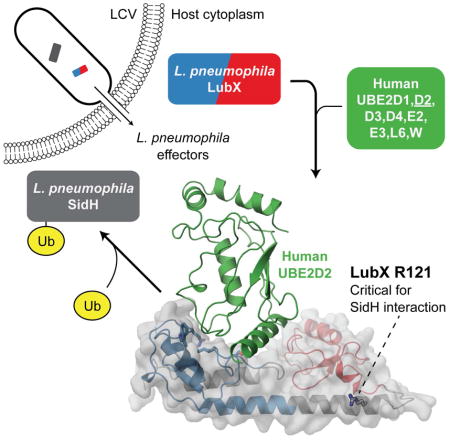
Summary
LubX, is part of Legionella pneumophila’s large arsenal of effectors that are translocated into the host cytosol during infection. Despite such unique features as the presence of two U-box motifs and its targeting of another effector SidH, the molecular basis of LubX’s activity remains poorly understood. Here we show that the N-terminal of LubX is able to activate an extended number of ubiquitin conjugating (E2) enzymes including UBE2W, UBEL6 and all tested members of UBE2D and UBE2E families. Crystal structures of LubX alone and in complex with UBE2D2 revealed drastic molecular diversification between the two U-box domains, with only the N-terminal U-box retaining E2 recognition features typical for its eukaryotic counterparts. Extensive mutagenesis followed by functional screening in a yeast model system captured functionally important LubX residues including Arg121 critical for interactions with SidH. Combined, this data provides a new molecular insight into function of this unique pathogenic factor.
Graphical Abstract
Introduction
Coevolution of bacterial and eukaryotic cells has led to the development of sophisticated multiprotein complexes that enable bacteria to deliver dedicated sets of proteins called bacterial effectors, to the eukaryotic host. Inside the host cell, these proteins are able to engage a diverse range of eukaryotic targets to ensure successful infection and immune system regulation. Amongst all bacterial pathogens studied to date, Legionella pneumophila, the causative agent of Legionnaires’ disease, maintains the largest arsenal of effectors, with over 300 proteins (Burstein et al., 2009; Huang et al., 2011; Lifshitz et al., 2013; Zhu et al., 2011) delivered to the host cell via the Dot/Icm type IVB translocation system (Segal et al., 1998; Vogel et al., 1998). Distributed throughout its genome, these Dot/Icm translocated substrates (DITS) represent a potential source of undiscovered molecular mechanisms that underpin the ability of this bacteria to colonize a broad spectrum of eukaryotic cell types, which ranges from human lung macrophages to diverse protozoan hosts (Fields, 1996).
While DITS and bacterial effectors in general, lack well-defined molecular signatures, there are examples whose function and activity can be gleaned from resemblance to typically eukaryote-specific functional domains. Genome analysis of several pathogenic Legionella strains (Cazalet et al., 2004; de Felipe et al., 2008; Lurie-Weinberger et al., 2010) has so far yielded the discovery of 102 eukaryotic-like proteins (ELPs) across a total of 72 strains. The ELPs identified are diverse and include both common protein-protein interaction motifs such as ankyrin repeats, leucine-rich repeats, coiled coils and also domains of specific functions including F-boxes and U-boxes (Cazalet et al., 2004; de Felipe et al., 2008; de Felipe et al., 2005). The presence of the latter two domains provided the first evidence that several DITS belong to the growing number of bacterial translocated proteins that interfere with ubiquitination (for review, see (Hubber et al., 2013)).
Ubiquitination is a eukaryote-specific post-translational modification (for review, see (Hershko and Ciechanover, 1998)) mediated by the sequential activity of ubiquitin activating (E1), ubiquitin conjugating (E2) and ubiquitin protein ligase (E3) enzymes. Ubiquitin chains on a substrate protein can communicate several possible signals, largely dependent on the length and topology of the ubiquitin chain. The most extensively characterized of these topologies is K48-linked polyubiquitin chains, which targets the substrate to the proteasome for degradation. Target specificity of ubiquitination is largely determined by E3 ubiquitin protein ligases and which have one of several E3 enzyme-specific domains. Single-subunit E3s have at least one HECT (homologous to E6-associated protein C-terminus), RING (really interesting new gene) or U-box domain that is responsible for direct interactions with the E2 enzyme. Alternatively, E3s can be part of multi-subunit complexes such as the SCF (Skp1-Cullin-F-box) complex.
Despite lacking complete ubiquitination machineries, pathogenic bacteria have developed many strategies to exploit ubiquitination processes via the translocated proteins that they deliver to the host cell. Several translocated proteins have been identified that mimic the activity of eukaryotic E3 ubiquitin ligases (Hicks and Galan, 2010); however, the manner in which they do so varies wildly. In several cases, the pathogenic E3 ubiquitin ligase does not resemble any previously known ubiquitination domains (Singer et al., 2008; Singer et al., 2013; Zhu et al., 2008). However, a growing number of pathogenic effectors have been shown to adopt structural folds that are well known as E3 structures. The NleG effectors found in attaching and effacing pathogens such as enterohaemorrhagic E. coli (EHEC) structurally mimic RING finger/U-box domains in spite of their low sequence homology (Wu et al., 2010). Similarly, AvrPtoB a type III secreted substrate from the tomato pathogen Pseudomonas syringae contains a functional U-box that is able to suppress programmed cell death in both plant and yeast cells (Abramovitch et al., 2006). L. pneumophila also possesses multiple methods of manipulation of host cell ubiquitination and has proteins with U-boxes and a family of F-box-containing proteins, several of which have been demonstrated to interact with the host ubiquitination machinery. For example, the F-box proteins LegU1 and LegAU13 can integrate with mammalian SCF complexes to direct ubiquitination in vitro (Ensminger and Isberg, 2010; Price et al., 2009).
While effectors are generally thought to target host proteins, the Legionella protein LubX/LegU2 was identified as the first example of an E3 ligase ‘metaeffector’, due to its ability to target another substrate of the Dot/Icm system, SidH (Kubori et al., 2010). LubX was also reported to target the human host kinase Clk1 for degradation (Kubori et al., 2008). LubX appears to be unique in that it contains U-box domains at both its N- and C-termini. Using a small panel of eight human E2 enzymes Kubori and colleagues showed that only the N-terminal portion of LubX is functional in activation of E2 ubiquitin-conjugating enzymes in vitro, whereas the C-terminal U-box motif was proposed to be involved in substrate interactions (Kubori et al., 2010), a function not previously reported for eukaryotic U-box domains. Further understanding of LubX function as a unique E3 ubiquitin protein ligase requires characterisation of its molecular structure. Accordingly, we used X-ray crystallography to determine the structure of individual U-box domains and full length LubX in complex with human UBE2D2 E2 enzyme. Informed by these data, we used a series of genetic and biochemical approaches to define critical LubX protein surfaces, residues, and other structural components necessary for E2 activation and metaeffector activity against SidH.
Results
Structures and in vitro activities of LubX U-box domains confirm their functional diversity
Residues 1-215 of LubX (Lpp2887) spanning both predicted U-box domains but lacking the last 25 amino acids containing the C-terminal translocation signal (Burstein et al., 2009; Huang et al., 2011; Lifshitz et al., 2013) was purified from E. coli and prepared for crystallization (see Materials and Methods for details). SDS-PAGE and gel filtration demonstrated accumulation of shorter fragments that corresponded to the approximate mass of the N- and C-terminal predicted U-box motifs, suggesting that LubX [1-215] is prone to cleavage by endogenous proteases. As such, LubX [1-117] and LubX [102-202] fragments were also purified and submitted for crystallization along with a LubX [1-215] fragment. Crystallization trials for both the N- and C-terminal fragments resulted in well-diffracting crystals. The structures of these LubX fragments were solved using selenomethionine-enriched protein crystals and the single-wavelength anomalous dispersion (SAD) phasing method and refined to 1.95 and 2.88 Å, respectively (Table 1). Interpretable electron density in the N- and C-terminal LubX fragment structures spanned residues 9 to 117 (Figure 1A) and residues 123 to 198, respectively (Figure 1B).
Table 1.
Crystallographic statistics.
| LubX U-box 1 | LubX U-box 2 wild-type | LubX U-box 2 Ile175Met | LubX-FL E2D2 complex | |
|---|---|---|---|---|
|
| ||||
| PDB code | 4WZ0 | 4XI1 | 4WZ2 | 4WZ3 |
|
| ||||
| Data collection | ||||
|
| ||||
| Space group | C2221 | P432 | P432 | P61 |
|
| ||||
| Cell dimensions | ||||
| a, b, c (Å) | 75.0, 90.4, 40.5 | 160.23, 160.23, 160.23 | 160.03, 160.03, 160.03 | 119.3, 119.3, 49.8 |
|
| ||||
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 120 |
|
| ||||
| Resolution (Å) | 50.0 – 1.95 | 20.0 – 2.98 | 40.0 – 3.40 | 40.0 – 2.70 |
|
| ||||
| Number of unique reflections | 10142 | 13171 | 9956 | 11243 |
|
| ||||
| Rmerge | 0.096 (0.418)a | 0.080 (0.623)b | 0.126 (0.649)c | 0.054 (0.665)d |
|
| ||||
| I/σI | 21.5 (2.94) | 23.6 (2.7) | 15.0 (2.97) | 20.05 (4.07) |
|
| ||||
| Completeness (%) | 97.5 (97.3) | 88.7 (92.7) | 98.6 (100) | 99.0 (99.8) |
|
| ||||
| Redundancy | 3.7 (3.9) | 5.7 (5.6) | 6.5 (6.6) | 5.0 (5.0) |
|
| ||||
| Refinement | ||||
|
| ||||
| Resolution (Å) | 50.0 – 1.95 | 20.0 – 2.88 | 40.0 – 3.41 | 40.0 – 2.70 |
|
| ||||
| No. of reflections: working, test | 9941, 995 | 13167, 659 | 9409, 948 | 10084, 484 |
|
| ||||
| Rwork/Rfree (%) | 23.4/27.8 | 16.2/21.0 | 17.8/21.6 | 16.7/22.3 |
|
| ||||
| Average B-factors | ||||
| Protein | 29.8 | 75.9 | 49.8 | 49.7 |
| Solvent | N/A | 70.5 | 41.0 | N/A |
| Water | 43.8 | 64.8 | 32.9 | 38.8 |
|
| ||||
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.009 | 0.009 | 0.005 | 0.005 |
| Bond angles (°) | 1.168 | 1.050 | 0.535 | 0.903 |
Values in parentheses refer to highest resolution shell of
1.98-1.95 Å,
3.03-2.98,
3.46-3.40,
2.75-2.70
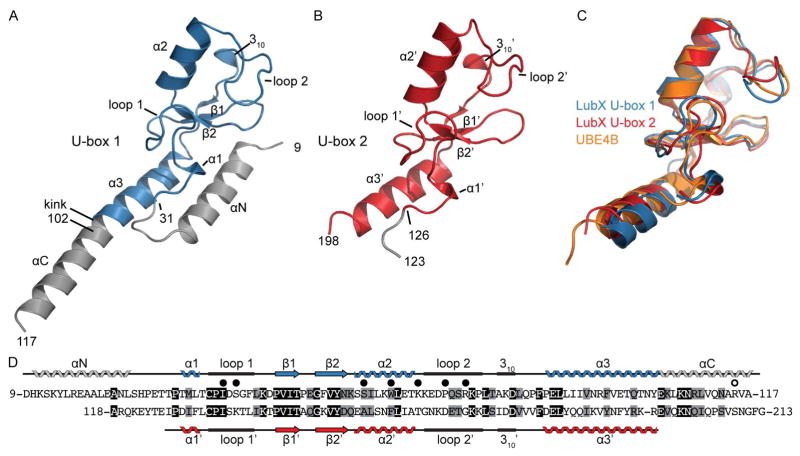
Figure 1. Structures of LubX U-box domains.
(A) 1.95 Å structure of LubX [9-117] (U-box 1). Residues highlighted in blue belong to the canonical U-box fold (B) 2.88 Å Structure of LubX [123-197] (U-box 2). (C) Superposition of crystal structures of LubX [9-117] and LubX [123-197] and UBE4B (PDB: 3L1Z). (D) Sequence alignment and secondary structure assignments of LubX U-box domains. Black and grey shading indicate identical and similar residues, respectively. Closed circles above alignment refer to LubX residues that form polar contacts or become >80% buried upon contact with UBE2D2. Open circle above alignment refers to residues important for interaction with SidH. See also Figure S1.
As predicted by primary sequence, the crystal structure of the N-terminal portion of LubX revealed that residues 31 to 102 adopted the typical fold of a eukaryotic U-box domain, consisting of a short α-helix (α1, residues 33-35), a central α-helix (α2, residues 58 to 67) set against a turn of a 310 helix (residues 80 to 82), two short β-strands forming an antiparallel β-sheet (residues 47 to 50 and 55 to 57), a C-terminal α-helix (α 3, residues 87-102) and finally, two prominent converging loops spanning residues 36 to 43 (loop 1), and 70 to 77 (loop 2). This region of LubX superimposes well with the U-box domains of eukaryotic ubiquitin ligases (i.e. UBE4B, PDB:3L1Z (Benirschke et al., 2010) and CHIP, PDB:2OXQ (Xu et al., 2008)) with root-mean-square deviations (RMSD) of 1.1 and 0.92 Å over 47 matching Cα atoms respectively (Figure 1C). Upstream of the N-terminal U-box domain, residues 11 to 25 of LubX formed an α-helix (αN) that packed against the C-terminal α3 helix, forming a platform for the N-terminal U-box domain (Figure 1A). An extensive survey of the 18 U-box structures in the Protein Data Bank (PDB) (Berman et al., 2000) (corresponding to 9 unique sequences) showed that they lack this additional structural element.
The crystal structure of the LubX C-terminal fragment (residues 126 to 198) also showed structural homology to eukaryotic U-boxes (Figure 1B). The LubX N- and C-terminal U-box domains superimposed with an RMSD of 1.34 Å over 48 Cα atoms, comprising all secondary structure elements of the U-box folds except for α3, which is rotated by ~10° to the equivalent helix in the N-terminal U-box (Figure 1C). In accordance with a previously defined nomenclature (Kubori et al., 2008; Kubori et al., 2010), the N- and C-terminal LubX U-boxes will hereafter be referred to as U-box 1 and 2, respectively.
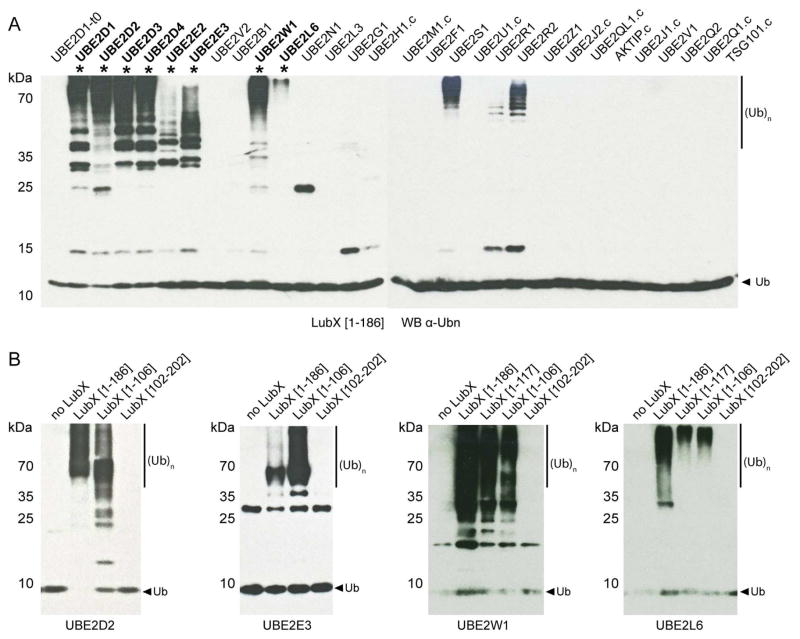
Typical of eukaryotic E3 ubiquitin protein ligases, LubX was previously shown to have in vitro poly-ubiquitination and auto-ubiquitination activities in the presence of human ubiquitin activating (E1) enzyme and ubiquitin conjugating (E2) UBE2D1 (UbcH5a) or UBE2D3 (UbcH5c) enzymes (Kubori et al., 2008; Kubori et al., 2010). The Ile39Ala mutation in U-box 1 abrogated LubX in vitro ubiquitination activity, while the equivalent mutation in U-box 2 (Ile134Ala) had no effect, suggesting that only U-box 1 is involved in interactions with human E2 enzymes (Kubori et al., 2008). However only 8 out of 37 predicted human E2s (Michelle et al., 2009) were tested for activity with LubX (Kubori et al., 2008). Thus, the in vitro ubiquitination activity of the LubX [1-186] fragment, which contained both U-box domains (but has better solubility than the full length protein), was tested against an expanded panel of 29 human E2 enzymes (Sheng et al., 2012) representing 13 out of 17 human E2 enzyme families (Michelle et al., 2009) (the remaining E2s were either not expressed or insoluble under our standard conditions, see Materials and Methods for details). The E2 enzymes that triggered LubX-specific accumulation of ubiquitinated protein species were further tested in the same assay but with LubX fragments corresponding to U-box 1 (LubX [1-106] or LubX [1-117]) or U-box 2 (LubX [102-202]) alone. According to our results (Figure 2A), eight human E2 conjugating enzymes, including the previously reported UBE2D1 and UBE2D3 and the newly identified UBE2D2, UBE2D4, UBE2E2, UBE2E3, UBE2W1 and UBE2L6 were able to support in vitro poly- and auto-ubiquitination activity of LubX. In each case, U-box 1 was both necessary and sufficient for the formation of polyubiquitin protein species, whereas U-box 2 was either unable to mediate polyubiquitination (Figure 2B) or was not clearly distinguishable from intrinsic E2 auto-ubiquitination activity (as in the case of UBE2S, UBE2R1 and UBE2R2, Figure S2). Thus, despite their close structural resemblance, LubX U-boxes 1 and 2 demonstrated drastically different functional properties, with only U-box 1 acting as an E3 ubiquitin ligase able to initiate interactions with a defined subset of E2 conjugating enzymes.
Figure 2. Autoubiquitination of LubX is Dependent on U-box 1.
(A) LubX [1-186] was tested for its ability to facilitate the formation of poly-ubiquitin chains in the presence of E1, ubiquitin, ATP and 29 different E2s. Ubiquitination activity was determined by SDS-PAGE and western blot analysis using anti-ubiquitin antibodies. Asterisks denote reactions with activity not attributable to E2 auto-ubiquitination (see Figure S2). (B) Representative auto-ubiquitination reactions for UBE2D2, UBE2E3, UBE2W and UBE2L6 with LubX [1-186] as well as U-box 1 and 2 domain fragments. Only LubX fragments containing U-box 1 exhibited auto-ubiquitination activity. See also Figure S2.
Structure of LubX in complex with the human UBE2D2 ubiquitin conjugating enzyme defines the U-box 1- E2 interface
LubX in vitro ubiquitination activity was particularly high in combination with the UBE2D2 human E2 enzyme (Figure 2). To reveal the structural basis for this activity, LubX [1-186] was co-crystallized with UBE2D2 and the complex structure was determined to 2.7 Å by the molecular replacement method (Table 1).
The model of the complex of LubX (residues 4 to 186) and UBE2D2 (residues 1 to 147) could be traced in unambiguous electron density (Figure 3A). In agreement with the isolated U-box fragment structures, this region of LubX featured two U-box domains connected by the long α-helix named αC. Both U-boxes retained largely the same conformations as in their isolated structures, except for changes in the conformation of U-box loops 1 and 2 (not shown). As implied by the model of U-box 1, the long α3/αC helix in context of this larger LubX fragment formed a platform against which both U-boxes were arranged. The position of U-box 2 relative to that of U-box 1 was twisted ~90° around the axis of inter-domain helix αC, with the loop (residues 120 to 125) connecting U-box 2 with αC helix adopting a turn conformation. This resulted in U-box 1 and U-box 2 positioned in a “front-to-back” arrangement, that is, the prominent converging loops of both U-boxes faced the same direction. Notably, the kink observed between α3 and αC in the structure of the isolated U-box 1 was not observed in the LubX [4-186]-UBE2D2 complex (Figure 1A and 3A), suggesting that this region may be conformationally mobile.
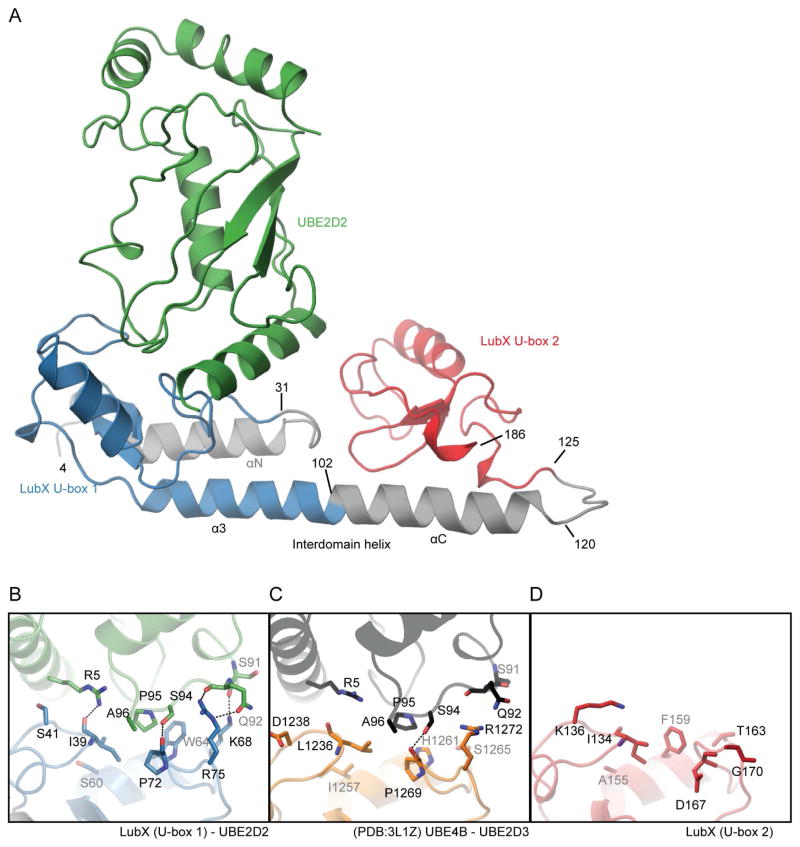
Figure 3. Structure of LubX -UBE2D2 complex.
(A) Overall architecture of LubX-UBE2D2 complex. LubX U-box 1 and 2 are colored blue and red, UBE2D2 green, respectively. Detailed comparison of U-box-E2 interactions. (B) LubX U-box1-UBED2, (C) UBE4B-UBE2D3 (PDB:3LIZ), (D) LubX U-box 2. Residues shown are those that form hydrogen bonds (dashed black line) or become >80% buried in the U-box-E2 interaction. See also Figure S4.
In full accordance with the LubX in vitro ubiquitination results (Figure 2), the interaction between LubX and UBE2D2 involves exclusively U-box 1 domain residues (Figure 3A). The U-box 1-UBE2D2 interface covered 607 Å2 and involved 18 LubX and 16 UBE2D2 residues. A comparison of the LubX-UBE2D2 complex structure with the human UBE4B U-box and UBE2D3 complex (PDB:3L1Z, (Benirschke et al., 2010)) highlighted significant similarity between bacterial and eukaryotic U-boxes in overall shape and location of E2 interaction surfaces. However, a detailed analysis of LubX interactions with E2 enzyme pointed to significant deviation in U-box 1 E2-interaction surface compared to equivalent regions in eukaryotic U-box domains (Figure 3B and C, see also structure based sequence alignments in Figure S4). While each of LubX residues contributing to interactions with UBE2D3 is conserved in subset of human U-box motifs (Figure S4), the overall composition of the U-box 1 E2-interaction surface appears to be unique to LubX and is not reproduced in any single human U-box domain. Furthermore, LubX-UBE2D2 interactions feature an expanded hydrogen bond network including three additional interactions between respective LubX-UBE2D3 residues Ile39-Arg5, Lys68-Ser91 and Arg75-Glu92, that do not have equivalents in structurally characterised human E2-U-box complexes such as UBE4B-UBE2D3.
With no evidence that U-box 2 can participate in E2 engagement, the specific structural compositions of the two U-boxes were examined for features that may explain this functional distinction. Of the 18 U-box 1 residues involved in interactions with UBE2D2, 15 showed non-conservative substitutions at the equivalent positions in U-box 2. In particular, the three U-box residues known to be critical for interactions with E2 and present in LubX U-box 1 (residues Ile39, Trp64, Pro72) are only partially conserved in U-box 2 (equivalent residues Ile134, Phe159, Asp167) (Figures 1D, 3D). The presence of Lys136 in U-box 2, which is structurally equivalent to Ser41 in U-box 1 (Figures 3B, 3D), may also prevent E2 association, significantly changing the physiochemical properties of the surface significantly and physically obscuring Ile134, which might otherwise fulfill its typical role in E3-E2 interactions in associating with α1 of the E2. These data show that although the U-box fold has been closely conserved, U-box 2 has significantly diverged from U-box 1 in residue composition throughout the solvent-exposed surface that formed the canonical E2 binding site in most U-boxes. These structural differences are consistent with the observed functional divergence between LubX U-box motifs and of the loss of E2-interaction capacity by U-box 2.
SidH is the primary target for LubX in Legionella proteome
The diverse nature of the two known LubX targets – SidH and host kinase Clk1 (Kubori et al., 2008; Kubori et al., 2010) and the large number (over 300) of DITS present in Legionella raised the possibility of LubX targeting multiple proteins, particularly among other DITS. To investigate this possibility, an unbiased screen for LubX targets within the Legionella proteome was performed using affinity purification coupled with mass spectrometry (AP-MS). To assess the repertoire of DITS expressed at each of these growth stages, the proteome of L. pneumophila str. Philadelphia-1 strain grown to exponential and post-exponential phase was profiled using high-resolution accurate mass spectrometry (see Supplemental Experimental Procedures). Details of all proteins identified in each growth phase are summarized in Table S1.
Between both Legionella proteome samples, 1498 out of 2942 possible proteins were identified at a false discovery rate (FDR) of 1% with 599 proteins detected in both growth stages. Among known Legionella DITS proteins, 77 and 99 were identified in exponential and post-exponential phases respectively, with only 32 DITS detected in both phases. Thus, combined exponential and post-exponential axenic grown Legionella lysates contained readily detectable amounts of close to the half of full DITS arsenal known for this bacterium. Based on this observation, a combined sample composed of equal amounts of exponential and post-exponential grown Legionella-clarified cell lysates was probed for potential LubX targets. Streptavidin-binding peptide (Keefe et al., 2001) tagged LubX [1-221] (Lpg2830) was used as a bait and proteins co-purified by streptavidin-based affinity were analyzed by mass spectrometry. SidH was the only DITS that co-precipitated with LubX (Table 2), confirming the highly specific nature of the interactions between these two proteins and suggesting that SidH is the primary interaction partner for LubX in the Legionella proteome under our experimental conditions. These experiments were also repeated with a LubX I45A, R121A and R121E mutants as outlined in the section below.
Table 2. Affinity purification-Mass spectrometry (AP-MS) with L. pneumophila lysate identifies SidH as the only strong candidate interactor for LubX.
SBP-tagged LubX was immobilized to magnetic sepharose-streptavidin beads and incubated with a 1:1 mixture of exponential and post-exponential lysates of Legionella pneumophila. Putative binding proteins were eluted with biotin and identified by mass spectrometry. Non LubX-specific background interactions were filtered out on the basis of GPM/X! Tandem expect score <−50, <2 unique peptides, between-replicate inconsistencies, or their appearance in the analyses of other SBP-tagged DITS proteins in similar or greater numbers. SidH was the only DITS identified as a specific interactor of LubX. Proteins NusA and RpoA are not translocated proteins and are therefore believed to be non-specific contaminants. See also Figure S3 and Table S1.
| LubX [1-221] | LubX [1-221].I45A | LubX [1-221].R121A | LubX [1-221].R121E | |||||
|---|---|---|---|---|---|---|---|---|
| Total peptides | Total peptides | Total peptides | Total peptides | |||||
| ID | Rep A | Rep B | Rep A | Rep B | Rep A | Rep B | Rep A | Rep B |
| LubX | 514 | 328 | 475 | 399 | 390 | 310 | 558 | 334 |
| SidH | 260 | 178 | 166 | 192 | ||||
| NusA | 51 | 41 | 33 | 20 | ||||
| RpsE | 9 | 14 | ||||||
Functional mapping of the LubX - SidH interaction interface
Empowered by this structural information, we set out to characterize the molecular basis of LubX interactions with SidH. For this, we established an in vivo functional screen using the budding yeast Saccharomyces cerevisiae. A previous study, in which individual DITS were expressed in S. cerevisiae, found that overexpression of SidH resulted in severe growth arrest (Heidtman et al., 2009). Based on the evidence that LubX ubiquitinates SidH leading to its degradation (Kubori et al., 2010), we predicted that co-expression of LubX and SidH would alleviate the toxic effect of SidH on yeast. Indeed, yeast cells co-overexpressing LubX and SidH demonstrated normal growth comparable to the control culture containing empty expression vector (Figure 4A). Notably, neither individual overexpression of LubX U-box 1 nor U-box 2 fragments was able to rescue SidH toxicity.
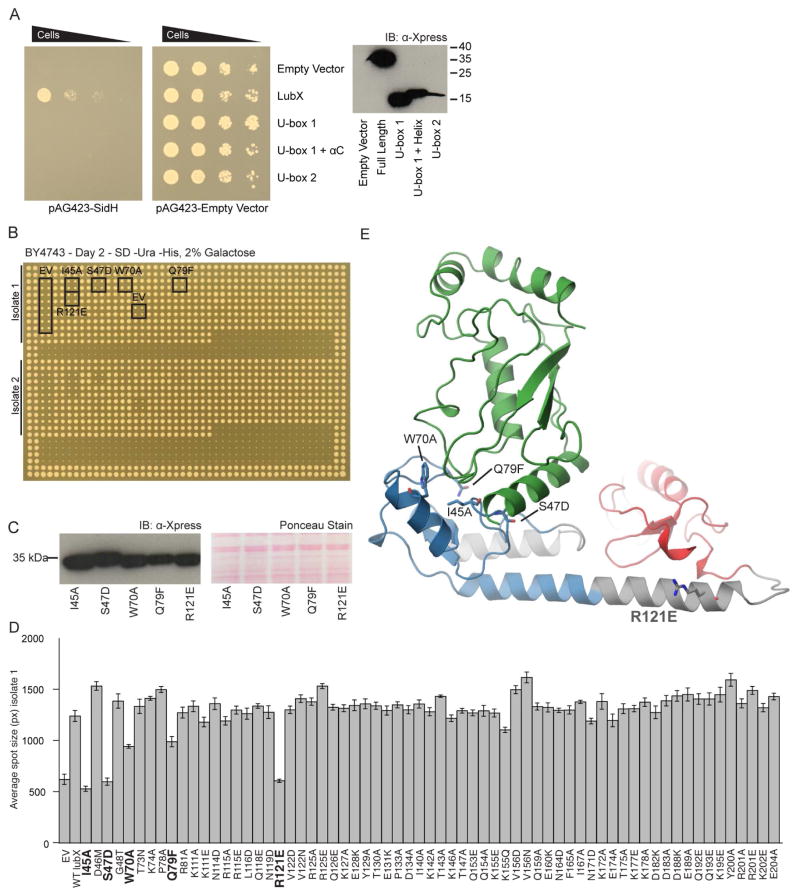
Figure 4. In vivo screening of LubX reveals domains and residues essential for effectual rescue of SidH toxicity.
(A) Spot dilutions of S. cerevisiae co-expressing SidH (lpg2829) and LubX (lpg2830), LubX U-box 1, U-box 1 plus αC helix or U-box 2 indicates full length LubX alleviates the toxicity of SidH. Rescue of SidH toxicity was not observed when using U-box 1, U-box 1 plus αC helix or U-box 2 domains. (B) Individual strains carrying SidH and LubX or a LubX mutants were arrayed in quadruplicate. 2 strains (‘isolate 1’ and ‘isolate 2’) of each mutant were assayed per plate. (C) Expression of mutant LubX strains with a significant reduction in ability to rescue SidH was validated by western blot. (D) Plate images were processed using SGAtools to obtain spot sizes. The graph shows data for one of the isolates. Mutations with a significant defect in their ability to rescue SidH are highlighted in bold. (E) The positions of LubX mutants with a significant reduction in their ability to rescue SidH are indicated on the LubX-UBE2D2 complex structure. See also Table S2.
In order to identify LubX surface-exposed residues are essential for the interaction with SidH, we prepared 65 LubX variants, each carrying individual residue mutations of 59 unique surface-exposed positions throughout the structure of LubX (Figure 3A), and tested the ability of these variants to alleviate SidH toxicity to yeast. For this, haploid yeast strains carrying individual LubX mutant were mated with a haploid yeast strain carrying a SidH-overexpressing plasmid or empty vector. The resulting diploid strains carrying both plasmids were selected by pinning onto medium selective for both plasmids and subsequently pinned onto protein expression-inducing medium. To quantify the growth of each resulting strain, the spot (pinned in quadruplicate) sizes of each diploid strain were measured and compared to the spot size of strains containing the empty vector control or wild-type LubX (Figure 4B).
According to this analysis, LubX variants carrying I45A, S47D, W70A or Q79F mutations were not able to rescue SidH-mediated toxicity (Figure 4B and D). These four residues localize to the LubX E2 interaction surface of U-box 1, suggesting that the observed effect is due to interruption of LubX-E2 interactions. In line with these observations, the substitution to alanine of the conserved isoleucine residue (Ile45 in Lpg2830 and Ile39 in lpp2887) at this surface was previously shown to result in complete loss of in vitro E3 ligase activity of LubX (Kubori et al., 2008) and several eukaryotic U-box E3 ligases (Andersen et al., 2004; Ohi et al., 2003). Unexpectedly, out of the remaining 61 LubX variants carrying single mutations of surface-exposed residues, only the R121E variant was defective in its ability to rescue SidH toxicity. The Arg121 residue localizes to the C-terminus of the αC helix connecting the two U-box domains (Figure 4D). Interestingly, LubX variants carrying individual substitutions of nearby residues, including Q118E, N119D, R125A/E, E189A, Q192E, Q193E, all retained the ability to rescue SidH toxicity. Furthermore, none of the mutations at the canonical E2 binding surface of U-box 2 (residues 140-143, 146, 147, 159-160, 164-165, and 167) had any detectable effect on LubX activity against SidH, suggesting that these residues may not play a critical role in LubX-SidH interactions. Furthermore, we tested these newly discovered mutations for their ability to bind to SidH by using the Streptavidin-binding peptide tagged LubX variants carrying R121E or R121A substitutions. These mutants lost their ability to co-precipitate SidH from Legionella lysate when used as baits in AP-MS experiment similar to the one described above (Table 2), despite still demonstrating robust in vitro ubiquitination activity in the presence of human E1 and UBE2D2 enzymes (Figure S3) in line with them containing intact U-box 1 domains. Taken together, this analysis suggests that the interaction between LubX and SidH are dissimilar from those characterized for canonical U-boxes and E2 ubiquitin conjugating enzymes and involves at least one LubX residue outside of the U-box 2 domain which has no known structural equivalent in its eukaryotic counterparts.
Discussion
The L. pneumophila translocated protein LubX mimics the function of a eukaryotic E3 ubiquitin ligase in order to specifically target another translocated protein, SidH, for degradation by the host proteasome (Kubori et al., 2008; Kubori et al., 2010). Here, we present the crystal structures of both the N- and C-terminal domains of LubX, which confirmed the presence of two distinct yet structurally similar domains reminiscent of eukaryotic U-boxes. Screening of LubX and its individual U-box domains against a panel of 29 ubiquitin conjugating E2 enzymes, representing 13 out of 17 E2 groups encoded by the human genome confirmed the ability of LubX U-box 1 to engage a defined subset of these enzymes for the formation of polyubiquitin protein species. In addition to the previously identified UBE2D1 and UBE2D3 E2 enzymes, we demonstrated that LubX U-box 1 is able to interact with UBE2D2 as well as the UBE2E family members and both UBE2W and UBE2L6 E2s. Unlike the UBE2D and UBE2E family enzymes, which are closely related in sequence, UBE2W and UBE2L6 stand out having only 30 to 40% identity to UBE2D and UBE2E family members. This new data shows that LubX is able to engage a significantly larger arsenal of host E2 enzymes than previously established and raises the possibility of this pathogenic E3 ligase having additional as-yet unidentified functions involving the diverse E2 enzymes identified in this study.
Characterization of a more comprehensive set of E2s able to support LubX activity also highlighted molecular features that may be responsible for LubX specificity for this particular subset of E2 enzymes. Our data shows that UBE2L3, or UBE2Q1 and Q2 E2 enzymes are not able to support in vitro ubiquitination activity of LubX despite their close similarity to UBE2L6 and UBE2W enzymes, respectively. Inability of UBE2L3 to support LubX activity is probably due to this E2 enzyme being HECT and RBR E3 specific (Wenzel et al., 2011). In case of UBE2Q1 and Q2 E2 enzymes, one possible explanation is that UBE2Q E2s feature an Asp residue (PDB: 1ZUO (Sheng et al., 2012)) at the position equivalent to Lys4 in UBE2D2 and UBE2W. This substitution would position this acidic residue in proximity to the hydrophobic patch of LubX around residue Ile39 which is critical for E2 interaction potentially causing surface charge clashes and disrupt the formation of Ile39-Arg5 hydrogen bond. The presence of Gly in UBE2Q1 and Q2 at the position equivalent to UBE2D2 Glu92 may also significantly reduce the ability for LubX to interact with these E2 enzymes.
The structure of LubX in complex with the human UBE2D2 E2 enzyme highlighted the role of the U-box 1 domain as the exclusive E2-interacting module within LubX. In addition it revealed that the LubX-E2 interface involves conserved UBE2D2 residues that are also critical for the interaction of this E2 enzyme with eukaryotic U-box domains. Specifically, the UBE2D2 residues Lys8, Phe62 and Pro95 located at the LubX interacting surface are conserved among all human E2 enzymes that are active with this E3. Pro95 of the Ser-Pro-Ala motif is an important determinant of specificity between human E2 enzymes and the eukaryotic U-box CHIP E3 (Soss et al., 2011; Xu et al., 2008). Notably, LubX and CHIP show similar profiles of E2 enzymes able to support their activity in vitro, all of which except for UBE2L6 harbor the requisite Ser-Pro-Ala motif required for binding. The ability to engage this latter E2, which features a Lys-Pro-Cys instead of the Ser-Pro-Ala motif indicates that LubX only partially relies on the latter motif in interactions with host E2 enzymes. Examination of the remainder of the LubX-E2 interface residues that play a significant role in the interaction with UBE2D2 shows that although LubX shares little similarity to any single known U-box domain, residues involved in the interaction can typically be found elsewhere in one or more eukaryotic U-boxes suggesting LubX may be optimized to interact with a broad range of host E2s.
Structural characterization of the LubX-UBE2D2 complex also highlighted significant differences between the U-box 1 and 2 domains. While the LubX U-box 2 domain follows the general architecture of the U-box fold, there are both significant changes in the biochemical makeup of its “E2 interaction” surface as well as more subtle structural differences including different conformations of the two key loops. Conformational changes in U-box 2 loop 1 and loop 2 in comparison with equivalent regions of U-box 1 may place residues of U-box 2 away from an orientation consistent with E2 binding.
The divergence of the LubX C-terminal domain from canonical U-box function was further emphasized by its role in interactions with SidH. Using a yeast-based assay that leverages the toxicity of heterologously expressed SidH and the ability for LubX co-expression to alleviate this toxicity we sought to better understand the nature and location of the LubX-SidH interaction. While the integrity of C-terminal domain encompassing U-box 2 domain was necessary for LubX function against SidH, extensive mutagenesis guided by the crystal structure of the LubX2-UBE2D2 complex revealed no evidence to support the direct involvement of U-box 2 in SidH recognition. Furthermore, the single residue that appears to be critical for LubX-SidH interactions, Arg121, localized to the end of the αC helix and is not part of the U-box fold. In addition, we verified the importance of this position for the physical interaction with SidH, by testing the abilities of wild type and I45A, R121A or R121E variants of LubX for their ability to co-precipitate the SidH from crude Legionella lysate. As expected, wild type LubX and LubX I45A co-precipitated SidH whereas R121A and R121E mutants (both of which were verified to be active as E3 ligases) did not. Notably, a LubX C-terminal fragment encompassing the U-box 2 motif that was previously reported (Kubori et al., 2010) to be required for binding to SidH also contained the sequence corresponding to αC helix and thus included the Arg121. Given the significant size of SidH (~ 253 kDa), it is likely that LubX-SidH interaction occurs over a large area with number of residues contributing to the interaction. Thus, the lack of LubX variants deficient in SidH interactions other than R121E LubX may be due to our single point mutation-focused analysis being inadequate to disrupt a potential large surface area of LubX-SidH interaction. Superimposition of our structures covering U-box 2 reveals that R121 may be well positioned to stabilize the observed conformation of LubX, providing a possible explanation for the apparent singular importance this residue. Interestingly, when we model Ub onto our co-crystal structure, non-covalently ‘backside bound’ ubiquitin that has been shown to greatly increase activation of Ub transfer (Buetow et al., 2015), is able to fit neatly into the space between UBE2D2 and U-box2 (data not shown) suggesting a new potential function for U-box2.
Our unbiased search of the L. pneumophila str. Philadelphia-1 proteome identified SidH as the primary target of LubX, however the possibility for functional interactions between LubX and other effectors not present in our lysate mix or that bind weakly to LubX remains. While the function of SidH during Legionella infection remains unknown, both ours and previously published data (Kubori et al., 2010) clearly show that SidH function requires tight regulation by LubX. In line with this observation, the expression of SidH in our yeast model eukaryotic system led to complete growth arrest that was alleviated by coexpression of LubX. These data, combined with the observation that many DITS exhibit a toxic phenotype similar to SidH when expressed in yeast (Heidtman et al., 2009), raise the possibility that L. pneumophila may be required to conduct self-regulation of its DITS functions in several other cases and that this requirement may be more frequent than previously thought. With over 300 DITS, further studies into the prevalence of DITS metaeffector activities are warranted. Understanding the dynamics of complex bacterial-host interactions such as those employed by Legionella will be critical to our detailed understanding of microbial disease.
Experimental Procedures
Protein purification
LubX orthologues from L. pneumophila str. Paris and L. pneumophila str. Philadelphia (Q5X159/Lpp2887 and Q5ZRQ0/Lpg2830 respectively) were cloned into either pET28-SBP-TEV (for proteins requiring SBP-tag) or p19MBP-L (no SBP-tag) as required, Human E2 expression constructs were obtained in pET28 (Sheng et al., 2012). BL21 (RIL) DE3 E. coli cultures were grown in LB or Studier media (without galactose) supplemented with the appropriate antibiotics and expression was induced with 0.4 mM IPTG. Cultures were harvested by centrifugation and pellets lysed by sonication on ice in 50 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM PMSF. All further purification was conducted at 4 °C. Lysate was clarified by ultracentrifugation at 17’000 × g for 30 minutes, and to this, 1–5 ml of Ni-NTA resin (Qiagen) was added and incubated with gentle rotation for 30 min. Resin was washed with 50 mM HEPES (pH 7.5), 150 mM NaCl and 30 mM imidazole and protein eluted with 50 mM HEPES (pH 7.5), 150 mM NaCl, 500 mM imidazole. Proteins were further concentrated using a centrifugal concentrator, flash frozen in liquid N2, and stored at −80 °C.
Crystallization and structure determination
Crystals of selenomethionine-substituted (SelMet) LubX [1-186], native LubX U-box 2, SelMet LubX U-box 2 mutant Ile175Met and SelMet LubX in complex with E2D2.C85S-Ub conjugate were grown at 23°C using hanging-drop vapor diffusion with the following protein concentrations plus reservoir solutions: ~30 mg/ml protein plus 0.25 M ammonium sulfate, 0.1 M imidazole pH 7.8 and 28% PEG 8K; 15 mg/ml protein and V8 protease (1:100 molar ratio protease:protein) plus 1.6 M ammonium sulfate, 0.1 M HEPES pH 7.5, 2% hexanediol; 15 mg/ml protein and 1.6 M ammonium sulfate, 0.1 M HEPES (pH 7.5), 2% hexanediol; 15 mg/ml of E2D2.(C85S)-Ub conjugate and an equimolar concentration of LubX, plus 0.2 M sodium tartrate, 0.1 M Tris-Cl (pH 8.5), 25% PEG3350. Crystals were cryoprotected with reservoir solution supplemented with 20% glycerol except for LubX U-box 2 mutant Ile175Met which was also supplemented with sodium chloride. SDS-PAGE of LubX [1-186] indicated degradation of the protein and residues 9-117 were identified in the electron density.
Diffraction data was collected at 100 K at beamline 19-ID at Structural Biology Center, Advanced Photon Source at wavelength 0.9794 Å (selenium peak) and reduced with HKL-3000 (Minor et al., 2006). LubX U-box 1 structure was solved first by SAD phasing using PHENIX.solve (Adams et al., 2010) which identified one SelMet site (residue 34), and followed by model building by PHENIX.autobuild. The structure of LubX-U-box 2 Ile175Met was determined by SAD phasing using PHENIX.solve, which identified 3 SelMet sites, and followed by model building by PHENIX.autobuild. This model of LubX U-box 2 Ile175Met was used to determine the structure of native LubX U-box 2; model refinement was focused on this fragment due to its higher resolution. Structure of LubX U-box 1 – UBE2D2 complex was determined by molecular replacement using as search models the structure of UBE2D2 from PDB 4DDI (Juang et al., 2012) and the structure of LubX U-box 1, using PHENIX.phaser. All structures were refined using PHENIX.refine. The final models include the following residues: LubX U-box 1 = 9-117; LubX U-box 2 (wild-type) = 123–198; LubX U-box 2 (Ile175Met) = 124–198; LubX U-box 1-UBE2D2 complex = 4–186 of LubX and 1-147 of E2D2 plus N-terminal Gly residue from the expression tag. All B-factors were refined as isotropic. All geometries were verified with PHENIX.refine and the RCSB PDB Validation server. Structure coordinates were deposited to the PDB under accession codes 4WZ0, 4WZ1, 4WZ2 and 4WZ3 for the LubX U-box 1, LubX U-box 2 (wild-type), LubX U-box 2 (Ile175 mutant) and LubX-UBE2D2 complex structures, respectively.
Structure analysis
Protein-protein interfaces were analyzed using PDBePISA (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html) (Krissinel and Henrick, 2007). For comparison of U-boxes, U-box 1 and U-box 2 were superposed based on the core central α-helix, 310 helix and β-sheets. Definitions for U-box boundaries were from PFAM PF04564 and publications for PDB 3L1Z (Benirschke et al., 2010), 2C2V (Zhang et al., 2005) and 2OXQ (Xu et al., 2008).
Ubiquitination Assays
Ubiquitination reactions were performed as described previously (Singer et al., 2008). Briefly, 4 μg of ubiquitin, 0.13 μg of E1 and 2 μg of E2 were incubated together with 2 μg of the specified fragment of LubX in 20 μl of reaction buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM ATP, 10 mM MgCl2, 0.5 mM DTT) at 25 °C for the time specified. Reactions were terminated by the addition of an equal volume of 2x Laemmli sample buffer (0.125 M Tris-HCl, pH 6.8, 20% (v/v) glycerol, 4% (w/v) SDS, 100 mM DTT, 0.004% (w/v) bromophenol blue). Reaction mixtures were separated by SDS-PAGE, transferred to nitrocellulose and probed with α-ubiquitin antibody (Millipore MAB1510).
Legionella pneumophila culture
A Legionella pneumophila str. Philadelphia-1 derived (Lp03) strain (Berger and Isberg, 1993; Rao et al., 2013) was patched on charcoal-N-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-yeast extract-thymidine (CYET) plates and used to inoculate overnight 37°C cultures in ACES-yeast extract-thymidine (AYET) broth (Berger and Isberg, 1993). Cultures were grown to either exponential phase (OD600 = 1.6) or post-exponential phase (cessation of growth, increase in pigmentation, and an increase in motility as visualized by light microscopy). Cells were harvested by centrifugation at 4°C at 5000 × g for 10 min.
Affinity Purification-Mass Spectrometry
Exponential and post-exponential Legionella cell pellets were resuspended in ice cold lysis/binding buffer (50 mM HEPES, (pH 8.0), 150 mM NaCl, 5 mM DTT, 5 mM EDTA 1x complete mini EDTA-free protease inhibitor cocktail (Roche)) at 1 × 109 cells/ml and sonicated on ice. Lysates were clarified by centrifugation at 21,000 × g for 10 minutes and stored at −80C until needed.
50 μl of Streptavidin Mag Sepharose beads bearing 6xHis-SBP baits were added to 1 ml of lysate depleted of excess endogenous biotin with streptavidin agarose and incubated with gentle agitation for 3 hrs. Beads were washed twice with binding buffer and transferred to a fresh tube during a final wash step. Bait and bound proteins were eluted using 100 μl of 100 mM ammonium bicarbonate (pH 8.5), 2.5 mM biotin. 1 μg of sequencing grade modified trypsin (Promega) was added to samples and incubated overnight at 37 °C. Digestion was terminated by addition of TFA to a final concentration of 0.5 %, and samples were further processed using OMIX C18 tips (Agilent) according to manufacturer’s instructions. Peptides were eluted with 95 % acetonitrile 0.1 % formic acid and dried to completion by vacuum centrifugation. Samples were resuspended in 0.1 % formic acid and analysed by mass spectrometry on an LTQ XL mass spectrometer. Peptide identifications were performed using GPM/X!Tandem. See Supplemental Experimental Procedures for details.
In vivo Yeast Screening
Spot dilutions
The ability of LubX, LubX U-box 1, U-box 1 plus αC and U-box 2 to rescue toxicity of SidH expression in yeast was assayed by spot dilution. BY4742 (MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) (Brachmann et al., 1998) was co-transformed with plasmids carrying sidH (lpg2829) Gateway cloned into pAG423-GAL-HA-ccdB and either lubX (lpg2830), one of its fragments in pYES2 NT/A, or empty vector pYES2 NT/A. Overnight cultures of each strain were grown in SD- uracil histidine, 2% glucose and subsequently diluted to OD600 of 1.0, 0.2, 0.04, 0.008, and spotted onto both SD –histidine, uracil + 2% galactose plates using the VP 407 AH pin tool (V&P scientific) and grown for 2 days at 30°C.
In vivo mutational characterization of LubX – SidH interaction
Characterization of the LubX – SidH interaction interface was performed using a high-density (1536) pinning assay, similar to the SGA procedure (Tong and Boone, 2006) with modifications (See Table S2 and Supplemental Experimental Procedures for full details and description of this procedure). Briefly, haploid S. cerevisiae strains BY4741 (MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) and BY4742 (Brachmann et al., 1998) carrying galactose-inducible pAG423GAL-HA-sidH (lpg2829) or empty vector control and pYES2 NT/A lubX (lpg2830), lubX mutants or empty vector control, respectively, were mated.. Resulting diploid strains carrying both plasmids were grown on SD –histidine, uracil + 2% galactose plates, imaged and quantified using SGAtools (Wagih et al., 2013)(http://sgatools.ccbr.utoronto.ca/). LubX mutants that failed to rescue the yeast toxicity of SidH were re-sequenced to confirm the identity of the mutations. Western blots were performed using α-Xpress antibodies (Life Technologies) to verify that the LubX mutants were expressed and stable.
Supplementary Material
Highlights.
LubX structures reveal two functionally diverse U-boxes connected by a central helix.
LubX U-box 1 demonstrates specificity toward UBE2D1, D2, D3, D4, E2, E3, L6 and W E2s.
In vivo yeast assay defines critical molecular determinants of LubX’s activity.
LubX R121 localised outside of the U-boxes is critical for LubX-SidH interaction.
Acknowledgments
Genomic DNA for Legionella pneumophila str. Paris was a generously provided by Carmen Buchrieser. Human E2 expression constructs were received as a generous gift from the S. Dhe-Paganon laboratory at the Structural Genomics Consortium. We also thank Adam Stein and Marianne Cuff at the Structural Biology Center, Advanced Photon Source, for X-ray diffraction data collection and processing, and Veronica Yim and Rosa Di Leo for cloning. This work was supported by the NIH grant GM094585 (to A.S. through the Midwest Center for Structural Genomics) and by the US Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357. A.W.E. is supported by a CIHR Operating Grant (MOP-13340), the University of Toronto Department of Molecular Genetics, and infrastructure grants from the Canada Foundation for Innovation and the Ontario Research Fund.
Footnotes
Author Contributions
Protein purification, crystallization and ubiquitination assays were performed by TS. X-ray data collection was performed by BN and analysed and interpreted by PS. All other experimental work was conducted and analysed by AQ and MU. All work was performed under the direction of AE and AS.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abramovitch RB, Janjusevic R, Stebbins CE, Martin GB. Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc Natl Acad Sci U S A. 2006;103:2851–2856. doi: 10.1073/pnas.0507892103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen P, Kragelund BB, Olsen AN, Larsen FH, Chua NH, Poulsen FM, Skriver K. Structure and biochemical function of a prototypical Arabidopsis U-box domain. The Journal of biological chemistry. 2004;279:40053–40061. doi: 10.1074/jbc.M405057200. [DOI] [PubMed] [Google Scholar]
- Benirschke RC, Thompson JR, Nomine Y, Wasielewski E, Juranic N, Macura S, Hatakeyama S, Nakayama KI, Botuyan MV, Mer G. Molecular basis for the association of human E4B U box ubiquitin ligase with E2-conjugating enzymes UbcH5c and Ubc4. Structure. 2010;18:955–965. doi: 10.1016/j.str.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic acids research. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Buetow L, Gabrielsen M, Anthony NG, Dou H, Patel A, Aitkenhead H, Sibbet GJ, Smith BO, Huang DT. Activation of a Primed RING E3-E2-Ubiquitin Complex by Non-Covalent Ubiquitin. Molecular cell. 2015 doi: 10.1016/j.molcel.2015.02.017. [DOI] [PubMed] [Google Scholar]
- Burstein D, Zusman T, Degtyar E, Viner R, Segal G, Pupko T. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 2009;5:e1000508. doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalet C, Rusniok C, Bruggemann H, Zidane N, Magnier A, Ma L, Tichit M, Jarraud S, Bouchier C, Vandenesch F, et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- de Felipe KS, Glover RT, Charpentier X, Anderson OR, Reyes M, Pericone CD, Shuman HA. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 2008;4:e1000117. doi: 10.1371/journal.ppat.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felipe KS, Pampou S, Jovanovic OS, Pericone CD, Ye SF, Kalachikov S, Shuman HA. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol. 2005;187:7716–7726. doi: 10.1128/JB.187.22.7716-7726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensminger AW, Isberg RR. E3 ubiquitin ligase activity and targeting of BAT3 by multiple Legionella pneumophila translocated substrates. Infect Immun. 2010;78:3905–3919. doi: 10.1128/IAI.00344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields BS. The molecular ecology of legionellae. Trends in Microbiology. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- Heidtman M, Chen EJ, Moy MY, Isberg RR. Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cellular Microbiology. 2009;11:230–248. doi: 10.1111/j.1462-5822.2008.01249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annual review of biochemistry. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hicks SW, Galan JE. Hijacking the host ubiquitin pathway: structural strategies of bacterial E3 ubiquitin ligases. Curr Opin Microbiol. 2010;13:41–46. doi: 10.1016/j.mib.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Boyd D, Amyot WM, Hempstead AD, Luo ZQ, O’Connor TJ, Chen C, Machner M, Montminy T, Isberg RR. The E Block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol. 2011;13:227–245. doi: 10.1111/j.1462-5822.2010.01531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubber A, Kubori T, Nagai H. Modulation of the ubiquitination machinery by Legionella. Current topics in microbiology and immunology. 2013;376:227–247. doi: 10.1007/82_2013_343. [DOI] [PubMed] [Google Scholar]
- Juang YC, Landry MC, Sanches M, Vittal V, Leung CC, Ceccarelli DF, Mateo AR, Pruneda JN, Mao DY, Szilard RK, et al. OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Molecular cell. 2012;45:384–397. doi: 10.1016/j.molcel.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefe AD, Wilson DS, Seelig B, Szostak JW. One-step purification of recombinant proteins using a nanomolar-affinity streptavidin-binding peptide, the SBP-Tag. Protein expression and purification. 2001;23:440–446. doi: 10.1006/prep.2001.1515. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of molecular biology. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Kubori T, Hyakutake A, Nagai H. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol Microbiol. 2008;67:1307–1319. doi: 10.1111/j.1365-2958.2008.06124.x. [DOI] [PubMed] [Google Scholar]
- Kubori T, Shinzawa N, Kanuka H, Nagai H. Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog. 2010;6:e1001216. doi: 10.1371/journal.ppat.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifshitz Z, Burstein D, Peeri M, Zusman T, Schwartz K, Shuman HA, Pupko T, Segal G. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc Natl Acad Sci U S A. 2013;110:E707–715. doi: 10.1073/pnas.1215278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurie-Weinberger MN, Gomez-Valero L, Merault N, Glockner G, Buchrieser C, Gophna U. The origins of eukaryotic-like proteins in Legionella pneumophila. International journal of medical microbiology : IJMM. 2010;300:470–481. doi: 10.1016/j.ijmm.2010.04.016. [DOI] [PubMed] [Google Scholar]
- Michelle C, Vourc’h P, Mignon L, Andres CR. What was the set of ubiquitin and ubiquitin-like conjugating enzymes in the eukaryote common ancestor? Journal of molecular evolution. 2009;68:616–628. doi: 10.1007/s00239-009-9225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta crystallographica Section D, Biological crystallography. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nature structural biology. 2003;10:250–255. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CT, Al-Khodor S, Al-Quadan T, Santic M, Habyarimana F, Kalia A, Kwaik YA. Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog. 2009;5:e1000704. doi: 10.1371/journal.ppat.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao C, Benhabib H, Ensminger AW. Phylogenetic reconstruction of the Legionella pneumophila Philadelphia-1 laboratory strains through comparative genomics. PLoS One. 2013;8:e64129. doi: 10.1371/journal.pone.0064129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G, Purcell M, Shuman HA. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y, Hong JH, Doherty R, Srikumar T, Shloush J, Avvakumov GV, Walker JR, Xue S, Neculai D, Wan JW, et al. A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen. Molecular & cellular proteomics : MCP. 2012;11:329–341. doi: 10.1074/mcp.O111.013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer AU, Rohde JR, Lam R, Skarina T, Kagan O, Dileo R, Chirgadze NY, Cuff ME, Joachimiak A, Tyers M, et al. Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nature structural & molecular biology. 2008;15:1293–1301. doi: 10.1038/nsmb.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer AU, Schulze S, Skarina T, Xu X, Cui H, Eschen-Lippold L, Egler M, Srikumar T, Raught B, Lee J, et al. A pathogen type III effector with a novel E3 ubiquitin ligase architecture. PLoS Pathog. 2013;9:e1003121. doi: 10.1371/journal.ppat.1003121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soss SE, Yue Y, Dhe-Paganon S, Chazin WJ. E2 conjugating enzyme selectivity and requirements for function of the E3 ubiquitin ligase CHIP. The Journal of biological chemistry. 2011;286:21277–21286. doi: 10.1074/jbc.M111.224006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- Wagih O, Usaj M, Baryshnikova A, VanderSluis B, Kuzmin E, Costanzo M, Myers CL, Andrews BJ, Boone CM, Parts L. SGAtools: one-stop analysis and visualization of array-based genetic interaction screens. Nucleic acids research. 2013;41:W591–596. doi: 10.1093/nar/gkt400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Skarina T, Yee A, Jobin MC, Dileo R, Semesi A, Fares C, Lemak A, Coombes BK, Arrowsmith CH, et al. NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases. PLoS Pathog. 2010;6:e1000960. doi: 10.1371/journal.ppat.1000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Kohli E, Devlin KI, Bold M, Nix JC, Misra S. Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC structural biology. 2008;8:26. doi: 10.1186/1472-6807-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Molecular cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Zhu W, Banga S, Tan Y, Zheng C, Stephenson R, Gately J, Luo ZQ. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS One. 2011;6:e17638. doi: 10.1371/journal.pone.0017638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Li H, Hu L, Wang J, Zhou Y, Pang Z, Liu L, Shao F. Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nature structural & molecular biology. 2008;15:1302–1308. doi: 10.1038/nsmb.1517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.