Abstract
Alkaline phosphatase (AP) is a key enzyme that enables marine phytoplankton to scavenge phosphorus (P) from dissolved organic phosphorus (DOP) when inorganic phosphate is scarce in the ocean. Yet how the AP gene has evolved in phytoplankton, particularly dinoflagellates, is poorly understood. We sequenced full-length AP genes and corresponding complementary DNA (cDNA) from 15 strains (10 species), representing four classes of the core dinoflagellate lineage, Gymnodiniales, Prorocentrales, Suessiales, and Gonyaulacales. Dinoflagellate AP gene sequences exhibited high variability, containing variable introns, pseudogenes, single nucleotide polymorphisms and consequent variations in amino acid sequence, indicative of gene duplication events and consistent with the “birth-and-death” model of gene evolution. Further sequence comparison showed that dinoflagellate APs likely belong to an atypical type AP (PhoAaty), which shares conserved motifs with counterparts in marine bacteria, cyanobacteria, green algae, haptophytes, and stramenopiles. Phylogenetic analysis suggested that PhoAaty probably originated from an ancestral gene in bacteria and evolved divergently in marine phytoplankton. Because variations in AP amino acid sequences may lead to differential subcellular localization and potentially different metal ion requirements, the multiple types of APs in algae may have resulted from selection for diversifying strategies to utilize DOP in the P variable marine environment.
Keywords: dinoflagellate, alkaline phosphatase, gene duplication, evolution, ecological strategy
Introduction
Alkaline phosphatase (AP) is a hydrolase [EC 3.1.3.1] that releases inorganic phosphate (Pi) from various types of phosphoester molecules. In the marine ecosystem, AP plays a critical role as it enables bacteria and phytoplankton to meet the P-nutrient requirement from dissolved organic phosphorus (DOP) when the energetically preferred dissolved inorganic phosphate (DIP) is scarce. As such, AP activity has widely been adopted as an indicator of DIP limitation (Kolowith et al., 2001; Dyhrman et al., 2007). Many studies have been carried out to identify and characterize the AP gene in marine microorganisms, leading to the categorization of three types of APs in marine prokaryotes, but APs in marine eukaryotic phytoplankton have remained largely uncategorized.
The three types of APs documented in prokaryotes, PhoAEC, PhoX, and PhoD, share weak sequence homology with each other and exhibit different metal ion requirements for their active sites, different subcellular localizations and various substrate preferences (Luo et al., 2009; Sebastián and Ammerman, 2009; White, 2009; Kathuria and Martiny, 2011). Typical AP, usually referred to as PhoAEC, was among the first isolated from E. coli and characterized as homodimeric phosphomonoesterase with phosphomonoesters as the major substrate but also showing lower activity on phosphate diesters (Zalatan et al., 2006). Yet many other enzymes (e.g., phosphodiesterase and phosphoglycerate mutases) with a similar mechanistic function as PhoAEC exist in a wide range of organisms. Because their folded protein structures share a conserved bimetal binding core (2Zn-Mg), PhoAEC and these other metalloenzymes are grouped into an AP superfamily, which has been adopted as a model to study the relationship between evolutionary driving forces and divergent enzymatic functions (Galperin and Jedrzejas, 2001; Zalatan et al., 2006). The promiscuity of PhoAEC for substrates (both monoester and diester) is suggested to have resulted from selective advantages in evolutionary optimization through gene duplication of an ancestral phosphohydrolase gene (Zalatan et al., 2006).
PhoD and PhoX are also able to hydrolyze both monoesters and diesters, and have been reported to be dependent on Ca and Ca-Fe, respectively (Zaheer et al., 2009; Kageyama et al., 2011; Sebastian and Ammerman, 2011; Yong et al., 2014). Contrary to the limited reports of PhoAEC amongst marine microbial communities (Luo et al., 2009, 2010), the Ca-dependent PhoX and PhoD appear to be much more common, presumably an adaption to Zn limitation in the ocean (Sebastian and Ammerman, 2011). Comparative genomic data reveals five different families of phosphatases in different cyanobacterial strains, amongst which there is an atypical PhoA-like AP, PhoAaty, so named because it is distantly related to PhoAEC (Ray et al., 1991; Moore et al., 2005; Orchard et al., 2009; Scanlan et al., 2009).
Growing genomic data provides evidence for the presence of different types of AP genes in eukaryotic phytoplankton (Armbrust et al., 2004; Dyhrman et al., 2012; Lin et al., 2012a, 2013), though only some of these have ever been (partially) characterized through gene isolation and expression. Putative phoX genes have been identified in the green algae, Volvox carteri and Chlamydomonas reinhardtii (Quisel et al., 1996; Hallmann, 1999; Moseley et al., 2006). Moreover, there are several novel (uncategorized) types of APs, e.g., EHAP1 of the haptophyte Emiliania huxleyi (Xu et al., 2006), an apparently Ca-dependent putative AP from the pelagophyte Aureoumbra lagunensis (Sun et al., 2012) and the diatom Phaeodactylum tricornutum (Bowler et al., 2008; Lin et al., 2013), and AP genes isolated from dinoflagellates (Lin et al., 2011, 2012a,b; Morey et al., 2011). Because of their highly divergent sequences, there is an increasing need for structural and functional classification of these APs. Besides, the evolutionary mechanism underlying the high variability of these diverse APs in marine phytoplankton needs to be characterized.
In this study, we attempted to classify APs in eukaryotic phytoplankton and gain an understanding of how the AP gene in this group of organisms has evolved, using dinoflagellates as a case study. We analyzed AP genes from 15 strains, representing four classes of the core dinoflagellates, Gymnodiniales Prorocentrales, Suessilaes, and Gonyaulacales. Existing AP sequences from both prokaryotic and eukaryotic phytoplankton were also collected from various public databases for phylogenetic analysis and sequence comparison. Our analyses indicated that dinoflagellate APs were only similar to PhoAaty-like APs reported in cyanobacteria, and that they have undergone gene duplication within species. We also discuss the evolutionary relationships amongst the multiple types of APs found in marine phytoplankton.
Materials and methods
Algal culture
Fifteen dinoflagellate strains used in this study (Table 1) were provided by the Center for Collections of Marine Bacteria and Phytoplankton, Xiamen University (CCMBP), Provasoli-Guillard National Center for Marine Algae and Microbiota (NCMA), and Jinan University. These strains were cultured in sterilized oceanic seawater (filtered through 0.22 μm pore size filters, 30 psu) enriched with the full nutrient regime of the L1 or f/2 medium (Guillard and Ryther, 1962; Guillard, 1975; Guillard and Hargraves, 1993) or the same nutrient regime except for the reduced phosphate concentration (2 μM), in a temperature controlled incubator at 15 or 20°C and under a 12:12 or 14:10 light dark cycle with a photon flux of 100 μE m−2 S−1. The identity of each strain was confirmed by amplifying the ITS ribosomal intergenic spacer region using 18ScomF-3end and com28SR1 primer pair (Wang et al., 2014).
Table 1.
Summary of the dinoflagellate AP characteristics.
| Class | Species | Gene (Protein) | ORF (bp) | Amino acid | Signal peptide | GPI anchor | Intron | Protein variants | Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| Gymnodiniales | Amphidinium carterae CCMP1314a | acaap (ACAAP) | 2115 | 704 | + | + | − | 1 | Extracellular |
| Karenia brevis CCMP2229a | kbrap (KBRAP) | 2061 | 686 | + | + | − | 6 | Extracellular | |
| Karenia mikimotoi C32-HK | kmiap (KMIAP) | 2043 | 680 | + | + | + | 1 | Cytoplasmic | |
| Prorocentrales | Prorocentrum minimum (Pavilard) Schiller 1933 CCMA15 | pmnap (PMNAP) | 2094 | 697 | + | + | + | 3 | Extracellular |
| Prorocentrum donghaienseb CCMA190 | pdoap (PDOAP) | 2127 | 709 | + | + | / | / | Extracellular | |
| Prorocentrum micans CCMA 143c | pmicap (PMICAP) | 1463 | 487 | / | / | / | 5 | / | |
| Suessiales | Symbiodinium sp. Clade E CCMA192 | symEap | 2004 | 667 | + | + | − | 1 | Extracellular |
| Symbiodinium kawagutii CCMP2468b Clade F | skaap (SKAAP) | 1725 | 574 | − | − | / | / | Extracellular | |
| Gonyaulacales | Alexandrium catenella (Clade IIC) ACHK-NTa | alecaap (ALECAAP) | 2181 | 726 | + | + | − | 1 | Extracellular |
| Alexandrium catenella (Clade IIC) ACHK-Tc | / | 1 | Extracellular | ||||||
| Alexandrium catenella (Clade IIC) ATDH02c | / | 1 | Extracellular | ||||||
| Alexandrium catenella (Clade IIC) ATMJ01c | / | 1 | Extracellular | ||||||
| Alexandrium catenella (Clade IIC) ATCI01c | ATCI01-ap | 2181/2193 | 726/730 | + | + | / | 6 | Extracellular | |
| Alexandrium tamarense (Clade IIB) CCAP1119/1 | ataap (ATAAP) | 2055/2160/2172/2184 | 684/719/723/727 | + | + | − | 36 | Extracellular, Cytoplasmic, Chloroplast, | |
| Alexandrium fundyense (Clade I) CCMP1719 | afuap (AlefuAP) | 2124/2136/2142/2166 | 707/711/713/721 | + | + | − | 11 | Plasma membrane |
Nucleic acid isolation and cDNA synthesis
Cells were collected from cultures during both the nutrient-replete exponential phase (genomic DNA extraction) and the phosphorus-depleted stationary phase (total RNA extraction) by centrifugation at 4000 × g for 15 min at 4°C. Cell pellets were re-suspended in DNA lysis buffer (10 mM Tris pH 8.0; 100 mM EDTA pH 8.0; 0.5% (w/v) SDS; 100 μg/ml proteinase K) for genomic DNA (gDNA) isolation and Tri-Reagent for total RNA isolation. Extraction of genomic DNA and total RNA was performed as previously reported (Zhang and Lin, 2005). The 1st-strand cDNA was synthesized from 300 ng of the total RNA using M-MLV reverse transcriptase (Promega, Madison, WI, USA) with a modified 454BT7-dT primer (Supplementary Table 1).
Amplification of the full-length alkaline phosphatase cDNA and genomic DNA (gDNA) sequences
Degenerate primers DinoAPNF4a/b and DinoAPNR6/7a/7b (Supplementary Table 1) were used to obtain a fragment of the AP gene from either the gDNA or cDNA of different dinoflagellate species following previously published PCR conditions (Lin et al., 2012a). Amplified fragments were cloned into a T-vector and sequenced using the Sanger method (at least 10 clones for each sample). Once this partial sequence was annotated as an alkaline phosphatase gene using BLASTX against the GenBank non-redundant protein database, gene specific primers were designed to amplify both the 5′ and 3′ cDNA ends using the rapid amplification of cDNA ends (RACE) technique. In general, the reverse primers were paired with DinoSL (a unique 22-nt spliced leader only exists at the 5′ end of the mature mRNA transcript in dinoflagellate) to perform the 5′ RACE, and the forward primers were paired with 454BT7 (the adaptor attached after dT used for cDNA synthesis) to perform the 3′ RACE (Zhang et al., 2007; Lin et al., 2011). Then, the specific primers (Supplementary Table 1) were designed based on the sequence of 5′ UTR and 3′ UTR regions to amplify full-length ORFs of each AP gene from cDNA and gDNA. All the sequences acquired in this study have been deposited in GenBank under the accession numbers KT274032-KT274169.
Analyses of introns, single nucleotide polymorphisms, and deduced protein sequences
Introns were detected by comparing the AP coding gene sequence of gDNA with the corresponding cDNA. Exon-intron boundaries were recorded in each case. In analyzing sequence polymorphisms, we took into account potential nucleotide bias caused by PCR and sequencing, and PCR products were purified and directly sequenced on the one hand and sequenced after cloning on the other (>10 clones per sample typically but >20 clones for P. minimum). Only sequences containing three or more mutations or those in which the same mutation occurred in more than two clones was accepted as a true sequence variant. Polymorphism analysis and Tajima's D-test for neutral mutation were conducted using DnaSP V 5.10.1 (Librado and Rozas, 2009).
A set of online programs was used to characterize the deduced protein as previously reported (Lin et al., 2011). Briefly, the signal peptide was predicted using SignalP V4.1 (Petersen et al., 2011). GPI-SOM (Fankhauser and Mäser, 2005) and PredGPI (Pierlenoi et al., 2008) were used to locate the GPI (glycosylphosphatidylinositol) anchor in the deduced protein sequence. The subcellular localization of AP was predicted using Cello (Yu et al., 2006) with the prediction in highest score presented in Table 1.
Phylogenetic analysis
Comparisons of amino acid sequences were performed between Dino-APs (alkaline phosphatase of dinoflagellates) and all reported phosphatases, including PhoAEC, PhoD, PhoX, phosphodiesterase, and other putatively assigned phosphatase genes identified in marine phytoplankton. Dino-APs were subjected to BLAST analysis against the GenBank, JGI (Joint Genome Institute) genome databases and a comprehensive algal EST datasets collected by our research group to retrieve potential homologs from marine phytoplankton. To examine the phylogenetic relationships between the Dino-APs and counterparts from other organisms, the deduced amino acid sequences from each strain were aligned with top hits in the BLAST analysis and other sequences from representative organisms. The alignment was run using MUSCLE (Edgar, 2004) and manually corrected. Conserved motifs were identified based on the sequence alignment and presented using WebLogo V3 (Crooks et al., 2004). Phylogenetic tree reconstruction was performed using Maximum-Likelihood (Guindon et al., 2010) and Neighbor-joining (NJ) with 1000 bootstraps on the Seaview V4.5.3 platform (Gouy et al., 2010). Bayesian analysis (Hueslenbeck and Ronquist, 2001) was carried out for 3,000,000 generations with burn-in at setting 250,000 (two independent runs were performed and the final standard deviation of split frequencies was lower than 0.01).
Results
Identification of introns in APs of Karenia mikimotoi and Prorocentrum minimum
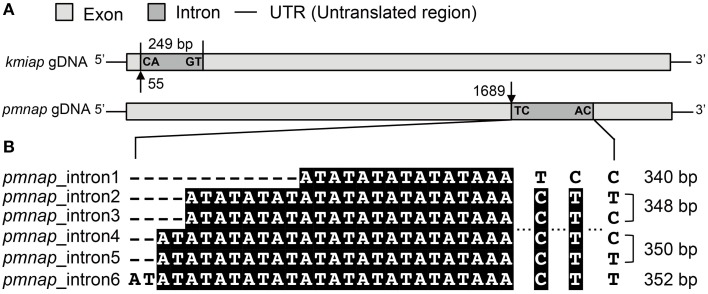
To examine the possible existence of introns, AP genes were amplified both from genomic DNA (gDNA) and cDNA from at least one representative strain in each family. Among the eight strains with both gDNA and cDNA analyzed, only APs in K. mikimotoi C32-HK (kmiap) and P. minimum CCMA 15 (pmnap) were found to contain introns (Table 1). The kmiap intron has CA as the donor site and GT as the acceptor site, while in pmnap, the donor site is TC and the acceptor site is AC (Figure 1A). By comparing the introns in pmnap and kmiap, no noticeable sequence similarity was identified. Unexpectedly, the intron in kmiap is translatable, encoding a peptide comprising 83 amino acids that had no BLASTP hits in GenBank. Moreover, at least six different variants of the introns were identified in pmnap based on the total of 21 full-length gDNA sequences aligned with the 28 full-length cDNA sequences (Figure 1B). The six intron variants with different lengths were classified by the combination of the repeat numbers of a (AT)n region and three sites of nucleotide substitution (T/C).
Figure 1.
Analysis of AP introns. (A) Schematic of the intron location and boundary sequences in kmiap and pmnap, the characters marked at the boundary of the intron represent donor site (left) and acceptor site (right), and the numbers with arrows indicate the nucleotide position of intron from the start codon. (B) Sequence comparison of six different variants of intron in pmnap, and the length for each intron variant was marked at the right.
Nucleotide substitution and apparent pseudogenization of AP genes in Prorocentrum spp.
By comparing full-length AP ORFs derived from cDNA (28 clones) with gDNA (21 clones), we found sequence discrepancies between the cDNA and the gDNA in P. minimum. This indicated either mRNA editing or differential recovery of paralogous sequences from the genomic DNA and the cDNA samples. In P. minimum, we identified only one type of ORF in pmnap cDNA while two different types in gDNA, with nucleotide substitutions observed in a total of 12 sites (Figure 2A, Supplementary Table 2). Except for a pyrimidine transitional substitution at one site, all the sites had transversional replacement. Half of those substitutions were synonymous. A 1463 bp AP gene fragment was obtained from P. micans genomic DNA, which also possessed multiple nucleotide substitutions. All of these 13 clone sequences could be translated into partial AP proteins of 487 amino acids in length, which can be grouped into five different variants resulting from 143 nucleotide substitutions (Supplementary Figure 1A).
Figure 2.
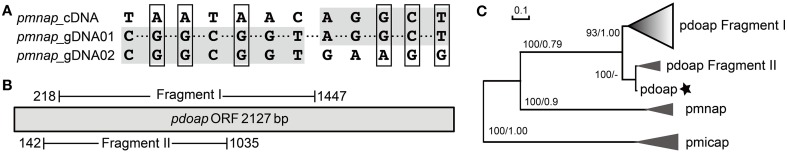
Sequence comparison of AP genes in Prorocentrum. (A) Nucleotide substitution of pmnap identified between gDNA and cDNA clones, shaded region represents sharing the same nucleotides and box indicates the nonsynonymous substitution. (B) Schematic of two different genomic fragments of pdoap. (C) Phylogenetic tree inferred from nucleotide sequences (common region) of AP gene in Prorocentrum. Prodo, P. donghaiense; Promini, P. minimum, support nodes shown as NJ/ML. Gradient shaded triangles represent the presence of pseudogenes and the asterisk represents the assembled pdoap ORF from transcriptomic data (intact tree refered to Supplementary Figure 1B).
A predicted full-length contig of the AP gene (pdoap) was assembled from a transcriptome dataset of P. donghaiense (Shi et al. unpublished data). The length of the deduced protein PDOAP was close to the PMNAP in P. minimum and the pairwise comparison between these two protein sequences showed 100% coverage, 87% identity, and E-value 0.0. However, PCR with different primer combinations consistently led to the recovery of two fragments from P. donghaiense genomic DNA, respectively sharing high similarity to two separate parts of pdoap. Fragment I was acquired using the specific primer pair prodoAPgF2/gR2 (32 clones) (Supplementary Table 1), and Fragment II using the Dino-AP degenerate primer pair DinoAPNF4b/NR6 (10 clones) (Figure 2B). Some of the Fragment I sequences (25 out of a total of 32 clones) appeared to be pseudogenes because they could not be translated into contiguous peptides due to one or more premature stop codons caused by nucleotide substitution/insertion, while the predicted peptide of Fragment II covered 57% of the predicted PDOAP with E-value 0.0. A phylogenetic tree inferred from the common nucleotide sequences shared by the assembled pdoap, pdoap fragments, pmnap, and pmicap showed that pdoap grouped together with pmnap, but distant from pmicap (Figure 2C, Supplementary Figure 1B). Moreover, pdoap fragment I dominated by pseudogene clones formed a separate branch from translatable fragment II and the assembled pdoap. All these results suggest that gene pseudogenization occurred after divergence of P. donghaiense and P. minimum.
High AP gene sequence diversity in Alexandrium spp.
To explore the conservation of AP gene sequences within a species, specific primers pairs AlexAPFLF/R (Supplementary Table 1) designed previously based on the 5′ and 3′ UTR region in A. catenella (ACHK-NT) (Lin et al., 2012a) were used to acquire the full length AP ORFs from three different Alexandrium species, including four additional strains from A. catenella (18S Clade IIC/28S Group IV), and one strain each from A. tamarense (18S Clade IIB/28S Group III) and A. fundyense (18S Clade I/28S Group I) (Table 1) (Wang et al., 2014). We successfully retrieved the complete ORFs from most of the tested Alexandrium spp.; however, the full-length AP gene in the strain A. tamarense CCAP1119/1 was obtained only by using specific primer pairs CCAP1119/1 APFLF/R designed based on the UTR region acquired as described in methodology (Supplementary Table 1).
In A. catenella, strain ACHK-NT and the other three strains (ACHK-T, ATDH02, and ATMJ01) share the same gene sequence (alecaap). Strain ATCI01 was the only exception in which a non-synonymous mutation occurred in 23 clones (variant I), and another five different AP variants occurred in one clone each (Table 2, Supplementary Table 3). Sequence comparison of the six different variants showed high nucleotide polymorphisms scattered along the whole length of the ORF region. Besides a 12-bp insertion, which was detected only in variant VI, there were 99 SNP sites, only one third of them being synonymous mutations (Table 2, Supplementary Table 3). Based on the nucleotide sequences of the six different variants, Tajima's D-value was calculated using DnaSP, which gave a value of 0.26 (p > 0.10), suggesting that those polymorphic sites were neutral mutations (Tajima, 1989). All the AP protein variants from A. catenella were predicted to be extracellular (Table 1).
Table 2.
Summary of nucleotide polymorphism in Alexandrium spp.
| Strain | nt variants | aa variants | Deletion (bp) | Total SNP sites (Syn/Nonsyn) | Tajima's D |
|---|---|---|---|---|---|
| Alexandrium catenella ATCI01 | 6 (gDNA) | 6 | 12 | 99 (34/65) | 0.26 (P > 0.10) |
| Alexandrium tamarense CCAP1119/1 | 2/37 (gDNA/cDNA) | 36 | 12/24/129 | 139 (39/100) | −2.75 (P < 0.001) |
| Alexandrium fundyense CCMP1719 | 6/18 (gDNA/cDNA) | 13 | 24/30/42 | 9 (3/6) | 2.46 (P < 0.05) |
nt, nucleotide; aa, amino acid.
In strain A. tamarense CCAP1119/1, among the 83 full-length clones we sequenced (18 for gDNA, 65 from cDNA), 139 nucleotide polymorphic sites and deletions in both gDNA and cDNA clones were observed, making the ataap highly polymorphic (Table 2, Figure 3A). There were three variants of deletions (12, 24, and 129 bp) when compared with the longest complete ORF (2184 bp) which was also the dominant variant in both gDNA and cDNA clone libraries. Overall 36 different ORF variants were identified considering both the gDNA and cDNA amplicons together. We noticed that the changes of amino acid sequences in different variants caused by 12 and 24 bp deletions resulted in the loss of different numbers of the “PTPA” repeat unit, which was completely absent in the variant with 129-bp deletions (Figure 3A). Correspondingly, the variants detected in CCAP1119/1 were predicted to have potentially different subcellular localization, extracellular in most cases but also cytoplasmic and chloroplast, which was exceptional compared with all other APs in Alexandrium spp. we tested (Table 1). With all the variants considered, the Tajima's D-value was −2.75 (p < 0.001), indicating that this gene has evolved under directional selection (Tajima, 1989).
Figure 3.
Sequence comparison showing intraspecific AP polymorphism and differences between closely related species of Alexandrium. (A) A. tamarense (strain CCAP1119/1); (B) “Alefu” represents A. fundyense (strain CCMP1719). The boxes mark the “PTPA” repeat motif with different repeat numbers in different variants. Asterisks indicate that this variant was present in both gDNA and cDNA clones.
In strain A. fundyense CCMP1719, a total of 15 gDNA and 53 cDNA full-length clones were obtained, also exhibiting highly polymorphic gene sequences. We identified six variants of afuap in gDNA clones and 18 variants in cDNA clones, only two of which were shared by both, which comprise 13 variants of protein sequences in total containing the “PTPA” repeat unit in different numbers as well (Figure 3B). The corresponding subcellular localization of all AFUAP variants was predicted to be in the plasma membrane, appearing to be unique among all detected dinoflagellate strains. Moreover, different from the result in ATCI01 and CCAP1119/1, the Tajima's D neutral test for alefuap was 2.46 (p < 0.05), suggesting evolution by balancing selection (Table 2).
Classification of dinoflagellate APs as an atypical AP (PhoAaty)
From the full-length AP genes from 10 dinoflagellate strains (including ORF fragments from P. micans) obtained in this study, and a predicted AP full-length ORF region retrieved from the transcriptome of P. donghaiense (Shi et al. unpublished) and the genome of S. kawagutii respectively (Lin et al. unpublished), in total we collected AP gene sequences from 15 dinoflagellate strains (three of them have been published in Lin et al., 2011, 2012a,b), representing four core dinoflagellate families (Table 1). Overall, all the deduced dinoflagellate AP proteins (Dino-APs) shared similar chemical characteristics and predicted protein structures, e.g., possessing a signal peptide and GPI anchor (Table 1), with that from S. kawagutii being the only exception.
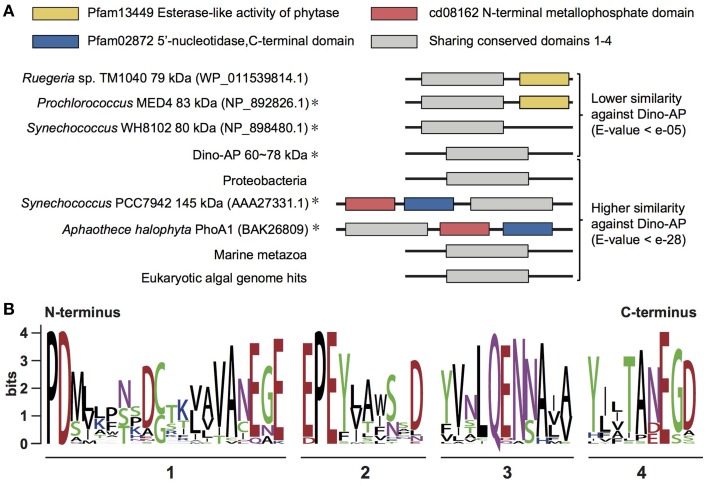
To determine which of the currently classified types of AP our Dino-APs belong to, pairwise sequence comparisons were carried out between the Dino-APs and all reported phosphatases. Overall, BLAST analysis results of Dino-APs revealed no recognizable functional domains in the sequences. The similarity between Dino-APs and typical PhoAEC was too low to identify the well-characterized PhoAEC domains in Dino-APs. Further comparison showed that Dino-APs only shared sequence similarity with partial sequences of a group of putative phosphatases identified in cyanobacteria (Figure 4A). BLASTP of Dino-APs against GenBank non-redundant protein database showed that the top100 hits (e-value < e−28) included three major groups, eukaryotic algae, marine animals, and bacteria (cyanobacteria and proteobacteria).
Figure 4.
Comparative structural analysis of AP in different organisms. (A) Sequence comparison of PhoAaty among different organisms and the section containing four conserved domains (adapted from Moore et al., 2005 and Scanlan et al., 2009). Representative sequence labeled with “asterisk” indicated this gene has been characterized partially. (B) WebLogo showing the consensus of amino acid sequences acquired from the alignment of four conserved domains (detail alignment shown in Supplementary Figure 2).
Despite the low sequence similarity among those putative APs (E-value e−05 to e−28), we were able to identify four conserved motifs from the amino acid alignment of all the AP gene sequences from dinoflagellates and other organisms (Figure 4B, Supplementary Figure 2). Although none of these four motifs fit any previously characterized domain data, there is a significant conservation of aspartic acid (D) and glutamic acid residues (E) in those motifs. Based on the presence of the common motifs in these APs, some of which have previously been assigned as an atypical PhoA type, we were led to classify these putative APs as PhoAaty to distinguish them from the typical PhoAEC and other phosphatases.
Phylogenetic analysis of phoAaty
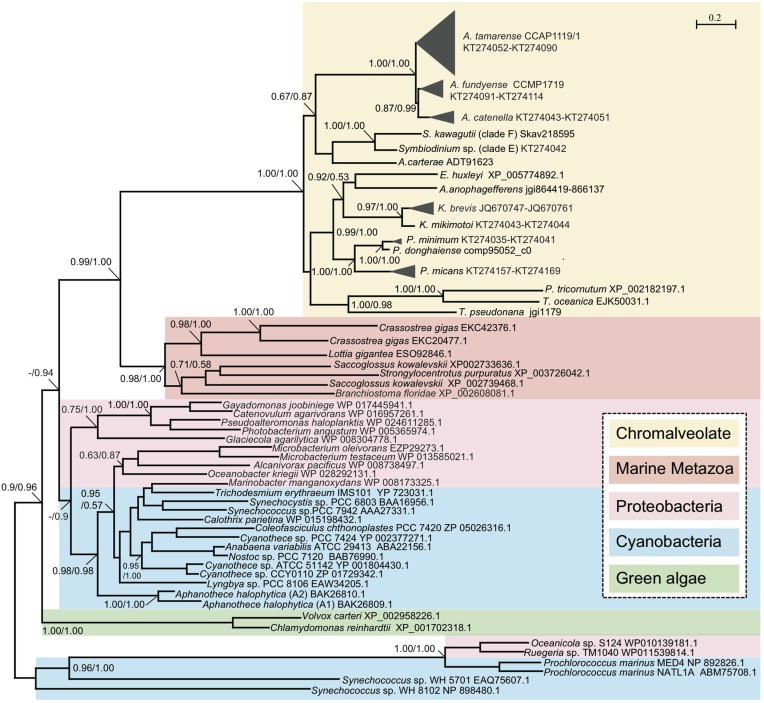
To gain an evolutionary perspective of these PhoAaty proteins, a phylogenetic tree was inferred based on the amino acid alignment of AP (unique variants) from dinoflagellates and representative species of other organisms. As shown in Figure 5, APs from cyanobacteria and proteobacteria formed a specific clade. With this clade as the root, two freshwater green algae formed a clade, clearly separated from a large clade comprising PhoAaty identified from dinoflagellates and other chromophytes (Chromalveolate clade), marine metazoans and bacteria. In the latter clade, APs from marine metazoans and chromalveolate respectively, formed monophyletic groups sister to each other, while cyanobacteria and proteobacteria inter-mingled as a separate group. In the Chromalveolate PhoAaty clade, diatoms formed a monophyletic lineage while dinoflagellates appeared to be paraphyletic due to the placement of the diatom group that separated AP of the Prorocentrum/Karenia clade from that of the Alexandrium/Amphidinium/Symbiodinium clade and the alliance of haptophytes and pelagophytes that separated the subclade of Karenia spp. from that of Prorocentrum spp. Within the dinoflagellate APs clusters, each genus (e.g., Alexandrium) formed a monophyletic clade and all variants in each species formed a monophyletic group. A similar tree topology for APs of Prorocentrum spp. was observed between the amino acid and nucleotide sequences (Figure 2C, Supplementary Figure 1).
Figure 5.
Phylogenetic tree inferred from amino acid sequences of PhoAaty in marine organisms. Tree topology shown as inferred from PhyML (Dayhoff model), and Bayesian analysis (BE) was also carried out based on Dayhoff model with similar tree topology. Support of nodes for most branches are shown as SH-like (Shimodaira-Hasegawa-like procedure) from ML and Bayesian posterior probability (ML/BE).
Discussion
Unusual introns and within-lineage duplication in dino-APs evolution
In contrast to the long-held notion that dinoflagellate genes have few introns (for review see Lin, 2011), fairly abundant introns have recently been reported in dinoflagellates (Bachvaroff and Place, 2008; Shoguchi et al., 2013). In this study, we detected introns in the AP gene in two out of the 12 dinoflagellates we examined, kmiap and pmnap, but their splice sites were unconventional. The translatable nature of the kmiap intron suggests that it may be functionally similar to Group II introns, a class of self-catalytic ribozymes containing an internal ORF which is capable of its own excision from precursor RNAs. This type of intron has been found only in bacteria and organellar genomes of fungi, plants, and protists (Rodríguez-Trelles et al., 2006; Roy and Gilbert, 2006).
Gene duplication is the major driving force in the evolution of functional diversification, and the duplicated genes (paralogs) can maintain the function of its parental gene, lose function due to dramatic sequence mutation (pseudogenization), perform partial function of the parental gene (subfunctionalization), or acquire and preserve a novel function (neofunctionalization) due to significant sequence divergence (Lynch and Conery, 2000; Zhang, 2003). The many variants of APs in each of the dinoflagellate strains examined in this study are a clear indication of gene duplication. The fact that the variants are grouped by species in the phylogenetic analyses further indicates that gene duplication occurs within a species. Given that Alexandrium is the latest-diverging genus of dinoflagellates (John et al., 2003), this result suggests that some of the gene duplications of Dino-APs might be quite recent. The clustering of pseudogenes in P. donghaiense suggests pseudogenization in the process or soon after gene duplication of Dino-APs. Moreover, the high sequence similarity with functional gene homologs (except for the internal stop codons or frame shifts), as is the case in P. donghaiense, suggests that these are young pseudogenes (Zhang, 2003).
Human AP is considered a multigene family derived from an original AP gene by gene duplication (Weiss et al., 1988). Comparably, multicopies of AP also occur in dinoflagellates and their monophyletic grouping within each species in the phylogenetic tree also suggests that these copies have also descended from a common ancestral gene. To describe the evolution of a multigene family, two different models have been proposed, concerted evolution and birth-and-death evolution (Nei and Rooney, 2005). In concerted evolution, paralogs of a gene within one species are closer to each other than to those in another species, even though gene duplication might have occurred before speciation. The monophyletic grouping of AP paralogs by species observed in Dino-APs is consistent with this model. However, the occurrence of pseudogenes and recently duplicated paralogs in dinoflagellate APs makes it fit the birth-and-death model better. In the birth-and-death model, new genes are generated by duplication, but subsequently whereas some of these will stay in the genome others are subject to deletion or inactivation during evolution. Therefore, most gene families that produce variable gene products are subject to birth-and-death evolution (Nei and Rooney, 2005). This model has also been invoked to explain the evolution of actin in dinoflagellates (Kim et al., 2011).
Sequence divergence of dino-APs and ecological implication
Duplicated genes may diverge and undergo functional differentiation (Kondrashov et al., 2002; Zhang, 2003). As the prototype AP, PhoAEC is an enzyme with promiscuous substrate specificity, which has a selective advantage during gene duplication to optimize the specificity and activity for a diverse type of substrates (Yoshikuni et al., 2006; Zalatan et al., 2006). Moreover, both PhoX and PhoD in bacteria exhibit promiscuous enzymatic activities on phosphomonoesters and phosphodiesters (Luo et al., 2009; Sebastian and Ammerman, 2011). It has been well documented that dinoflagellates are able to utilize different types of DOP, each as a sole P source (Huang et al., 2005; Oh et al., 2010), suggesting that Dino-APs are also promiscuous. Substrate promiscuity has clear advantages for versatility in acquiring P from different forms of DOP, and might be an evolutionary adaptation to the abundant and chemically diverse DOP in the ocean (Paytan and McLaughlin, 2007).
It is interesting to observe that the AP genes of different Alexandrium catenella strains (most of them isolated from East China Sea) share the same sequence, except that in strain ATCI01, which was isolated from the South China Sea (Wang et al., 2014). A. tamarense (strain CCAP1119-1 from England) and A. fundyense (strain CCMP1719 from the Gulf of Maine) exhibit high AP sequence variations. The Tajima' D-values for these AP sequences were totally different in these three geographic regions (China Seas, Bay of Tamar Esturary, England, and the Gulf of Maine, USA), which indicate that the evolution of AP in Alexandrium spp. is probably driven by habitat-specific environmental conditions. As an example, it has been thought that the independent loss and duplication of the intestinal AP in vertebrate lineages are driven by their respective substrate landscapes dictated by their different gut microbiomes and diets (Yang et al., 2012). Furthermore, our results also showed that deduced protein variants in Alexandrium spp. might have differential subcellular localizations as computationally predicted, A. catenella in the extracellular space, A. fundyense in the plasma membrane, and A. tamarense in three possible subcellular compartments. These differential subcellular distributions of APs would enable the different dinoflagellate species to utilize different DOP sources (e.g., intracellular and extracellular). Therefore, the diverging sequences of different Dino-AP potentially have significant ecological implications (e.g., utilizing different sources of DOP) (Lin et al., 2012a).
PhoAaty as a phylogenetically distinct type of AP in marine microorganisms
Sequence comparisons and phylogenetic analyses of Dino-APs indicated that they comprise a type of AP–PhoAaty similar to those reported in several bacteria. Although there is not yet sufficient biochemical data of PhoAaty (i.e., metal ion requirement, substrate specificity) for us to ascribe the observed sequence divergence to potential enzymatic structural and functional diversification, we can obtain some insights from the relatively well-characterized counterparts. As confirmed in PhoAEC, the metal-binding site shows a preference for histidine (H) and aspartic acid residues (D), consistent with what has been observed in most Zn-binding proteins (Galperin and Jedrzejas, 2001; Patel et al., 2007). PhoAaty in different species shares conserved motifs, in which there is a significant conservation of aspartic acid (D) and glutamic acid residues (E) that have been found to be responsible for Ca-binding in PhoX. Besides, a highly conserved Ca-binding “proline-aspartic acid” motif is shared by PhoAaty, PhoX, and phosphotriesterase (Wu et al., 2007; Zaheer et al., 2009; Yong et al., 2014).
The high sequence divergence observed in PhoAaty may be attributed to serial gene duplications followed by divergent evolution between each species. As observed in the phylogenetic analyses (Figure 5), PhoAaty in green algae was placed in a separate cluster from the other eukaryotic algae, and the PhoAaty from dinoflagellates, haptophytes, and stramenopiles were all mingled together. This coincides with the different endosymbiosis histories of these algae: green algae emerged from primary endosymbiosis and the rest from secondary endosymbiosis or tertiary replacement (Falkowski et al., 2004). It is interesting to note that AP from Karenia spp. is close to that from E. huxleyi while the plastid of Karenia and other Kareniacea lineages arose as a result of tertiary replacement from a haptophyte origin (Keeling et al., 2004; Tirichine and Bowler, 2011; Jackson et al., 2013). HGT has been considered as a significant driver of gene innovation in both dinoflagellates (Wisecaver et al., 2013) and diatoms (Bowler et al., 2008). Based on both the sequence comparison and phylogenetic analysis, PhoAaty in the chromalveolate algae likely arose through HGT from a common ancestor. Similarly, it has been suggested that PhoX has undergone horizontal gene transfer among different marine bacteria (Zaheer et al., 2009; Kathuria and Martiny, 2011).
The remarkably low sequence similarity between the identified multiple types of APs in different algal lineages qualifies this group of enzymes as Non-homologous ISofunctional Enzymes (NISE), which refers to enzymes that lack detectable sequence similarity but catalyze the same biochemical reactions (Omelchenko et al., 2010). The pressure of adapting to the fluctuating nutrient availability in marine environments is likely to trigger the convergent evolution of a group of NISE toward APs to hydrolyze phosphoesters from different ancestral genes. Over-representation of NISE, such as in the case of antioxidant analogous enzymes, may imply a powerful selective pressure from rapid oxygenation, since the best-known superoxide dismutase (SOD) enzymes include four distinct structural types with different metal ion requirements (Omelchenko et al., 2010). Similarly, multiple types of APs also exhibit different metal requirements, with most characterized APs in marine microorganisms apparently having adopted Ca as a cofactor instead of a typical Zn/Mg type (Luo et al., 2009; Sun et al., 2012; Lin et al., 2013). The adoption of Ca may simultaneously require Fe as the cofactor partner, as found in PhoX (Yong et al., 2014). Although whether Ca/Fe is the only pair that replaces Zn/Mg remains to be further studied, the differential divalent metal cation requirements of multiple types of APs represents an interesting case of adaptive evolution in organisms living in the oceanic habitat where Zn bioavailability is often limited (Sebastián and Ammerman, 2009; Kathuria and Martiny, 2011).
Concluding remarks
In this study, we found introns, pseudogenes, and presence of multiple copies of AP in dinoflagellates indicating that the evolution of APs in dinoflagellates has been driven by gene duplication and fits the birth-and-death model. The Tajima's D-value differed amongst the three geographic species of Alexandrium predictive of differential subcellular localizations amongst the various deduced proteins, implying APs in these species have evolved under different environmental selection pressure. Phylogenetic analysis showed that Dino-APs, as well as putative APs from other eukaryotic phytoplankton, might be classified as an atypical type of AP, PhoAaty, which is prevalent in marine microorganisms and shares a common ancestral gene. We found evidence that environmental stress and endosymbiosis (plastid acquisition) might have contributed to the divergent evolution of PhoAaty. On this basis, we propose that multiple types of APs in marine ecosystems could be considered a group of NISE, sharing weak sequence similarity and different metal ion requirements while performing the same phosphoester hydrolyzing function. Adaption to the heterogeneous and limited availability of metal ions and phosphate in the ocean (Moore et al., 2013), by owning multiple types of AP would provide a competitive advantage to marine phytoplankton living in different environments.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by National Natural Science Foundation of China grants NSFC41176091 and NSFC41330959 (to SL) and China Postdoctoral Science Foundation Grant 2013M531546 (to XL). We thank the reviewers for their constructive comments and Ed. J. Carpenter from Romberg Tiburon Center of SFSU for English proof reading, which led to substantial improvement of this manuscript.
Glossary
Abbreviations
- AP
alkaline phosphatase
- Dino-AP
dinoflagellate AP
- PhoAEC
typical E. coli-type AP
- PhoAaty
atypical type of PhoA.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00868
References
- Armbrust E. V., Berges J. A., Bowler C., Green B. R., Martinez D., Putnam N. H., et al. (2004). The genome of the diatom Thalassioria pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. 10.1126/science.1101156 [DOI] [PubMed] [Google Scholar]
- Bachvaroff T. R., Place A. R. (2008). From stop to start: tandem gene arrangement, copy number and trans-splicing sites in the dinoflagellate Amphidinium carterae. PLoS ONE 3:e2929. 10.1371/journal.pone.0002929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C., Allen A. E., Badger J. H., Grimwood J., Jabbari K., Kuo A., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. 10.1038/nature07410 [DOI] [PubMed] [Google Scholar]
- Crooks G. E., Hon G., Chandonia J.-M., Brenner S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyhrman S. T., Ammerman J. W., Van Mooy B. A. S. (2007). Microbes and the marine phosphorus cycle. Oceanography 20, 110–116. 10.5670/oceanog.2007.54 [DOI] [Google Scholar]
- Dyhrman S. T., Jenkins B. D., Rynearson T. A., Saito M. A., Mercier M. L., Alexander H., et al. (2012). The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response. PLoS ONE 7:e33768. 10.1371/journal.pone.0033768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowski P. G., Katz M. E., Knoll A. H., Quigg A., Raven J. A., Schofield O., et al. (2004). The evolution of modern eukaryotic phytoplankton. Science 305, 354–360. 10.1126/science.1095964 [DOI] [PubMed] [Google Scholar]
- Fankhauser N., Mäser P. (2005). Identification of GPI anchor attachment signals by a Kohonen self-organizing map. Bioinformatics 21, 1846–1852. 10.1093/bioinformatics/bti299 [DOI] [PubMed] [Google Scholar]
- Galperin M. Y., Jedrzejas M. J. (2001). Conserved core structure and active site residues in alkaline phosphatase superfamily enzymes. Proteins 45, 318–324. 10.1002/prot.1152 [DOI] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- Guillard R. R. L. (1975). Culture of phytoplankton for feeding marine invertebrates, in Culture of Marine Invertebrate Animals, eds Smith W. L., Chanley M. H. (New York, NY: Plenum Press; ), 26–60. 10.1007/978-1-4615-8714-9_3 [DOI] [Google Scholar]
- Guillard R. R. L., Hargraves P. E. (1993). Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia 32, 234–236. 10.2216/i0031-8884-32-3-234.1 [DOI] [Google Scholar]
- Guillard R. R. L., Ryther J. H. (1962). Studies of marine planktonic diatoms: I. Cyclotella nana Hustedt and Detonula confervacea Cleve. Can. J. Microbiol. 8, 229–239. 10.1139/m62-029 [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hallmann A. (1999). Enzymes in the extracellular matrix of Volvox: an inducible, calcium-dependent phosphatase with a modular composition. J. Biol. Chem. 274, 1691–1697. 10.1074/jbc.274.3.1691 [DOI] [PubMed] [Google Scholar]
- Huang B., Ou L., Hong H., Luo H., Wang D. (2005). Bioavailability of dissolved organic phosphorus compounds to typical harmful dinoflagellate Prorocentrum donghaiense Lu. Mar. Pollut. Bull. 51, 838–844. 10.1016/j.marpolbul.2005.02.035 [DOI] [PubMed] [Google Scholar]
- Hueslenbeck J. P., Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Jackson C. J., Gornik S. G., Waller R. F. (2013). A tertiary plastid gains RNA editing in its new host. Mol. Biol. Evol. 30, 788–792. 10.1093/molbev/mss270 [DOI] [PubMed] [Google Scholar]
- John U., Fensome R. A., Medlin L. K. (2003). The application of a molecular clock based on molecular sequences and the fossil record to explain biogeographic distributions within the Alexandrium tamarense “species complex” (Dinophyceae). Mol. Biol. Evol. 20, 1015–1027. 10.1093/molbev/msg105 [DOI] [PubMed] [Google Scholar]
- Kageyama H., Tripathi K., Rai A. K., Cha-Um S., Waditee-Sirisattha R., Takabe T. (2011). An alkaline phosphatase/phosphodiesterase, PhoD, induced by salt stress and secreted out of the cells of Aphanothece halophytica, a halotolerant cyanobacterium. Appl. Environ. Microbiol. 77, 5178–5183. 10.1128/AEM.00667-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathuria S., Martiny A. C. (2011). Prevalence of a calcium-based alkaline phosphatase associated with the marine cyanobacterium Prochlorococcus and other ocean bacteria. Environ. Microbiol. 13, 74–83. 10.1111/j.1462-2920.2010.02310.x [DOI] [PubMed] [Google Scholar]
- Keeling P. J., Archibald J. M., Fast N. M., Palmer J. D. (2004). Comment on “The evolution of modern eukaryotic phytoplankton.” Science 306, 2191. 10.1126/science.1103879 [DOI] [PubMed] [Google Scholar]
- Kim S., Bachvaroff T. R., Handy S. M., Delwiche C. F. (2011). Dynamics of actin evolution in dinoflagellates. Mol. Biol. Evol. 28, 1469–1480. 10.1093/molbev/msq332 [DOI] [PubMed] [Google Scholar]
- Kolowith L. C., Ingall E. D., Benner R. (2001). Composition and cycling of marine organic phosphorus. Limnol. Oceanogr. 46, 309–321. 10.4319/lo.2001.46.2.0309 [DOI] [Google Scholar]
- Kondrashov F. A., Rogozin I. B., Wolf Y. I., Koonin E. V. (2002). Selection in the evolution of gene duplications. Genome Biol. 3:8. 10.1186/gb-2002-3-2-research0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P., Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- Lin S. (2011). Genomic understanding of dinoflagellates. Res. Microbiol. 162, 551–569. 10.1016/j.resmic.2011.04.006 [DOI] [PubMed] [Google Scholar]
- Lin H. Y., Shih C. Y., Liu H. C., Chang J., Chen Y. L., Chen Y. R., et al. (2013). Identification and characterization of an extracellular alkaline phosphatase in the marine diatom Phaeodactylum tricornutum. Mar. Biotechnol. 15, 425–436. 10.1007/s10126-013-9494-3 [DOI] [PubMed] [Google Scholar]
- Lin X., Zhang H., Cui Y., Lin S. (2012a). High sequence variability, diverse subcellular localizations, and ecological implications of alkaline phosphatase in dinoflagellates and other eukaryotic phytoplankton. Front. Microbiol. 3:235. 10.3389/fmicb.2012.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Zhang H., Huang B., Lin S. (2011). Alkaline phosphatase gene sequence and transcriptional regulation by phosphate limitation in Amphidinium carterae (Dinophyceae). J. Phycol. 47, 1110–1120. 10.1111/j.1529-8817.2011.01038.x [DOI] [PubMed] [Google Scholar]
- Lin X., Zhang H., Huang B., Lin S. (2012b). Alkaline phosphatase gene sequence characteristics and transcriptional regulation by phosphate limitation in Karenia brevis (Dinophyceae). Harmful Algae 17, 14–24. 10.1016/j.hal.2012.02.005 [DOI] [Google Scholar]
- Luo H., Benner R., Long R. A., Hu J. (2009). Subcellular localization of marine bacterial alkaline phosphatases. Proc. Natl. Acad. Sci. U.S.A. 106, 21219–21223. 10.1073/pnas.0907586106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Guo Y. C., Deng J. Y., Wei H. P., Zhang Z. P., Leng Y., et al. (2010). Characterization of a monomeric heat-labile classical alkaline phosphatase from Anabaena sp. PCC7120. Biochemistry (Moscow) 75, 655–664. 10.1134/S0006297910050172 [DOI] [PubMed] [Google Scholar]
- Lynch M., Conery J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. 10.1126/science.290.5494.1151 [DOI] [PubMed] [Google Scholar]
- Moore C., Mills M., Arrigo K., Berman-Frank I., Bopp L., Boyd P., et al. (2013). Processes and patterns of oceanic nutrient limitation. Nat. Geosci. 6, 701–710. 10.1038/ngeo176522615968 [DOI] [Google Scholar]
- Moore I. R., Ostrowski M., Scanlan D. J., Feren K., Sweetsir I. (2005). Ecotypic variation in phosphorus-acquisition mechanisms within marine picocyanobacteria. Aquat. Microb. Ecol. 39, 257–269. 10.3354/ame039257 [DOI] [Google Scholar]
- Morey J. S., Monroe E. A., Kinney A. L., Beal M., Johnson J. G., Hitchcock G. L., et al. (2011). Transcriptomic response of the red tide dinoflagellate, Karenia brevis, to nitrogen and phosphorus depletion and addition. BMC Genomics 12:346. 10.1186/1471-2164-12-346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J. L., Chang C. W., Grossman A. R. (2006). Genome-based approaches to understanding phosphorus deprivation responses and PSR1 control in Chlamydomonas reinhardtii. Eukaryotic Cell 5, 26–44. 10.1128/EC.5.1.26-44.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Rooney A. P. (2005). Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 39, 121–152. 10.1146/annurev.genet.39.073003.112240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. J., Kwon H. K., Noh I. H., Yang H. S. (2010). Dissolved organic phosphorus utilization and alkaline phosphatase activity of the dinoflagellate Gymnodinium impudicum isolated from the South Sea of Korea. Ocean Sci. 45, 171–178. 10.1007/s12601-010-0015-2 [DOI] [Google Scholar]
- Omelchenko M. V., Galperin M. Y., Wolf Y. I., Koonin E. V. (2010). Research Non-homologous isofunctional enzymes: a systematic analysis of alternative solutions in enzyme evolution. Biol. Direct 5:31. 10.1186/1745-6150-5-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard E. D., Webb E. A., Dyhrman S. T. (2009). Molecular analysis of the phosphorus starvation response in Trichodemium spp. Environ. Microbiol. 11, 2400–2411. 10.1111/j.1462-2920.2009.01968.x [DOI] [PubMed] [Google Scholar]
- Patel K., Kumar A., Durani S. (2007). Analysis of the structural consensus of the zinc coordination centers of metalloprotein structures. Biochim. Biophys. Acta 1774, 1247–1253. 10.1016/j.bbapap.2007.07.010 [DOI] [PubMed] [Google Scholar]
- Paytan A., McLaughlin K. (2007). The oceanic phosphorus cycle. Chem. Rev. 107, 563–576. 10.1021/cr0503613 [DOI] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., Von Heijne G., Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Pierlenoi A., Martelli P. L., Casadio R. (2008). PredGPI: a GPI-anchor predictor. BMC Bioinformatics 9:392. 10.1186/1471-2105-9-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quisel J. D., Wykoff D. D., Grossman A. R. (1996). Biochemical characterization of the extracellular phosphatases produced by phosphorus-deprived Chlamydomonas reinhardtii. Plant Physiol. 111, 839–848. 10.1104/pp.111.3.839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray J. M., Bhaya D., Block M. A., Grossman A. R. (1991). Isolation, transcription, and inactivation of the gene for an atypical alkaline phosphatase of Synechococcus sp. strain PCC 7942. J. Bacteriol. 173, 4297–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Trelles F., Tarrío R., Ayala F. J. (2006). Origins and evolution of spliceosomal introns. Annu. Rev. Genet. 40, 47–76. 10.1146/annurev.genet.40.110405.090625 [DOI] [PubMed] [Google Scholar]
- Roy S. W., Gilbert W. (2006). The evolution of spliceosomal introns: patterns, puzzles and progress. Nat. Rev. Genet. 7, 211–221. 10.1038/nrg1807 [DOI] [PubMed] [Google Scholar]
- Scanlan D. J., Ostrowski M., Mazard S., Dufresne A., Garczarek L., Hess W. R., et al. (2009). Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 73, 249–299. 10.1128/MMBR.00035-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian M., Ammerman J. W. (2011). Role of the phosphatase PhoX in the phosphorus metabolism of the marine bacterium Ruegeria pomeroyi DSS−3. Envrion. Microbiol Rep. 3, 535–542. 10.1111/j.1758-2229.2011.00253.x [DOI] [PubMed] [Google Scholar]
- Sebastián M., Ammerman J. W. (2009). The alkaline phosphatase PhoX is more widely distributed in marine bacteria than the classical PhoA. ISME J. 3, 563–572. 10.1038/ismej.2009.10 [DOI] [PubMed] [Google Scholar]
- Shoguchi E., Shinzato C., Kawashima T., Gyoja F., Mungpakdee S., Koyanagi R., et al. (2013). Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 23, 1399–1408. 10.1016/j.cub.2013.05.062 [DOI] [PubMed] [Google Scholar]
- Sun M. M., Sun J., Qiu J. W., Jing H., Liu H. (2012). Characterization of the proteomic profiles of the brown tide alga Aureoumbra lagunensis under phosphate- and nitrogen-limiting conditions and of its phosphate limitation-specific protein with alkaline phosphatase activity. Appl. Environ. Microbiol. 78, 2025–2033. 10.1128/AEM.05755-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L., Bowler C. (2011). Decoding algal genomes: tracing back the history of photosynthetic life on Earth. Plant J. 66, 45–57. 10.1111/j.1365-313X.2011.04540.x [DOI] [PubMed] [Google Scholar]
- Wang L., Zhuang Y., Zhang H., Lin X., Lin S. (2014). DNA barcoding species in Alexandrium tamarense complex using ITS and proposing designation of five species. Harmful Algae 31, 100–113. 10.1016/j.hal.2013.10.013 [DOI] [PubMed] [Google Scholar]
- Weiss M. J., Ray K., Henthorn P. S., Lamb B., Kadesch T., Harris H. (1988). Structure of the human liver/bone/kidney alkaline phosphatase gene. J. Biol. Chem. 263, 12002–12010. [PubMed] [Google Scholar]
- White A. E. (2009). New insights into bacterial acquisition of phosphorus in the surface ocean. Proc. Natl. Acad. Sci. U.S.A. 106, 21013–21014. 10.1073/pnas.0912475107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisecaver J. H., Brosnahan M. L., Hackett J. D. (2013). Horizontal gene transfer is a significant driver of gene innovation in dinoflagellates. Genome Biol. Evol. 5, 2368–2381. 10.1093/gbe/evt179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. R., Shien J. H., Shieh H. K., Hu C. C., Gong S. R., Chen L. Y., et al. (2007). Cloning of the gene and characterization of the enzymatic properties of the monomeric alkaline phosphatase (PhoX) from Pasteurella multocida strain X-73. FEMS Microbiol. Lett. 7, 113–120. 10.1111/j.1574-6968.2006.00542.x [DOI] [PubMed] [Google Scholar]
- Xu Y., Wahlund T. M., Feng L., Shaked Y., Morel F. M. M. (2006). A novel alkaline phosphatase in the Coccolithophore Emiliania huxleyi (Prymnesiophyceae) and its regulation by phosphorus. J. Phycol. 42, 835–844. 10.1111/j.1529-8817.2006.00243.x [DOI] [Google Scholar]
- Yang Y., Wandler A. M., Postlethwait J. H., Guillemin K. (2012). Dynamic evolution of the LPS-detoxifying enzyme intestinal alkaline phosphatase in zebrafish and other vertebrates. Front. Immunol. 3:314. 10.3389/fimmu.2012.00314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong S. C., Roversi P., Lillington J., Rodriguez F., Krehenbrink M., Zeldin O. B., et al. (2014). A complex iron-calcium cofactor catalyzing phosphotransfer chemistry. Science 345, 1170–1173. 10.1126/science.1254237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikuni Y., Ferrin T. E., Keasling J. D. (2006). Designed divergent evolution of enzyme function. Nature 440, 1078–1082. 10.1038/nature04607 [DOI] [PubMed] [Google Scholar]
- Yu C. S., Chen Y. C., Lu C. H., Hwang J. K. (2006). Prediction of protein subcellular localization. Proteins 64, 643–651. 10.1002/prot.21018 [DOI] [PubMed] [Google Scholar]
- Zaheer R., Morton R., Proudfoot M., Yakunin A., Finan T. M. (2009). Genetic and biochemical properties of an alkaline phosphatase PhoX family protein found in many bacteria. Environ. Microbiol. 11, 1572–1587. 10.1111/j.1462-2920.2009.01885.x [DOI] [PubMed] [Google Scholar]
- Zalatan J. G., Fenn T. D., Brunger A. T., Herschlag D. (2006). Structural and functional comparisons of nucleotide pyrophosphatase/phosphodiesterase and alkaline phosphatase: implications for mechanism and evolution. Biochemistry 45, 9788–9803. 10.1021/bi060847t [DOI] [PubMed] [Google Scholar]
- Zhang H., Hou Y., Miranda L., Campbell D. A., Sturm N. R., Gaasterland T., et al. (2007). Spliced leader RNA trans-splicing in dinoflagellates. Proc. Natl. Acad. Sci. U.S.A. 104, 4618–4623. 10.1073/pnas.0700258104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Lin S. (2005). Development of a cob-18S rRNA gene real-time PCR assay for quantifying Pfiesteria shumwayae in the natural environment. Appl. Environ. Microb. 71, 7053–7063. 10.1128/AEM.71.11.7053-7063.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. (2003). Evolution by gene duplication: an update. Trends Ecol. Evol. 18, 292–298. 10.1016/S0169-5347(03)00033-8 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.