Abstract
The molecular motors kinesin and dynein drive bidirectional motility along microtubules (MTs) in most eukaryotic cells. Land plants, however, are a notable exception, because they contain a large number of kinesins but lack cytoplasmic dynein, the foremost processive retrograde transporter. It remains unclear how plants achieve retrograde cargo transport without dynein. Here, we have analysed the motility of the six members of minus-end-directed kinesin-14 motors in the moss Physcomitrella patens and found that none are processive as native dimers. However, when artificially clustered into as little as dimer of dimers, the type-VI kinesin-14 (a homologue of Arabidopsis KCBP (kinesin-like calmodulin binding protein)) exhibited highly processive and fast motility (up to 0.6 μm s−1). Multiple kin14-VI dimers attached to liposomes also induced transport of this membrane cargo over several microns. Consistent with these results, in vivo observations of green fluorescent protein-tagged kin14-VI in moss cells revealed fluorescent punctae that moved processively towards the minus-ends of the cytoplasmic MTs. These data suggest that clustering of a kinesin-14 motor serves as a dynein-independent mechanism for retrograde transport in plants.
Organelle transport in plant cells has generally been considered to be actin and myosin dependent (for example cytoplasmic streaming)1. However, MT-based motility has also been observed in some plant systems and is plausibly dependent on kinesin, another class of cytoskeletal motor1–4. Kinesins constitute a large superfamily, the founding member of which (kinesin-1) forms homodimers that take many steps along a MT towards the plus end before dissociating. Such processive movement allows this kinesin to function efficiently in the long distance anterograde transport of cargo5–8. Within the kinesin superfamily, the kinesin-14 motors are distinct from other kinesin families in that they display minus-end-directed movement, and are therefore potential retrograde transporters9. Recently Kar3, an atypical kinesin-14 present in budding yeast, was shown to move processively towards minus-ends via heterodimerization with a non-motor subunit10,11. However, none of the animal or plant kinesin-14s characterized to date, which form homodimers, have shown fast and processive motility. The best-studied protein is Ncd, the sole kinesin-14 member in Drosophila, which exhibits short residency times that coincide with the length of time it takes to bind and hydrolyse ATP12. Ncd is required for mitotic and meiotic spindle MT cross-linking, but is sequestered in the nucleus in interphase13,14; thus, it is unlikely that Ncd plays a major role in cargo transport in the interphase cytoplasm. In plants, kinesin-14 genes have been heavily duplicated, and there are 21 and 15 genes in the seed plant Arabidopsis and the moss Physcomitrella patens, respectively15,16. Some kinesin-14 members appear to have non-mitotic roles, such as KCBP in trichome morphogenesis17 and the KCA/KAC kinesin in positioning of the chloroplast in the cytoplasm18,19. It is unknown whether any members of the kinesin-14 subfamily in plants are capable of processive motility and/or are involved in MT-based cargo transport.
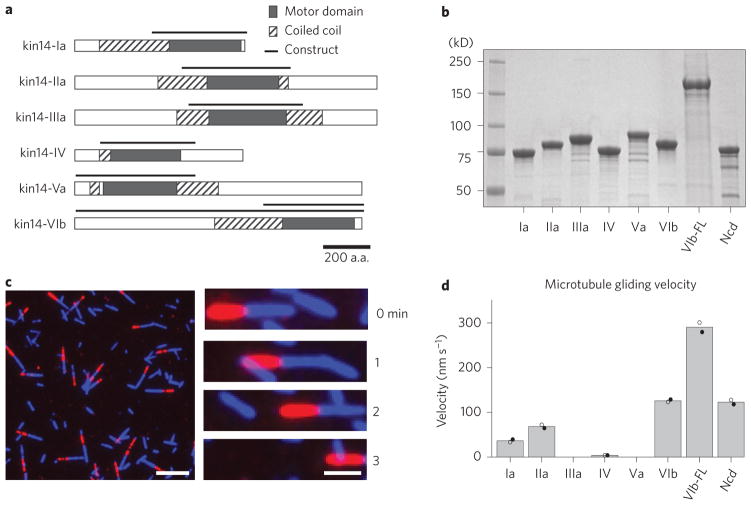
Fifteen kinesin-14s of P. patens are further subdivided into six subgroups, based on the amino acid sequence similarity of the motor and the adjacent neck domains (Fig. 1a)15. Within these subgroups, the amino acid sequences are very similar to each other (for example kin14-Ia and kin14-Ib share nearly 87% sequence identity) and are therefore thought to function redundantly, as was previously shown for the kin14-V proteins18. On the other hand, the lengths, sequences and domain organization are markedly different between the subgroups (Fig. 1a). To test whether any of the kinesin-14 motors show processive minus-end-directed motility, we selected one representative member from each of the six protein subgroups for biochemical analysis. Characterization of kinesins has generally been achieved with truncated constructs in which the neck and motor domains are included. We therefore engineered truncations of P. patens kinesin-14s fused with an N-terminal green fluorescent protein (GFP) (Fig. 1b). Gel filtration chromatography showed that they eluted at a similar fraction to a dimeric Ncd motor construct (236–700 a.a., tagged with GFP), suggesting that they are also dimeric (Supplementary Fig. 1b).
Figure 1. Four kinesin-14 subgroup members exhibit minus-end-directed motor activity.
a, Gene maps of the six subgroups of kinesin-14s in Physcomitrella patens. b, Coomassie blue staining after SDS-PAGE of the purified proteins used in the motility assays (FL stands for full length). c, An example of a gliding assay used to determine motor velocity and directionality (in this case kin14-Ia). MT minus-ends are labelled in red. Scale bar, 5 μm (left) or 2 μm (right). d, Gliding velocities. Bars represent the mean velocity of two independent experiments utilizing different protein purifications; solid and filled circles show the results from each preparation. Each circle represents the mean velocity of at least 100 motile MTs.
The purified proteins were assayed for motility in a MT gliding assay, in which motors were adhered to a cover glass and then MTs and ATP were added to the reaction chamber. Four of the six truncated chimeras translocated MTs with velocities ranging from 4 to 130 nm s−1 (Fig. 1d and Supplementary Movie 1). The fastest motor (kin14-VIb) showed a gliding velocity similar to Ncd and KCBP20. Kin14-IIIa did not translocate MTs along the glass surface, although they bound to MTs in an ATP-dependent manner in a sedimentation assay (Supplementary Fig. 1c). Kin14-Va did not efficiently bind to MTs, consistent with its Arabidopsis orthologue (Supplementary Fig. 1c)18. To determine the directionality of the moving MTs, we also performed a gliding assay with polarity marked MTs (Fig. 1c). As the MTs predominantly moved with their plus-ends leading (Supplementary Fig. 1a), we concluded that the four motile kinesin-14 subgroups are all minus-end-directed motors.
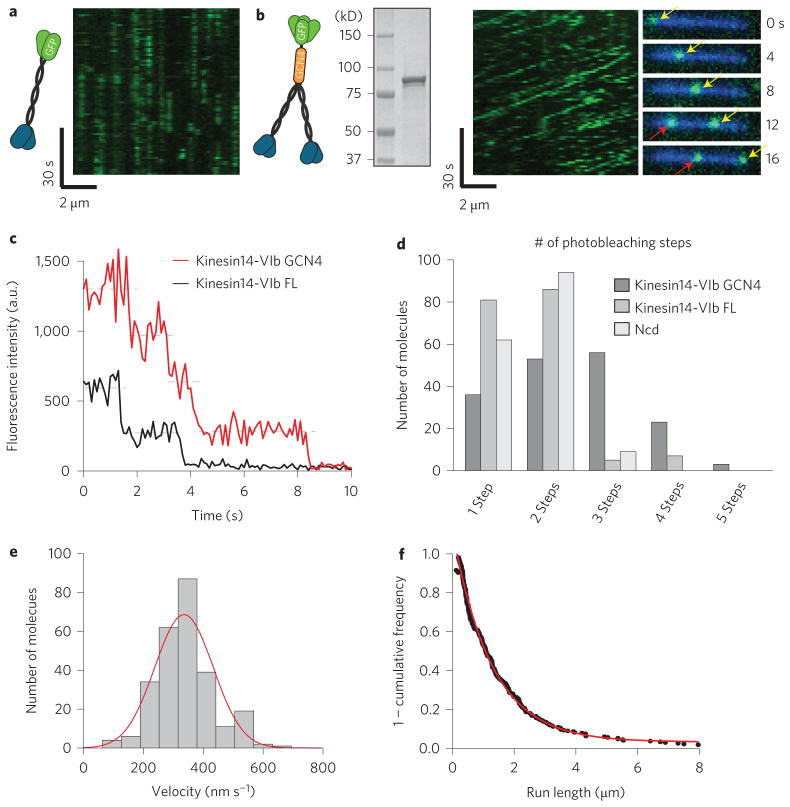
We asked whether any of the active motors might also be processive in a single-molecule motility assay. This assay involves attaching MTs to a coverslip and then adding low levels of GFP-tagged kinesin to examine the interactions of single motors with MTs. We performed this assay with high (2 mM) and low (10 μM) concentrations of ATP, but did not observe processive motion for any construct (Fig. 2a shows a representative kymograph for kin14-VIb).
Figure 2. Artificially tetramerized kin14-VIb showed processive motility.
a, A kymograph for dimeric kin14-VIb motors reveals no processive movement. b, A kymograph for the GFP-kin14-VIb GCN4 tetramer construct exhibits clear processive movement (diagonal lines) and long run lengths. Movie frames show two separate GFP spots (arrows), moving along a MT (blue). c, Representative traces of the photobleaching of kin14-VIb FL and the kin14-VIb GCN4 tetramer. d, Quantitation of the number of photobleaching steps. e, Velocity histogram of the kin14-VIb GCN4 tetramer with mean of 336 ± 97 nm s−1 (mean ± s.d., n=267). f, 1 – cumulative frequency for run lengths of the GCN4 tetramer construct, which were fitted to an exponential yielding a fit parameter of λ = 1.27±0.03 μm (error was determined from goodness of fit parameters; R2 = 0.995, n=267 particles).
We measured gliding velocity as a function of surface motor density for kin14-VIb (Supplementary Fig. 2a). The velocity was insensitive to a wide range of surface densities. However, at low surface densities (less than approximately 1 molecule per μm2), MTs attached to the surface but no longer exhibited unidirectional motion. The gliding velocity decreased slightly for the two highest surface density measurements, indicating that the motors can interfere with each other when present in sufficiently high numbers.
We then wondered whether the full-length proteins might contain some additional domain that confers processivity. We therefore expressed full-length versions of the two fastest motors (kin14-IIa and kin14-VIb). While we failed to purify full-length kin14-IIa after several attempts, we successfully purified the full-length kin14-VIb (kin14-VIb FL) (Fig. 1b). Kin14-VIb FL showed minus-end-directed MT gliding activity, and the gliding velocity was faster than the truncated form (~300 nm s−1; Fig. 1d). To test whether the increase in velocity is due to higher ATPase rate of the FL, we measured the steady-state ATPase activity of truncated and FL constructs (Supplementary Fig. 2b). The truncated kin14-VIb had a similar rate to those previously reported for Ncd21,22. However, the rate was ~10-fold lower for the FL. A likely interpretation of this data is that the full-length protein in solution is in an auto-inhibitory form with a low ATPase rate, as is well documented for kinesin-1 and other anterograde kinesin motors23. We speculate that the overall protein size affects its velocity, as is the case for Ncd22. Although it translocated MTs at a faster rate, the full-length dimeric proteins were still non-processive in the single-molecule motility assay (Supplementary Fig. 2c). On rare occasions, we observed a GFP particle that moved processively (Supplementary Fig. 2c). However, these motile particles were significantly brighter than the non-motile particles, suggesting that they are small aggregates and not single native dimers.
These results suggested that a small cohort of kin14-VIb motors could potentially move processively. We therefore engineered the ‘dimer of dimers’ construct by introducing the coding sequence for a GCN4 parallel tetramer motif 24 into the N-terminus (Fig. 2b). To confirm that the kin14-VIb GCN4 construct is indeed a tetramer, we measured its photobleaching characteristics (Fig. 2c,d). Figure 2c compares representative traces for kin14-VIb GCN4 and dimeric kin14-VIb FL, which showed four- and two-step photobleaching processes, respectively. The observed photobleaching step distribution of kin14-VIb GCN4 (n = 199) was distinct from that of dimeric Ncd or kin14-VIb FL (Fig. 2d), and was more consistent with that of other tetrameric proteins analysed in a similar manner25. Prevalence of three or fewer bleaching steps over four steps in the tetramer is probably due to GFP photo-inactivation prior to image acquisition, as is commonly reported26. Taken together we conclude, as expected, that kin14-VIb GCN4 is tetrameric, while kin14-VIb FL, like Ncd, is dimeric.
In the single-molecule motility assay, kin14-VIb GCN4 exhibited frequent processive motility (Fig. 2b and Supplementary Movie 2). The velocity was 336 ± 97 nm s−1, which was significantly higher than its gliding velocity of 125 nm s−1 (Fig. 1d). Kin14-VIb GCN4 exhibited long run lengths of 1.27 ± 0.03 μm, (exponential fit parameter ± error of fit, R2 = 0.995), which is comparable to the run length of human kinesin-1 processivity in vitro (Fig. 2f )27. When we attached the GCN4 tetramer motif to kin-14IIa, the second fastest kin14 (Fig. 1d), we did not observe processive motion (Supplementary Fig. 2d). We therefore focused on kin-14VIb for further characterization.
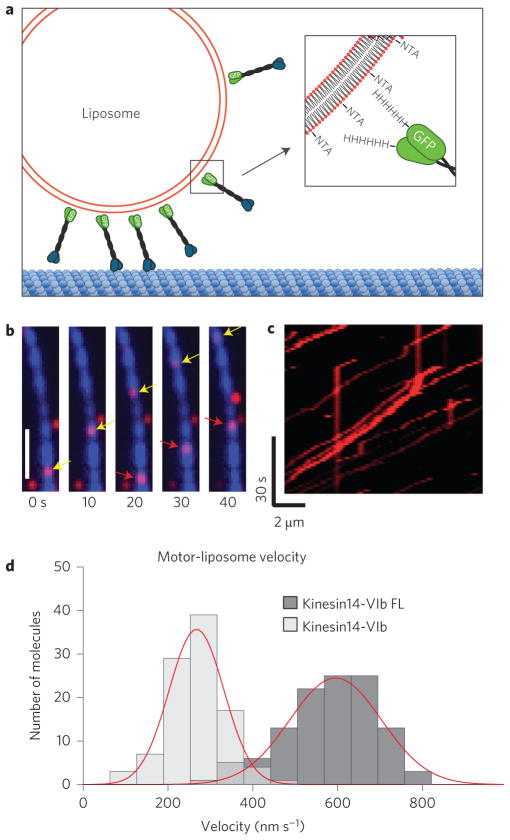
As an alternative in vitro clustering assay, we adhered multiple kin14-VIb motors to liposomes. To this end, we assembled liposomes with incorporated DOGS-NTA-Ni lipids, such that multiple histidine-tagged kin14-VIb proteins could bind to the liposomes (Fig. 3a and Supplementary Fig. 2e). The liposomes were transported along MTs for long distances (Fig. 3b,c and Supplementary Movie 3). Some moving liposomes switched between MT tracks, indicating that multiple motors were indeed present on their surface. The velocities (266 ± 69 nm s−1 for kin14-VIb and 597 ± 134 nm s−1 for kin14-VIb FL; Fig. 3d) were twice as fast as the gliding velocities, and the FL transport length was several microns, which is comparable to mammalian dynein28 (Supplementary Fig. 2f).
Figure 3. Kin14-VIb transports liposomes along MTs.
a, The attachment of histidine-tagged motor to Ni-NTA lipids incorporated into liposomes. b, Transport of liposomes. Two separate motor-coated fluorescently-labelled liposomes (red, indicated by arrows) can be seen moving along a Cy5-labelled MT (blue). Bar, 2 μm. c, A kymograph shows long run lengths of kin14-VIb-coated liposomes. d, Velocity histograms for liposomes coated with kin14-VIb (266 ± 69 nm s−1; mean±s.d., n = 100 particles) and kin14-VIb FL (597 ± 134 nm s−1; mean±s.d., n = 116 particles).
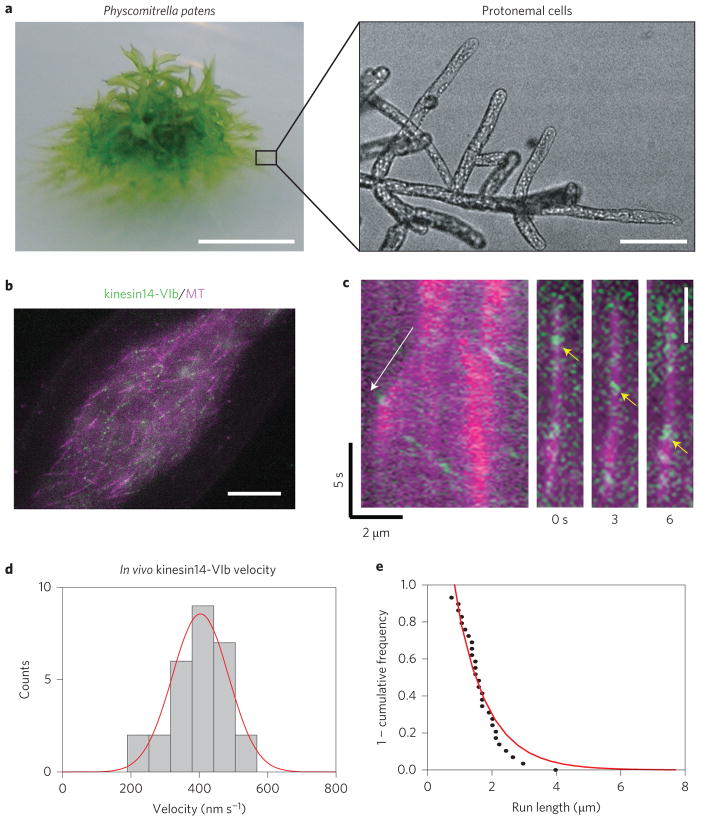
Finally, we tested whether native kin14-VIb moves processively in vivo. Taking advantage of the very high frequency of homologous recombination in P. patens29, we tagged the Citrine (a GFP variant) gene to the N-terminus of the endogenous kin14-VIb gene without inserting any other sequences such as selection markers; the established transgenic moss lines expressed these tagged motors under the control of the endogenous promoter and 3′ UTR sequences at the native locus (Supplementary Fig. 3a–c). We observed interphase cells in the protonemal tissue (Fig. 4a) using oblique illumination fluorescence microscopy; this technique has been used successfully to visualize endoplasmic MTs as well as the associated γ-tubulin-Citrine with minimum interference by autofluorescence derived from the chloroplast7. For Citrine-kin14-VIb, we observed discrete fluorescent punctate along the MTs, many of which showed unidirectional motility (Fig. 4b,c and Supplementary Movie 4). Because only the motors and MTs were visualized by fluorescence, we could not ascertain whether or not the Citrine-kin14-VIb were bound to and moving a cargo. The mean velocity of this movement was 413 ± 18 nm s−1 (mean ± s.d.; n = 29), which was comparable to the in vitro velocity, and the run length was 1.01 ± 0.31 μm (exponential fit parameter ± error of fit; R2 = 0.942; n = 26) (Fig. 4d,e). Run lengths shorter than those that were observed in the liposome assay might reflect the fact that MT binding of this kinesin can be negatively regulated by calmodulin binding proximal to the motor region20. In some instances, we could identify the polarity of the MT along which the Citrine-kin14-VIb moved. The signals moved away from the dynamic MT plus-end in all 12 cases analysed, indicating that the Citrine-kin14-VIb moved towards minus-ends (Fig. 4c). Under the same imaging condition, the fluorescence intensity of the Citrine-kin14-VIb punctae was about half of γ-tubulin-Citrine and slightly higher than γ-tubulin complex protein 4 (GCP4) tagged with Citrine in punctae corresponding to individual γ-tubulin ring complexes (Supplementary Fig. 3d; both were expressed from endogenous promoters). Assuming that 13–15 molecules of γ-tubulin and 2–4 molecules of GCP4 are present in the γ-tubulin ring complex of P. patens, as has been estimated for animal and yeast cells30,31, then it is likely that the Citrine-kin14-VIb punctae consist of more than one but relatively few dimeric kin14-VIb motor proteins. Thus, we conclude that a small number of kin14-VIb motors, clustered into a diffraction-limited spot, canmove processively along the MT towards minus-ends in moss cells.
Figure 4. Minus-end-directed motility of kin14-VIb clusters in vivo.
a, Protonemal cells were imaged in this study. Scale bars, 5 mm (left) and 100 μm (right). b, The endoplasm close to the cell cortex was observed with oblique illumination fluorescence microscopy (green; Citrine-kin14-VIb, magenta; mCherry-tubulin). Note that most of the MTs visualized in this area are single MTs, not bundles7. Scale bar, 5 μm. c, Citrine-kin14-VIb signals moved away from the growing plus-end (white arrow in the kymograph). (Right) An example of Citrine movement (yellow arrows). Scale bar, 2 μm. d, The velocity of Citrine-kin14-VIb motility (n = 29). Static signals were not counted. e, 1 – cumulative frequency for run lengths of Citrine-kin14-VIb which were fit to an exponential yielding a fit parameter of λ = 1.01 ± 0.31 μm (error was determined from goodness-of-fit parameters; R2=0.942, n = 26 particles).
In conclusion, we have obtained evidence that the collective actions of as few as two non-processive kinesin-14s achieve retrograde transport in vitro and most likely in plant cells as well. Based on its rapid speed of transport (up to 600 nm s−1; the fastest ever reported for a kinesin-14 motor) and high processivity achieved upon motor clustering, we propose that kin14-VIb acts as a cargo transport motor in plants, serving an analogous role to cytoplasmic dynein in animal and fungal cells. In the protonemal cells, we recently observed retrograde motility of newly generated MTs along the existing MTs7 and MT-dependent translocation of the nucleus8. In the latter process, the responsible anterograde kinesin was identified and the involvement of a minus-end-directed transporter was suggested8. Thus, MT-dependent bidirectional transport is present in moss cells in addition to the actomyosin-based mechanism. Kin14-VIb would be a candidate retrograde motor in these processes. Previous loss-of-function and gain-of-function analyses suggested a variety of roles of kin14-VI in seed plants, such as trichome morphogenesis, pollen tube growth, organelle positioning and cell division17,32,33. The versatility might be explained if kin14-VI associates with and transports various cargoes. A proof for this model would require identifying a specific cargo for kin14-VI in vivo.
This study also reveals similarities between kin14-VIb and other types of motors. Ncd was recently found to move with run lengths greater than 1 μm when clustered on a DNA scaffold in vitro34. Therefore, other subgroup members of kinesin-14, even ones not involved in cargo transport, can potentially become processive in vivo in a similar manner to kin14-VIb. As for anterograde motors, kinesin-3 (Unc-104/KIF1), which is responsible for long-distance transport of neuronal vesicles, transports liposomes in vitro only after motor clustering on the liposome surface35,36. In this instance, processivity is probably achieved by promoting the association of two monomeric kinesin-3 motors into a dimer, which is the highly processive form of the motor35,36. Similarly myosin VI, an actin-based motor, is monomeric and non-processive, but small clusters of this motor can efficiently produce cargo transport37. Thus, the cargo-dependent clustering may be a widely utilized mechanism for cytoskeletal motors to produce long-distance transport along cytoskeletal polymers.
Methods
Constructs
PCR primers for constructing the plasmids for protein expression and transgenic line generation are listed in Supplementary Table 1. EGFP, mGFP (in vitro) or Citrine (in vivo) was attached. The information on the linker sequences is also available in the footnote to Supplementary Table 1.
Protein purification
Plasmids containing the coding sequences for each of the GFP-kinesin-14 motors were transformed and expressed in BL21-AI cells. Expression was induced by addition of 0.2% arabinose and 0.2 mM IPTG and were left overnight at 18 °C. Cells were pelleted and harvested in lysis buffer (25 mM MOPS pH 7.0, 2 mM MgCl2, 250 mM NaCl, 30 mM imidazole, 5 mM β-mercaptoethanol, 5% sucrose), and lysed by the EmulsiFlex homogenizer in the presence of protease inhibitors. After lysis, the extract was loaded onto a Ni-NTA column, washed with additional lysis buffer, and then eluted by increasing the imidazole concentration to 400 mM. Proteins were dialysed for 4 h to remove imidazole and then snap frozen in liquid nitrogen and stored at −80 °C.
Motility assays
The standard assay buffer contained 25 mM MOPS pH 7.0, 75 mM KCl, 2 mM MgCl2, 1 mM EGTA. Addition of the PCA/PCD/Trolox oxygen scavenging system38 was used in all in vitro microscopy experiments. Purified motor proteins were subjected to a ‘bind-and-release’ reaction, in order to select for active motors, in all motility assays used in this study. Motors were first bound to MTs for 10 min in the absence of ATP. The reaction was then layered over a 60% glycerol cushion in 1× assay buffer, supplemented with 20 μM taxol, and centrifuged at 80,000 rpm in a TLA 100 rotor for 10 min. The supernatant was discarded and the pellet was washed and resuspended in 1× assay buffer plus added KCl (to a final concentration of 150 mM) with 20 μM taxol and 5 mM ATP and left to incubate for 5 min. The solution was recentrifuged at 80,000 rpm for 10 min and the supernatant (containing motors released from the MTs with ATP) was kept on ice and used in the motility assays. Fractions at each step were analysed by SDS-PAGE and Coomassie blue staining (Supplementary Fig. 1c). For gliding assays, motors were added to a microscope slide flow chamber (~10 μl in volume) made with an untreated glass coverslip, a microscope slide and double-stick tape. After a brief incubation (2–5 min), the flow chamber was washed with assay buffer containing 1 mM casein, followed by incubation with the motility buffer containing casein, 2 mM ATP and polymerized Cy5-labelled MTs. For single-molecule assays27, 5mg ml−1 biotin-BSA was added to a flow chamber made with acid-washed coverslips and allowed to incubate for 2–3 min. The flow chamber was washed and supplied with 0.5 mg ml−1 streptavidin (another 2–3 min incubation). The flow chamber was then washed with the assay buffer containing 1 mg ml−1 casein, followed by a 5 min incubation with labelled MTs (10% Cy5-labelled tubulin and 10% biotin-labelled tubulin). Finally, kinesin motors in the assay buffer (with 2 mM ATP, 20 μM taxol and 1 mg ml−1 casein) were added. Polarity-marked MTs were made by preparing Alexa 561-labelled MT seeds, blocking the minus-ends with NEM-treated tubulin, and allowing a mixture of unlabelled and Cy5-labelled tubulin (10%) to polymerize exclusively at the plus-end. For the surface density titration, the same attachment chemistry was used as in the photobleaching experiments (see below). The surface density was successively titrated by preparing coverslips with the same adhered antibody concentration followed by incubating serial dilutions of GFP motor. For very low concentrations of motor, the surface could be determined directly by counting the number of clearly visible GFP particles in a defined area. The higher surface densities (which were too high to accurately count motors individually) were estimated based on the added motor concentration and extrapolation from the surface densities that were counted directly.
In vitro microscopy instrumentation and analysis
All in vitro motility assays were performed at room temperature (~23 °C) using total internal reflection fluorescence (TIRF) microscopy on a Nikon Eclipse Ti microscope fitted with a 100× (1.45 NA) objective and an Andor iXON EMCCD camera. The acquisition software was Micromanager39, and the analyses of velocities and run lengths were performed using Fiji. Briefly, we made kymographs for a series of motile MTs (or translocating particles) in a field of view. Velocity was then determined from these kymographs based on the acquisition parameters and the known pixel size of the microscope camera. Run lengths were also determined from kymographs of processively translocating particles. 1 – cumulative frequency distribution was plotted against run length40 and fit to a single exponential.
Liposome preparation and motility assay
Liposomes were prepared by dissolving lipids (79.7% POPC, 10% POPS, 10% DOGS-NTA-Ni, 0.3% rhodamine PE (Avanti)) in chloroform, drying them under a constant stream of N2 and desiccating in a vacuum for at least 1 h, and then resuspending in 1× assay buffer. To make liposomes of uniform size, the solution was extruded through polycarbonate filters with a 200 nm pore size. Motors were then added to liposomes (with at least 10-fold molar excess of motor to DOGS-NTA-Ni lipids) and incubated on ice for at least 1 h.
Photobleaching
For the photobleaching assay, HEPES pH 7.5 was substituted in the assay buffer for MOPS, because the higher pH gives a brighter GFP signal and reduces blinking. Flow chambers, made with acid-washed coverslips, were incubated with 0.1 mg ml−1 protein G (Sigma Chemical Co.) for ~3 min. The flow chamber was then washed, followed by a 3-min incubation with an antibody against 6x histidine (0.1 mg ml−1; Roche). After a wash, soluble motor was added at an appropriate concentration to achieve a surface coating of no more than ~1 particle per μm2. The GFP intensity was plotted in time and sequences were analysed with custom-designed Matlab software. In Fig. 2d, ~200 traces were observed and scored according to the number of discernible bleaching events (~20% of traces were rejected due to noise).
ATPase measurement
Steady-state ATP turnover was determined by a standard PK/LDH coupled assay. We diluted the standard assay buffer to 0.5× (12.5 mM MOPS pH 7.0, 37.5 mM KCl, 1 mMMgCl2, 0.5 mM EGTA) in order to decrease the ionic strength, which can interfere with motor–MT interactions. Final concentrations of motor were 100 nM for kin14-VIb and 225 nM for kin14-VIb FL and the MT concentration was 20 μM (which we verified was sufficiently high for maximal MT-stimulated ATP turnover). Concentrations for the other components of the assay were as follows: MgATP (2 mM), NADH (0.2 mM), phosphoenol pyruvate (1 mM), pyruvate kinase (0.01 U), lactate dehydrogenase (0.03 U), taxol (20 μM). Absorbance at 340 nm was continuously measured in an Eppendorf Spectrophotometer (UV-Vis BioSpectrometer) to determine the rate of ATP turnover.
Moss lines
Transgenic lines were selected by the conventional PEG-mediated transformation41,42 with some modification (see Supplementary Fig. 3a; detailed in Yamada et al.43). Briefly, a plasmid in which the Citrine gene was flanked by ~1-kb 5′-UTR and N-terminal sequences of the kin14-VIb was constructed. After linearization, it was co-transformed into the mCherry-tubulin-expressing line with a circular plasmid with the hygromycin-resistant gene cassette. The transformants were selected by hygromycin (2 weeks), followed by transferring to the drug-free medium to allow cells to proliferate without the circular plasmid (10 d). In the end, we selected 200 transgenic lines that were no longer hygromycin resistant, and assessed the integration of Citrine at the N-terminus of kin14-VIb by PCR and immunoblotting (Supplementary Fig. 3b,c). We obtained two independent Citrinekin14-VIb replacement lines, which grew normally on the regular culture medium.
In vivo microscopy
Protonemata, the tissue containing actively dividing cells, were imaged in this study, following the method described in Nakaoka et al.7 Briefly, cells were homogenized and plated on the cellophane-coated BCDAT agar medium for 5–7 d. Protonemal cells and 20 μl distilled water were placed on a slide glass and covered by a coverslip. Extra water was wiped out. A TIRF microscope (Nikon Ti; 100× 1.49 NA lens) with a GEMINI split view (Hamamatsu) and an EMCCD camera Evolve (Roper) or iXON (DU888E; Andor) was used with oblique illumination fluorescence. The endoplasm of the sub-apical cells (or in rare cases apical cells) that was most closely located to the coverslip was in focus. Imaging was performed at 24–25 °C. The microscope was controlled by Micromanager software39 or Nikon’s NIS-Elements. The velocity of moving particles was measured based on kymographs.
Supplementary Material
Acknowledgments
We are grateful to S. Ross and L. Chang (Nikon USA) for providing microscopes at MBL.W. Huynh provided invaluable advice in establishing the liposome assay, and A. Jain assisted with the photobleaching experiment. We also thank M. Bezanilla (University of Massachusetts Amherst) and Tomoko Nishiyama (Nagoya University) for help with microscopy and helpful discussion, Y. Nakaoka for moss lines, and M. Nishina and R. Inaba for technical assistance. This work was supported by the Human Frontier Science Program, the James A. and Faith Miller Memorial Fund (MBL), the Laura and Arthur Colwin Endowed Summer Research Fellowship Fund (MBL), the TORAY Science Foundation, Grants-in-Aid for Scientific Research (15K14540, MEXT) (G.G.), and the NIH (38499) (R.D.V.).
Footnotes
Author contributions
E.J., M.Y., R.D.V. and G.G. designed the research; E.J., M.Y. and G.G. performed experiments; E.J., M.Y., R.D.V. and G.G. analysed data; E.J., R.D.V. and G.G. wrote the paper.
Supplementary information is available online.
Competing interests
The authors declare no competing financial interests.
References
- 1.Shimmen T, Yokota E. Cytoplasmic streaming in plants. Curr Opin Cell Biol. 2004;16:68–72. doi: 10.1016/j.ceb.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Cai G, Cresti M. Are kinesins required for organelle trafficking in plant cells? Front Plant Sci. 2012;3:170. doi: 10.3389/fpls.2012.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakaoka Y, Kimura A, Tani T, Goshima G. Cytoplasmic nucleation and atypical branching nucleation generate endoplasmic microtubules in Physcomitrella patens. Plant Cell. 2015;27:228–242. doi: 10.1105/tpc.114.134817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miki T, Nishina M, Goshima G. RNAi screening identifies the armadillo repeat-containing kinesins responsible for microtubule-dependent nuclear positioning in Physcomitrella patens. Plant Cell Physiol. 2015;56:737–749. doi: 10.1093/pcp/pcv002. [DOI] [PubMed] [Google Scholar]
- 5.Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nature Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 6.Hancock WO. Bidirectional cargo transport: moving beyond tug of war. Nature Rev Mol Cell Biol. 2014;15:615–628. doi: 10.1038/nrm3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawrence CJ, Morris NR, Meagher RB, Dawe RK. Dyneins have run their course in plant lineage. Traffic. 2001;2:362–363. doi: 10.1034/j.1600-0854.2001.25020508.x. [DOI] [PubMed] [Google Scholar]
- 8.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 9.Endow SA. Determinants of molecular motor directionality. Nature Cell Biol. 1999;1:E163–E167. doi: 10.1038/14113. [DOI] [PubMed] [Google Scholar]
- 10.Mieck C, et al. Non-catalytic motor domains enable processive movement and functional diversification of the kinesin-14 Kar3. eLife. 2015;4:e04489. doi: 10.7554/eLife.04489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hepperla AJ, et al. Minus-end-directed kinesin-14 motors align antiparallel microtubules to control metaphase spindle length. Dev Cell. 2014;31:61–72. doi: 10.1016/j.devcel.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Case RB, Pierce DW, Hom-Booher N, Hart CL, Vale RD. The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell. 1997;90:959–966. doi: 10.1016/s0092-8674(00)80360-8. [DOI] [PubMed] [Google Scholar]
- 13.Hatsumi M, Endow SA. Mutants of the microtubule motor protein, nonclaret disjunctional, affect spindle structure and chromosome movement in meiosis and mitosis. J Cell Sci. 1992;101:547–559. doi: 10.1242/jcs.101.3.547. [DOI] [PubMed] [Google Scholar]
- 14.Goshima G, Vale RD. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol Biol Cell. 2005;16:3896–3907. doi: 10.1091/mbc.E05-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen Z, Collatos AR, Bibeau JP, Furt F, Vidali L. Phylogenetic analysis of the kinesin superfamily from Physcomitrella. Front Plant Sci. 2012;3:230. doi: 10.3389/fpls.2012.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy AS, Day IS. Kinesins in the Arabidopsis genome: a comparative analysis among eukaryotes. BMC Genomics. 2001;2:2. doi: 10.1186/1471-2164-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oppenheimer DG, et al. Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc Natl Acad Sci USA. 1997;94:6261–6266. doi: 10.1073/pnas.94.12.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suetsugu N, et al. The KAC family of kinesin-like proteins is essential for the association of chloroplasts with the plasma membrane in land plants. Plant Cell Physiol. 2012;53:1854–1865. doi: 10.1093/pcp/pcs133. [DOI] [PubMed] [Google Scholar]
- 19.Suetsugu N, et al. Two kinesin-like proteins mediate actin-based chloroplast movement in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107:8860–8865. doi: 10.1073/pnas.0912773107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song H, Golovkin M, Reddy AS, Endow SA. In vitro motility of AtKCBP, a calmodulin-binding kinesin protein of Arabidopsis. Proc Natl Acad Sci USA. 1997;94:322–327. doi: 10.1073/pnas.94.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pechatnikova E, Taylor EW. Kinetic mechanism of monomeric non-claret disjunctional protein (Ncd) ATPase. J Biol Chem. 1997;272:30735–30740. doi: 10.1074/jbc.272.49.30735. [DOI] [PubMed] [Google Scholar]
- 22.Endres NF, Yoshioka C, Milligan RA, Vale RD. A lever-arm rotation drives motility of the minus-end-directed kinesin Ncd. Nature. 2006;439:875–878. doi: 10.1038/nature04320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verhey KJ, Hammond JW. Traffic control: regulation of kinesin motors. Nature Rev Mol Cell Biol. 2009;10:765–777. doi: 10.1038/nrm2782. [DOI] [PubMed] [Google Scholar]
- 24.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 25.Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nature Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hines KE. Inferring subunit stoichiometry from single molecule photobleaching. J Gen Physiol. 2013;141:737–746. doi: 10.1085/jgp.201310988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vale RD, et al. Direct observation of single kinesin molecules moving along microtubules. Nature. 1996;380:451–453. doi: 10.1038/380451a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 2014;345:337–341. doi: 10.1126/science.1254198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cove D. The moss Physcomitrella patens. Annu Rev Genet. 2005;39:339–358. doi: 10.1146/annurev.genet.39.073003.110214. [DOI] [PubMed] [Google Scholar]
- 30.Choi YK, Liu P, Sze SK, Dai C, Qi RZ. CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J Cell Biol. 2010;191:1089–1095. doi: 10.1083/jcb.201007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA. Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 2010;466:879–882. doi: 10.1038/nature09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vos JW, Safadi F, Reddy AS, Hepler PK. The kinesin-like calmodulin binding protein is differentially involved in cell division. Plant Cell. 2000;12:979–990. doi: 10.1105/tpc.12.6.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazzaro MD, Marom EY, Reddy AS. Polarized cell growth, organelle motility, and cytoskeletal organization in conifer pollen tube tips are regulated by KCBP, the calmodulin-binding kinesin. Planta. 2013;238:587–597. doi: 10.1007/s00425-013-1919-8. [DOI] [PubMed] [Google Scholar]
- 34.Furuta K, et al. Measuring collective transport by defined numbers of processive and nonprocessive kinesin motors. Proc Natl Acad Sci USA. 2013;110:501–506. doi: 10.1073/pnas.1201390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomishige M, Klopfenstein DR, Vale RD. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 2002;297:2263–2267. doi: 10.1126/science.1073386. [DOI] [PubMed] [Google Scholar]
- 36.Klopfenstein DR, Tomishige M, Stuurman N, Vale RD. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 2002;109:347–358. doi: 10.1016/s0092-8674(02)00708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sivaramakrishnan S, Spudich JA. Coupled myosin VI motors facilitate unidirectional movement on an F-actin network. J Cell Biol. 2009;187:53–60. doi: 10.1083/jcb.200906133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using μManager. Curr Protoc Mol Biol. 2010;92:14.20.1–14.20.17. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bieling P, et al. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450:1100–1105. doi: 10.1038/nature06386. [DOI] [PubMed] [Google Scholar]
- 41.Nakaoka Y, et al. An inducible RNA interference system in Physcomitrella patens reveals a dominant role of augmin in phragmoplast microtubule generation. Plant Cell. 2012;24:1478–1493. doi: 10.1105/tpc.112.098509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miki T, Naito H, Nishina M, Goshima G. Endogenous localizome identifies 43 mitotic kinesins in a plant cell. Proc Natl Acad Sci USA. 2014;111:E1053–E1061. doi: 10.1073/pnas.1311243111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamada M, Miki T, Goshima G. Imaging mitosis in the moss Physcomitrella patens. Methods Mol Biol. doi: 10.1007/978-1-4939-3542-0_17. in the press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.