Abstract
Spermatogonial stem cells (SSCs) undergo self-renewal divisions to provide the foundation for spermatogenesis. Although Rb1 deficiency is reportedly essential for SSC self-renewal, its mechanism has remained unknown. Here we report that Rb1 is critical for cell cycle progression and protection of SSCs from DNA double-strand breaks (DSBs). Cultured SSCs depleted of Cdkn1b proliferated poorly and showed diminished expression of CDK4 and RB1, thereby leading to hypophosphorylation of RB1. Rb1 deficiency induced cell cycle arrest and apoptosis in cultured SSCs, which expressed markers for DNA DSBs. This DNA damage is caused by increased E2F1 activity, the depletion of which decreased DNA DSBs caused by Rb1 deficiency. Depletion of Cdkn1a and Bbc3, which were upregulated by Trp53, rescued Rb1-deficient cells from undergoing cell cycle arrest and apoptosis. These results suggest that the CDKN1B-RB1-E2F1 pathway is essential for SSC self-renewal and protects SSCs against genomic damage.
Keywords: Apoptosis, Cell cycle arrest, Genomic damage, Rb1, Trp53
Spermatogenesis is a unique process that produces mature gametes and consists of three phases: amplification of spermatogonia, genetic diversification in spermatocytes and maturation of spermatids to spermatozoa to acquire mobility [1, 2]. Because parental genetic information must be preserved faithfully for the next generation, germ cells are thought to maintain a stable genetic integrity compared with somatic cells. Although little is known regarding the mechanism, germ cells may protect their pristine genome through different strategies. Of the different spermatogenic cell types, spermatogonial stem cells (SSCs) are the only cell type with self-renewal potential. These cells continuously undergo mitotic divisions and produce enormous amounts of committed progenitors during their lifetime. SSCs are thought to consist of ~5–10% of Asingle (As) spermatogonia [3, 4]. Since SSCs are the founder population of spermatogenesis, the maintenance of SSC genetic integrity is of utmost importance. Furthermore, the accumulation of genetic abnormalities in SSCs can lead to the suppression of spermatogenesis and thus infertility.
Despite their apparent biological importance, SSC analysis has been challenging because of their low frequency in the testis and the lack of SSC-specific markers. However, SSC culture techniques have allowed us to study SSCs in vitro [5]. SSCs can be cultured for long periods of time in the presence of glial cell line-derived neurotrophic factor (GDNF) and fibroblast growth factor 2 (FGF2). Cultured SSCs, designated as germline stem (GS) cells, proliferate in vitro as grape-like clusters of spermatogonia on mouse embryonic fibroblasts (MEFs). These cells initiate spermatogenesis upon introduction into seminiferous tubules of infertile testes. One of the most important findings from culture studies was the stable genetic and epigenetic integrity of SSCs [6]. GS cells were shown to maintain a normal number of chromosomes and androgenetic imprinting patterns despite 2 years of consecutive cultures. This result was unexpected given that many cultured cells undergo senescence and exhibit karyotype abnormalities and abnormal DNA methylation. Although factors involved in the maintenance of genetic integrity have not been identified, these results confirmed that replication of genetic information in SSCs proceeds with higher fidelity.
Our understanding of the signaling pathway of self-renewal factors, however, has improved. GDNF is known to activate HRAS via Src family kinase molecules [7, 8], and cells transfected with activated Hras undergo self-renewal division without exogenous cytokines [7]. Activation of HRAS increases the expression of Ccnd2 and Ccne1. Overexpression (OE) of both Ccnd2 and Ccne1 in GS cells allows cytokine-free self-renewal in a manner similar to Hras-transfected cells. Although these cells are known to have SSC activity based on a transplantation assay, both types of GS cells produced germ cell tumors in recipient mice, suggesting that excessive stimulation of self-renewal triggers tumor formation. Hras and Ccnd2 play similar roles in humans, because human germ cell tumors show enhanced expression of Hras and Ccnd2 [9, 10]. While these previous studies revealed the critical role of G1/S cyclins in self-renewal, how they regulate the G1/S transition in SSCs remains unknown. Cyclins bind to cyclin-dependent kinase (CDK) and phosphorylate RB1 [11]. RB1 phosphorylation causes changes in cell cycle-related genes, including E2F1 activation. Understanding the dynamics of these molecules is a prerequisite for clarifying the link between cytokine signaling and self-renewal.
Two recent studies have addressed the function of Rb1 in SSCs. One study showed that Rb1 deficiency caused progressive loss of GFRA1-positive (GFRA1+) As spermatogonia when the Rb1 gene was deleted by Cre driven by the Ddx4 promoter [12]. The Ddx4 promoter became active during embryonic development at ~15.5 days post coitum (dpc). Rb1-deficient As spermatogonia incorporated 5-bromo-2-deoxyuridine (BrdU) at a similar rate as controls and did not show increased apoptosis. Notably, As spermatogonia generated no As spermatogonia, only Apaired (Apr) spermatogonia upon Rb1 loss. In contrast, another group suggested that SSCs do not form in Ddx4-Cre Rb1flox/– mice and reported that the loss of Rb1 influenced SSC maturation from gonocytes [13]. When Stra8- or Neurog3-Cre transgenic mice that express Cre in undifferentiated spermatogonia were used to delete Rb1, these mice showed normal spermatogenesis, suggesting that Rb1 may play a role in the transition of gonocytes to SSCs. Although SSC self-renewal was shown to be repressed in pup testis cells, this study involved small interfering RNA (siRNA)-mediated partial knockdown (KD), and this conclusion does not agree with the observation that germ cells, which were suggestive of SSCs, were present in mature Ddx4-Cre Rb1flox/– mice. Thus, although both studies showed the critical role of Rb1 in male germline cells, they reached different conclusions regarding the role of Rb1. Moreover, they did not address how molecules involved in the G1/S transition regulate Rb1, and the precise function and the mechanism of action of Rb1 in postnatal SSCs remain elusive.
In this study, we extended our previous observations and analyzed the molecular mechanism of the G1/S transition in GS cells. We found that depletion of the CDK inhibitor (CDKI) Cdkn1b decreased CDK4 and RB1 levels in GS cells. Moreover, we found that Rb1 deficiency induced DNA double-strand breaks (DSBs) in GS cells and that Rb1-deficient GS cells were arrested at the G2/M phase and underwent apoptosis. This cell cycle suppression and apoptosis could be rescued by depletion of Trp53, which protects the genome. Thus, the network involving Trp53 and Rb1 governs the genetic integrity and maintenance of SSCs.
Materials and Methods
Animals and transplantation
Rb1flox/flox mice were obtained from the NCI Mouse Models of Human Cancers Consortium (NCI Mouse Repository, Frederick, MD, USA) [14]. These mice were crossed with R26R female mice to introduce the LacZ reporter construct for Cre-mediated deletion [15] (The Jackson Laboratory, Bar Harbor, ME, USA). For production of Ddx4-Cre Rb1flox/– mice, Rb1flox/flox mice were mated with Ddx4-Cre transgenic mice (The Jackson Laboratory). The genotypes of the mice were examined by polymerase chain reaction (PCR) with the primers listed in Supplementary Table 1 (online only). For deletion of Rb1in vitro, dissociated testis cells were exposed to adenovirus expressing Cre (AxCANCre, RIKEN BRC, Tsukuba, Japan) at a density of 1 × 106 cells/9.5 cm2, as described previously [16]. After an overnight incubation, the virus was removed on the next day, and cells were used for transplantation. The multiplicities of infection (MOIs) were adjusted to 2.0. For transplantation, testis cells were dissociated into a single-cell suspension using a two-step enzymatic digestion with collagenase type IV and trypsin (Sigma, St Louis, MO, USA), as described previously [17]. Cells were transplanted into seminiferous tubules of WBB6F1-W/Wv (designated W) mice (Japan SLC, Hamamatsu, Japan) through the efferent duct [17]. For allogeneic transplantation, recipient mice were treated with anti-CD4 antibody, as described previously [18]. Approximately 4 μl could be introduced into each testis, which filled 75–85% of the seminiferous tubules. The Institutional Animal Care and Use Committee of Kyoto University approved all animal experimentation protocols.
Cell culture
GS cells were established from B6/Tg14 (act-EGFP-OsbY01) mice (a gift from Dr M Okabe, Osaka University, Japan) or B6-TgR (ROSA26)26Sor (ROSA; The Jackson Laboratory) mice that were backcrossed to a DBA/2 background for at least 7 generations [5, 19]. GS cells from Trp53 KO mice were previously described [20]. We also derived Rb1flox/flox GS cells from 4- to 5-day-old Rb1flox/flox mice, which were produced by mating F1 offspring that resulted from crossing of Rb1flox/+ mice in a B6 background to ICR mice. AxCANCre was added to these cells at MOIs of 2.0 to produce Rb1 KO GS cells. AxCANLacZ (RIKEN BRC) was used as a control. The conditions of GS cell culture using StemPro-34 SFM (Invitrogen, Carlsbad, CA, USA) were described previously [5]. The growth factors used were 10 ng/ml human FGF2 and 15 ng/ml rat GDNF (Peprotech, Rocky Hill, NJ, USA). The cells were maintained on mitomycin C (Sigma)-treated MEFs.
Gene expression analyses
Total RNA was recovered using TRIzol reagent (Invitrogen), and first-strand cDNA was produced using a Verso cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) for reverse transcription (RT)-PCR. For real-time PCR, a StepOnePlusTM Real-Time PCR System (Applied Biosystems, Warrington, UK) and FastStart Universal SYBR Green Master Mix (Roche Applied Science, Mannheim, Germany) were used according to the manufacturers’ protocols. Transcript levels were normalized relative to those of Hprt. The real-time PCR conditions were 95 C for 10 min, 40 cycles at 95 C for 15 sec, and then 60 C for 1 min. Each PCR was performed at least in triplicate, and gene expression levels were determined with at least three biological repeats. For RT-PCR, the PCR conditions were 95 C for 5 min followed by 30 cycles at 94 C for 30 sec, 60 C for 30 sec and 72 C for 30 sec. The primers used for PCR are listed in Supplementary Table 1.
Lentivirus infection
For gene OE experiments, PSM-human Rb1 (Addgene, Cambridge, MA, USA), human papillomavirus E7 (Addgene), mouse Bbc3 (transOMIC technologies, Huntsville, AL, USA), mouse Cdkn1a (Addgene) and mouse Cdkn2a (p16; Addgene) were cloned into the CSII-EF-IRES2-Venus vector. For E2f1 OE, mouse E2f1 cDNA containing the full-length open reading frame was amplified by PCR and cloned into the CSII-EF-IRES2-Venus vector. Lentivirus particles were produced by transient transfection of 293T cells, and GS cells or testes cells were transfected as described previously [21]. CSII-EF-MCS-IRES2-Venus was used as a control. Titers of the virus were determined by transfecting 293T cells, and the MOIs were adjusted to 4.0. For gene KD experiments, KD vectors for Cdk4, Cdk6, Cdkn1b, E2f1 and Bbc3 were obtained from Open Biosystems (Huntsville, AL, USA). For Trp53 KD, we used pSicoR Trp53 and its control vector pSicoR (Addgene). For Cdkn1a KD, we used pFUGWH1-Cdkn1a and its control vector pFUGWH1 (a gift from Dr S Temple, Neural Stem Cell Institute, Rensselaer, NY, USA). A mixture of lentivirus particles was used to transfect GS cells from Rb1flox/flox or ROSA mice. pLKO1-Scramble shRNA was used as a control (Addgene). The lentivirus titer was determined using a Lenti-X p24 Rapid Titer Kit (Clontech, Mountain View, CA, USA). KD vectors from Open Biosystems are listed in Supplementary Table 2 (online only).
Analyses of recipient testes
Recipient mice were killed at the indicated time points, and their testes were analyzed by staining for β-galactosidase with 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal; Wako Pure Chemical Industries, Osaka, Japan) [19]. A germ cell cluster was defined as a colony when it occupied the entire basal surface of the tubule and was longer than 0.1 mm. For histological analysis, paraffin-embedded sections were stained with hematoxylin and eosin. The number of tubules with spermatogenesis, as defined by the presence of multiple layers of germ cells in the entire circumference of the tubules, was recorded for one section from each testis.
Western blotting
Samples were separated by SDS-PAGE and transferred onto Hybond-P membranes (GE Healthcare, Piscataway, NJ, USA). The membranes were incubated with primary antibodies. The antibodies used in the experiments are shown in Supplementary Table 3 (online only).
Southern blotting
Genomic DNA was digested with PstI and transferred and hybridized with the exon 18 probe [22], as described previously [16]. The PCR product was subsequently cloned into the pGEMT easy vector (Promega, Madison, WI, USA). The plasmid was then digested with EcoRI to produce a 492-bp fragment, which was used as a hybridization probe. Band intensity was quantified using the ImageJ 1.43r software (US National Institutes of Health, Bethesda, MD, USA).
Flow cytometry
For analysis of cell surface markers, GS cells were stained in phosphate-buffered saline (PBS)/1% fetal bovine serum (FBS) using the antibodies listed in Supplementary Table 3. For cell cycle analysis, GS cells were incubated with Hoechst 33342 (12.5 μg/ml; Sigma) for 30 min at 37 C and suspended in PBS/1% FBS. For quantification of cell cycle phases using combined propidium iodide (PI) and BrdU staining, GS cells were cultured in the presence of BrdU (50 μM; BD Biosciences, San Jose, CA, USA) for 6 h. After fixation by ethanol, cells were treated with HCl (2N) for 20 min at room temperature. The cells were then incubated with Alexa 647-conjugated anti-BrdU antibody for 30 min and treated with 5 μg/ml PI and 100 μg/ml RNaseA for 30 min before analysis. Stained cells were analyzed using a FACSCalibur (BD Biosciences).
Apoptosis assay
To perform a terminal deoxynucleotidyl transferase biotin-dUTP nick-end labeling (TUNEL) assay, a single cell suspension was concentrated on glass slides by centrifugation with Cytospin 4 (Thermo Electron, Altrincham, UK). After fixation in 4% paraformaldehyde for 1 h, cells were labeled using an In Situ Cell Death Detection Kit, TMR red (Roche Applied Science), according to the manufacturer’s protocol. The nuclei were counterstained with Hoechst 33342 (2 μg/ml; Sigma) to determine the percentage of TUNEL-positive nuclei relative to the total number of Hoechst 33342-stained nuclei. Apoptotic cells were quantified by collecting images of stained cells using the Photoshop software (Adobe Systems, San Jose, CA, USA).
Karyotype analysis
Cells were incubated with colcemid solution (60 ng/ml; KaryoMAX; Invitrogen) for 1 h, recovered by trypsin and treated with 75 mM KCl for 7 min. Metaphase spreads were prepared using a standard procedure after fixing the cells with methanol/acetic acid (3:1). The slides were stained with Hoechst 33342 (Sigma).
Comet assay
A comet assay was performed using a CometAssay Kit (Trevigen, Gaithersburg, MD, USA) according to the manufacturer’s protocol. Briefly, GS cells were cultured for 2 weeks after adenovirus infection and suspended in PBS (2 × 105 cells/ml). The cell suspension was mixed with CometAaay LM Agarose (Trevigen) at a ratio of 1:10, and 50 μl was transferred to a CometSlide (Trevigen) for immobilization. After incubation in lysis solution, cells were treated with an alkaline solution for 30 min followed by electrophoresis. DNA was stained with SYBR Green I (TaKaRa Bio, Otsu, Japan). DNA damage was quantified by measuring the length of the visible comet tail. Cells with a ratio greater than or equal to 2 were counted as positive for DNA damage. Figures show the percentage of cells positive for DNA damage per treatment.
Immunostaining
Testes samples were fixed in 4% paraformaldehyde for 2 h and then frozen in Tissue-Tek O.C.T. Compound (Sakura Finetechnical, Tokyo, Japan). For immunostaining, samples were treated with 0.1% Triton-X in PBS. After immersion in blocking buffer (0.1% Tween 20, 3% bovine serum albumin and 10% goat or donkey serum in PBS), samples were incubated with primary antibodies at 4 C overnight. Secondary antibodies were incubated for 1 h at room temperature. Samples were counterstained with Hoechst 33342 (Sigma). Images were collected using a confocal microscope (Fluoview FV1000D; Olympus, Tokyo, Japan). The antibodies used are listed in Supplementary Table 3.
Statistical analyses
The results are presented as means ± SEM. Significant differences between means for single comparisons were determined using the Student’s t-test. Multiple comparison analyses were performed using analysis of variance followed by Tukey’s honestly significant differences test.
Results
Depletion of Cdk4 impairs GS cell proliferation
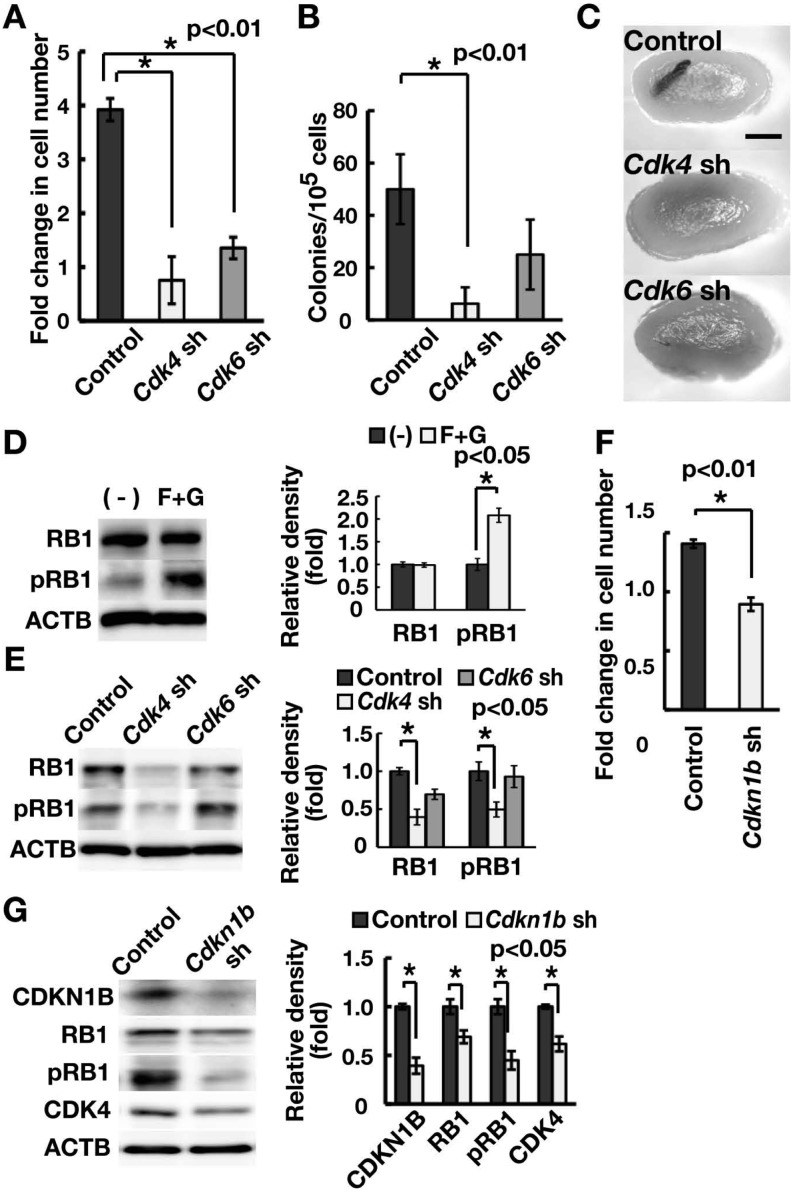
To examine the mechanism of the G1/S transition in GS cells, we first analyzed the role of Cdk4 and Cdk6, which are regulated by G1/S cyclins to drive the cell cycle. We transduced GS cells with lentiviruses expressing short hairpin RNA (shRNA) against Cdk4 or Cdk6. GS cells that had been depleted of either Cdk4 or Cdk6 proliferated more slowly compared to non-depleted controls (Fig. 1A; Supplementary Fig. 1A, online only). Since the SSC frequency in GS cell cultures is low (1–2%) [5], we confirmed the effect of gene depletion on SSCs based on germ cell transplantation into the seminiferous tubules of congenitally infertile W mice that lack endogenous spermatogenesis [23]. Analysis of the recipient testes showed that depletion of Cdk4, but not Cdk6, decreases SSC activity (Figs. 1B and C). The numbers of colonies generated by GS cells expressing shRNA against Cdk4, Cdk6 and the control luciferase gene were 6.3 ± 6.3, 25.0 ± 13.4 and 50.0 ± 13.4 per 105 cells (n = 8), respectively, suggesting that Cdk4 is responsible for the G1/S transition in SSCs.
Fig. 1.
Regulation of RB1 in spermatogonia. (A) Suppression of GS cell proliferation through depletion of Cdk4 or Cdk6. GS cells were infected with the indicated lentivirus expressing shRNA and passaged 4 days after transfection. Cell recovery was determined 6 days after passage (n = 4; MOI = 10). (B) Colony counts. Cells were transplanted 2 days after transfection. Results of two experiments (n = 8; MOI = 10). (C) Macroscopic appearance of recipient testes that received transplantation of GS cells depleted of Cdk4 or Cdk6. (D) Western blot analysis of RB1. GS cells were cultured without cytokines for 3 days and stimulated with FGF2 and GDNF. Cells were recovered 1 h after cytokine stimulation. Note the relative increase in phosphorylated RB1 (pRB1) caused by cytokine stimulation. The graph shows relative band intensity (n = 3). (E) Western blot analysis of RB1 following depletion of Cdk4 or Cdk6. Cells were recovered 4 days after transfection (MOI = 10). The graph shows relative band intensity (n = 3). (F) Suppression of GS cell proliferation through depletion of Cdkn1b. GS cells were infected with the indicated lentivrus, and cells were recovered 4 days after transfection (n = 4; MOI = 4). (G) Western blot analysis of cell cycle-related proteins following depletion of Cdkn1b. Cells were recovered 4 days after transfection (MOI = 4). The graph shows relative band intensity (n = 3). Bar = 1 mm (C).
We next examined the involvement of Rb1 family genes, the major target of the CDK4-CCND complex. RB1 was phosphorylated upon addition of GDNF and FGF2 (Fig. 1D), which promote GS cell proliferation. When we analyzed RB1 expression levels using Western blotting, Cdk4 depletion significantly decreased both the phosphorylated form and the total amount of RB1 (Fig. 1E).
We then examined the impact of Cdkn1b depletion on RB1 phosphorylation because CDKN1B is involved in stabilizing the CDK4/6-CCND complex in somatic cells [11]. In spermatogonia, we previously found (using serial transplantation) that SSC self-renewal is decreased in Cdkn1b knockout (KO) SSCs [24]. Consistently, Cdkn1b depletion inhibited GS cell proliferation and reduced the levels of RB1 and CDK4 in GS cells (Figs. 1F and G). These results suggested that Cdk4 and Cdkn1b play critical roles in regulating RB1 function in GS cells.
Rb1 deficiency impairs SSC activity
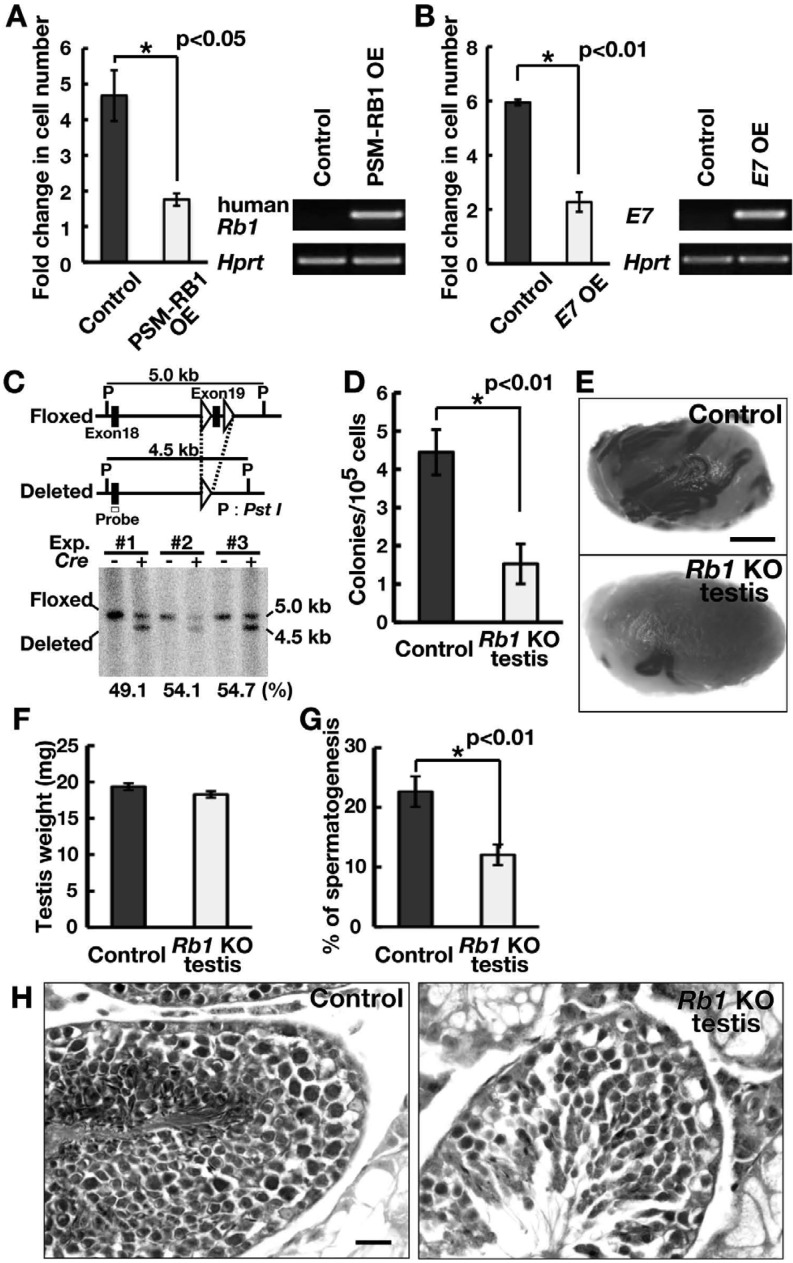
To examine the function of Rb1 in SSCs, we first used phosphorylation site-mutated RB1 (PSM-RB1), a constitutive active mutant of RB1 [25]. PSM-RB1 has been shown to cause G1 arrest and to interfere with S phase progression. We transfected GS cells with a lentivirus expressing PSM-RB1 and found that PSM-RB1 significantly reduced the GS cell number (Fig. 2A), suggesting that excessive activation of RB1 abrogates self-renewal of SSCs. To examine whether diminished RB1 influences self-renewal, we next used human papillomavirus E7 oncoprotein, which induces degradation of RB1 through the ubiquitin-proteasome pathway [26]. E7 OE significantly lowered GS cell proliferation compared with the control (Fig. 2B). These results suggested that appropriate activity levels of RB1 are important for driving self-renewal division.
Fig. 2.
Functional analysis of Rb1 in self-renewal division. (A) Suppression of GS cell proliferation following transfection of constitutively active RB1 (PSM-RB1). GS cells were infected with the indicated lentivirus and passaged 4 days after transfection. Cell recovery was determined 6 days after passage (n = 3; MOI = 10). OE was confirmed by RT-PCR 2 days after transfection. (B) Suppression of GS cell proliferation following transfection of E7. GS cells were infected with the indicated lentivirus and passaged 4 days after transfection. Cell recovery was determined 6 days after passage (n = 3; MOI = 10). OE was confirmed by RT-PCR 2 days after transfection. (C) Conditional mutant mice used in the experiment. Exon 19 of the Rb1 gene was deleted using Cre-mediated recombination. The indicated probe was used for Southern blot analysis. (D) Colony counts. Results of three experiments (n = 12). (E) Macroscopic appearance of recipient testis. (F) Testicular weight (n = 8). (G) Tubules with spermatogenesis, defined as the presence of multiple layers of germ cells in the entire circumference of the tubules. At least 169 tubules were counted (n = 7). (H) Histological appearance of recipient testis. Bar = 1 mm (E), 20 μm (H).
Based on these results, we used Rb1 floxed mice to confirm the role of Rb1 function on freshly isolated SSCs [14]. These mice were mated with R26R reporter mice to visualize the colonization of donor cells [15]. Testis cells were collected from 9-day-old pup testes and dissociated into single cells. Littermates were used as controls. The cells were then exposed to an AxCANCre overnight in vitro [16]. Southern blot analysis of the Cre-infected cells revealed that the deletion efficiency of Rb1 was 49.1–54.7% (with an average of 52.6%) at the time of cell recovery (Fig. 2C). The recovered cells were transplanted into W mice. Three separate experiments were performed involving 12 testes for each cell type.
Analysis of the recipient testes showed that the numbers of colonies produced by Rb1 KO and control testis cells were 1.5 ± 0.5 and 4.4 ± 0.6 (n = 12) per 105 cells, respectively (Fig. 2D and E), and the difference was significant. Although the weights of the recipient testes were comparable (Fig. 2F), histological analysis of the recipient testes revealed that the number of tubules with spermatogenesis significantly decreased in testes transplanted with Rb1 KO testis cells (Fig. 2G and H). These results suggest that Rb1 is required for SSC activity.
Rb1 deficiency increases apoptosis and cell cycle progression
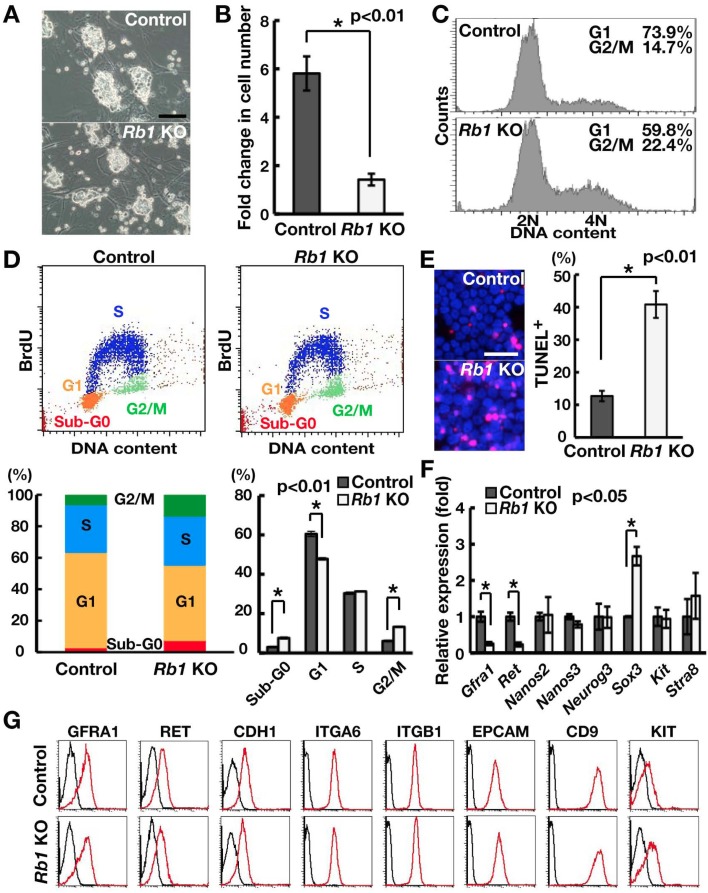
To characterize the mechanism of Rb1 function in SSC activity, we derived GS cells from Rb1flox/flox mice and examined their response upon Cre-mediated Rb1 deletion (Fig. 3A). Although we observed no apparent changes in colony morphology upon Cre-mediated gene deletion, we noted that CRE exposure significantly reduced cell recovery (Fig. 3B). Due to the close association of Rb1 with cell cycle progression, we performed flow cytometry to examine the impact of Rb1 deficiency on cell cycle progression (Fig. 3C). Analysis using Hoechst 33342 showed that Rb1 deficiency increases the number of GS cells at the G2/M phase of the cell cycle, and this is accompanied by a relative decrease in the G1 phase. Further analysis using BrdU staining confirmed these results (Fig. 3D). Since we observed an increase in the sub-G0 population by flow cytometry (Fig. 3D), we performed TUNEL staining to directly examine apoptosis levels (Fig. 3E). Our results showed that the frequency of TUNEL-positive (TUNEL+) cells increased significantly upon Rb1 deletion compared with the control.
Fig. 3.
Increased apoptosis and abnormal cell cycle progression of Rb1 KO GS cells. (A) Appearance of Rb1 KO GS cells. (B) Impaired proliferation of Rb1 KO GS cells (n = 3). Cells were recovered 2 weeks after AxCANCre transfection. (C) Analysis of cell cycle distribution using Hoechst 33342. Cells were analyzed 2 weeks after AxCANCre transfection. (D) Quantification of cell cycle phases by combined propidium iodide and BrdU staining. Cells were analyzed 2 weeks after AxCANCre transfection (n = 3). (E) Quantification of apoptotic cells using TUNEL staining (n = 3). At least 273 cells were counted 1 week after AxCANCre transfection. (F) Real-time PCR analysis of spermatogonia markers (n = 6). (G) Flow cytometric analysis of cell surface markers. Bar = 50 μm (A, E).
Because loss of SSC activity can occur due to increased differentiating divisions, we also examined the expression of several spermatogonia markers. Real-time PCR analysis showed that Rb1 deficiency significantly decreased the expression of Gfra1 and Ret, both of which comprise a GDNF receptor (Fig. 3F) [27]. However, we observed no apparent changes in cell surface marker expression using flow cytometry (Fig. 3G). We also detected increased expression of differentiation markers at the mRNA level. However, only Sox3 was expressed more strongly in Rb1 KO GS cells, and the remaining differentiation markers were not affected by Rb1 deficiency.
E2f1 OE decreases SSC activity
To characterize the molecules underlying the phenotype of Rb1 KO GS cells, we first analyzed the role of E2f1. E2F1 is released from RB1 when RB1 is phosphorylated by the CDK-cyclin complex in somatic cells [28]. In spermatogonia, previous studies using transgenic mice showed that E2f1 OE in vivo induces apoptosis of spermatogonia and severe testicular atrophy [29]. E2f1 KO mice also showed a gradual loss of spermatogenesis, which is thought to be caused by depletion of SSCs [30, 31].
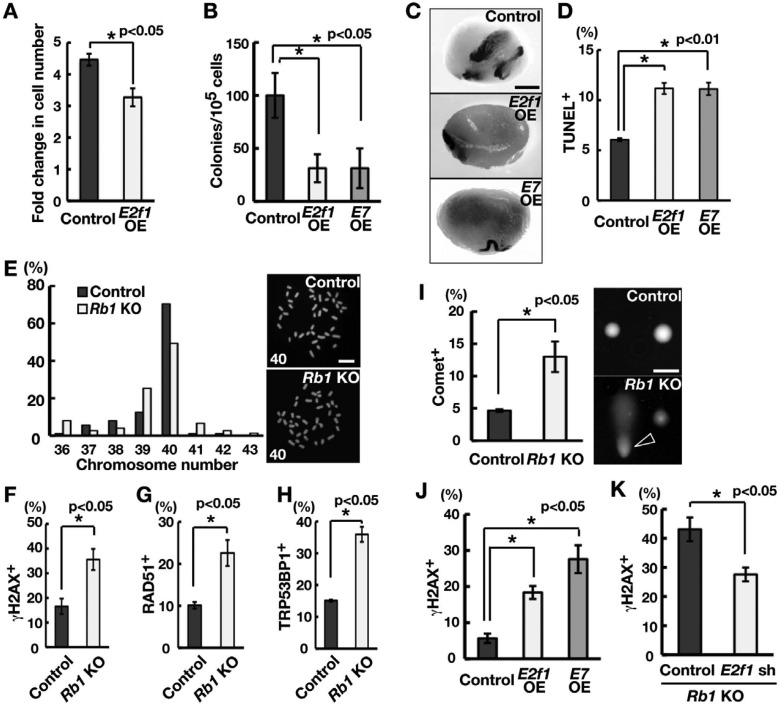
To examine the role of E2f1 in SSCs, we used GS cells. Transfection experiments showed that E2f1 OE reduced the number of GS cells (Fig. 4A; Supplementary Fig. 1B, online only). To directly examine whether E2f1 OE has any impact on SSCs, we transplanted E2f1-transfected GS cells into the seminiferous tubules of W mice. Analysis of the recipient mice showed that the numbers of colonies generated by E2f1 OE and control GS cells were 31.3 ± 13.2 and 100. 0 ± 21.1 per 105 cells (n = 8), respectively (Figs. 4B and C). These results indicate that excessive E2f1 activity reduces the SSC concentration in GS cell cultures.
Fig. 4.
Induction of DNA DSBs by E2F1 activation. (A) Suppression of GS cell proliferation following transfection of E2f1. GS cells were infected with the indicated lentivirus and passaged 4 days after transfection. Cell recovery was determined 6 days after passage (n = 3; MOI = 10). (B) Colony counts. Results of two experiments (n = 8). (C) Macroscopic appearance of recipient testis. (D) Quantification of apoptotic cells following E2f1 or E7 OE using TUNEL staining. At least 536 cells were counted 10 days after AxCANCre transfection. Results of four experiments (MOI = 10). (E) Karyotype analysis and metaphase spread of GS cells. Cells were analyzed 2 weeks after AxCANCre transfection (n = 88 for control; n = 75 for KO). The number in the metaphase spread indicates the chromosome number. (F–H) Quantification of γH2AX- (F), RAD51- (G) and TRP53BP1 (H)-positive cells of Rb1 KO GS cells. Cells were recovered 2 weeks after AxCANCre transfection (n =3). At least 338 (F), 401 (G) and 149 (H) cells were counted. Representative pictures are shown in Supplementary Fig. 2 (online only). (I) Comet assay. Data are expressed as the percent damage compared with the control sample. The arrowhead indicates a cell with DNA damage. (J) Immunocytochemistry of GS cells transfected with E2f1 or E7 for γH2AX. Cells were recovered 10 days after transfection (n = 3). At least 304 cells were counted. (K) Suppression of γH2AX staining following depletion of E2f1. Cells were co-transfected with AxCANCre and a lentivirus expressing shRNA against E2f1 and recovered 6 days after transfection (n = 3; MOI = 4). At least 252 cells were counted. Bar = 1 mm (C), 50 μm (E, I).
To understand the function of E2F1, we examined the apoptosis level of E2f1-transfected GS cells. TUNEL staining showed that E2f1 OE increases apoptosis of GS cells within 10 days (Fig. 4D). E2F1 is known to cause apoptosis through several pathways [32] and is considered a strong regulator of apoptosis after DNA damage in many cancers [32]. Therefore, although Rb1 KO GS cells contained a normal chromosome number (Fig. 4E), Rb1 deficiency may induce DNA DSBs by activating E2F1. To test this hypothesis, we performed immunocytochemistry of Rb1 KO GS cells. Analysis of Rb1 KO GS cells showed enhanced staining of DNA DSB markers, including γH2AX, RAD51 and TRP53BP1 (Figs. 4F–H; Supplementary Figs. 2A–C, online only). To further confirm whether the increase in γH2AX staining corresponded to DNA damage, we subjected Rb1 KO GS cells to a comet assay, which revealed increases in tail DNA content in Rb1 KO GS cells (Fig. 4I).
Increased γH2AX staining was also observed in E2f1-transfected GS cells (Fig. 4J). Because E2F1 is activated by Rb1 deficiency, we depleted E2f1 in Rb1 KO GS cells to examine whether E2f1 is required for DNA DSBs. Immunocytochemical staining showed that depletion of E2f1 could suppress increases in γH2AX staining in Rb1 KO GS cells (Fig. 4K; Supplementary Fig. 1A). These results suggested that activation of E2F1 by Rb1 deficiency causes DNA DSBs and apoptosis, thereby reducing SSC activity.
Rb1 deficiency activates Trp53 and increases pro-apoptotic gene expression
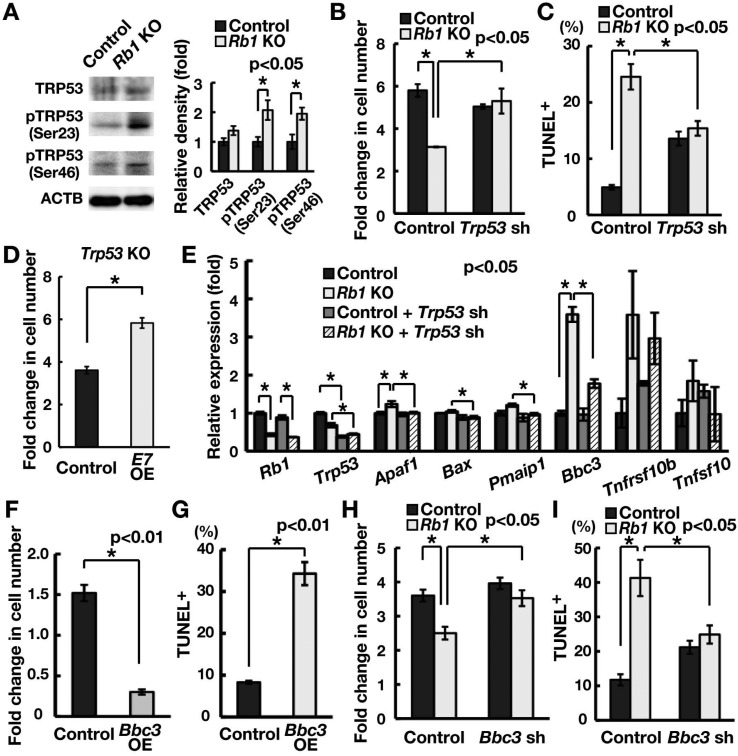
We next examined how Rb1 deficiency causes apoptosis and cell cycle arrest. We hypothesized that DNA damage induced by E2f1 OE activates TRP53. We first performed Western blot to examine TRP53 levels (Fig. 5A). Although Rb1 deficiency did not change the amount of total TRP53, it induced phosphorylation at Ser23 and Ser46, both of which have been implicated in the DNA damage response [33, 34]. Consistent with our hypothesis, depletion of Trp53 rescued the reduced recovery of GS cells upon Rb1 deletion (Fig. 5B). Because this result suggested activation of the TRP53-mediated apoptosis pathway, we examined the impact of Trp53 depletion on Rb1 KO GS cells and found that depletion of Trp53 by shRNA significantly suppressed apoptosis of Rb1 KO GS cells (Fig. 5C). Although combined deficiency of Trp53 and Rb1 can cause transformation of somatic cells [35], we found no clear evidence of transformed GS cells after depletion of Trp53. However, when we transfected Trp53 KO GS cells with E7, cells proliferated more actively (Fig. 5D), suggesting that Rb1 suppresses the proliferation of Trp53 KO GS cells.
Fig. 5.
Increased Bbc3 expression causes apoptosis of GS cells upon Rb1 deletion. (A) Western blot analysis of TRP53. Cells were recovered 3 days after AxCANCre transfection. The graph shows relative band intensity (n = 3). Note the relative increase in phosphorylated TRP53 (pTRP53) in Rb1 KO GS cells. (B) Increased recovery of Rb1 KO GS cells following depletion of Trp53. Cells were transfected with AxCANCre and a lentivirus expressing shRNA against Trp53 and recovered 1 week after infection (n = 3; MOI = 4). (C) Quantification of apoptotic cells using TUNEL staining following depletion of Trp53 (n = 3). Cells were transfected with AxCANCre and a lentivirus expressing shRNA against Trp53 (MOI = 4) and recovered 1 week after infection. At least 545 cells were counted in three experiments. (D) Increased cell recovery of E7-transfected Trp53 KO GS cells (n = 3; MOI = 10). Cells were transfected with a lentivirus expressing E7 and passaged 4 days after transfection. Cells were recovered 6 days after passage. (E) Real-time PCR analysis of apoptosis-related genes following Rb1 gene deletion. Cells were transfected with AxCANCre and a lentivirus expressing shRNA against Trp53, and recovered 3 days after infection (n = 3; MOI = 4). (F) Reduced GS cell recovery upon Bbc3 OE. GS cells were transfected with a lentivirus expressing Bbc3. Cells were recovered 4 days after transfection (n = 3; MOI = 10). (G) Quantification of apoptotic cells after Bbc3 OE using TUNEL staining (n = 3; MOI = 10). At least 239 cells were counted 4 days after transfection. (H) Increased recovery of Rb1 KO GS cells following depletion of Bbc3. Cells were transfected with AxCANCre and a lentivirus expressing shRNA against Bbc3 and recovered 1 week after infection (n = 6; MOI = 10). (I) Quantification of apoptosis of Rb1 KO cells using TUNEL staining after Bbc3 depletion. Cells were transfected with AxCANCre and a lentivirus expressing shRNA against Bbc3 and recovered 1 week after infection (n = 3; MOI = 10). At least 270 cells were counted.
To examine the mechanism of Trp53-induced apoptosis, we examined the expression of apoptosis-related genes that are regulated by Trp53. Our screening using real-time PCR revealed that Rb1 KO GS cells showed increased expression of Bbc3 (Fig. 5E). Although the level of Tnfrsf10b increased slightly, the increase was not significant. The increase in Bbc3 was inhibited by Trp53 depletion (Fig. 5E), confirming that it was activated by Trp53. To examine the function of Bbc3, we transfected Bbc3 into GS cells. Bbc3 OE decreased cell recovery and increased the number of cells undergoing apoptosis (Fig. 5F and G; Supplementary Fig. 1B). In contrast, Bbc3 depletion increased the cell recovery of Rb1 KO GS cells and reduced apoptosis levels (Fig. 5H and I; Supplementary Fig. 1A). These results suggested that upregulation of Bbc3 is responsible for the increased apoptosis caused by Rb1 deficiency.
Induction of Cdkn1a blocks cell cycle progression following induction of Rb1 deficiency
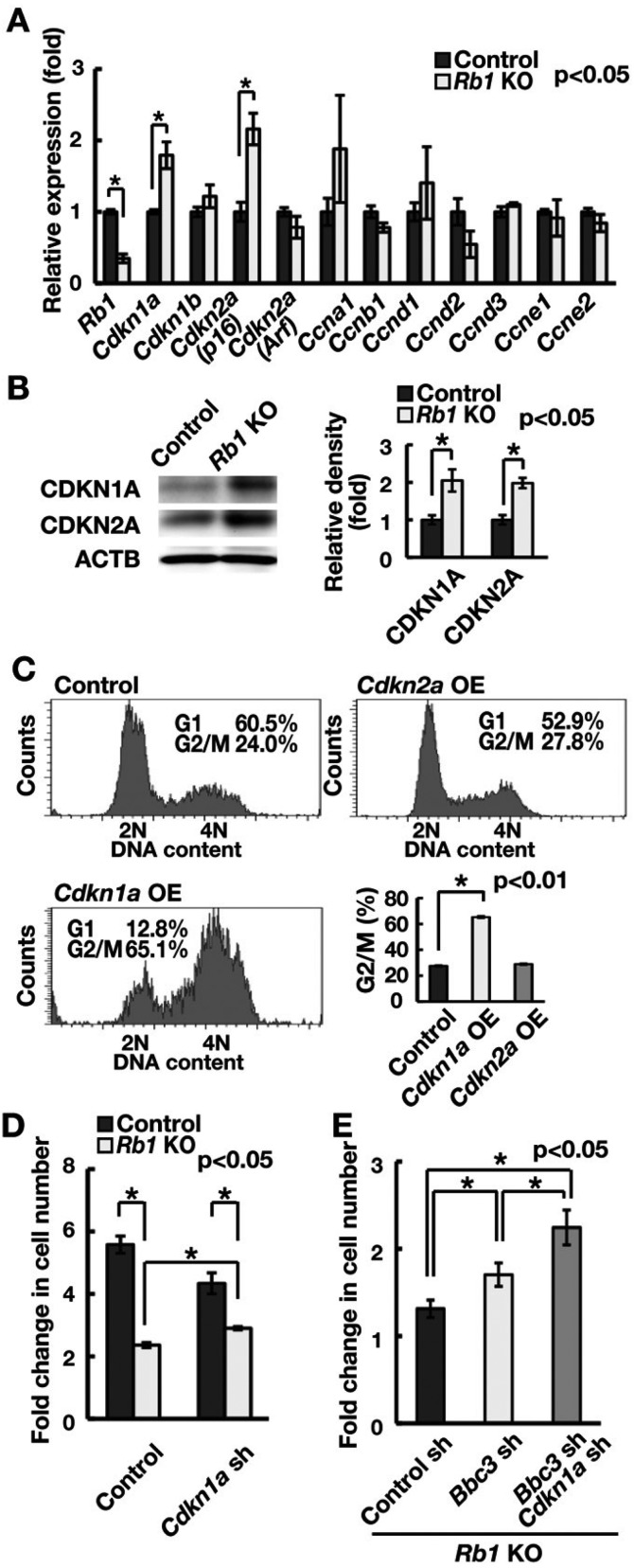
We next examined cell cycle arrest in Rb1 KO GS cells. When the expression of several cell cycle-related genes was examined using real-time PCR, no apparent changes were observed in cyclin expression (Fig. 6A). However, Rb1 KO GS cells showed increased expression of CDKIs Cdkn1a and Cdkn2a (p16), suggesting that upregulation of Cdkn1a and Cdkn2a (p16) caused cell cycle arrest. No increase in the levels of Cdkn2a (Arf), an important mediator of TRP53-dependent apoptosis induced by E2F1, was observed. Western blot analysis confirmed the increased expression of CDKN1A and CDKN2A (Fig. 6B). To explore whether Cdkn1a or Cdkn2a blocks the cell cycle of GS cells, GS cells were transfected with a lentivirus vector expressing either Cdkn1a or Cdkn2a. When Cdkn1a was transfected into GS cells, a significant number of cells were arrested at the G2/M phase (Fig. 6C; Supplementary Fig. 1B). Although Cdkn2a depletion similarly induced cell cycle arrest, its effect was not significant.
Fig. 6.
Suppression of the cell cycle by Cdkn1a induction following induction of Rb1 deficiency. (A) Real-time PCR analysis of cell cycle-related genes following Rb1 gene deletion. Cells were recovered 2 weeks after AxCANCre transfection (n = 3). (B) Western blot analysis of CDKI proteins following Rb1 gene deletion. Cells were recovered 2 weeks after AxCANCre transfection. The graph shows relative band intensity (n = 3). (C) Cell cycle analysis by Hoechst 33342 following Cdkn1a or Cdkn2a OE (MOI = 10). (D) Increased cell recovery of Rb1 KO GS cells following depletion of Cdkn1a. Cells were transfected with AxCANCre and a lentivirus expressing shRNA against Cdkn1a and recovered 1 week after infection (n = 3; MOI = 4). (E) Increased cell recovery of Rb1 KO GS cells following double depletion of Bbc3 and Cdkn1a. Cells were transfected with AxCANCre and a lentivirus expressing shRNA against the indicated genes and recovered 1 week after infection (n = 8; MOI = 4).
Because Cdkn1a is a Trp53 downstream gene, this result suggested that activation of the Trp53-Cdkn1a pathway blocks cell cycle progression in Rb1 KO GS cells. We examined this hypothesis by depleting Cdkn1a in Rb1 KO GS cells. Analysis of the cell count after transfection showed that depletion of Cdkn1a increased the cell recovery after induction of Rb1 deficiency (Fig. 6D; Supplementary Fig. 1A). Furthermore, double depletion of Bbc3 and Cdkn1a synergistically improved cell recovery upon induction of Rb1 deficiency (Fig. 6E). These results suggested that induction of Trp53 activation reduced cell recovery upon induction of Rb1 deficiency by upregulating Bbc3 and Cdkn1a levels.
Increased DNA damage in undifferentiated spermatogonia of Ddx4-Cre Rb1flox/– mice
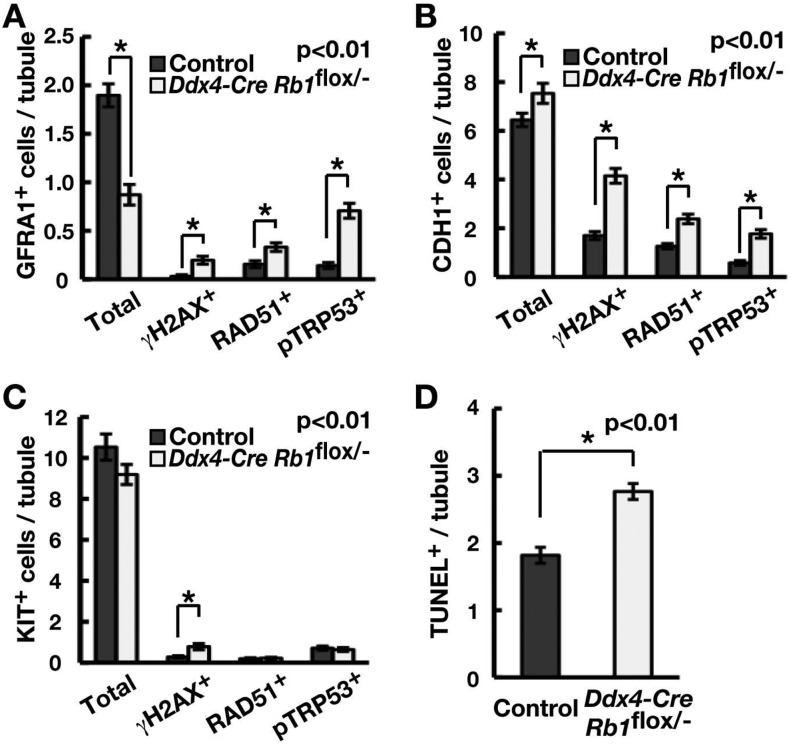
In the final set of experiments, we analyzed undifferentiated spermatogonia in 10-day-old Ddx4-Cre Rb1flox/– mice to examine the mechanism by which spermatogonia undergo depletion after prenatal loss of Rb1. We performed double immunohistochemistry using GFRA1, CDH1 and KIT. GFRA1 is highly expressed in As and Apr spermatogonia and gradually decreases in Aaligned(Aal) spermatogonia [36]. CDH1 is expressed in the total undifferentiated spermatogonia population [37], while KIT is expressed in differentiating spermatogonia.
The number of GFRA1+ cells was significantly decreased in Ddx4-Cre Rb1flox/– mice, as reported in a recent study [12], and this was accompanied by increased expression of γH2AX, RAD51 and pTRP53 (Fig. 7A; Supplementary Fig. 3A, online only). In contrast, CDH1-positive (CDH1+) cells were significantly increased in Ddx4-Cre Rb1flox/– mice, as reported previously [12], despite the increased amount of DNA DSB markers and pTRP53 (Fig. 7B; Supplementary Fig. 3B, online only). The number of KIT-expressing cells showed no significant changes, but γH2AX-positive (γH2AX+) cells significantly increased (Fig. 7C; Supplementary Fig. 3C, online only). As expected based on immunohistochemistry, the number of TUNEL+ cells significantly increased in Ddx4-Cre Rb1flox/– mice (Fig. 7D; Supplementary Fig. 3D, online only). These results suggest that prenatal loss of Rb1in vivo induces DNA damage and Trp53 activation, thereby increasing apoptosis and infertility in Ddx4-Cre Rb1flox/– mice.
Fig. 7.
Analysis of undifferentiated spermatogonia in Ddx4-Cre Rb1flox/– mice. (A–C) Immunohistochemical analysis of γH2AX, RAD51 or pTRP53 expression in GFRA1+ (A), CDH1+ (B) or KIT+ (C) cells in the 10-day-old Ddx4-Cre Rb1flox/– mouse testis. At least 50 tubules were counted in three testes. (D) TUNEL staining of the 10-day-old Ddx4-Cre Rb1flox/– mouse testis. At least 50 tubules were counted in three testes. Representative pictures are shown in Supplementary Fig. 3 (online only).
Discussion
This study was performed to increase our understanding of the G1/S transition in GS cells. We were interested in this process because we found that loss of Cdkn1b decreased self-renewal division and that Ccnd2 acts downstream of HRAS to promote self-renewal division [7, 24]. CDKN1B is known to suppress the cell cycle but also to play a critical role in forming the CDK4/6-CCND complex, whose primary target is RB1 [11]. Although Rb1 has been implicated in SSC self-renewal, the roles of Rb1 in SSCs remain unclear. Therefore, understanding the relationship between these molecules would provide valuable information on the mechanism of self-renewal division.
We first found that Cdk4 is responsible for SSC activity. Although depletion of Cdk4 or Cdk6 suppressed GS cell proliferation, only Cdk4 depletion showed significant effects on SSC activity. Reduced proliferation of Cdk4-depleted spermatogonia was consistent with the previous observation that Cdk4 KO mice show decreased spermatogenesis and gradual loss of fertility [38]. Although SSC activity was not examined in that study, the results of our transplantation assay confirmed that Cdk4 plays a role in SSCs. Since we previously demonstrated that self-renewal of SSCs by cytokines is mediated by Hras and Ccnd2 [7], these results suggest that CDK4 combines with CCND2 to drive SSC self-renewal.
A major target of the CDK4 kinase is RB1. RB1 phosphorylation was significantly reduced by depletion of Cdk4, but not Cdk6. Our analysis further showed that Cdkn1b depletion not only reduced GS cell proliferation but also RB1 and its phosphorylation levels. In many cases, RB1 dephosphorylation is associated with a reduction in RB1 [39]. Although kinase inhibitory protein (KIP) OE suppresses cell progression, a low level of KIP proteins is also known to be important for formation of the CDK4-CCND complex [11]. Because the amount of CDK4 protein is diminished by Cdkn1b depletion, the RB1 phosphorylation level is significantly decreased by poor CDK4-CCND complex formation and reduced CDK4 levels. However, note that Cdk4 and Cdkn1b may not function in a similar manner. In fact, unlike Cdk4 KO mice that have small testes, Cdkn1b KO mice have enlarged testes and are fertile [40]. The possibility exists that other KIP family molecules, such as CDKN1C, are involved in RB1 phosphorylation. However, our results strongly suggest that Cdkn1b is one of the major drivers of cell cycle progression in SSCs.
We also showed that Rb1 deficiency reduces colony formation after transplantation. Transfection of E7, which binds to RB1 and promotes its ubiquitin-mediated degradation [26], also resulted in poor colonization. These results are consistent with previous findings showing that transient depletion of Rb1 by siRNA in primary cultures of undifferentiated spermatogonia decreases colony formation after transplantation [13]. However, a critical difference between the current study and the previous study was the colony morphology. Rb1 depletion by siRNA produced large clumps of donor cells, and the cells were suggested to be malignant because colonies of them invaded interstitial cells. These changes occurred despite transient transfection of siRNA against Rb1, which suggested a strong growth inhibitory function of Rb1. Contrary to these observations, we did not observe apparent cancerous lesions in the current study. Although the cause of these discrepancies remains unknown, Rb1 inhibition by siRNA in the previous study may have influenced additional off-target genes and caused tumor-like cell formation. Alternatively, the differences may have been caused by the differences in the genetic backgrounds (C57BL/6 (B6) vs. 129/B6). Thus, although both studies support the importance of Rb1 in SSC self-renewal, the reasons for these discrepancies remain unclear.
Using GS cells, we examined the mechanism of reduced SSC activity. Although we observed increased expression of Sox3, whether SSCs were fully committed to differentiation was not clear because other differentiating spermatogonia marker levels did not change significantly. Rb1 deficiency induced DNA DSBs, which was confirmed based on increased expression of DNA damage proteins and a comet assay. Increased DNA damage protein expression was also found when E2f1 was overexpressed in GS cells. Because E2f1 depletion could decrease γH2AX expression in Rb1 KO GS cells, we speculate that Rb1 plays a critical role in genomic stability via regulation of E2F1. Although a direct causal relationship between RB1 and E2F1 has not been completely clarified in this study, the involvement of E2f1 in spermatogonial proliferation was previously reported; E2f1 deficiency results in spermatogonia depletion [30, 31], while E2f1 OE also impairs spermatogenesis [29]. However, how these changes occur in vivo and whether they influence SSCs remain unclear. Our results showed that E2f1 OE suppresses cell cycle progression by inducing DNA damage, which likely causes reduced colony formation. E2F1 stimulates DNA damage by activating DNA replication at inappropriate times [41]. Although E2F1 is known to support an apoptotic DNA damage response in Trp53-dependent and Trp53-independent pathways [42], partial rescue of Rb1 KO cells by Trp53 depletion suggests that DNA damage caused by the RB-E2F1 pathway activates Trp53. Based on these observations, we analyzed Trp53 downstream genes to explain the Rb1 KO GS cell phenotype.
We observed that an important characteristic of the Rb1 KO GS cell phenotype is cell cycle arrest, which was unexpected because Rb1 was previously shown to suppress the cell cycle progression of undifferentiated spermatogonia by increasing the expression of Ccnd2 and Ccne1 [13]. However, we found that GS cells upregulate Cdkn1a and Cdkn2a upon induction of Rb1 deficiency. Although Cdkn1a deficiency does not influence SSC self-renewal in vivo [24], Cdkn1a OE occurs in Atm deficient GS cells and inhibits their proliferation [43]. However, the role of Cdkn2a in SSCs remains unclear because no apparent reproductive phenotype has been reported in Cdkn2a KO mice. When we examined the impact of these CDKIs, suppression of Cdkn1a (but not Cdkn2a) resulted in increased cell recovery, suggesting that Cdkn1a is responsible for cell cycle arrest upon Rb1 deletion. Because depletion of Trp53, which suppresses Cdkn1a, also improved cell recovery, activation of the Trp53-Cdkn1a pathway is likely responsible for suppressing the cell cycle arrest that occurs upon induction of Rb1 deficiency.
Increased apoptosis was another feature of Rb1 KO GS cells. Our analysis revealed that Bbc3 is upregulated in Rb1 KO GS cells. Bbc3 OE caused increased apoptosis, while its depletion increased cell survival. This increase in apoptosis may explain why we observed no apparent chromosomal abnormalities in Rb1 KO GS cells. Genomic damage was also reported in both mouse and human embryonic stem (ES) cells that were deficient in Rb1. However, while we did not observe an increase in chromosome number, both mouse and human ES cells showed a high incidence of chromosomal abnormalities [44, 45]. In particular, human ES cells showed increased 4N cells, but Cdkn1a KD did not affect the genomic instability. Mouse ES cells may have adapted to the complete loss of function of Rb family members. In contrast to ES cells that often show trisomy in chromosome 8 or 11 [46, 47], GS cells are relatively stable as far as chromosomal abnormalities are concerned. We previously showed that GS cells maintain euploidy for at least 2 years [6], which is suggestive of stable genetic integrity. GS cells may be more susceptible to DNA damage, and those that have experienced severe damage may be eliminated by apoptosis instead of acquiring additional mutations before the occurrence of apparent chromosomal aberrations. Because ES cells do not show severe checkpoint responses [48], they likely easily acquire more damage, leading to increased chromosome abnormalities.
Increased DNA damage proteins in Ddx4-Cre Rb1flox/– mice showed that DNA damage was caused by Rb1 deficiency in vivo. Although the number of GFRA1+ spermatogonia decreased, an increased number of CDH1+ spermatogonia was observed. This result is in contrast to our observation of GS cells that showed apoptosis and blocked cell cycle progression. This difference may have occurred because Ddx4-Cre expression occurs prenatally at ~15.5 dpc [49]. The lack of Rb1 during the prenatal period likely influenced the maturation of germ cells into SSCs because transplantation experiments revealed that a dramatic increase in the SSC pool size occurs postnatally [50]. A previous study showed that Rb1 deficiency causes a delay in cell cycle arrest and increases in Cdkn1b and Cdkn2b CDKIs in gonocytes [51]. Based on these results, it is possible that the lack of cell cycle arrest in undifferentiated spermatogonia of Ddx4-Cre Rb1flox/– mice is caused by the developmental defect of gonocytes that prevents them from acquiring the ability to respond to the Trp53-induced damage response. Future experiments are required to confirm this hypothesis.
Overall, our results support the critical roles of the CDKN1B-CDK4/CCND2-RB1-E2F1 pathway in regulating SSC self-renewal (Supplementary Fig. 4, online only). Our analysis of cell cycle progression led to the identification of regulatory functions of Rb in genomic damage. Loss of Rb1 results in activation of Trp53, which induces Bbc3 and Cdkn1a, compromising self-renewal division. However, several questions remain to be addressed. First, the effect of Rb1 on SSC fate commitment remains unclear. The frequency of self-renewal and differentiating division is believed to occur at the same frequency in SSCs in a steady state [52]. In Ddx4-Cre Rb1flox/– mice, As spermatogonia exclusively differentiated into Apr spermatogonia and did not self-renew to form new As spermatogonia [12]. Although we observed increased expression of Sox3, we were not able to obtain evidence for the role of Rb1 in SSC differentiation. Second, the effect of E2F1 on genetic integrity has not been characterized. Although E2F1 is responsible, determining how activation of E2F1 leads to DNA damage in SSCs requires further study, and the molecular targets of E2F1 need to be identified. Finally, how E2F1 is regulated in SSCs remains unknown. Because E2f1 deficiency leads to spermatogonia depletion in E2f1 KO mice, the activity of E2F1 requires tight regulation to assure genetic integrity and cell cycle progression. In addition to regulation by Rb1, other molecules may control or modify E2F1 function, such as Myc and Foxo, which are known to influence SSCs. Thus, characterizing the molecular networks surrounding Rb1 will increase our understanding of how SSCs determine the choice between self-renewal and differentiating divisions and how germ cells maintain genetic integrity.
Supplementary
Acknowledgments
We thank Ms Y Ogata for technical assistance.
References
- 1.de Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J Androl 2000; 21: 776–798. [PubMed] [Google Scholar]
- 2.Meistrich ML, van Beek MEAB. Spermatogonial stem cells. In: Desjardins C, Ewing LL (eds.), Cell and Molecular Biology of the Testis. New York: Oxford University Press; 1993: 266–295.
- 3.Nagano M, Avarbock MR, Brinster RL. Pattern and kinetics of mouse donor spermatogonial stem cell colonization in recipient testes. Biol Reprod 1999; 60: 1429–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogawa T, Ohmura M, Yumura Y, Sawada H, Kubota Y. Expansion of murine spermatogonial stem cells through serial transplantation. Biol Reprod 2003; 68: 316–322. [DOI] [PubMed] [Google Scholar]
- 5.Kanatsu-Shinohara M, Ogonuki N, Inoue K, Miki H, Ogura A, Toyokuni S, Shinohara T. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod 2003; 69: 612–616. [DOI] [PubMed] [Google Scholar]
- 6.Kanatsu-Shinohara M, Ogonuki N, Iwano T, Lee J, Kazuki Y, Inoue K, Miki H, Takehashi M, Toyokuni S, Shinkai Y, Oshimura M, Ishino F, Ogura A, Shinohara T. Genetic and epigenetic properties of mouse male germline stem cells during long-term culture. Development 2005; 132: 4155–4163. [DOI] [PubMed] [Google Scholar]
- 7.Lee J, Kanatsu-Shinohara M, Morimoto H, Kazuki Y, Takashima S, Oshimura M, Toyokuni S, Shinohara T. Genetic reconstruction of mouse spermatogonial stem cell self-renewal in vitro by Ras-cyclin D2 activation. Cell Stem Cell 2009; 5: 76–86. [DOI] [PubMed] [Google Scholar]
- 8.Oatley JM, Avarbock MR, Brinster RL. Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J Biol Chem 2007; 282: 25842–25851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartkova J, Rajpert-De Meyts E, Skakkebaek NE, Lukas J, Bartek J. Deregulation of the G1/S-phase control in human testicular germ cell tumours. APMIS 2003; 111: 252–265, discussion :265–266. [DOI] [PubMed] [Google Scholar]
- 10.Goriely A, Hansen RM, Taylor IB, Olesen IA, Jacobsen GK, McGowan SJ, Pfeifer SP, McVean GA, Rajpert-De Meyts E, Wilkie AO. Activating mutations in FGFR3 and HRAS reveal a shared genetic origin for congenital disorders and testicular tumors. Nat Genet 2009; 41: 1247–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinberg RA. pRB and control of the cell cycle clock. In: Weinberg RA (ed.), The Biology of Cancer. New York: Garland Science; 2007: 255–306.
- 12.Hu YC, de Rooij DG, Page DC. Tumor suppressor gene Rb is required for self-renewal of spermatogonial stem cells in mice. Proc Natl Acad Sci USA 2013; 110: 12685–12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang QE, Gwost I, Oatley MJ, Oatley JM. Retinoblastoma protein (RB1) controls fate determination in stem cells and progenitors of the mouse male germline. Biol Reprod 2013; 89: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev 2000; 14: 994–1004. [PMC free article] [PubMed] [Google Scholar]
- 15.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 1999; 21: 70–71. [DOI] [PubMed] [Google Scholar]
- 16.Takehashi M, Kanatsu-Shinohara M, Inoue K, Ogonuki N, Miki H, Toyokuni S, Ogura A, Shinohara T. Adenovirus-mediated gene delivery into mouse spermatogonial stem cells. Proc Natl Acad Sci USA 2007; 104: 2596–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogawa T, Aréchaga JM, Avarbock MR, Brinster RL. Transplantation of testis germinal cells into mouse seminiferous tubules. Int J Dev Biol 1997; 41: 111–122. [PubMed] [Google Scholar]
- 18.Kanatsu-Shinohara M, Ogonuki N, Inoue K, Ogura A, Toyokuni S, Honjo T, Shinohara T. Allogeneic offspring produced by male germ line stem cell transplantation into infertile mouse testis. Biol Reprod 2003; 68: 167–173. [DOI] [PubMed] [Google Scholar]
- 19.Kanatsu-Shinohara M, Inoue K, Ogonuki N, Morimoto H, Ogura A, Shinohara T. Serum- and feeder-free culture of mouse germline stem cells. Biol Reprod 2011; 84: 97–105. [DOI] [PubMed] [Google Scholar]
- 20.Kanatsu-Shinohara M, Inoue K, Lee J, Yoshimoto M, Ogonuki N, Miki H, Baba S, Kato T, Kazuki Y, Toyokuni S, Toyoshima M, Niwa O, Oshimura M, Heike T, Nakahata T, Ishino F, Ogura A, Shinohara T. Generation of pluripotent stem cells from neonatal mouse testis. Cell 2004; 119: 1001–1012. [DOI] [PubMed] [Google Scholar]
- 21.Kanatsu-Shinohara M, Muneto T, Lee J, Takenaka M, Chuma S, Nakatsuji N, Horiuchi T, Shinohara T. Long-term culture of male germline stem cells from hamster testes. Biol Reprod 2008; 78: 611–617. [DOI] [PubMed] [Google Scholar]
- 22.Vooijs M, van der Valk M, te Riele H, Berns A. Flp-mediated tissue-specific inactivation of the retinoblastoma tumor suppressor gene in the mouse. Oncogene 1998; 17: 1–12. [DOI] [PubMed] [Google Scholar]
- 23.Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proc Natl Acad Sci USA 1994; 91: 11298–11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanatsu-Shinohara M, Takashima S, Shinohara T. Transmission distortion by loss of p21 or p27 cyclin-dependent kinase inhibitors following competitive spermatogonial transplantation. Proc Natl Acad Sci USA 2010; 107: 6210–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knudsen KE, Weber E, Arden KC, Cavenee WK, Feramisco JR, Knudsen ES. The retinoblastoma tumor suppressor inhibits cellular proliferation through two distinct mechanisms: inhibition of cell cycle progression and induction of cell death. Oncogene 1999; 18: 5239–5245. [DOI] [PubMed] [Google Scholar]
- 26.Boyer SN, Wazer DE, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res 1996; 56: 4620–4624. [PubMed] [Google Scholar]
- 27.Sariola H, Saarma M. Novel functions and signalling pathways for GDNF. J Cell Sci 2003; 116: 3855–3862. [DOI] [PubMed] [Google Scholar]
- 28.Müller H, Helin K. The E2F transcription factors: key regulators of cell proliferation. Biochim Biophys Acta 2000; 1470: M1–M12. [DOI] [PubMed] [Google Scholar]
- 29.Agger K, Santoni-Rugiu E, Holmberg C, Karlström O, Helin K. Conditional E2F1 activation in transgenic mice causes testicular atrophy and dysplasia mimicking human CIS. Oncogene 2005; 24: 780–789. [DOI] [PubMed] [Google Scholar]
- 30.Hoja MR, Liu JG, Mohammadieh M, Kvist U, Yuan L. E2F1 deficiency impairs murine spermatogenesis and augments testicular degeneration in SCP3-nullizygous mice. Cell Death Differ 2004; 11: 354–356. [DOI] [PubMed] [Google Scholar]
- 31.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson NJ. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell 1996; 85: 537–548. [DOI] [PubMed] [Google Scholar]
- 32.Pützer BM, Engelmann D. E2F1 apoptosis counterattacked: evil strikes back. Trends Mol Med 2013; 19: 89–98. [DOI] [PubMed] [Google Scholar]
- 33.Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, Taya Y. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 2000; 102: 849–862. [DOI] [PubMed] [Google Scholar]
- 34.Wu Z, Earle J, Saito S, Anderson CW, Appella E, Xu Y. Mutation of mouse p53 Ser23 and the response to DNA damage. Mol Cell Biol 2002; 22: 2441–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 2003; 4: 181–189. [DOI] [PubMed] [Google Scholar]
- 36.Grisanti L, Falciatori I, Grasso M, Dovere L, Fera S, Muciaccia B, Fuso A, Berno V, Boitani C, Stefanini M, Vicini E. Identification of spermatogonial stem cell subsets by morphological analysis and prospective isolation. Stem Cells 2009; 27: 3043–3052. [DOI] [PubMed] [Google Scholar]
- 37.Tokuda M, Kadokawa Y, Kurahashi H, Marunouchi T. CDH1 is a specific marker for undifferentiated spermatogonia in mouse testes. Biol Reprod 2007; 76: 130–141. [DOI] [PubMed] [Google Scholar]
- 38.Rane SG, Dubus P, Mettus RV, Galbreath EJ, Boden G, Reddy EP, Barbacid M. Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in beta-islet cell hyperplasia. Nat Genet 1999; 22: 44–52. [DOI] [PubMed] [Google Scholar]
- 39.Broude EV, Swift ME, Vivo C, Chang BD, Davis BM, Kalurupalle S, Blagosklonny MV, Roninson IB. p21(Waf1/Cip1/Sdi1) mediates retinoblastoma protein degradation. Oncogene 2007; 26: 6954–6958. [DOI] [PubMed] [Google Scholar]
- 40.Beumer TL, Kiyokawa H, Roepers-Gajadien HL, van den Bos LA, Lock TM, Gademan IS, Rutgers DH, Koff A, de Rooij DG. Regulatory role of p27kip1 in the mouse and human testis. Endocrinology 1999; 140: 1834–1840. [DOI] [PubMed] [Google Scholar]
- 41.Pickering MT, Kowalik TF. Rb inactivation leads to E2F1-mediated DNA double-strand break accumulation. Oncogene 2006; 25: 746–755. [DOI] [PubMed] [Google Scholar]
- 42.Croxton R, Ma Y, Song L, Haura EB, Cress WD. Direct repression of the Mcl-1 promoter by E2F1. Oncogene 2002; 21: 1359–1369. [DOI] [PubMed] [Google Scholar]
- 43.Takubo K, Ohmura M, Azuma M, Nagamatsu G, Yamada W, Arai F, Hirao A, Suda T. Stem cell defects in ATM-deficient undifferentiated spermatogonia through DNA damage-induced cell-cycle arrest. Cell Stem Cell 2008; 2: 170–182. [DOI] [PubMed] [Google Scholar]
- 44.Conklin JF, Baker J, Sage J. The RB family is required for the self-renewal and survival of human embryonic stem cells. Nat Commun 2012; 3: 1244. [DOI] [PubMed] [Google Scholar]
- 45.Zheng L, Flesken-Nikitin A, Chen PL, Lee WH. Deficiency of Retinoblastoma gene in mouse embryonic stem cells leads to genetic instability. Cancer Res 2002; 62: 2498–2502. [PubMed] [Google Scholar]
- 46.Liu X, Wu H, Loring J, Hormuzdi S, Disteche CM, Bornstein P, Jaenisch R. Trisomy eight in ES cells is a common potential problem in gene targeting and interferes with germ line transmission. Dev Dyn 1997; 209: 85–91. [DOI] [PubMed] [Google Scholar]
- 47.Longo L, Bygrave A, Grosveld FG, Pandolfi PP. The chromosome make-up of mouse embryonic stem cells is predictive of somatic and germ cell chimaerism. Transgenic Res 1997; 6: 321–328. [DOI] [PubMed] [Google Scholar]
- 48.Aladjem MI, Spike BT, Rodewald LW, Hope TJ, Klemm M, Jaenisch R, Wahl GM. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr Biol 1998; 8: 145–155. [DOI] [PubMed] [Google Scholar]
- 49.Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 2007; 45: 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shinohara T, Orwig KE, Avarbock MR, Brinster RL. Remodeling of the postnatal mouse testis is accompanied by dramatic changes in stem cell number and niche accessibility. Proc Natl Acad Sci USA 2001; 98: 6186–6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spiller CM, Wilhelm D, Koopman P. Retinoblastoma 1 protein modulates XY germ cell entry into G1/G0 arrest during fetal development in mice. Biol Reprod 2010; 82: 433–443. [DOI] [PubMed] [Google Scholar]
- 52.de Rooij DG. Proliferation and differentiation of spermatogonial stem cells. Reproduction 2001; 121: 347–354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.