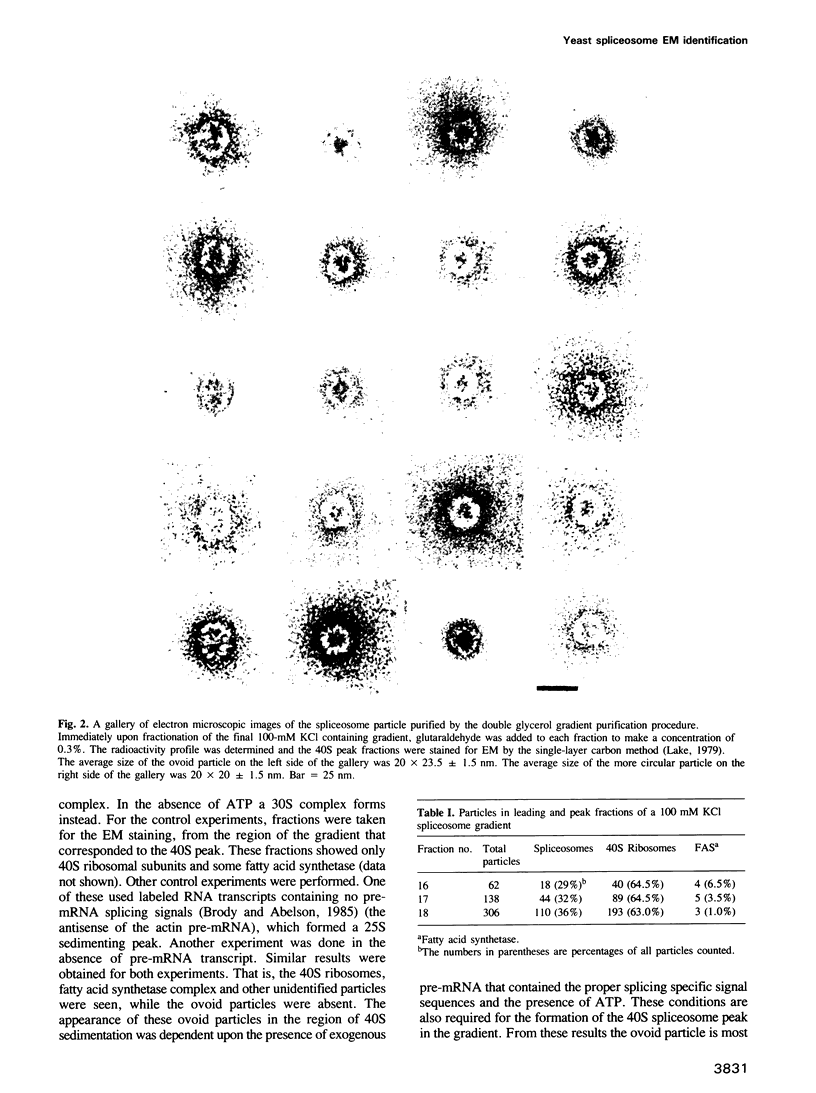
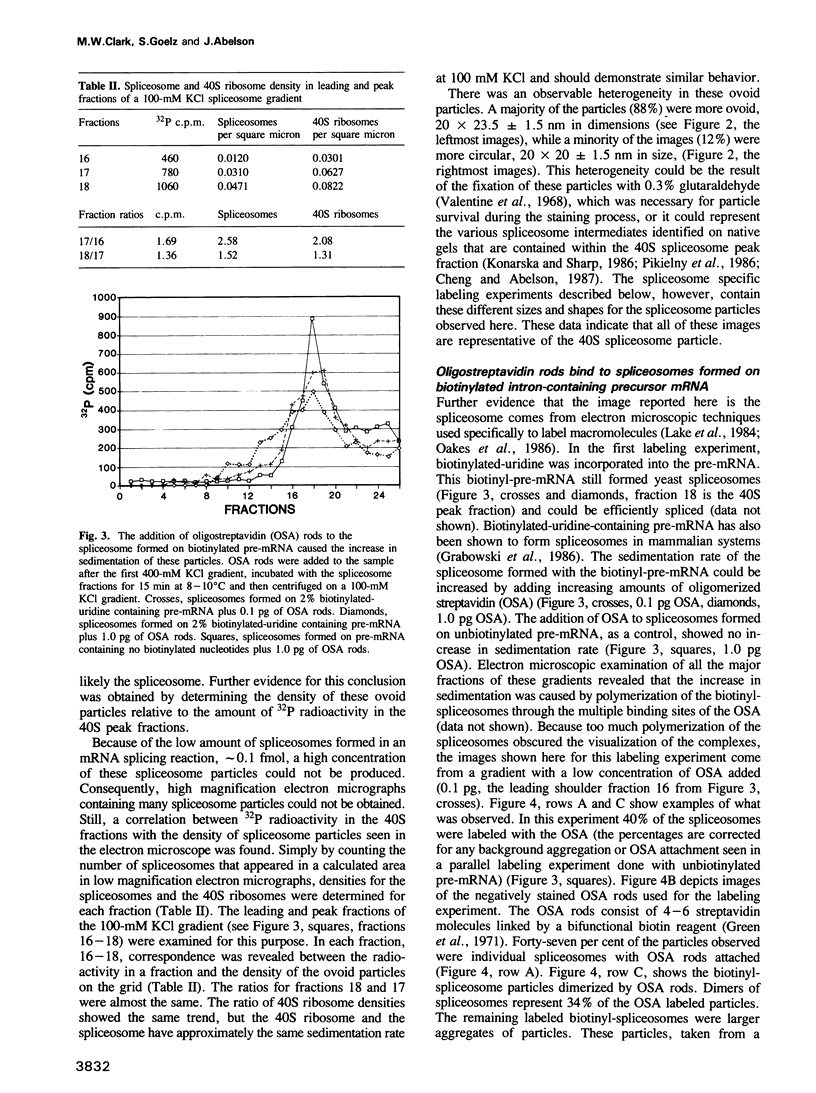
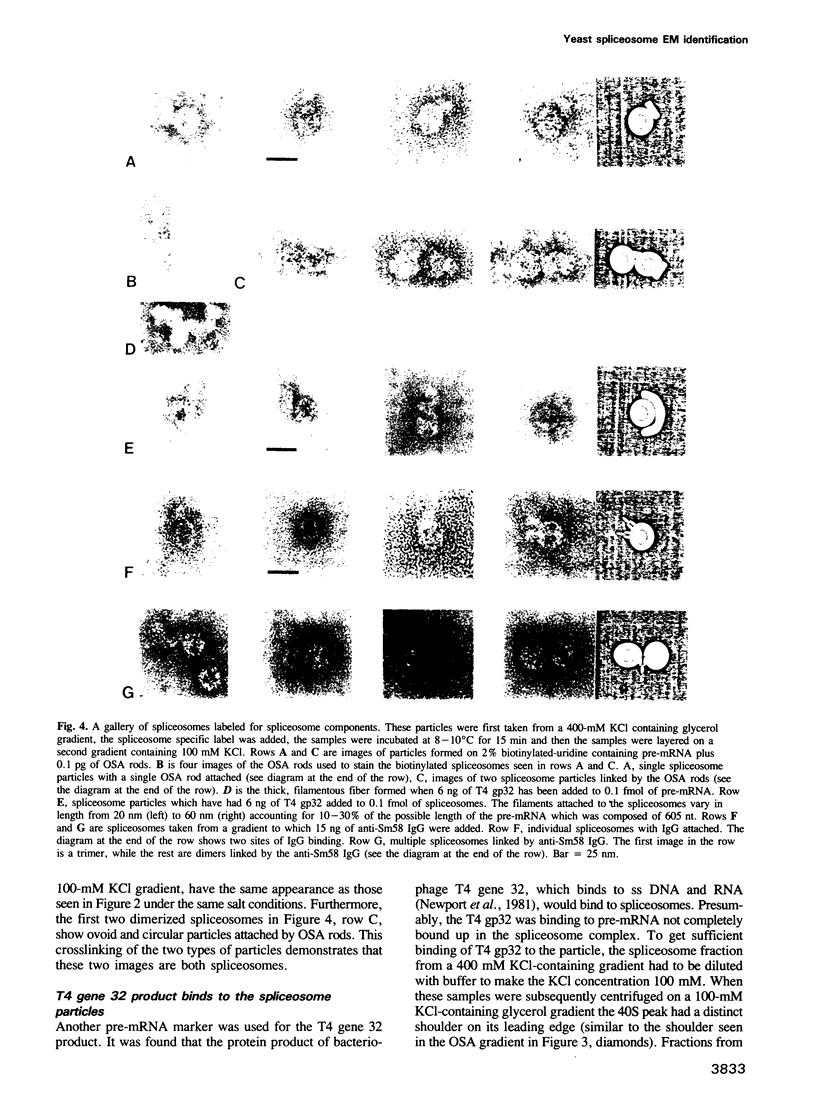
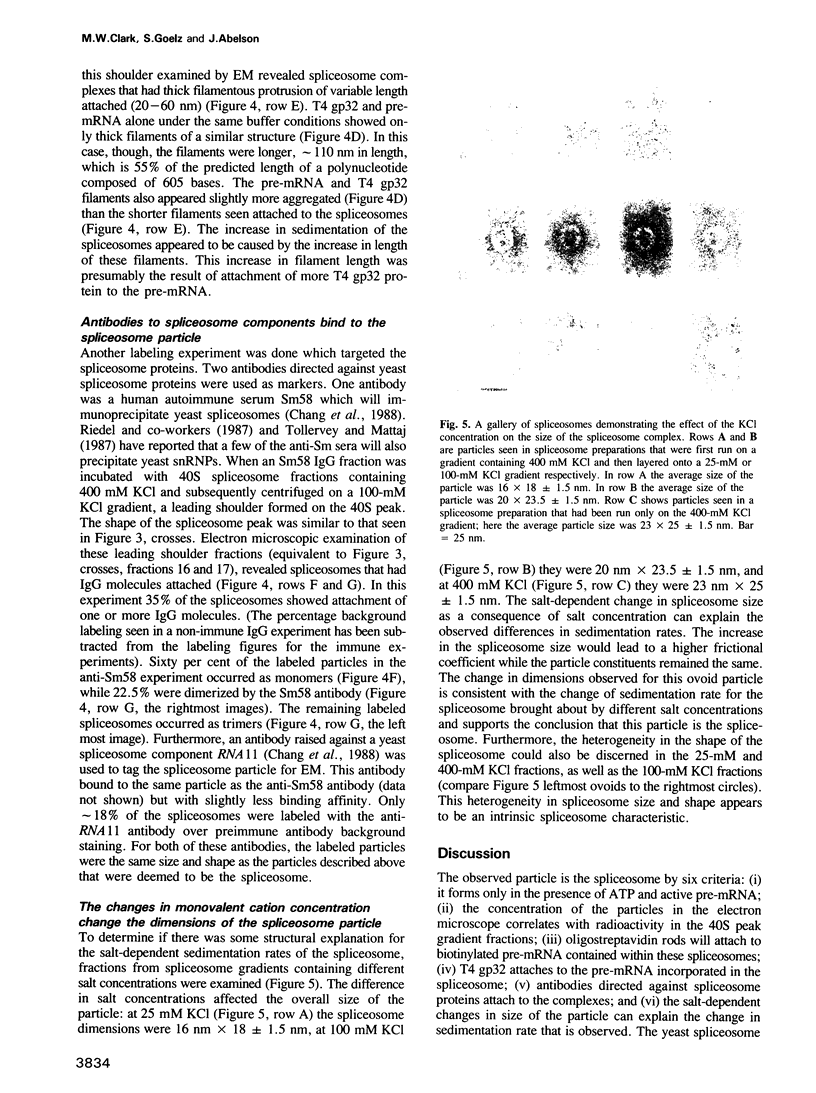
Abstract
We have partially purified the yeast spliceosome by differential sedimentation in glycerol gradients. By electron microscopy we have identified a particle in these fractions that is the spliceosome. In 100 mM KCl buffer, the yeast spliceosome is an ovoid disc with the dimensions of 20 x 23.5 nm with a central indentation. To verify that these ovoid particles were spliceosomes, specific labels were used to tag them. These tagged spliceosomes were then identified in the electron microscope. The salt dependent shift of sedimentation rate for the spliceosome can be explained by a change in size of the particle.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beyer A. L., Bouton A. H., Miller O. L., Jr Correlation of hnRNP structure and nascent transcript cleavage. Cell. 1981 Oct;26(2 Pt 2):155–165. doi: 10.1016/0092-8674(81)90299-3. [DOI] [PubMed] [Google Scholar]
- Beyer A. L., Miller O. L., Jr, McKnight S. L. Ribonucleoprotein structure in nascent hnRNA is nonrandom and sequence-dependent. Cell. 1980 May;20(1):75–84. doi: 10.1016/0092-8674(80)90236-6. [DOI] [PubMed] [Google Scholar]
- Beyer A. L., Osheim Y. N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988 Jun;2(6):754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Bindereif A., Green M. R. An ordered pathway of snRNP binding during mammalian pre-mRNA splicing complex assembly. EMBO J. 1987 Aug;6(8):2415–2424. doi: 10.1002/j.1460-2075.1987.tb02520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D. L., Chabot B., Steitz J. A. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985 Oct;42(3):737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- Black D. L., Steitz J. A. Pre-mRNA splicing in vitro requires intact U4/U6 small nuclear ribonucleoprotein. Cell. 1986 Aug 29;46(5):697–704. doi: 10.1016/0092-8674(86)90345-4. [DOI] [PubMed] [Google Scholar]
- Brody E., Abelson J. The "spliceosome": yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science. 1985 May 24;228(4702):963–967. doi: 10.1126/science.3890181. [DOI] [PubMed] [Google Scholar]
- Chabot B., Steitz J. A. Multiple interactions between the splicing substrate and small nuclear ribonucleoproteins in spliceosomes. Mol Cell Biol. 1987 Jan;7(1):281–293. doi: 10.1128/mcb.7.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T. H., Clark M. W., Lustig A. J., Cusick M. E., Abelson J. RNA11 protein is associated with the yeast spliceosome and is localized in the periphery of the cell nucleus. Mol Cell Biol. 1988 Jun;8(6):2379–2393. doi: 10.1128/mcb.8.6.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. C., Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987 Nov;1(9):1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Grabowski P. J., Seiler S. R., Sharp P. A. A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell. 1985 Aug;42(1):345–353. doi: 10.1016/s0092-8674(85)80130-6. [DOI] [PubMed] [Google Scholar]
- Grabowski P. J., Sharp P. A. Affinity chromatography of splicing complexes: U2, U5, and U4 + U6 small nuclear ribonucleoprotein particles in the spliceosome. Science. 1986 Sep 19;233(4770):1294–1299. doi: 10.1126/science.3638792. [DOI] [PubMed] [Google Scholar]
- Green N. M., Konieczny L., Toms E. J., Valentine R. C. The use of bifunctional biotinyl compounds to determine the arrangement of subunits in avidin. Biochem J. 1971 Dec;125(3):781–791. doi: 10.1042/bj1250781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967 May;93(5):1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J., Vidali G., Boffa L. C., Allfrey V. G. Characterization of the non-histone nuclear proteins associated with rapidly labeled heterogeneous nuclear RNA. J Biol Chem. 1977 Oct 25;252(20):7307–7322. [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986 Sep 12;46(6):845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- Lake J. A., Henderson E., Oakes M., Clark M. W. Eocytes: a new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3786–3790. doi: 10.1073/pnas.81.12.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. A. Practical aspects of immune electron microscopy. Methods Enzymol. 1979;61:250–257. doi: 10.1016/0076-6879(79)61014-5. [DOI] [PubMed] [Google Scholar]
- LeStourgeon W. M., Beyer A. L., Christensen M. E., Walker B. W., Poupore S. M., Daniels L. P. The packaging proteins of core hnRNP particles and the maintenance of proliferative cell states. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):885–898. doi: 10.1101/sqb.1978.042.01.090. [DOI] [PubMed] [Google Scholar]
- Lin R. J., Lustig A. J., Abelson J. Splicing of yeast nuclear pre-mRNA in vitro requires a functional 40S spliceosome and several extrinsic factors. Genes Dev. 1987 Mar;1(1):7–18. doi: 10.1101/gad.1.1.7. [DOI] [PubMed] [Google Scholar]
- Lin R. J., Newman A. J., Cheng S. C., Abelson J. Yeast mRNA splicing in vitro. J Biol Chem. 1985 Nov 25;260(27):14780–14792. [PubMed] [Google Scholar]
- Lossky M., Anderson G. J., Jackson S. P., Beggs J. Identification of a yeast snRNP protein and detection of snRNP-snRNP interactions. Cell. 1987 Dec 24;51(6):1019–1026. doi: 10.1016/0092-8674(87)90588-5. [DOI] [PubMed] [Google Scholar]
- Lustig A. J., Lin R. J., Abelson J. The yeast RNA gene products are essential for mRNA splicing in vitro. Cell. 1986 Dec 26;47(6):953–963. doi: 10.1016/0092-8674(86)90810-x. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987 Feb 19;325(6106):673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- Newport J. W., Lonberg N., Kowalczykowski S. C., von Hippel P. H. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. II. Specificity of binding to DNA and RNA. J Mol Biol. 1981 Jan 5;145(1):105–121. doi: 10.1016/0022-2836(81)90336-3. [DOI] [PubMed] [Google Scholar]
- Oakes M. I., Clark M. W., Henderson E., Lake J. A. DNA hybridization electron microscopy: ribosomal RNA nucleotides 1392-1407 are exposed in the cleft of the small subunit. Proc Natl Acad Sci U S A. 1986 Jan;83(2):275–279. doi: 10.1073/pnas.83.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim Y. N., Miller O. L., Jr, Beyer A. L. RNP particles at splice junction sequences on Drosophila chorion transcripts. Cell. 1985 Nov;43(1):143–151. doi: 10.1016/0092-8674(85)90019-4. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Rymond B. C., Rosbash M. Electrophoresis of ribonucleoproteins reveals an ordered assembly pathway of yeast splicing complexes. 1986 Nov 27-Dec 3Nature. 324(6095):341–345. doi: 10.1038/324341a0. [DOI] [PubMed] [Google Scholar]
- Reed R., Griffith J., Maniatis T. Purification and visualization of native spliceosomes. Cell. 1988 Jun 17;53(6):949–961. doi: 10.1016/s0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- Riedel N., Wolin S., Guthrie C. A subset of yeast snRNA's contains functional binding sites for the highly conserved Sm antigen. Science. 1987 Jan 16;235(4786):328–331. doi: 10.1126/science.2948278. [DOI] [PubMed] [Google Scholar]
- Rosbash M., Harris P. K., Woolford J. L., Jr, Teem J. L. The effect of temperature-sensitive RNA mutants on the transcription products from cloned ribosomal protein genes of yeast. Cell. 1981 Jun;24(3):679–686. doi: 10.1016/0092-8674(81)90094-5. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. Splicing of messenger RNA precursors. Science. 1987 Feb 13;235(4790):766–771. doi: 10.1126/science.3544217. [DOI] [PubMed] [Google Scholar]
- Soltyk A., Tropak M., Friesen J. D. Isolation and characterization of the RNA2+, RNA4+, and RNA11+ genes of Saccharomyces cerevisiae. J Bacteriol. 1984 Dec;160(3):1093–1100. doi: 10.1128/jb.160.3.1093-1100.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoops J. K., Awad E. S., Arslanian M. J., Gunsberg S., Wakil S. J., Oliver R. M. Studies on the yeast fatty acid synthetase. Subunit composition and structural organization of a large multifunctional enzyme complex. J Biol Chem. 1978 Jun 25;253(12):4464–4475. [PubMed] [Google Scholar]
- Thomas J. O., Glowacka S. K., Szer W. Structure of complexes between a major protein of heterogeneous nuclear ribonucleoprotein particles and polyribonucleotides. J Mol Biol. 1983 Dec 25;171(4):439–455. doi: 10.1016/0022-2836(83)90039-6. [DOI] [PubMed] [Google Scholar]
- Tollervey D., Mattaj I. W. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J. 1987 Feb;6(2):469–476. doi: 10.1002/j.1460-2075.1987.tb04777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine R. C., Shapiro B. M., Stadtman E. R. Regulation of glutamine synthetase. XII. Electron microscopy of the enzyme from Escherichia coli. Biochemistry. 1968 Jun;7(6):2143–2152. doi: 10.1021/bi00846a017. [DOI] [PubMed] [Google Scholar]
- Vasiliev V. D. Morphology of the ribosomal 30S subparticle according to electron microscopic data. Acta Biol Med Ger. 1974;33(5-6):779–793. [PubMed] [Google Scholar]
- Vijayraghavan U., Parker R., Tamm J., Iimura Y., Rossi J., Abelson J., Guthrie C. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986 Jul;5(7):1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooley J., Chung S. Y., Wall J., Lestourgeon W. Architecture of pre-messenger, nuclear ribonucleoprotein monoparticles. Biophys J. 1986 Jan;49(1):17–19. doi: 10.1016/S0006-3495(86)83575-5. [DOI] [PMC free article] [PubMed] [Google Scholar]






