Abstract
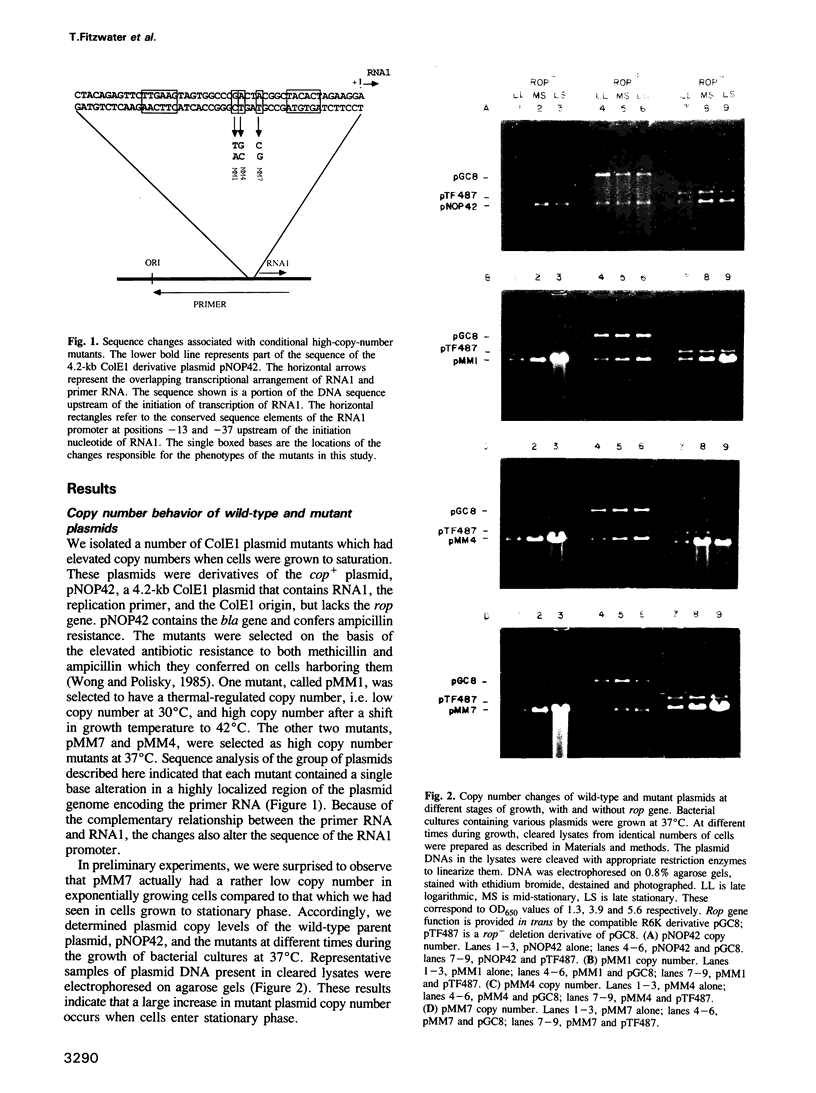
We describe three independently isolated copy number mutants of a plasmid ColE1 derivative which undergo temperature- and growth-phase-dependent DNA amplification in Escherichia coli. These mutants have single base-pair alterations in a highly localized region of the plasmid genome encoding the replication primer RNA. The mutations map immediately upstream of the RNA1 transcript, altering the sequence between conserved elements of the RNA1 promoter. These mutants have 2- to 4-fold increased copy number relative to wild-type plasmids in exponential growth at 37 degrees C but undergo 20-fold amplification of copy number relative to wild-type when cells enter stationary phase. Cells containing these plasmids grow with normal kinetics at 37 degrees C but grow poorly at 42 degrees C. The poor growth is associated with high-level plasmid amplification. Both the temperature and growth phase plasmid DNA amplification are suppressed if the ColE1 rop gene product is provided in trans from a compatible plasmid. Analysis of steady-state RNA1 levels indicates that DNA amplification occurs in the presence of RNA1 made by the mutant plasmid. Thus, the DNA amplification of the copy mutant is not due to an inability to synthesize RNA1. Using an in vitro transcription system containing RNase H, we show that mutant primer processing by RNase H is resistant to levels of the replication initiation inhibitor RNA1 that inhibit wild-type primer processing. The defect in inhibition appears not to be at the level of association of RNA1 with nascent primer. These results indicate that mutant plasmid amplification is due to the ability of its primer precursor transcripts to serve as substrates for RNase H despite the presence of RNA1.
Full text
PDF


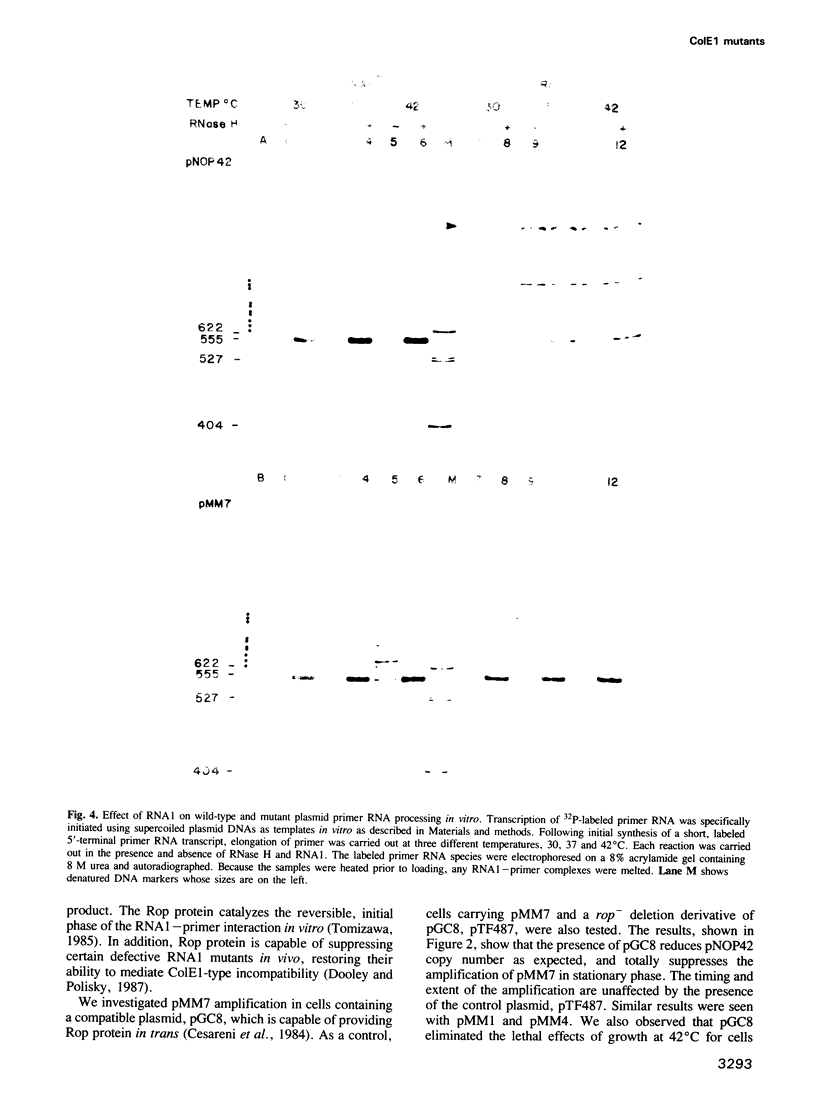



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Castagnoli L., Lacatena R. M., Cesareni G. Analysis of dominant copy number mutants of the plasmid pMB1. Nucleic Acids Res. 1985 Jul 25;13(14):5353–5367. doi: 10.1093/nar/13.14.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesareni G., Cornelissen M., Lacatena R. M., Castagnoli L. Control of pMB1 replication: inhibition of primer formation by Rop requires RNA1. EMBO J. 1984 Jun;3(6):1365–1369. doi: 10.1002/j.1460-2075.1984.tb01978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesareni G., Muesing M. A., Polisky B. Control of ColE1 DNA replication: the rop gene product negatively affects transcription from the replication primer promoter. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6313–6317. doi: 10.1073/pnas.79.20.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley T. P., Polisky B. Suppression of ColE1 RNA-RNA mismatch mutations in vivo by the ColE1 Rop protein. Plasmid. 1987 Jul;18(1):24–34. doi: 10.1016/0147-619x(87)90075-8. [DOI] [PubMed] [Google Scholar]
- Frey J., Timmis K. N. ColD-derived cloning vectors that autoamplify in the stationary phase of bacterial growth. Gene. 1985;35(1-2):103–111. doi: 10.1016/0378-1119(85)90162-3. [DOI] [PubMed] [Google Scholar]
- Itoh T., Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A. 1980 May;77(5):2450–2454. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacatena R. M., Banner D. W., Castagnoli L., Cesareni G. Control of initiation of pMB1 replication: purified Rop protein and RNA I affect primer formation in vitro. Cell. 1984 Jul;37(3):1009–1014. doi: 10.1016/0092-8674(84)90435-5. [DOI] [PubMed] [Google Scholar]
- Lacatena R. M., Cesareni G. Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature. 1981 Dec 17;294(5842):623–626. doi: 10.1038/294623a0. [DOI] [PubMed] [Google Scholar]
- Lin-Chao S., Bremer H. Activities of the RNAI and RNAII promoters of plasmid pBR322. J Bacteriol. 1987 Mar;169(3):1217–1222. doi: 10.1128/jb.169.3.1217-1222.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masukata H., Tomizawa J. Control of primer formation for ColE1 plasmid replication: conformational change of the primer transcript. Cell. 1986 Jan 17;44(1):125–136. doi: 10.1016/0092-8674(86)90491-5. [DOI] [PubMed] [Google Scholar]
- Masukata H., Tomizawa J. Effects of point mutations on formation and structure of the RNA primer for ColE1 DNA replication. Cell. 1984 Feb;36(2):513–522. doi: 10.1016/0092-8674(84)90244-7. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M., Oka A. The structure of a transcriptional unit on colicin E1 plasmid. Eur J Biochem. 1979 Jul;97(2):435–443. doi: 10.1111/j.1432-1033.1979.tb13131.x. [DOI] [PubMed] [Google Scholar]
- Moser D. R., Moser C. D., Sinn E., Campbell J. L. Suppressors of a temperature-sensitive copy-number mutation in plasmid NTP1. Mol Gen Genet. 1983;192(1-2):95–100. doi: 10.1007/BF00327652. [DOI] [PubMed] [Google Scholar]
- Muesing M., Tamm J., Shepard H. M., Polisky B. A single base-pair alteration is responsible for the DNA overproduction phenotype of a plasmid copy-number mutant. Cell. 1981 Apr;24(1):235–242. doi: 10.1016/0092-8674(81)90519-5. [DOI] [PubMed] [Google Scholar]
- Som T., Tomizawa J. Regulatory regions of ColE1 that are involved in determination of plasmid copy number. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3232–3236. doi: 10.1073/pnas.80.11.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stueber D., Bujard H. Transcription from efficient promoters can interfere with plasmid replication and diminish expression of plasmid specified genes. EMBO J. 1982;1(11):1399–1404. doi: 10.1002/j.1460-2075.1982.tb01329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm J., Polisky B. Characterization of the ColE1 primer-RNA1 complex: analysis of a domain of ColE1 RNA1 necessary for its interaction with primer RNA. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2257–2261. doi: 10.1073/pnas.82.8.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm J., Polisky B. Structural analysis of RNA molecules involved in plasmid copy number control. Nucleic Acids Res. 1983 Sep 24;11(18):6381–6397. doi: 10.1093/nar/11.18.6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomcsányi T., Apirion D. Processing enzyme ribonuclease E specifically cleaves RNA I. An inhibitor of primer formation in plasmid DNA synthesis. J Mol Biol. 1985 Oct 20;185(4):713–720. doi: 10.1016/0022-2836(85)90056-7. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. I., Itoh T. The importance of RNA secondary structure in CoIE1 primer formation. Cell. 1982 Dec;31(3 Pt 2):575–583. doi: 10.1016/0092-8674(82)90313-0. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: initial interaction of RNA I and the primer transcript is reversible. Cell. 1985 Mar;40(3):527–535. doi: 10.1016/0092-8674(85)90201-6. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell. 1984 Oct;38(3):861–870. doi: 10.1016/0092-8674(84)90281-2. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T. Plasmid ColE1 incompatibility determined by interaction of RNA I with primer transcript. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6096–6100. doi: 10.1073/pnas.78.10.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T., Selzer G., Som T. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1421–1425. doi: 10.1073/pnas.78.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J., Som T. Control of ColE1 plasmid replication: enhancement of binding of RNA I to the primer transcript by the Rom protein. Cell. 1984 Oct;38(3):871–878. doi: 10.1016/0092-8674(84)90282-4. [DOI] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Wong E. M., Muesing M. A., Polisky B. Temperature-sensitive copy number mutants of CoIE1 are located in an untranslated region of the plasmid genome. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3570–3574. doi: 10.1073/pnas.79.11.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E. M., Polisky B. Alternative conformations of the ColE1 replication primer modulate its interaction with RNA I. Cell. 1985 Oct;42(3):959–966. doi: 10.1016/0092-8674(85)90292-2. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]