Abstract
NF-κB is generally believed to be pro-tumorigenic. Here, we report a tumor-suppressive function for NF-κB1, the prototypical member of NF-κB. While NF-κB1 down-regulation is associated with high lung cancer risk in humans and poor patient survival, NF-κB1 deficient mice are more vulnerable to lung tumorigenesis induced by the smoke carcinogen, urethane. Notably, the tumor suppressive function of NF-κB1 is independent of its classical role as an NF-κB factor, but instead through stabilization of the Tpl2 kinase. NF-κB1 deficient tumors exhibit “normal” NF-κB activity, but a decreased protein level of Tpl2. Reconstitution of Tpl2 or the NF-κB1 p105, but not p50 (the processed product of p105), inhibits the tumorigenicity of NF-κB1 deficient lung tumor cells. Remarkably, Tpl2 knockout mice resemble NF-κB1 knockouts in urethane-induced lung tumorigenesis. Mechanistic studies indicate that p105/Tpl2 signaling is required for suppressing urethane-induced lung damage and inflammation, and activating mutations of the K-Ras oncogene. These studies reveal an unexpected, NF-κB-independent but Tpl2-depenednt role of NF-κB1 in lung tumor suppression. These studies also reveal a previously unexplored role of p105/Tpl2 signaling in lung homeostasis.
Keywords: lung cancer, NF-κB1, p105, p50, Tpl2
Introduction
NF-κB is not a single protein, but a collection of five structurally-related inducible transcription factors that are able to bind to κb site-containing promoters to regulate gene expression (1). Upon the molecular identification in 1990 of NF-κB1 and RelA (also known as p65), two prototypical members of NF-κB, as the homologs of the avian retroviral oncoprotein v-Rel, a role for NF-κB in tumorigenesis was highly expected (2). Since then, the importance of NF-κB in tumor biology has been extensively investigated and is now widely appreciated (1–3). Although oncogenic NF-κB mutations are relatively rare and mainly occur in certain hematological malignancies, constitutive activation of NF-κB has been detected in almost all tumors, either hematological or solid (4, 5). More importantly, aberrant NF-κB activation has been suggested to be involved in all steps of tumor development, from initiation to promotion to progression, as well as resistance to cancer therapies (1, 3). Mechanistic studies indicate that NF-κB contributes to tumor pathogenesis both intrinsically and extrinsically. Within precancerous or cancerous cells, activated NF-κB induces a large range of genes not only to promote malignant cell survival, proliferation and metastasis but also to induce angiogenesis and tumorigenic inflammation (1, 3). Moreover, NF-κB activated in nontumor cells, particularly immune cells, also contributes to tumor pathogenesis indirectly through establishing a tumorigenic microenvironment (1, 3).
Intriguingly, the link between NF-κB and lung cancer has been established only quite recently, although lung cancer is the leading cause of cancer deaths worldwide (6–9). NF-κB can be activated by tobacco smoking, the major risk factor that accounts for approximately 87% of lung cancer cases (10, 11). Accordingly, NF-κB is persistently activated in human lung cancers, and aberrant NF-κB activation is associated with lung tumor progression and poor patient survival (12). In vitro cell line and in vivo animal studies involving NF-κB inhibition suggest that persistent NF-κB activation is one important mechanism underlying lung cancer development and therapy resistance (13–17).
Those studies, however, mainly focus on RelA, and their conclusions on the tumorigenic role of NF-κB in lung and other cancers are largely derived from the anti-tumor effects of NF-κB inhibition by knockout/knockdown of the NF-κB activator IKK, overexpression of the NF-κB inhibitor IκBα, or administration of IKK inhibitors (12–17). IKK and IκBα have many NF-κB-independent functions that are also implicated in tumorigenesis (18, 19). Furthermore, they control the activation of other members of the NF-κB family, besides RelA (1). Currently, it remains largely unknown whether and how individual NF-κB members are involved in lung and other cancers. Given the functional complexities of NF-κB members, addressing this question is of importance. It will not only advance our understanding of NF-κB’s role in tumorigenesis but also set up a basis for us to target NF-κB for cancer therapy. This is particularly true for NF-κB1. NF-κB1 protein exists as two forms, p105 and p50, the precursor and mature forms, respectively (1). NF-κB1 p50 is believed to be the most important functional partner of RelA, and the RelA/p50 heterodimer is often referred to as NF-κB. On the other hand, NF-κB1 p105 has two different functions related to NF-κB: serving as an inhibitor of NF-κB and the precursor of p50 (1). In addition, NF-κB1 p105 also has an NF-κB-independent function: binding to and stabilizing Tpl2 (also known as Cot), a kinase that was originally identified as a proto-oncoprotein (20, 21).
Using lung cancer as a model, here we demonstrate an unexpected tumor suppressive function for NF-κB1. Remarkably, NF-κB1 suppresses lung tumorigenesis independently of its NF-κB function but instead through p105 stabilization of Tpl2. The non-canonical p105/Tpl2 signaling pathway is required for maintaining pulmonary homeostasis under inflammation and oncogenic stress. Genetic deficiency of either p105 or Tpl2 makes animals susceptible to lung damage and inflammation, activating mutation of the K-Ras oncogene, and subsequent lung carcinogenesis in response to tobacco carcinogens. In addition to advancing our understanding of lung cancer and the complex functions of NF-κB1, these studies provide the first line of evidence demonstrating that the Tpl2 proto-oncoprotein may exert a tumor suppressive effect.
Results
Repression of NF-κB1 expression is involved in human lung cancer
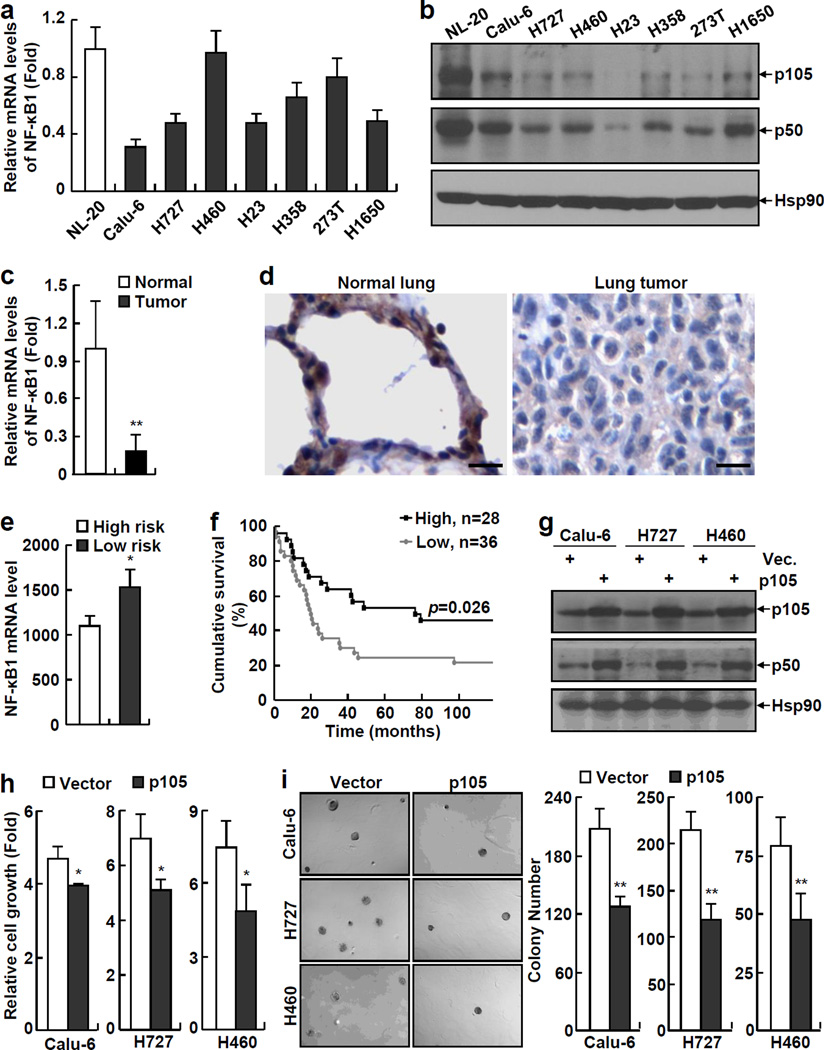
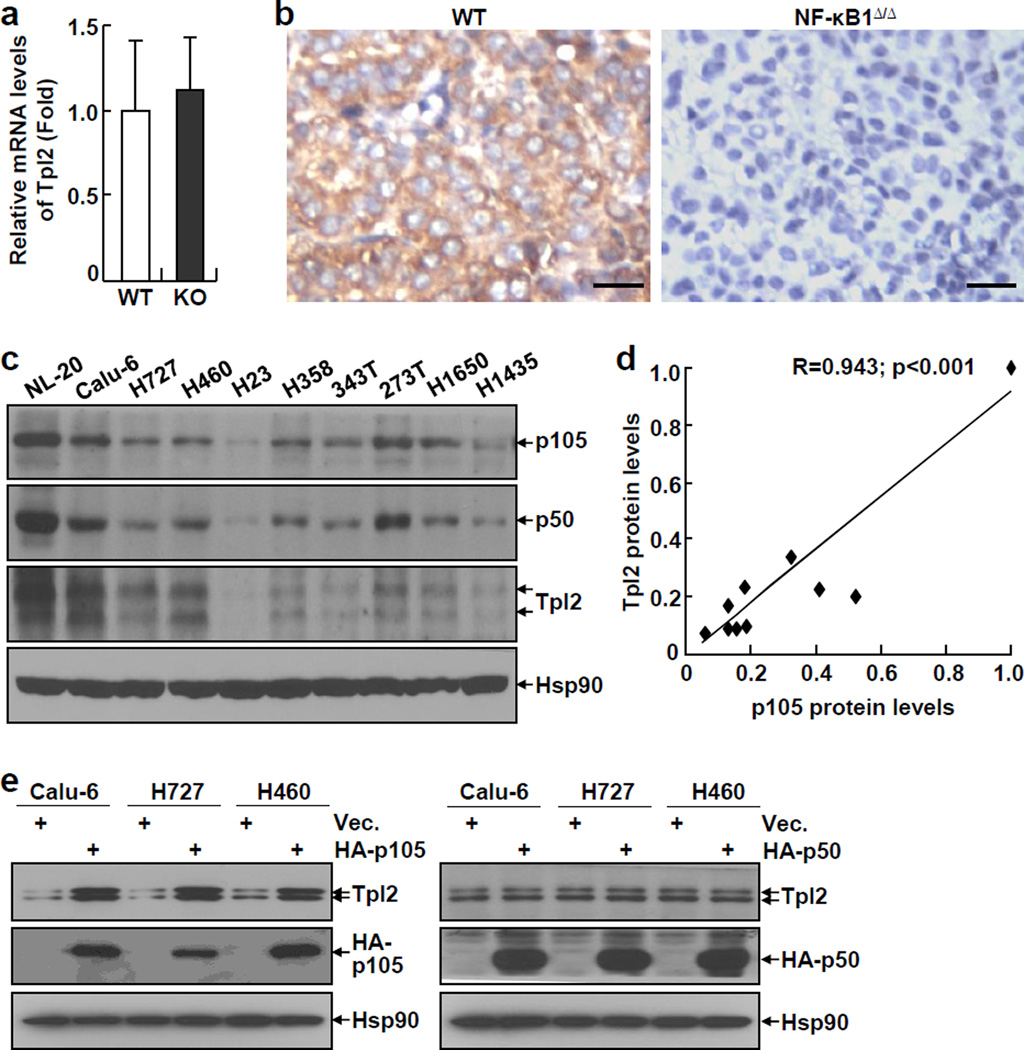
Although recent studies have suggested RelA as a promoter of lung cancer, the potential roles of other members of the NF-κB family in this deadliest form of human cancer have not yet been examined. To address this important issue, we initially examined the expression levels of NF-κB1, the most important functional partner of RelA, in a large range of human lung cancer cell lines. Surprisingly, most human lung cancer cell lines we examined expressed significantly lower levels of NF-κB1 mRNA, in comparison to the normal human lung epithelial cell line NL-20 (Figure 1a). In line with the decreased mRNA expression, the protein levels of both NF-κB1 p105 and p50 were significantly lower in those human lung cancer cell lines (Figure 1b, and Supplemental Figure S1). For simplicity, hereinafter, these cells are referred to as NF-κB1low lung cancer cells.
Figure 1.
NF-κB1 expression is decreased in human lung cancer cells and its re-expression inhibits the tumorigenicities of human lung cancer cells. (a) Real-time PCR assays showing decreased expression of NF-κB1 mRNA in human lung cancer cell lines. Normal human lung epithelial cell line NL-20 was used as a control. Data shown are means ± standard deviation (SD) (n = 3). (b) Immunoblotting (IB) assays showing decreased expression of NF-κB1 protein (p105 and p50) in human lung cancer cell lines. Hsp90 was used as a loading control. (c) Real-time PCR assays showing decreased expression of NF-κB1 mRNA in human primary lung tumor tissues. Normal control tissues from the same patients were used as controls. Data shown are means ± SD (n = 10; **, p < 0.01). (d) Immunohistochemical (IHC) analysis showing decreased expression of NF-κB1 protein in human primary lung tumor tissues. Scale bar: 20 µm. (e) Gene array assays showing an association between down-regulation of NF-κB1 mRNA and high risk of lung cancer in humans (The clinicopathological characteristics of patients used for the gene array were listed in the supplemental Table S1). (f) Gene array assays showing an association between low NF-κB1 mRNA expression and poor survival of patients with lung cancer. (g) IB assays confirming the expression of p105 and p50 proteins in NF-κB1 human lung cancer stable cell lines. (h) Cell growth assays showing decreased growth rate of NF-κB1 human lung cancer stable cell lines. Cells were cultured 3 days before growth assays. Data shown are means ± SD (n ≥ 3; *, p < 0.05). (i) Soft agar colony formation assays showing decreased anchorage-independent growth of NF-κB1 human lung cancer stable cell lines. Data shown are means ± SD (n ≥ 3; **, p < 0.01).
To test the clinical relevance of this finding, we examined the mRNA expression levels of NF-κB1 in human lung tumor tissues freshly isolated from patients and their matched normal control tissues from the same patients. Indeed, the mRNA expression level of NF-κB1 was significantly decreased in human primary lung tumor samples compared to their matched normal controls (Figure 1c). Immuno-histochemistry (IHC) staining using an antibody that can recognize both p105 and p50 proteins indicated the protein expression of NF-κB1 was also repressed in human primary lung cancer tissues (Figure 1d). Our gene array analysis further indicated that the down-regulation of NF-κB1 is associated with high risk of lung cancer in humans and poor patient survival (Figures 1e and 1f). These studies suggested that NF-κB1 repression is a clinically relevant event of human lung cancer.
To directly examine whether the repression of NF-κB1 expression contributes to the pathogenesis of human lung cancer, we reconstituted NF-κB1 cDNA, which expresses both p105 and p50 proteins, into NF-κB1low lung cancer cells (Figure 1g). Interestingly, NF-κB1 reconstitution alone was sufficient to block the growth of those NF-κB1low lung cancer cells in culture (Figure 1h). Moreover, NF-κB1 reconstitution also suppressed the anchorage-independent growth of those cancer cells (Figure 1i). On the other hand, further knockdown of NF-κB1 increased the growth of those cancer cells (Supplemental Figure S2). These data suggested that NF-κB1 down-regulation is one important mechanism underlying human lung cancer pathogenesis.
NF-κB1 knockout mice are prone to lung tumorigenesis induced by the smoke carcinogen urethane
To systemically investigate the role of NF-κB1 repression in lung cancer, we employed a well-accepted and widely-used mouse model of lung cancer induced by the tobacco carcinogen urethane. Urethane alone is sufficient to induce lung carcinogenesis in mice, and importantly, urethane-induced lung cancers in mice faithfully recapitulate human lung cancers, and in particular adenocarcinomas associated with tobacco smoking (22). We initially examined whether NF-κB1 deficiency in mice makes animals more sensitive to urethane-induced lung tumorigenesis.
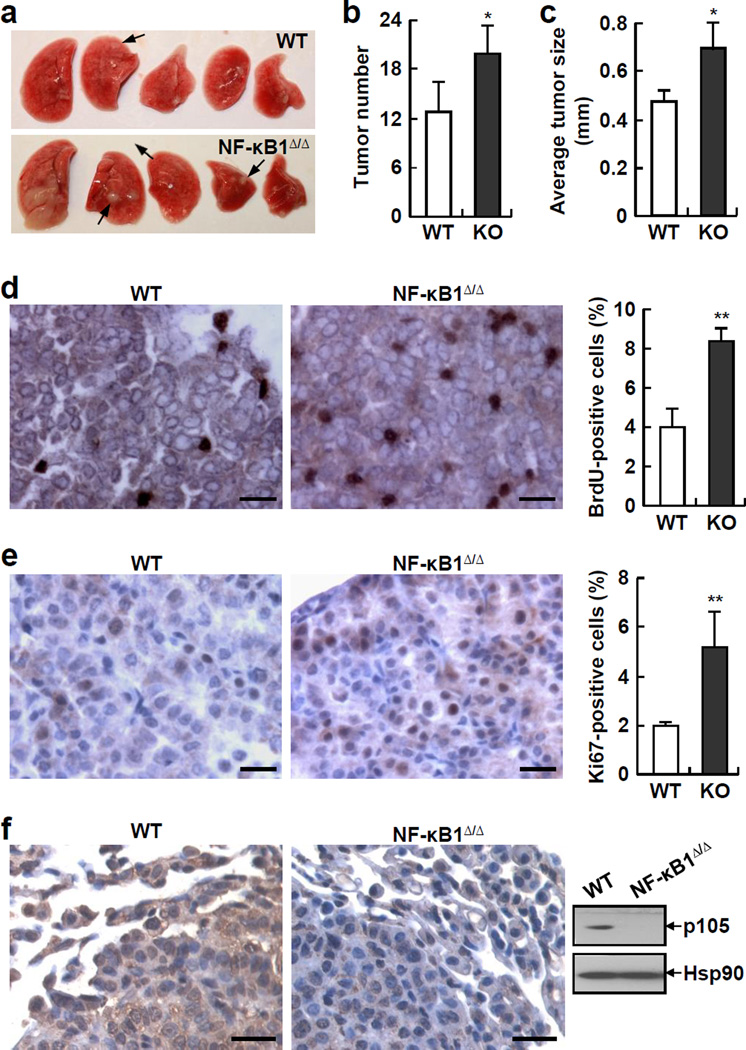
Consistent with previous findings that NF-κB1 is dispensable for lung development and function (23), NF-κB1 knockout (NF-κB1Δ/Δ) mice did not show apparent abnormalities in lung size or morphology (Figure 2a, and data not shown). Notably, NF-κB1Δ/Δ mice developed significantly more lung tumors than wild type (WT) mice after exposure to urethane (Figures 2a and 2b). Moreover, the tumors in NF-κB1Δ/Δ mice were bigger than those in WT mice, as evidenced by a significant increase in the average size of tumors (Figure 2c). Consistent with this observation, both BrdU and Ki-67 proliferation assays showed that in comparison with WT tumors, NF-κB1Δ/Δ tumors showed an increased cell proliferation rate (Figures 2d and 2e). Of note, the absence of NF-κB1 in the tumors from urethane-treated NF-κB1Δ/Δ mice was validated by IHC and immunoblotting (IB) assays (Figure 2f). Our IHC assays also showed that NF-κB1Δ/Δ mice had significantly more atypical adenomatous hyperplasia (AAH), adenomas (AD) and adenocarcinomas (AC) in their lungs (Supplemental Figure S3). These data clearly indicated that NF-κB1 deficiency contributes to both lung tumor development and progression in a urethane-induced lung tumor model.
Figure 2.
NF-κB1 knockout (NF-κB1Δ/Δ) mice are more susceptible to urethane-induced lung tumorigenesis than wild type (WT) mice. (a) Lung tissues from urethane-treated NF-κB1Δ/Δ mice and WT mice. Representative tumors are indicated by arrows. The large white nodules in the most left lung tissue of NF-kB1Δ/Δ mice are not tumors but actually damages associated with increased inflammation (see Figure 3). (b) Increased lung tumor multiplicities in urethane-treated NF-κB1Δ/Δ mice. Data shown are means ± SD (n ≥ 6; *, p < 0.05). (c) Increased average size of lung tumors in urethane-treated NF-κB1Δ/Δ mice. Data shown are means ± SD (n ≥ 5; *, p < 0.05). (d) BrdU labeling showing increased proliferation rate of lung tumors in urethane-treated NF-κB1Δ/Δ mice. Scale bar: 20 µm. BrdU-positive cells were also counted and represented as the percentage of total cells. Data shown are means ± SD (n ≥ 5; **, p < 0.01). (e) Ki-67 IHC staining showing increased proliferation rate of lung tumors in urethane-treated NF-κB1Δ/Δ mice. Scale bar: 20 µm. Ki-67-positive cells were also counted and represented as the percentage of total cells. Data shown are means ± SD (n ≥ 5; **, p < 0.01). (f) IHC staining and IB assays showing absence of NF-κB1 proteins in lung tumors from urethane-treated NF-κB1Δ/Δ mice. Scale bar: 20 µm.
Increased lung tumorigenesis in urethane-treated NF-κB1 knockout mice is associated with augmented lung damage, pro-tumorigenic inflammation and oncogenic K-Ras mutation
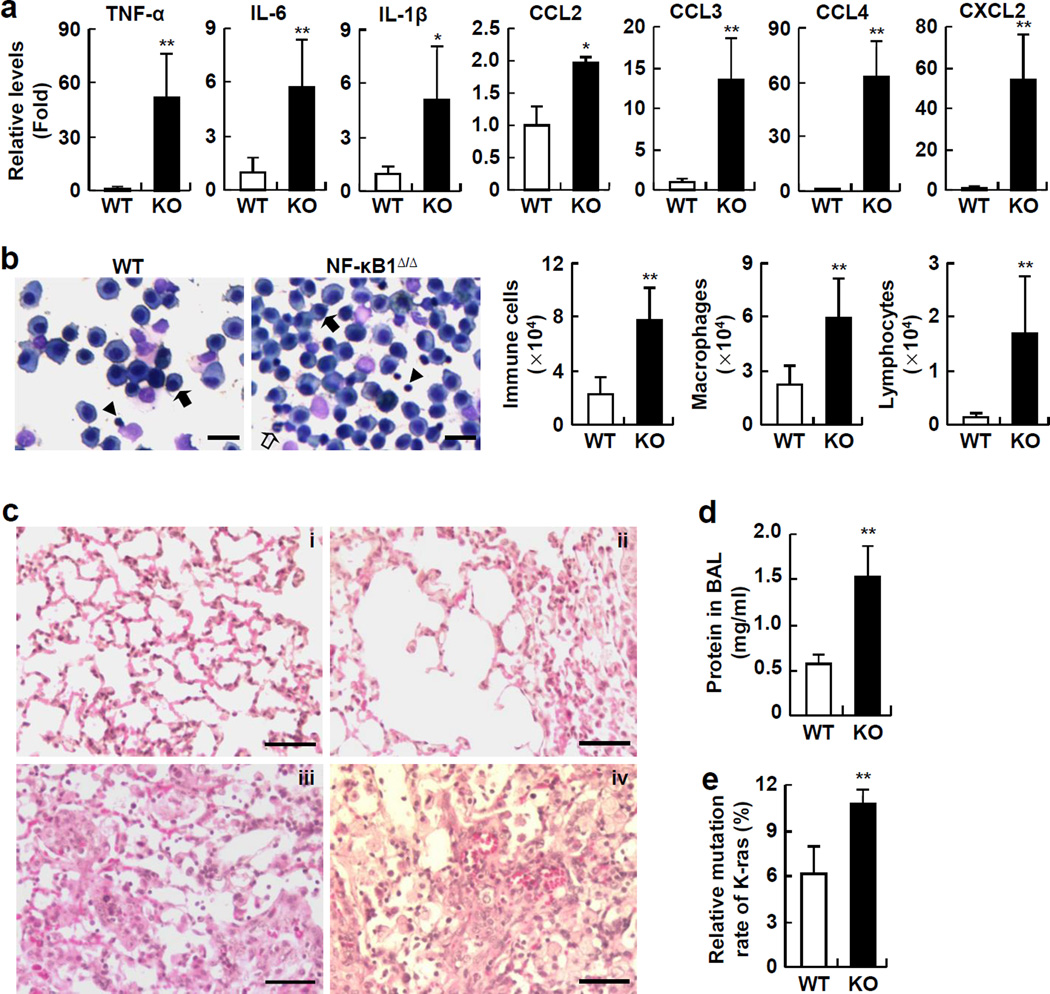
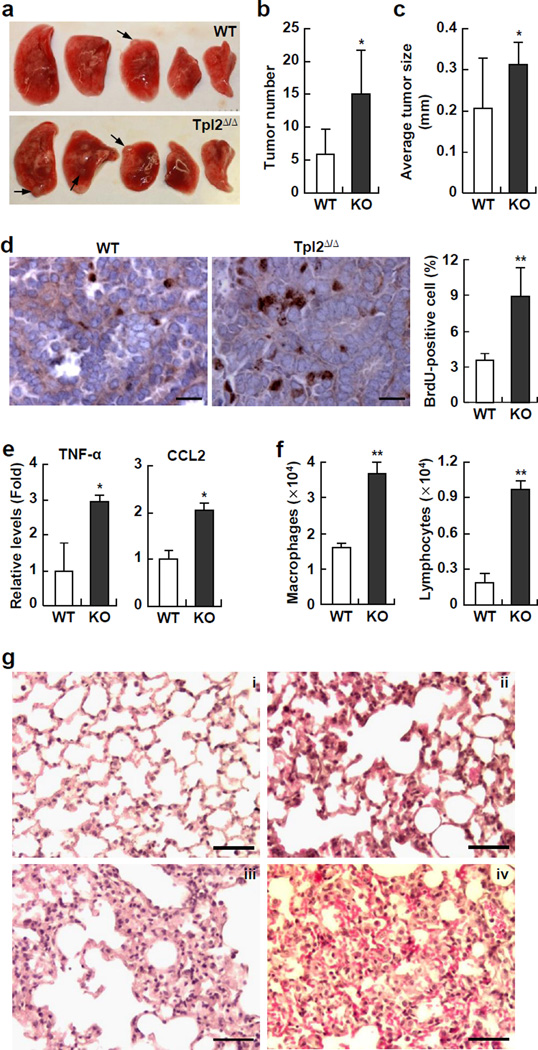
To investigate the mechanisms underlying the increased lung tumorigenesis in NF-κB1Δ/Δ mice, we examined the pulmonary inflammatory responses to urethane in NF-κB1Δ/Δ mice and WT mice, because recent studies indicated that pulmonary inflammation plays a critical role in the initiation and progression of lung cancer (24, 25). Interestingly, we found that several pro-inflammatory cytokines that can promote tumor initiation and progression, such as tumor necrosis factor-alpha (TNF-α), interleukin-1beta (IL-1β), and interleukin 6 (IL-6), were significantly higher in the lungs of NF-κB1Δ/Δ mice compared to WT mice (Figure 3a). In addition, the immune cell-attractive chemokines CCL2, CCL3, CCL4 and CXCL2, were also significantly increased in the lungs of urethane-treated NF-κB1Δ/Δ mice (Figure 3a). Consistently, significantly more immune cells were represented in bronchoalveolar lavage fluid (BALF) from NF-κB1Δ/Δ mice (Figure 3b). Further analysis indicated that lymphocytes and macrophages were increased in the BALF of NF-κB1Δ/Δ mice. These data suggested that NF-κB1 restricts urethane-induced pro-tumorigenic inflammation.
Figure 3.
NF-κB1Δ/Δ mice are more susceptible to urethane-induced lung damage, pro-tumorigenic inflammation and K-Ras oncogenic mutation than WT mice. (a) Real-time PCR assays showing increased expressions of pro-inflammatory cytokines and chemokines in lungs of urethane-treated NF-κB1Δ/Δ mice. Data shown are means ± SD (n ≥ 5; *, p < 0.05; **, p < 0.01). (b) Hema 3 staining assays showing increase in total immune cells, macrophages, and lymphocytes, in BALF from urethane-treated NF-κB1Δ/Δ mice. Representative macrophages and neutrophils and lymphocytes were indicated by the filled arrows, open arrows and arrowheads, respectively. Scale bar: 20 µm. Data shown are means ± SD (n ≥ 5; **, p < 0.01). (c) Histological analysis showing severe lung injury in urethane-treated NF-κB1Δ/Δ mice. Minor histological changes in the lungs of urethane-treated WT mice (i). Severe epithelial cell death, protein leak, thickened alveoli, perivascular edema and hemorrhage in the lungs of urethane-treated NF-κB1Δ/Δ mice (ii–iv). Scale bar: 50 µm. (d) BCATM protein assay showing higher protein concentration in BALF from urethane-treated NF-κB1Δ/Δ mice. Data shown are means ± SD (n = 5; **, p < 0.01). (e) Genomic sequencing showing increased K-Ras mutation frequency in the lungs of urethane-treated NF-κB1Δ/Δ mice. Data shown are means ± SD (n = 4; **, p < 0.01).
In association with the increased pulmonary inflammation in urethane-treated NF-κB1Δ/Δ mice, much more serious lung injury was observed in these mice (Figure 3c). Before urethane treatment, the lungs of NF-κB1Δ/Δ mice were normal and displayed the same morphology and histology as those of WT mice (data not shown). However, exposure to urethane caused significantly enhanced alveolar thickening and congestion, loss of integrity of the alveolar capillary membrane, as well as perivascular edema and hemorrhage, leading to air space enlargement, protein leak, and red blood cell extravasation in the lungs of NF-κB1Δ/Δ mice compared to WT mice (Figure 3c). In further support of these observations, an increased protein concentration was detected in the lung lavage fluid of urethane-treated NF-κB1Δ/Δ mice (Figure 3d). These data suggested that NF-κB1 protects lungs from urethane-induced pre-neoplastic injury.
One of the important roles of local inflammation and tissue damage is to establish a microenvironment that facilitates induction of oncogenic mutations and subsequent tumor initiation and progression. Thus, we compared the mutation frequency of the K-Ras oncogene in lung epithelium, an early event involved in lung tumorigenesis induced by urethane (26). We found that about 6% of lung epithelial cells in urethane-treated WT mice harbored K-Ras activating mutations (Figure 3e). However, the same urethane treatment resulted in K-Ras mutations in over 10% of lung epithelial cells of NF-κB1Δ/Δ mice. Collectively, these data suggested that NF-κB1 prevents urethane-induced K-Ras oncogenic mutations and lung tumorigenesis through maintaining pulmonary homeostasis.
NF-κB1 suppresses lung cancers independent of its NF-κB activity
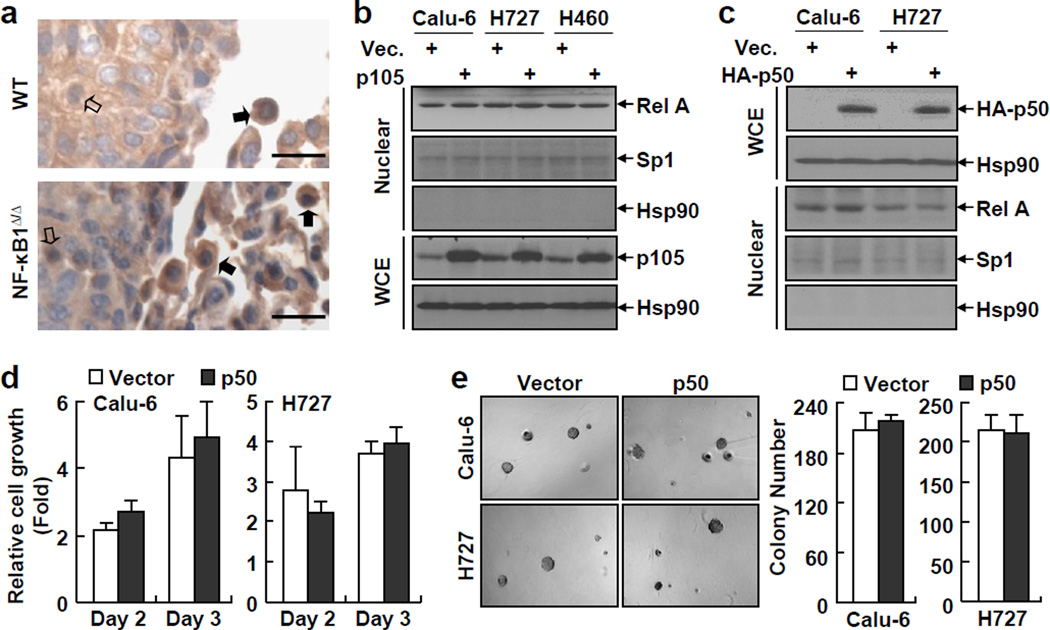
The human and mouse studies above clearly demonstrated a lung tumor suppressive role for NF-κB1. Thus, it is both interesting and important to determine the molecular mechanisms by which NF-κB1 suppresses lung cancer. First, we examined whether the tumor suppressive role of NF-κB1 involves its NF-κB inhibition function, since its precursor form, p105, can act as an inhibitor of NF-κB through sequestering NF-κB in the cytoplasm (1). Given the tumor promoting role of RelA in lung and other cancers (Supplemental Figure S4), we examined its nuclear expression levels in murine NF-κB1Δ/Δ or WT lung tumors. However, those murine tumors showed no difference in the nuclear expression of RelA (Figure 4a). As a matter of fact, the immune cells surrounding or within the murine NF-κB1Δ/Δ or WT lung tumors also exhibited comparable expressions of nuclear RelA. Consistently, p105 re-expression had no obvious effect on RelA’s nuclear expression in human NF-κB1low lung cancer cells (Figure 4b). These data suggested that the tumor suppressive role of NF-κB1 does not involve the NF-κB inhibitor function of p105.
Figure 4.
RelA activation in lung cancer cells is independent of NF-κB1 and expression of exogenous p50 has no effect on the tumorigenicities of NF-κB1low lung cancer cells. (a) IHC staining showing a similar RelA nuclear expression in NF-κB1Δ/Δ tumors and WT tumors from urethane-treated mice. Scale bar: 20 µm. Representatives of tumor cells and immune cells with positive staining of nuclear RelA are indicated by open and filled arrows, respectively. (b) IB assays showing no change in RelA nuclear expression in p105 human lung cancer stable cell lines. (c) IB assays confirming expression of exogenous HA-tagged p50 in human lung cancer stable cell lines and showing no change in RelA nuclear expression in these stable cell lines. (d) Cell growth assays showing no change in the growth of p50 human lung cancer stable cell lines. Data shown are means ± SD (n ≥ 3). (e) Soft agar colony formation assays showing no change in anchorage-independent growth of p50 human lung cancer stable cell lines. Data shown are means ± SD (n ≥ 3).
Next, we examined whether the tumor suppressive role of NF-κB1 is mediated by its mature form, p50. To do so, we stably expressed exogenous p50 to a level equal to the physiological level in human lung cancer cell lines in which endogenous NF-κB1 (both p105 and p50) is repressed (Figure 4c). As expected, re-expression of p50 did not affect the nuclear expression of RelA in those NF-κB1low lung cancer cells (Figure 4c). In addition, p50 re-expression had no effect on the tumorigenicities of those cancer cells, as evidenced by our cell proliferation and soft-agar colony formation assays (Figures 4d and 4e). Given our findings that p105/p50 re-expression is sufficient to suppress the tumorigenicities of NF-κB1low lung cancer cells (Figures 1g-1i), these data together suggested that the lung tumor suppressive role of NF-κB1 is mediated by its precursor form, p105, but independent of its NF-κB activities (p50 generation and p105 inhibition of NF-κB).
NF-κB1 p105 is required for the stabilization of the Tpl2 kinase in lung cancer cells
Previous studies suggested that p105, but not p50, is required for the stabilization of the Tpl2 kinase in murine macrophage under physiological conditions (21). Thus, we examined the mRNA and protein expression levels of Tpl2 in NF-κB1Δ/Δ or WT lung tumors from urethane-treated mice. While the mRNA expression levels of Tpl2 were comparable in these tumors, Tpl2 proteins could be detected only in WT tumors, but not in NF-κB1Δ/Δ tumors (Figures 5a and 5b). Similarly, we found that although there was no association between mRNA expression levels of NF-κB1 and Tpl2 (Supplemental Figure S5), their protein expression levels were highly associated with each other in human lung cancer cell lines (Figures 5c and 5d). These data suggested that NF-κB1 p105 is required for the protein stabilization of Tpl2 in both murine and human lung cancer cells.
Figure 5.
NF-κB1 p105 stabilizes Tpl2 in lung cancer cells. (a) Real-time PCR showing a similar expression of Tpl2 mRNA in NF-κB1Δ/Δ tumors and WT tumors from urethane-treated mice. Data shown are means ± SD (n ≥ 3). (b) IHC staining showing no Tpl2 protein in NF-κB1Δ/Δ tumors from urethane-treated mice. Scale bar: 20 µm. (c) IB assays showing the protein expression levels of p105, p50 and Tpl2 in human lung cancer cell lines. (d) Quantitation of IB assays in C showing a positive correlation in the protein expression levels of NF-κB1 and Tpl2 in human lung cancer cell lines. (e) IB assays showing restoration of Tpl2 protein expression in human lung cancer cell lines stably expressing HA-p105, but not HA-p50.
To confirm the role of NF-κB1 p105 in Tpl2 stabilization in lung cancer cells, we checked whether p105 re-expression could re-store the expression of Tpl2 proteins in human NF-κB1low lung cancer cell lines. In this regard, we took advantage of human NF-κB1low lung cancer cell lines stably expressing p105, p50, or an empty vector, which had already been generated (see above Figures 1 and 4). As expected, re-expression of p105, but not p50, increased expression levels of endogenous Tpl2 proteins in human NF-κB1low lung cancer cell lines (Figure 5e), although both p105 and p50 did not affect the mRNA expression levels of Tpl2 in those same cells (Supplemental Figure S6). These data indicated that NF-κB1 p105 stabilizes Tpl2 protein in lung cancer cells.
Reconstitution of Tpl2 inhibits the tumorigenicities of NF-κB1low lung cancer cells
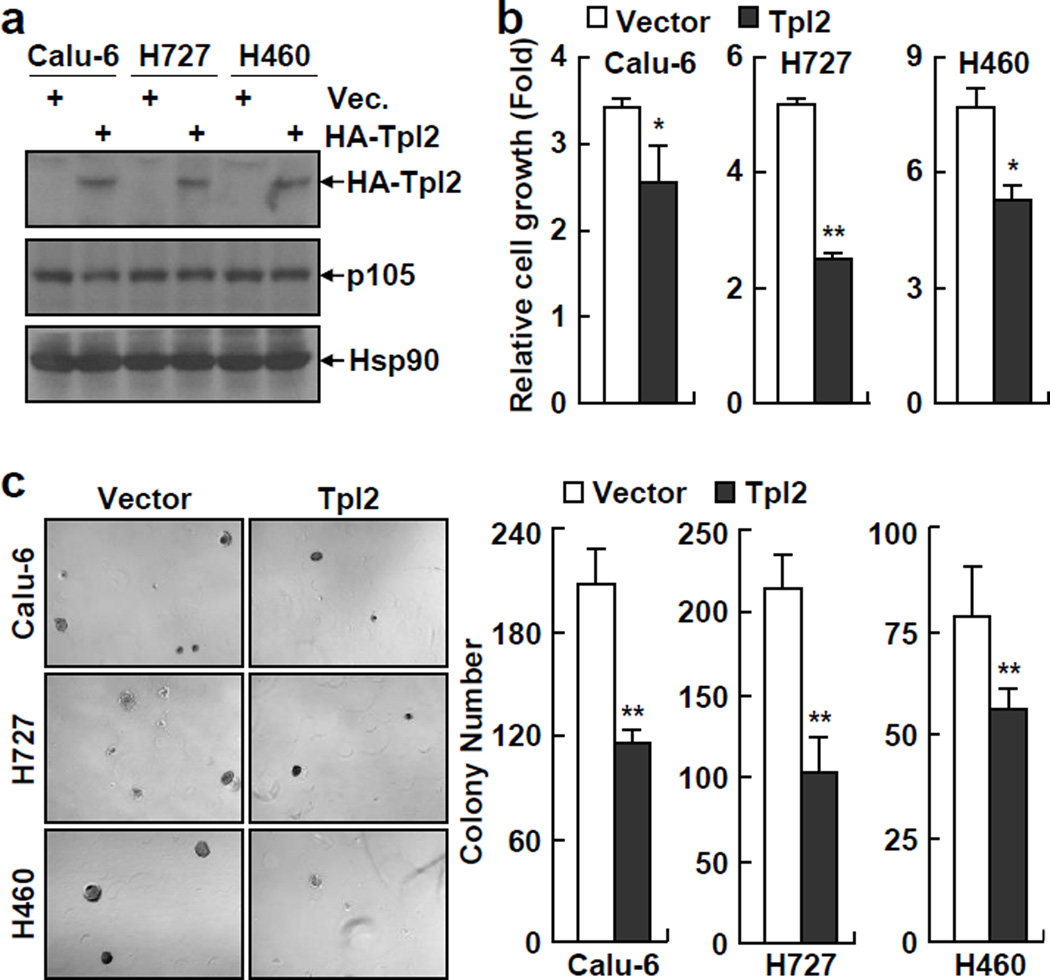
Although Tpl2 was originally identified as a proto-oncoprotein (27, 28), we examined whether p105 stabilization of Tpl2 protein accounts for the lung tumor suppressive function of NF-κB1, as our recent studies demonstrate that STAT3, a well-known tumor promoter that is often referred as an oncoprotein, actually suppresses lung cancer initiation when specifically knocked out from lung epithelial cells (29). In fact, a tumor suppressive role of Tpl2 has been suggested in colon cancer and T-cell lymphoma models (30, 31). Thus, we reconstituted Tpl2 into NF-κB1low lung cancer cells in which endogenous Tpl2 is disrupted due to p105 deficiency (Figure 6a). Consistent with the fact that Tpl2 functions as a downstream target of p105, Tpl2 reconstitution did not affect expression of p105 in those lung cancer cells (Figure 6a). However, interestingly enough, Tpl2 reconstitution blocked the growth in culture and colony formation in soft agar of those NF-κB1low lung cancer cells, exactly the same as p105 re-expression did (Figures 6b and 6c). On the other hand, knockdown of Tpl2 increased the growth of those cancer cells (Supplemental Figure S7). These data suggested that the lung tumor suppressive function of NF-κB1 is attributed to p105 stabilization of Tpl2. These data also suggested a lung tumor suppressive function for the Tpl2 proto-oncoprotein.
Figure 6.
Tpl2 expression blocks the tumorigenicities of human NF-κB1low lung cancer cells. (a) IB assays confirming the expression of exogenous HA-tagged Tpl2 in human lung cancer stable cell lines. (b) Cell growth assays showing decreased growth rate of Tpl2 human lung cancer stable cell lines. Cells were cultured 3 days before growth assays. Data shown are means ± SD (n ≥ 3; *, p < 0.05; **, p < 0.01). (c) Soft agar colony formation assays showing decreased anchorage-independent growth of Tpl2 human lung cancer stable cell lines. Data shown are means ± SD (n ≥ 3; **, p < 0.01).
Tpl2 knockout mice resemble NF-κB1 knockouts in urethane-induced lung tumorigenesis
To further validate whether Tpl2 works downstream of NF-κB1 p105 in lung cancer suppression, we examined whether Tpl2 knockout (Tpl2Δ/Δ) mice, like NF-κB1 knockouts, are prone to urethane-induced lung tumorigenesis. Indeed, Tpl2Δ/Δ knockout mice, like NF-κB1Δ/Δ mice, developed more and overall bigger lung tumors and have significant more AAH, AD and AC in their lungs than WT mice after exposure to urethane (Figures 7a–7c, and Supplemental Figure S8). Moreover, Tpl2Δ/Δ tumors, like NF-κB1Δ/Δ ones, showed an increased cell proliferation rate (Figure 7d). In agreement with our human lung cancer cell line studies showing that Tpl2 did not affect NF-κB1 expression, Tpl2Δ/Δ tumors expressed a similar level of NF-κB1 proteins compared to wild type tumors (Supplemental Figure S9). These data further support that Tpl2 works downstream of p105 in suppressing lung cancer.
Figure 7.
Tpl2 knockout (Tpl2Δ/Δ) mice are more susceptible to urethane-induced lung damage, pro-tumorigenic inflammation and tumorigenesis than WT mice. (a) Lung tissues from urethane-treated Tpl2Δ/Δ mice and WT mice. Representative tumors are indicated by arrows. (b) Increased lung tumor multiplicities in urethane-treated Tpl2Δ/Δ mice. Data shown are means ± SD (n ≥ 6; *, p < 0.05). (c) Increased average size of lung tumors in urethane-treated Tpl2Δ/Δ mice. Data shown are means ± SD (n ≥ 5; *, p < 0.05). (d) BrdU labeling showing increased proliferation rate of lung tumors in urethane-treated Tpl2Δ/Δ mice. Scale bar: 20 µm. BrdU-positive cells were also counted and represented as the percentage of total cells. Data shown are means ± SD (n ≥ 5; **, p < 0.01). (e) Real-time PCR assays showing increased expressions of TNF-α and CCL2 in the lungs of urethane-treated Tpl2Δ/Δ mice. Data shown are means ± SD (n ≥ 5; *, p < 0.05). (f) Hema 3 staining assays indicating increase in macrophages and lymphocytes in BALF from urethane-treated Tpl2Δ/Δ mice. Data shown are means ± SD (n ≥ 5; **, p < 0.01). (g) Histological analysis showing severe lung injury in urethane-treated Tpl2Δ/Δ mice. Severe epithelial cell death, protein leak, thickened alveoli, perivascular edema and hemorrhage in the lungs of urethane-treated Tpl2Δ/Δ mice were shown. Scale bar: 50 µm. i, WT mice; ii–iv, Tpl2Δ/Δ mice.
Remarkably, Tpl2Δ/Δ mice, like NF-κB1Δ/Δ mice, had a significantly elevated inflammation and injury in their lungs after exposure to urethane. The pro-inflammatory cytokine TNF-α and immune cell-attractive chemokine CCL2 were significantly increased in the lungs of Tpl2Δ/Δ mice, compared to WT mice (Figure 7e). Consistently, more inflammatory cells, particularly macrophages and lymphocytes, were found in the BALF from Tpl2Δ/Δ mice (Figure 7f). Moreover, Tpl2Δ/Δ mice showed more severe lung damage, as evidenced by enhanced alveolar thickening and congestion, loss of integrity of the alveolar capillary membrane, air space enlargement, protein leak, and red blood cell extravasation in their lungs (Figure 7g). These data clearly indicated that Tpl2Δ/Δ mice phenocopy NF-κB1 knockouts in urethane-induced lung tumorigenesis. All together, these studies strongly suggested that NF-κB1 restricts lung inflammation, maintains pulmonary homeostasis under oncogenic stress, and suppresses lung tumorigenesis through p105 stabilization of Tpl2, but independent of its NF-κB-related activities.
Discussion
Lung cancer accounts for approximately 1.2 million deaths annually worldwide, and roughly 85% of the patients with lung cancer die of the disease within five years (9). A better understanding of the mechanisms underlying lung cancer development is desperately needed to design new effective therapies for this deadliest cancer. Recent studies suggest that NF-κB, particularly its prototypical member RelA, is involved in lung cancer development. RelA activation in lung cancer is associated with disease progression and poor patient survival (12). On the other hand, genetic deletion of RelA significantly, although not completely, blocks lung tumorigenesis induced by the onogenic form of K-Ras in a mouse model (16). Thus, RelA provides a novel target for lung cancer therapy. However, like other transcription factors, RelA is believed to be undruggable. Most currently available inhibitors of NF-κB indirectly target its upstream activating kinase IKK, which activates not only RelA but also all other members of the NF-κB family, including NF-κB1, another prototypical NF-κB member and most important functional partner of RelA (1, 32). Given the functional complex of NF-κB, it is thus paramount important and interesting to delineate the role of NF-κB1 and other NF-κB members in lung cancer.
Astonishingly, both animal and human studies showed that in sharp contrast to RelA, NF-κB1 actually suppresses lung tumorigenesis. Genetic deletion of NF-κB1 makes animals more sensitive to lung cancer induction. Compared to wild type mice, NF-κB1 deficient mice developed much more and larger lung tumors after the same treatment of the smoke carcinogen urethane (Figure 2). Similarly, the expression of NF-κB1 is repressed in human lung cancer cell lines and primary tissues, and low expression of NF-κB1 is associated with high risk of lung cancer in humans and poor patient survival (Figure 1). More importantly, NF-κB1 re-expression in those lung cancer cells inhibits their tumorigenicity (Figure 1). These data indicate that NF-κB1 suppresses both initiation and progression of lung cancer. Of note, this is the first evidence directly showing a tumor suppressive role for a prototypical NF-κB member.
Remarkably, the lung tumor suppressive function of NF-κB1 is independent of its two NF-κB-related activities: generation of p50 and NF-κB inhibition by p105. As a matter of fact, the two NF-κB-related activities of NF-κB1 play a minor, if any, role in NF-κB activation in lung cancer cells. Although p105 re-expression reverses the tumor phenotypes of NF-κB1low lung cancer cells (Figure 1), it fails to block persistent RelA activation in those malignant cells (Figure 4), suggesting that p105 does not exhibit NF-κB inhibitor function in lung cancer cells. Conversely, p50 re-expression has no effect on the tumorigenicity of NF-κB1low lung cancer cells (Figure 4). This is the first evidence showing that NF-κB1 p50 is dispensable for cancer development. It also suggests that RelA can exert tumor promoting role in the absence of its most important functional partner, p50. Collectively, these findings significantly improve our understanding of the complex function of NF-κB, and further suggest that NF-κB1 exerts its tumor suppressive role in lung cancer via NF-κB-independent function of p105.
Our mechanistic studies indicate that p105 suppresses lung cancer initiation and progression through stabilization of the Tpl2 kinase. Human and mouse lung cancers defective in NF-κB1 are also defective in the expression of Tpl2 protein (Figure 5). Re-expression of p105, but not p50, restores Tpl2 protein expression in those NF-κB1low lung cancer cells, although both p105 and p50 do not affect the RNA expression level of Tpl2. More importantly, re-expression of Tpl2, like re-expression of p105, inhibits the tumorigenicity of NF-κB1low lung cancer cells (Figures 1 versus 6), and Tpl2 knockout mice faithfully recapitulate NF-κB1 knockouts in urethane-induced lung tumorigenesis (Figures 2 and 3 versus Figure 7).
Interestingly, our studies suggest that the lung tumor suppressive functions of the noncanonical p105/Tpl2 signaling pathway involve its different abilities in regulating the growth of normal and malignant lung cells. It suppresses lung cancer induction through maintaining lung epithelial survival and homeostasis under inflammatory and oncogenic stress, whereas it inhibits lung cancer progression via blocking cancer cell growth (Figures 1–7). In addition, the p105/Tpl2 signaling pathway also suppresses activation of immune cells, particularly macrophages and lymphocytes, restricting inflammation under oncogenic stress. In association with an increase in the incidence and size of lung cancer, p105 or Tpl2 knockout mice display an increased pulmonary inflammation and injury in response to urethane (Figures 2, 3 and 7). Lung damage and inflammation enhance each other and facilitate K-Ras activating mutations (Figure 3). Activated K-Ras in turn induces and exacerbates lung inflammation and damage (Figure 3). In this way, lung damage, inflammation and K-Ras mutations function as extrinsic and/or intrinsic forces driving lung cancer initiation and progression. Loss of the p105/Tpl2 signaling in cancer cells also directly contributes to lung cancer growth. It seems that the intrinsic p105/Tpl2 signaling within lung cancer cells activates the mitogen-activated protein kinase kinase 7 (MKK7) to activate the c-Jun N-terminal kinase (JNK), which in turn induces the activation of the tumor suppressor p53, resulting in proliferation inhibition and apoptosis induction of cancer cells (33, 34). In support of this, lung cancer cells defective in p105 or Tpl2 have higher proliferation rates, and re-expression of p105 or Tpl2 suppresses growth of those lung cancer cells (Figures 1 and 6).
In summary, the data presented here demonstrate that in contrast to the tumor promoting role of RelA, NF-κB1 suppresses lung cancer development and progression. The tumor suppressive function of NF-κB1 is mediated by p105 stabilization of Tpl2, but independent of its NF-κB function. On one hand, the non-canonical p105/Tpl2 signaling pathway contributes to lung cell survival and pulmonary homeostasis under inflammation and oncogenic stress, therefore suppressing lung tumorigenesis. On the other hand, the p105/Tpl2 signaling pathway within lung cancer cells directly inhibits cancer cell growth. Moreover, the p105/Tpl2 signaling pathway also limits immune cell activation and restricts pulmonary inflammation under oncogenic stress. These findings describe a previously unidentified role of the p105/Tpl2 signaling pathway in lung cancer suppression, and therefore significantly improve our understanding of this deadliest human cancer. These findings also help us understand the complex functions of NF-κB.
Materials and Methods
Illumina Microarray Analysis
The gene array assay was performed essentially as previously described (35). Briefly, total RNAs were extracted from formalin fixed paraffin embedded (FFPE) human lung cancer tissues. The integrity of the RNAs was examined by using the Agilent platform (2100 Bioanalyzer; Agilent Technologies, Palo Alto, CA). High quality total RNA was labeled using Illumina TotalPrep RNA Amplification kit from Ambion and hybridized on the Illumina Whole-Genome DASL Assay (HT-12 V4 Bead Chip platform) according to the manufacturer’s instructions. The studies were approved by the University of Pittsburgh Institutional Review Board (IRB).
Animals
NF-κB1Δ/Δ mice and Tpl2Δ/Δ mice under C57BL/6 background were from Jackson Laboratory (Bar Harbor, ME, USA) and Dr. Philip N Tsichlis (Tufts University, Boston, MA, USA). Since C57BL/6 mice are resistant to urethane-induced lung tumorigenesis (15), those mice were backcrossed to WT FVB/N mice (Jackson Laboratory, Bar Harbor, ME, USA) for more than ten generations for pure FVB/N background. All animals were housed under specific pathogen-free conditions at the Hillman Cancer Center of the University of Pittsburgh Cancer Institute. Animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Pittsburgh.
Lung Carcinogenesis
For urethane induction of lung tumors, six to eight week old NF-κB1Δ/Δ mice Tpl2Δ/Δ mice, or wild type FVB/N mice were intraperitoneally (i.p.) injected with urethane (1 mg/g body weight, Sigma-Aldrich, St Louis, MO, USA) one time per week for six weeks. Six weeks post-urethane treatment, all mice were sacrificed for lung tumor examinations.
Bronchioalveolar Lavage (BAL)
Mice were sacrificed, and their lungs were lavaged three times with phosphate buffered saline (PBS). The recovered BAL fluids (BALF) were centrifuged. Cells from BALF were visualized by Hema3-staining. BALF total cell counts were determined by using a grid hemocytometer, and numbers of different leukocytes were obtained by counting at least 400 cells on Hema3-stained cytocentrifuge slides. Supernatants of BALF were subjected to the Bicinchoninic Acid (BCA)TM protein assay (Thermo Scientific, Rockford, IL, USA) for protein quantitation.
Lung Tumor Enumeration
Surface tumors in mouse lungs were counted by three blinded readers under a dissecting microscope. Tumor diameters were determined by microcalipers.
Histology and Immunohistochemistry (IHC)
Mouse and human lung tissues were fixed in formalin, embedded in paraffin, and cut into 5¼m-thick sections. Sections were stained with H&E, or subjected to IHC as previously described (29, 36). The following primary antibodies were used: anti-p105/50 (ab32360)(Abcam, Cambridge, MA, USA), anti-Tpl2 (sc-720) and anti-Ki-67 (sc-7846)(Santa Cruz Biotechnology, Dallas, TX, USA).
BrdU Labeling
Mice were i.p. injected with 50 mg/kg BrdU (Sigma-Aldrich, St Louis, MO, USA) 24 hrs prior to sacrifice. Mouse lung tissue sections were stained with anti-BrdU (Sigma-aldrich, St Louis, MO, USA). More than 3000 cells per lung were counted in randomly selected fields. BrdU labeling index was calculated as the percentage of labeled cells per total cells counted as described previously (37).
Quantitative Polymerase Chain Reaction (qPCR) Analysis
Mouse lung tissues, BAL cells, or type II epithelial cells were subjected to RNA extraction, RNA reverse transcription and real-time PCR as described (28–40). The expression levels of the indicated genes were normalized to that of ribosomal 18S within the same cells. Primer pairs used for qPCR are listed in the supplemental Table S2.
Isolation of Lung Type II Epithelial Cells
The detailed protocol has been described before (29). Briefly, crude lung cell suspensions were prepared by intratracheal instillation of Dispase and agarose followed by mechanical disaggregation of the lungs. Crude cell suspensions were purified by negative selection using a biotinylated-antibody, streptavidin-coated biomagnetic particle system. Cell purities were determined by Pap staining and confirmed ultrastructurally. Purified ATII cells were cultured on fibronectin-coated chamber slides and maintained for up to 5 days in DMEM with 10% fetal bovine serum (FBS).
K-Ras Sequencing
The exon 3 of the K-Ras gene was amplified by PCR using AccuPrime Pfx polymerase (Invitrogen, Grand Island, NY, USA) for DNA sequencing. The mutation frequencies of K-Ras in type II lung epithelial cells were determined by the ratios of the area of the mutated nucleotide versus that of wild type nucleotide of K-Ras in the sequencing map as described before (29).
Retroviral Transduction and Generation of Stable cDNA Transfectants
All the experiments involving virus were performed under Biohazard Safety Level BSL 2+ conditions. HEK293T cells were transfected with the GFP-expressing retroviral vector pCLXSN(GFP) alone or vector simultaneously expressing p105, p50 or Tpl2 using FuGENE 6 (Roche Applied Science) followed by viral supernatant collection and infection of human lung cancer cell lines as described (41). Stable transfectants were obtained by GFP sorting.
Colony Formation Assays
Soft agar assays were performed as previously described (42). Briefly, cell suspensions in culture medium containing 0.6% SeaPlaque low melting agarose were plated on the top of 1% agarose in culture medium. Colony growth was scored after 21 days of cell incubation. All the colony formation assays presented in this study were repeated in at least 3 independent experiments.
Immunoblotting (IB) Analysis
Whole cell extracts (WCE) were prepared by lysing cells in radioimmune precipitation assay buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.25% (w/v) sodium deoxycholate, 1% (v/v) Nonidet P-40, 1 mM dithiothreitol) supplemented with 1 mM phenylmethylsulfonyl fluoride. Cell nuclear extracts were prepared using hypertonic buffer (20 mM HEPES, pH 8.0, 1 mM EDTA, 20% (v/v) glycerol, 0.1% (v/v) Triton X-100, and 400 mM NaCl) after the cytoplasmic fraction was extracted by the hypotonic buffer (20 mM HEPES, pH 8.0, 10 mM KCl, 1 mM MgCl2, 0.1% (v/v) Triton X-100, and 20% (v/v) glycerol). The purity of the nuclear fractions was confirmed by detecting expression of Sp1 (nuclear marker), but not Hsp90 (cytoplasm marker). The whole cell lysates and nuclear extracts were subjected to SDS-PAGE and immunoblotting using the indicated antibodies as described previously (43).
Statistical Analysis
Data were reported as mean ± standard deviation (SD). The Student’s t test (two tailed) was used to assess significance of differences between two groups, and p values < 0.05 and 0.01 were considered statistically significant and highly statistically significant, respectively (44). Log-rank tests were used to compare overall survival (OS) between high and low NF-κB1 expression (45).
Supplementary Material
Acknowledgments
We thank Dr. Philip N Tsichlis for providing us the Tpl2Δ/Δ mice. This study was supported in part by the National Institute of Health (NIH)/National Cancer Institute (NCI) grants R01 CA172090, R21 CA175252, R21 CA189703, P30 CA047904, P50 CA090440-Lung Cancer Developmental Research Award, as well as the American Lung Association (ALA) Lung Cancer Discovery Award and American Cancer Society (ACS) Fellowship PF-12-081-01-TBG.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Fu J, Xiao G. NF-κB and cancer: a paradigm of Yin-Yang. Am J Cancer Res. 2011;1:192–221. [PMC free article] [PubMed] [Google Scholar]
- 2.Gilmore TD. The Rel/NF-κB/IκB signal transduction pathway and cancer. Cancer Treat Res. 2003;115:241–265. [PubMed] [Google Scholar]
- 3.Karin M, Greten FR. NF-κB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 4.Xiao G, Rabson AB, Young W, Qing G, Qu Z. Alternative pathways of NF-κB activation: a double-edged sword in health and disease. Cytokine Growth Factor Rev. 2006;17:281–293. doi: 10.1016/j.cytogfr.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Rayet B, Gélinas C. Aberrant rel/nfκb genes and activity in human cancer. Oncogene. 1999;18:6938–6947. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 6.Wong KK, Jacks T, Dranoff G. NF-κB fans the flames of lung carcinogenesis. Cancer Prev Res (Phila) 2010;3:403–405. doi: 10.1158/1940-6207.CAPR-10-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai Z, Tchou-Wong KM, Rom WN. NF-κB in lung tumorigenesis. Cancers (Basel) 2011;3:4258–4268. doi: 10.3390/cancers3044258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W, Li Z, Bai L, Lin Y. NF-κB in lung cancer, a carcinogenesis mediator and a prevention and therapy target. Front Biosci (Landmark Ed) 2011;16:1172–1185. doi: 10.2741/3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 10.Wen J, Fu JH, Zhang W, Guo M. Lung carcinoma signaling pathways activated by smoking. Chin J Cancer. 2011;30:551–558. doi: 10.5732/cjc.011.10059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hecht SS. Cigarette smoking and lung cancer: chemical mechanisms and approaches to prevention. Lancet Oncol. 2002;3:461–469. doi: 10.1016/s1470-2045(02)00815-x. [DOI] [PubMed] [Google Scholar]
- 12.Zhang D, Jin X, Wang F, Wang S, Deng C, Gao Z, et al. Combined prognostic value of both RelA and IκB-α expression in human non-small cell lung cancer. Ann Surg Oncol. 2007;14:3581–3592. doi: 10.1245/s10434-007-9560-z. [DOI] [PubMed] [Google Scholar]
- 13.Bassères DS, Ebbs A, Cogswell PC, Baldwin AS. IKK is a therapeutic target in KRAS-Induced lung cancer with disrupted p53 activity. Genes Cancer. 2014;5:41–55. doi: 10.18632/genesandcancer.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, et al. Requirement for NF-κB signaling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stathopoulos GT, Sherrill TP, Cheng DS, Scoggins RM, Han W, Polosukhin VV, et al. Epithelial NF-κB activation promotes urethane-induced lung carcinogenesis. Proc Natl Acad Sci USA. 2007;104:18514–18519. doi: 10.1073/pnas.0705316104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bassères DS, Ebbs A, Levantini E, Baldwin AS. Requirement of the NF-κB subunit p65/RelA for K-Ras-induced lung tumorigenesis. Cancer Res. 2010;70:3537–3546. doi: 10.1158/0008-5472.CAN-09-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones DR, Broad RM, Madrid LV, Baldwin AS, Jr, Mayo MW. Inhibition of NF-κB sensitizes non-small cell lung cancer cells to chemotherapy-induced apoptosis. Ann Thorac Surg. 2000;70:930–936. doi: 10.1016/s0003-4975(00)01635-0. [DOI] [PubMed] [Google Scholar]
- 18.Chariot A. The NF-κB-independent functions of IKK subunits in immunity and cancer. Trends Cell Biol. 2009;19:404–413. doi: 10.1016/j.tcb.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Mulero MC, Ferres-Marco D, Islam A, Margalef P, Pecoraro M, Toll A, et al. Chromatin-bound IκBα regulates a subset of polycomb target genes in differentiation and cancer. Cancer Cell. 2013;24:151–166. doi: 10.1016/j.ccr.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belich MP, Salmerón A, Johnston LH, Ley SC. TPL-2 kinase regulates the proteolysis of the NF-κB-inhibitory protein NF-κB1 p105. Nature. 1999;397:363–368. doi: 10.1038/16946. [DOI] [PubMed] [Google Scholar]
- 21.Waterfield MR, Zhang M, Norman LP, Sun SC. NF-κB1/p105 regulates lipopolysaccharide-stimulated MAP kinase signaling by governing the stability and function of the Tpl2 kinase. Mol Cell. 2003;11:685–694. doi: 10.1016/s1097-2765(03)00070-4. [DOI] [PubMed] [Google Scholar]
- 22.Tuveson DA, Jacks T. Modeling human lung cancer in mice: similarities and shortcomings. Oncogene. 1999;18:5318–5324. doi: 10.1038/sj.onc.1203107. [DOI] [PubMed] [Google Scholar]
- 23.Sha WC, Liou HC, Tuomanen EI, Baltimore D. Targeted disruption of the p50 subunit of NF-κB leads to multifocal defects in immune responses. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 24.Dougan M, Li D, Neuberg D, Mihm M, Googe P, Wong KK, et al. A dual role for the immune response in a mouse model of inflammation-associated lung cancer. J Clin Invest. 2011;121:2436–2446. doi: 10.1172/JCI44796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu Z, Sun F, Zhou J, Li L, Shapiro SD, Xiao G. Interleukin-6 prevents the initiation but enhances the progression of lung cancer. Cancer Res. 2015 doi: 10.1158/0008-5472.CAN-14-3042. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ichikawa T, Yano Y, Uchida M, Otani S, Hagiwara K, Yano T. The activation of K-ras gene at an early stage of lung tumorigenesis in mice. Cancer Lett. 1996;107:165–170. doi: 10.1016/0304-3835(96)04351-0. [DOI] [PubMed] [Google Scholar]
- 27.Aoki M, Akiyama T, Miyoshi J, Toyoshima K. Identification and characterization of protein products of the cot oncogene with serine kinase activity. Oncogene. 1991;6:1515–1519. [PubMed] [Google Scholar]
- 28.Patriotis C, Makris A, Bear SE, Tsichlis PN. Tumor progression locus 2 (Tpl-2) encodes a protein kinase involved in the progression of rodent T-cell lymphomas and in T-cell activation. Proc Natl Acad Sci USA. 1993;90:2251–2255. doi: 10.1073/pnas.90.6.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou J, Qu Z, Yan S, Sun F, Whitsett JA, Shapiro SD, et al. Differential roles of STAT3 in the initiation and growth of lung cancer. Oncogene. 2014 doi: 10.1038/onc.2014.318. advance online publication 6 October 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serebrennikova OB, Tsatsanis C, Mao C, Gounaris E, Ren W, Siracusa LD, et al. Tpl2 ablation promotes intestinal inflammation and tumorigenesis in Apcmin mice by inhibiting IL-10 secretion and regulatory T-cell generation. Proc Natl Acad Sci USA. 2012;109:E1082–E1091. doi: 10.1073/pnas.1115098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsatsanis C, Vaporidi K, Zacharioudaki V, Androulidaki A, Sykulev Y, Margioris AN, et al. Tpl2 and ERK transduce antiproliferative T cell receptor signals and inhibit transformation of chronically stimulated T cells. Proc Natl Acad Sci USA. 2008;105:2987–2992. doi: 10.1073/pnas.0708381104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilmore TD, Herscovitch M. Inhibitors of NF-κB signaling: 785 and counting. Oncogene. 2006;25:6887–6899. doi: 10.1038/sj.onc.1209982. [DOI] [PubMed] [Google Scholar]
- 33.Gkirtzimanaki K, Gkouskou KK, Oleksiewicz U, Nikolaidis G, Vyrla D, Liontos M, et al. TPL2 kinase is a suppressor of lung carcinogenesis. Proc Natl Acad Sci USA. 2013;110:E1470–E1479. doi: 10.1073/pnas.1215938110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schramek D, Kotsinas A, Meixner A, Wada T, Elling U, Pospisilik JA, et al. The stress kinase MKK7 couples oncogenic stress to p53 stability and tumor suppression. Nat Genet. 2011;43:212–219. doi: 10.1038/ng.767. [DOI] [PubMed] [Google Scholar]
- 35.Qing G, Qu Z, Xiao G. Endoproteolytic processing of C-terminally truncated NF-κB2 precursors at κB-containing promoters. Proc Natl Acad Sci USA. 2007;104:5324–5329. doi: 10.1073/pnas.0609914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun F, Xiao Y, Qu Z. Oncovirus KSHV represses tumor suppressor PDLIM2 to persistently activate NF-κB and STAT3 transcription factors for tumorigenesis and tumor maintenance. J Biol Chem. 2015;290:7362–7368. doi: 10.1074/jbc.C115.637918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qu Z, Sun D, Young W. Lithium promotes neural precursor cell proliferation: evidence for the involvement of the non-canonical GSK-3β-NF-AT signaling. Cell Biosci. 2011;1:18. doi: 10.1186/2045-3701-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qu Z, Fu J, Ma H, Zhou J, Jin M, Mapara MY, Grusby MJ, Xiao G. PDLIM2 restricts Th1 and Th17 differentiation and prevents autoimmune disease. Cell Biosci. 2012;2:23. doi: 10.1186/2045-3701-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan P, Fu J, Qu Z, Li S, Tanaka T, Grusby MJ, et al. PDLIM2 suppresses HTLV-I Tax-mediated tumorigenesis by targeting Tax into the nuclear matrix for proteasomal degradation. Blood. 2009;113:4370–4380. doi: 10.1182/blood-2008-10-185660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu J, Qu Z, Yan P, Ishikawa C, Aqeilan RI, Rabson AB, et al. The tumor suppressor gene WWOX links the canonical and noncanonical NF-κB pathways in HTLV-I Tax-mediated tumorigenesis. Blood. 2011;117:1652–1661. doi: 10.1182/blood-2010-08-303073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu J, Yan P, Li S, Qu Z, Xiao G. Molecular determinants of PDLIM2 in suppressing HTLV-I Tax-mediated tumorigenesis. Onocogene. 2010;29:6499–6507. doi: 10.1038/onc.2010.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qu Z, Yan P, Fu J, Jiang J, Grusby MJ, Smithgall TE, et al. DNA methylation-dependent repression of PDLIM2 in colon cancer and its role as a potential therapeutic target. Cancer Res. 2010;70:1766–1772. doi: 10.1158/0008-5472.CAN-09-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qing G, Qu Z, Xiao G. Stabilization of basally translated NF-κB-inducing kinase (NIK) protein functions as a molecular switch of processing of NF-κB2 p100. J Biol Chem. 2005;280:40578–40582. doi: 10.1074/jbc.M508776200. [DOI] [PubMed] [Google Scholar]
- 44.Qu Z, Fu J, Yan P, Hu J, Cheng SY, Xiao G. Epigenetic repression of PDLIM2: implications for the biology and treatment of breast cancer. J Biol Chem. 2010;285:11786–11792. doi: 10.1074/jbc.M109.086561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stabile LP, Dacic S, Land SR, Lenzner DE, Dhir R, Acquafondata M, et al. Combined analysis of estrogen receptor beta-1 and progesterone receptor expression identifies lung cancer patients with poor outcome. Clin Cancer Res. 2011;17:154–164. doi: 10.1158/1078-0432.CCR-10-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.