcriptome variation across individuals and tissues. Tissues exhibit characteristic transcriptional signatures that show stability in postmortem samples. These signatures are dominated by a relatively small number of genes—which is most clearly seen in blood—though few are exclusive to a particular tissue and vary more across tissues than individuals. Genes exhibiting high interindividual expression variation include disease candidates associated with sex, ethnicity, and age. Primary transcription is the major driver of cellular specificity, with splicing playing mostly a complementary role; except for the brain, which exhibits a more divergent splicing program. Variation in splicing, despite its stochasticity, may play in contrast a comparatively greater role in defining individual phenotypes.
Gene expression is the key determinant of cellular phenotype, and genome-wide expression analysis has been a mainstay of genomics and biomedical research, providing insights into the molecular events underlying human biology and disease. Whereas expression data sets from tissues/primary cells (1, 2) and individuals (3) have accumulated over recent years, only limited expression data sets have allowed analysis across tissues and individuals simultaneously (4). The Genotype-Tissue Expression Project (GTEx) is developing such a resource (5, 6), collecting multiple “nondiseased” tissues sampled from recently deceased human donors. We analyzed the GTEx pilot data freeze (6), which comprised RNA sequencing (RNA-seq) from 1641 samples from 175 individuals representing 43 sites: 29 solid organ tissues, 11 brain subregions, whole blood, and two cell lines: Epstein-Barr virus–transformed lymphocytes (LCL) and cultured fibroblasts from skin [table S1 and (7)].
The identification and characterization of genetic variants that are associated with gene expression are extensively discussed in (6). Here we use the GTEx data to investigate the patterns of transcriptome variation across individuals and tissues and how these patterns associate with human phenotypes. RNA-seq performed on the GTEx pilot samples produced an average of 80 million paired-end mapped reads per sample (fig. S1) (7, 8). We used the mapped reads to quantify gene expression using Gencode V12 annotation (9), which includes 20,110 protein-coding genes (PCGs) and 11,790 long noncoding RNAs (lncRNAs). Comparison with microarray-based quantification for a subset of 736 samples showed concordance between the two technologies (average correlation coefficient = 0.83, fig. S2). At the threshold defined for expression quantitative trait loci (eQTL) analysis [reads per kilobase per million mapped reads (RPKM) > 0.1, see (7)], at which 88% of PCGs and 71% of lncRNAs are detected in at least one sample, the distribution of gene expression across tissues is U-shaped and complementary between PCGs (generally ubiquitously expressed) and lncRNAs (typically tissue-specific or not expressed, Fig. 1A).
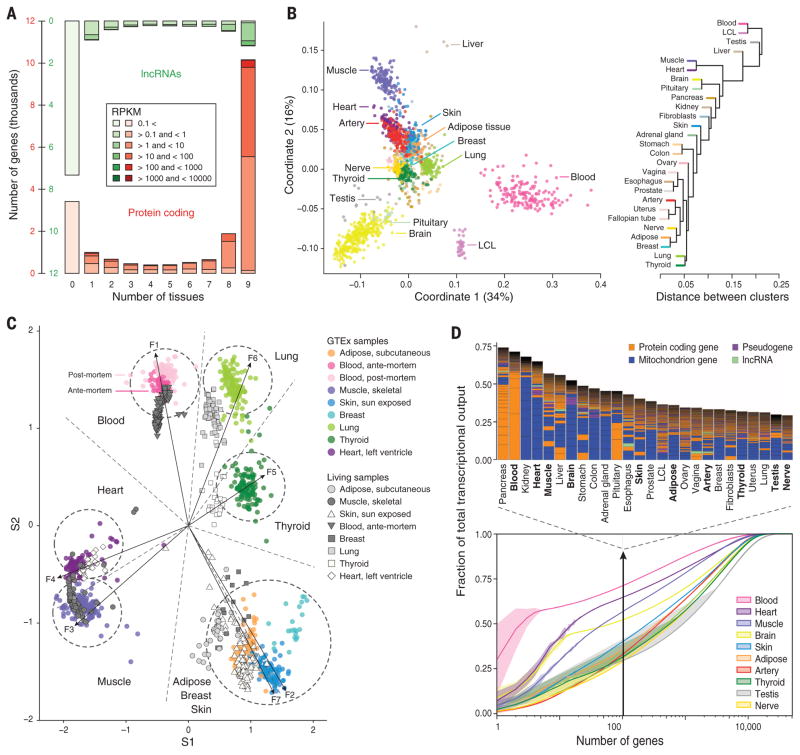
Fig. 1. The GTEx multitissue transcriptome.
(A) Gene expression levels and number of tissues in which genes are expressed (>0.1 RPKM in at least 80% of the samples). RPKMs are averaged over all genes expressed in a given number of tissues. (B) Sample and tissue similarity on the basis of gene expression profiles. Left: Multidimensional scaling Right: Tissue hierarchical clustering. (C) Expression values from eight GTEx tissues (colored circles) plotted radially along seven metagenes extracted from expression data. Antemortem samples curated from the Gene Expression Omnibus (GEO) cluster strongly with GTEx tissues. (D) Transcriptome complexity. Bottom: Cumulative distribution of the average fraction of total transcription contributed by genes when sorted from most-to-least expressed in each tissue (x axis). Lines represent mean values across samples of the same tissue, and lighter-color surfaces around the mean represent dispersion calculated as the standard deviation divided by the cumulative sum of all means.Top: Biological type and relative contribution to total transcription of the hundred most expressed genes. Height of the bars is proportional to the fraction that these genes contribute to total transcription.
Tissues show a characteristic transcriptional signature, as revealed by multidimensional scaling, of both PCG and lncRNA expression (figs. 1B, S3, and S4), with individual phenotypes contributing little (fig. S5). The primary separation, as observed in prior studies (10), is between nonsolid (blood) and solid tissues and, within solid tissues, brain is the most distinct. Brain subregions are not well differentiated, with the exception of cerebellum (fig. S6). Postmortem ischemia appears to have little impact on the characteristic tissue transcriptional signatures, as previously noted (11). In a comparison of 798 GTEx samples with 609 “nondis-eased” samples obtained from living (surgical) donors (table S2), we found that GTEx samples clustered with surgical samples of the same tissue type (Fig. 1C and table S3) (12).
Tissue transcription is generally dominated by the expression of a relatively small number of genes. Indeed, we found that for most tissues, about 50% of the transcription is accounted for by a few hundred genes (13). In many tissues, the bulk of transcription is of mitochondrial origin (Fig. 1D and table S4) (14). In kidney, for instance, a highly aerobic tissue with many mitochondria, a median of 51% (>65% in some samples) of the transcriptional output is from the mitochondria (fig. S7). Other tissues show nuclear-dominated expression; in blood, for example, three hemoglobin genes contribute more than 60% to total transcription. Genes related to lipid metabolism in pancreas, actin in muscle, and thyroglobulin in thyroid are other examples of nuclear genes contributing disproportionally to tissue-specific transcription. Because RNA samples are generally sequenced to the same depth, in tissues where a few genes dominate expression, fewer RNA-seq reads are comparatively available to estimate the expression of the remaining genes, decreasing the power to estimate expression variation. These tissues—i.e., blood, muscle, and heart (Fig. 1E)—are, consequently, those with less power to detect eQTLs (6). Because most eQTL analyses are performed on easily accessible samples, such as blood, this highlights the relevance of the GTEx multitissue approach.
Although thousands of genes are differentially expressed between tissues (fig. S8) or show tissue-preferential expression (fig. S9 and table S5), fewer than 200 genes are expressed exclusively in a given tissue (figs. S10 and S11 and tables S6 and S7, A to E). The vast majority (~ 95%) are exclusive to testis and many are lncRNAs. This may reflect low-level basal transcription common to all cell types or result from general tissue heterogeneity, with few primary cell types being specific to a given tissue.
Expression of repetitive elements also recapitulates tissue type (table S8 and fig. S12A). We identified 3046 PCGs whose expression, in at least one tissue, was correlated with the expression of the closest repeat element (on average 2827 base pairs away, fig. S12B). In about half of these cases, the repeat was also significantly coexpressed with other repeats of its same family (table S8 and fig. S13). LncRNA expression can be regulated by specific repeat families (15), and we found evidence that testis-specific expression could be regulated by endogenous retrovirus L repeats (ERVL and ERVL-MaLR) (fig. S12C).
Using linear mixed models, we found that variation in gene expression is far greater among tissues (47% of total variance in gene expression) than among individuals (4% of total variance, Fig. 2A and table S9), and very similar for PCGs and lncRNAs when controlling for gene expression (Fig. 2A). Genes that show high expression variance across individuals and low variance across tissues include genes on the sex chromosomes, as well as autosomal genes, such as the RHD gene that determines Rh blood group.
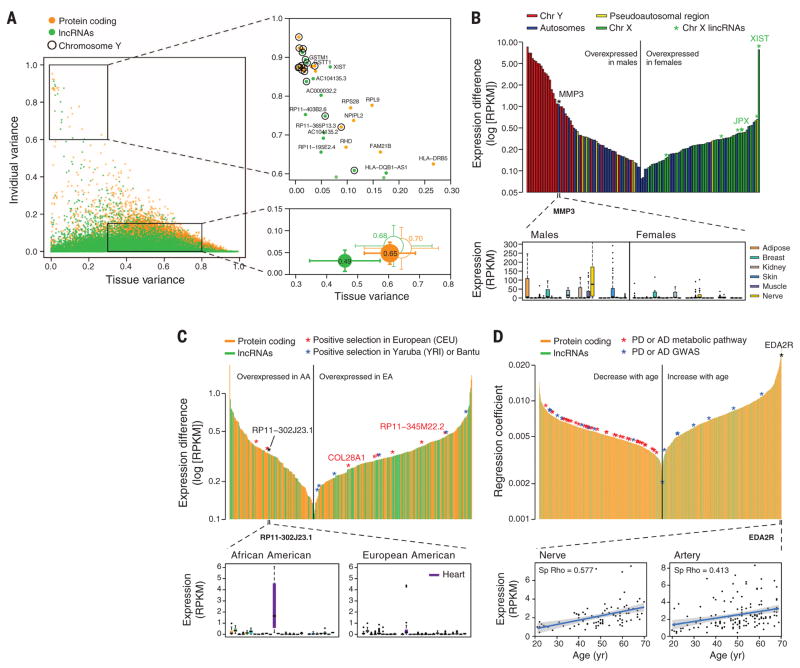
Fig. 2. Gene expression across tissues and individuals.
(A) Left: Contribution of tissue and individual to gene expression variation of PCGs and lncRNAs. Bottom right: Mean ± SD over all genes (filled circles) and over genes with similar expression levels in PCGs and lncRNAs (unfilled circles). Circle size is proportional to the sum of tissue and individual variation, and segment length corresponds to 0.5 SD. Top right: genes with high individual variation and low tissue variation. (B) Sex differentially expressed genes. Top: differentially expressed genes (FDR < 0.05) sorted according to expression differences between males and females. Genes in the Y chromosome are sorted according to the expression in males. Bottom: MMP3 gene expression in males and females. (C) Genes differentially expressed with ethnicity. Top: differentially expressed genes (FDR < 0.05) between African Americans (AA) and European Americans (EA) sorted according to expression differences. A few of these genes lie in regions reported to be under positive selection in similar populations. Bottom: expression of RP11-302J23.1. (D) Genes differentially expressed with age. Top: Genes sorted according to the regression coefficient. Bottom: expression of EDAR2 gene in nerve and artery as a function of age. Shaded area around the regression line represents 95% confidence interval.
We identified 92 PCGs and 43 lncRNAs with global sex-biased expression [false discovery rate (FDR) < 0.05, Fig. 2B and table S10]. Genes over-expressed in males are predominantly located on the Y chromosome. Conversely, many genes on the X chromosome are overexpressed in females, suggesting that more genes might escape X inactivation than previously described (16). Among these, we found XIST and JPX, known to participate in X inactivation, as well as the lncRNAs RP11-309M23.1 and RP13-216E22.4, the expression of which shows enrichment in the nucleus in female cell lines from ENCODE (17) and hence could be candidates to also participate in X inactivation (fig. S14) (16). Among autosomal PCGs, MMP3, linked to susceptibility to coronary heart disease [Online Mendelian Inheritance in Man (OMIM) no. 614466] and more prevalent in males, shows the strongest expression bias (Fig. 2B).
We detected 221 PCGs and 153 lncRNAs globally differentially expressed between individuals of European and African-American ancestry (FDR < 0.05, Fig. 2C and table S11). There is a slight enrichment of lncRNAs (P < 1 × 10−6), among which we identified the RP11-302J23.1 gene, highly expressed in cardiac tissue in African Americans only, and located in a region that harbors weak associations to heart disease (18). Additionally, some genes showing differential expression by ethnicity lie in genomic regions under positive selection in European or sub-Saharan African populations (Fig. 2C and fig. S15).
Finally, we detected 1993 genes that globally change expression with age (FDR < 0.05, Fig. 2D and table S12). Genes that decrease expression are enriched in functions and pathways related to neurodegenerative diseases such as Parkinson’s and Alzheimer’s diseases, among which eight harbor single-nucleotide polymorphisms (SNPs) for these diseases identified from genome-wide association studies (P < 0.05). Among the genes that increase expression with age is EDA2R, whose ligand, EDA, has been associated with age-related phenotypes (19).
We also identified 753 genes with tissue specific sex-biased expression (FDR < 0.05, table S13) predominantly in breast tissue (92%), and 31 genes with tissue-specific ethnicity-biased expression, many in the skin (FDR < 0.05, Table 1 and table S14). Among the sex-differentially expressed genes, five show biased expression specifically in heart and are of interest given the differing prevalence of cardiovascular disease between males and females. One of these genes, PLEKHA7 (fig. S15C), contains SNPs associated with risk for cardiovascular disease.
Table 1. Genes with sex-biased and ethnicity-biased expression in GTEx tissues.
Differentially expressed genes between males (M) and females (F) and between African American (AA) and European American (EA) in those tissues with at least 10 samples per group. Median fold change (on autosomal genes) was calculated for tissues with more than two significant genes.
| Sex | Ethnicity | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||||||
| No. of samples |
No. of genes |
Med. fold change |
No. of samples |
No. of genes |
Med. fold change |
|||||||||||
|
|
|
|||||||||||||||
| M | F | All genes |
PCGs | lncRNA | Y chrom. |
X chrom. |
Pseudo- autosomal region |
Autosomal | AA | EA | All genes |
PCGs | lncRNA | |||
| Adipose | 76 | 37 | 45 | 29 | 16 | 28 | 4 | 1 | 12 | 2.8 | 17 | 95 | 0 | 0 | 0 | – |
| Artery | 88 | 57 | 40 | 21 | 19 | 29 | 2 | 0 | 9 | 2.3 | 21 | 121 | 3 | 0 | 0 | 2.9 |
| Blood | 99 | 57 | 20 | 12 | 8 | 18 | 1 | 0 | 1 | – | 24 | 129 | 0 | 0 | 0 | – |
| Breast | 14 | 13 | 762 | 567 | 195 | 26 | 21 | 0 | 715 | 4.2 | 5 | 22 | – | – | – | – |
| Esophagus | 27 | 11 | 23 | 14 | 9 | 22 | 1 | 0 | 0 | – | 5 | 33 | – | – | – | – |
| Heart | 74 | 34 | 27 | 15 | 12 | 23 | 1 | 0 | 3 | 3 | 11 | 95 | 2 | 1 | 1 | – |
| LCL | 26 | 13 | 23 | 11 | 12 | 21 | 1 | 1 | 0 | – | 9 | 30 | – | – | – | – |
| Lung | 76 | 43 | 34 | 17 | 17 | 31 | 1 | 0 | 2 | – | 14 | 104 | 4 | 3 | 1 | 3 |
| Muscle | 87 | 51 | 42 | 27 | 15 | 24 | 6 | 0 | 12 | 2.7 | 18 | 117 | 2 | 2 | 0 | – |
| Nerve | 54 | 34 | 38 | 24 | 14 | 25 | 5 | 1 | 7 | 2.3 | 13 | 73 | 2 | 1 | 1 | – |
| Skin | 76 | 43 | 47 | 32 | 15 | 31 | 5 | 0 | 11 | 2.3 | 14 | 103 | 14 | 13 | 1 | 2.9 |
| Thyroid | 65 | 40 | 42 | 25 | 17 | 27 | 5 | 1 | 9 | 1.9 | 13 | 90 | 13 | 7 | 6 | 3.5 |
| No. of samples | 942 | 566 | 172 | 1307 | 35 | 27 | 8 | |||||||||
| No. of individuals | 111 | 64 | 847 | 628 | 219 | 41 | 34 | 1 | 771 | 24 | 148 | |||||
Overall, tissue specificity is likely to be driven by the concerted expression of multiple genes. Thus, we performed sex-based differential analysis of coexpression networks. We identified 42 coexpression modules in males and 46 in females (fig. S16). Among male-specific modules, we found one related to spermatid differentiation and development (FDR = 9.0 × 10−4, fig. S16B), and among female-specific modules, we found one related to epidermis and ectoderm development (FDR = 4.6 × 10−14, fig. S16C). Differential network expression, therefore, distinguishes differences between male and females not well captured by analysis of individual genes.
Split-mapped RNA-seq reads predict about 87,000 novel junctions with very strong support (fig. S17). These tend to be more tissue specific, detected in fewer samples, and less conserved than previously annotated junctions (only 2.6% of novel junctions can be detected as orthologous in mouse, compared to 65% for annotated junctions). Multidimensional scaling based on exon inclusion levels again largely recapitulates tissue type (Fig. 3A). However, samples from brain cluster as the primary out-group, supporting the existence of a distinct splicing program in the brain (20). Furthermore, preferential gene expression of RNA-binding proteins and both differential and preferential exon inclusion are enriched in the brain (figs. S18 and S19 and table S15). We found very few exons exclusively included or excluded in a given tissue (fig. S20 and table S16), 40% of which show exclusive inclusion in the brain. We also found that micro-exons (<15 bp) are overwhelmingly used in the brain compared to other tissues (Wilcoxon test, P < 1 × 10−7, Fig. 3B). This pattern is not obvious in short exons longer than 15 bp (P = 0.3, fig. S21). This observed brain-specific splicing pattern may result from differential splicing in the cerebellum, because expression clustering of the brain regions reveals a general up-regulation of RNA-binding proteins specifically in the cerebellum (Fig. 3C). This is also the brain region exhibiting the largest proportion of novel splicing events (fig. S22).
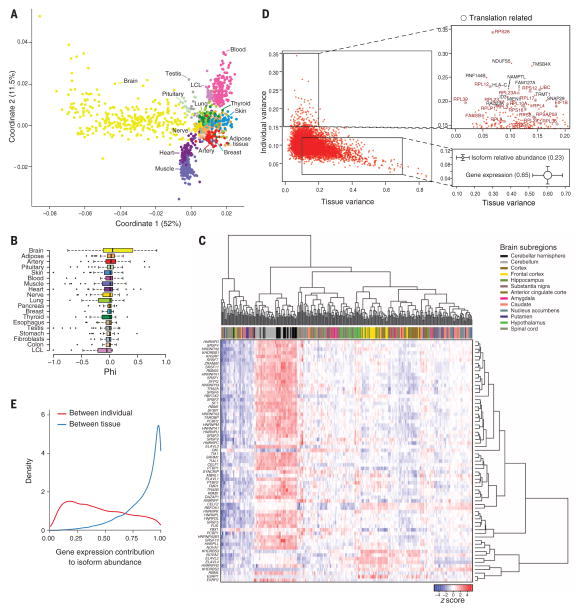
Fig. 3. Splicing across tissue and individuals.
(A) Multidimensional scaling of all samples on the basis of exon inclusion levels (Percent spliced in, PSI). (B) Microexon inclusion across tissues.Values of tissue exon inclusion close to 1 (−1) indicate that the microexon is included (excluded), in nearly all samples from the tissue, and excluded (included) in nearly all samples from the rest of the tissues.Tissues are sorted according to tissue exon inclusion (phi) median value. (C) Clustering of brain samples on the basis of the normalized expression levels of 67 RNA binding proteins involved in splicing. The order of samples and genes is obtained by biclustering the expression matrix. (D) Left: Contribution of tissue and individual to splicing variation in PCGs. Bottom right: Mean ± SD of individual and tissue contributions to splicing and to gene expression variation. Circle size is proportional to the sum of tissue and individual variation and segment length corresponds to 0.5 SD. Top right: Genes with high splicing variation across individuals. (E) Contribution of gene expression to the between-individual and between-tissue variation in isoform abundance
In contrast to gene expression, variation of splicing, measured either from relative isoform abundance or exon inclusion, is similar across tissues and across individuals, but exhibits a much larger proportion of residual unexplained variation (Fig. 3D, fig. S23, and table S17). This could arise from nonadditive interactions between individuals and tissues, but might also reflect stochastic, nonfunctional fluctuations that are more common in splicing than in expression (21). Among the genes that show high interindividual splicing variability, we found an enrichment of ribosomal proteins and genes related to translation and protein biosynthesis (Fig. 3D and table S18). Higher variability between individuals may also partially reflect an effect of ischemic time on splicing, which we observed when clustering samples by exon inclusion within each tissue (fig. S24).
The abundance of splicing isoforms reflects the actions of both primary transcription and posttranscriptional processing—mostly alternative splicing. To determine the relative contribution of each process, we estimated the proportion of variance in isoform abundance that can be simply explained by variance in gene expression. We found that gene expression explains only 45% of the variance between individuals, but 84% of the variance between tissues (Fig. 3E and fig. S25). This strongly suggests that primary transcription is the main driver of cellular specificity, with splicing playing a complementary role. Although this may be unexpected, given the magnitude of the effect, it is consistent with recent findings of low proteomic support for alternatively spliced isoforms (22) and few shifts in major protein isoforms across cell types (table S19) (23).
Overall, our results underscore the value of monitoring the transcriptome of multiple tissues and individuals in order to understand tissue-specific transcriptional regulation and to uncover the transcriptional determinants of human phenotypic variation and disease susceptibility.
Supplementary Material
Acknowledgments
We acknowledge and thank the donors and their families for their generous gifts of organ donation for transplantation and tissue donations for the GTEx research study. We thank the Genomics Platform at the Broad Institute for data generation; L. Gaffney for help with figures; E. Gelfand and C. Trowbridge for project support and members of the Analysis Working Group for feedback; and D. MacArthur, J. Maller, and B. Neale for critical reading of the manuscript. The primary and processed data used to generate the analyses presented here are available in the following locations: All primary sequence files are deposited in and available from dbGaP (phs000424.v3.p1); gene and transcript quantifications are available on the GTEx Portal (www.gtexportal.org). The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health (http://commonfund.nih.gov/GTEx). Additional funds were provided by the National Cancer Institute (NCI); National Human Genome Research Institute; National Heart, Lung, and Blood Institute; National Institute on Drug Abuse; National Institute of Mental Health; and National Institute of Neurological Disorders and Stroke. This work was supported by the following grants and contracts from the United States National Institutes of Health: contract HHSN261200800001E (Leidos Prime contract with NCI); contracts 10XS170 [National Disease Research Interchange (NDRI)], 10XS171 (Roswell Park Cancer Institute), 10X172 (Science Care Inc.), and 12ST1039 (IDOX); contract 10ST1035 (Van Andel Institute); contract HHSN268201000029C (Broad Institute); R01 DA006227-17 (University of Miami Brain Bank); R01 MH090941 (University of Geneva), European Research Council, Swiss National Science Foundation, and Louis-Jeantet Foundation to E.T.D.; R01 MH090936 (University of North Carolina–Chapel Hill); and grants BIO2011-26205 from the Spanish Ministerio de Ciencia e Innovación (MICINN), 2014 SGR 464 and 2014 SGR 1319 from the Generalitat de Catalunya, and 294653 from the European Research Council–European Commission.
Footnotes
REFERENCES AND NOTES
- 1.FANTOM Consortium and the RIKEN PMI and CLST (DGT) et al . Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ENCODE Project Consortium. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lappalainen T, et al. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grundberg E, et al. Nat Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lonsdale TJ, et al. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The GTEx Consortium Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Materials and methods are available in the supplementary materials on Science Online.
- 8.DeLuca DS, et al. Bioinformatics. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrow J, et al. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lukk M, et al. Nat Biotechnol. 2010;28:322–324. doi: 10.1038/nbt0410-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birdsill AC, Walker DG, Lue L, Sue LI, Beach TG. Cell Tissue Bank. 2011;12:311–318. doi: 10.1007/s10561-010-9210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Proc Natl Acad Sci USA. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carninci P, et al. Genome Res. 2000;10:1617–1630. doi: 10.1101/gr.145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly RD, Mahmud A, McKenzie M, Trounce IA, St John JC. Nucleic Acids Res. 2012;40:10124–10138. doi: 10.1093/nar/gks770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelley D, Rinn J. Genome Biol. 2012;13:R107. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrel L, Willard HF. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 17.Djebali S, et al. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Regitz-Zagrosek V, Seeland U. Wien Med Wochenschr. 2011;161:109. doi: 10.1007/s10354-011-0892-8. [DOI] [PubMed] [Google Scholar]
- 19.Yan M, et al. Science. 2000;290:523–527. doi: 10.1126/science.290.5491.523. [DOI] [PubMed] [Google Scholar]
- 20.Yeo G, Holste D, Kreiman G, Burge CB. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickrell JK, Pai AA, Gilad Y, Pritchard JK. PLOS Genet. 2010;6:e1001236. doi: 10.1371/journal.pgen.1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ezkurdia I, et al. Mol Biol Evol. 2012;29:2265–2283. doi: 10.1093/molbev/mss100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzàlez-Porta M, Frankish A, Rung J, Harrow J, Brazma A. Genome Biol. 2013;14:R70. doi: 10.1186/gb-2013-14-7-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.