Abstract
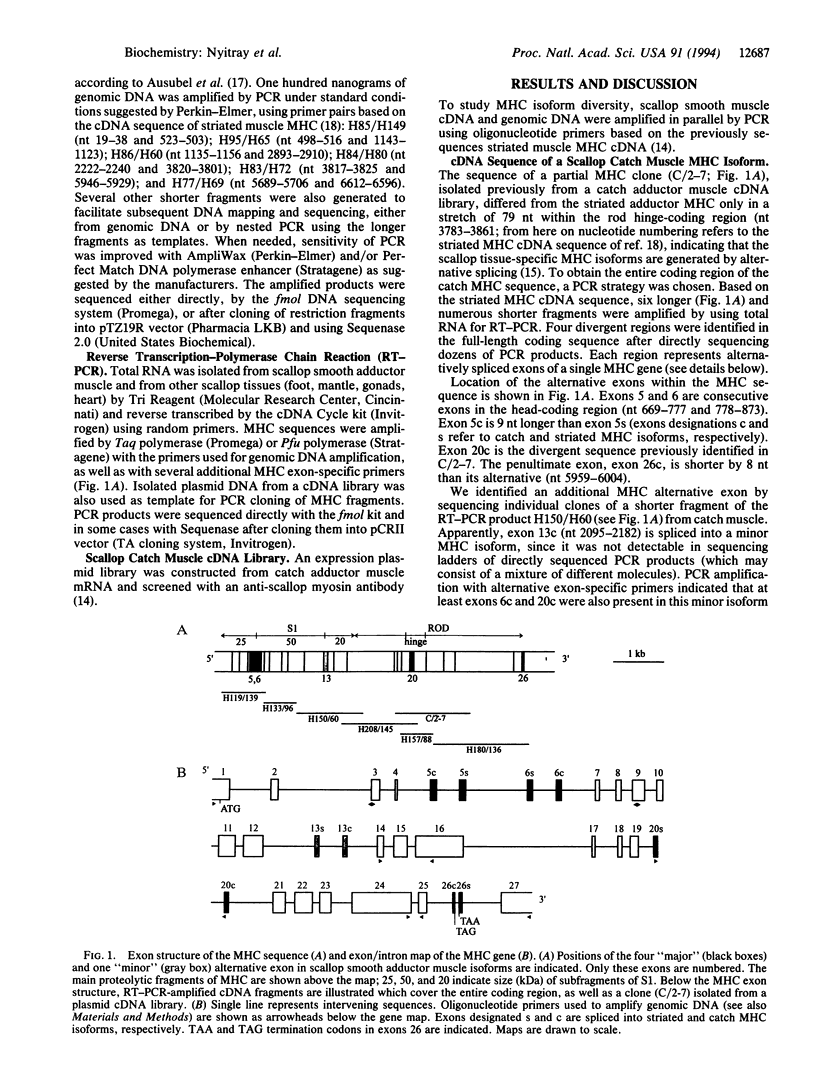
We report here that the catch and striated adductor muscle myosin heavy-chain (MHC) isoforms of scallop (Argopecten irradians, previously Aequipecten irradians) are generated by alternative RNA splicing from a single gene. Scallop catch muscle cDNA and genomic DNA were amplified by PCR using primers based on the previously sequenced scallop striated muscle MHC cDNA. Mapping of the exon/intron borders and sequencing of a full-length catch muscle MHC in overlapping fragments revealed that the 24-kb gene encodes the MHC polypeptide in 27 exons and that four sets of tandem exon pairs are alternatively spliced into a striated and a catch MHC isoform. An additional alternative exon was identified in catch cDNA and is apparently spliced into a minor MHC isoform. The striated muscle-specific isoform is not expressed in other tissues, whereas the catch-type isoforms were also detected in various smooth muscles, but not in the striated one. Of the alternative exons, exons 5 and 6 encode part of the ATP-binding region and the 25-kDa/50-kDa proteolytic junction; exon 13 encodes part of one of the actin-binding regions and extends to the active site; exon 20 encodes the middle of the rod hinge region; exon 26 in the striated-specific sequence starts with the stop codon, whereas the catch-specific exon codes for an additional 10 residues. Differences between the alternative exons presumably determine the lower ATPase activity of smooth muscle myosin, contribute to the different structure of the striated and smooth muscle thick filaments, and may also be important for the molecular mechanism of the catch phenomenon.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Babij P., Periasamy M. Myosin heavy chain isoform diversity in smooth muscle is produced by differential RNA processing. J Mol Biol. 1989 Dec 5;210(3):673–679. doi: 10.1016/0022-2836(89)90142-3. [DOI] [PubMed] [Google Scholar]
- Babij P. Tissue-specific and developmentally regulated alternative splicing of a visceral isoform of smooth muscle myosin heavy chain. Nucleic Acids Res. 1993 Mar 25;21(6):1467–1471. doi: 10.1093/nar/21.6.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani L., Cohen C. Myosin rod phosphorylation and the catch state of molluscan muscles. Science. 1987 Jan 16;235(4786):334–337. doi: 10.1126/science.3026049. [DOI] [PubMed] [Google Scholar]
- Cheney R. E., Riley M. A., Mooseker M. S. Phylogenetic analysis of the myosin superfamily. Cell Motil Cytoskeleton. 1993;24(4):215–223. doi: 10.1002/cm.970240402. [DOI] [PubMed] [Google Scholar]
- Collier V. L., Kronert W. A., O'Donnell P. T., Edwards K. A., Bernstein S. I. Alternative myosin hinge regions are utilized in a tissue-specific fashion that correlates with muscle contraction speed. Genes Dev. 1990 Jun;4(6):885–895. doi: 10.1101/gad.4.6.885. [DOI] [PubMed] [Google Scholar]
- Csizmadia A. M., Bonet-Kerrache A., Nyitray L., Mornet D. Purification and properties of caldesmon-like protein from molluscan smooth muscle. Comp Biochem Physiol Biochem Mol Biol. 1994 May;108(1):59–63. doi: 10.1016/0305-0491(94)90165-1. [DOI] [PubMed] [Google Scholar]
- Dibb N. J., Maruyama I. N., Krause M., Karn J. Sequence analysis of the complete Caenorhabditis elegans myosin heavy chain gene family. J Mol Biol. 1989 Feb 5;205(3):603–613. doi: 10.1016/0022-2836(89)90229-5. [DOI] [PubMed] [Google Scholar]
- George E. L., Ober M. B., Emerson C. P., Jr Functional domains of the Drosophila melanogaster muscle myosin heavy-chain gene are encoded by alternatively spliced exons. Mol Cell Biol. 1989 Jul;9(7):2957–2974. doi: 10.1128/mcb.9.7.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson H. V., Spudich J. A. Molecular evolution of the myosin family: relationships derived from comparisons of amino acid sequences. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):659–663. doi: 10.1073/pnas.90.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes T. R., Block S. M., White B. T., Spudich J. A. Movement of myosin fragments in vitro: domains involved in force production. Cell. 1987 Mar 27;48(6):953–963. doi: 10.1016/0092-8674(87)90704-5. [DOI] [PubMed] [Google Scholar]
- Kelley C. A., Takahashi M., Yu J. H., Adelstein R. S. An insert of seven amino acids confers functional differences between smooth muscle myosins from the intestines and vasculature. J Biol Chem. 1993 Jun 15;268(17):12848–12854. [PubMed] [Google Scholar]
- Kerwin B. A., Yount R. G. Photolabeling evidence for calcium-induced conformational changes at the ATP binding site of scallop myosin. Proc Natl Acad Sci U S A. 1993 Jan 1;90(1):35–39. doi: 10.1073/pnas.90.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketchum A. S., Stewart C. T., Stewart M., Kiehart D. P. Complete sequence of the Drosophila nonmuscle myosin heavy-chain transcript: conserved sequences in the myosin tail and differential splicing in the 5' untranslated sequence. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6316–6320. doi: 10.1073/pnas.87.16.6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S., Morita F. Smooth muscle of scallop adductor contains at least two kinds of myosin. J Biochem. 1981 Sep;90(3):673–681. doi: 10.1093/oxfordjournals.jbchem.a133522. [DOI] [PubMed] [Google Scholar]
- Mahdavi V., Periasamy M., Nadal-Ginard B. Molecular characterization of two myosin heavy chain genes expressed in the adult heart. Nature. 1982 Jun 24;297(5868):659–664. doi: 10.1038/297659a0. [DOI] [PubMed] [Google Scholar]
- McLachlan A. D., Karn J. Periodic charge distributions in the myosin rod amino acid sequence match cross-bridge spacings in muscle. Nature. 1982 Sep 16;299(5880):226–231. doi: 10.1038/299226a0. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai R., Kuro-o M., Babij P., Periasamy M. Identification of two types of smooth muscle myosin heavy chain isoforms by cDNA cloning and immunoblot analysis. J Biol Chem. 1989 Jun 15;264(17):9734–9737. [PubMed] [Google Scholar]
- Nguyen H. T., Gubits R. M., Wydro R. M., Nadal-Ginard B. Sarcomeric myosin heavy chain is coded by a highly conserved multigene family. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5230–5234. doi: 10.1073/pnas.79.17.5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyitray L., Goodwin E. B., Szent-Gyorgyi A. G. Nucleotide sequence of full length cDNA for a scallop striated muscle myosin heavy chain. Nucleic Acids Res. 1990 Dec 11;18(23):7158–7158. doi: 10.1093/nar/18.23.7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyitray L., Goodwin E. B., Szent-Györgyi A. G. Complete primary structure of a scallop striated muscle myosin heavy chain. Sequence comparison with other heavy chains reveals regions that might be critical for regulation. J Biol Chem. 1991 Oct 5;266(28):18469–18476. [PubMed] [Google Scholar]
- Rayment I., Rypniewski W. R., Schmidt-Bäse K., Smith R., Tomchick D. R., Benning M. M., Winkelmann D. A., Wesenberg G., Holden H. M. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993 Jul 2;261(5117):50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Sohma H., Yazawa M., Morita F. Phosphorylation of regulatory light chain a (RLC-a) in smooth muscle myosin of scallop, Patinopecten yessoensis. J Biochem. 1985 Aug;98(2):569–572. doi: 10.1093/oxfordjournals.jbchem.a135311. [DOI] [PubMed] [Google Scholar]
- Strehler E. E., Strehler-Page M. A., Perriard J. C., Periasamy M., Nadal-Ginard B. Complete nucleotide and encoded amino acid sequence of a mammalian myosin heavy chain gene. Evidence against intron-dependent evolution of the rod. J Mol Biol. 1986 Aug 5;190(3):291–317. doi: 10.1016/0022-2836(86)90003-3. [DOI] [PubMed] [Google Scholar]
- Szent-Györgyi A. G., Cohen C., Kendrick-Jones J. Paramyosin and the filaments of molluscan "catch" muscles. II. Native filaments: isolation and characterization. J Mol Biol. 1971 Mar 14;56(2):239–258. doi: 10.1016/0022-2836(71)90462-1. [DOI] [PubMed] [Google Scholar]
- Szent-Györgyi A. G., Szentkiralyi E. M., Kendrick-Jonas J. The light chains of scallop myosin as regulatory subunits. J Mol Biol. 1973 Feb 25;74(2):179–203. doi: 10.1016/0022-2836(73)90106-x. [DOI] [PubMed] [Google Scholar]
- Takahashi M., Kawamoto S., Adelstein R. S. Evidence for inserted sequences in the head region of nonmuscle myosin specific to the nervous system. Cloning of the cDNA encoding the myosin heavy chain-B isoform of vertebrate nonmuscle myosin. J Biol Chem. 1992 Sep 5;267(25):17864–17871. [PubMed] [Google Scholar]
- Ueno H., Harrington W. F. Local melting in the subfragment-2 region of myosin in activated muscle and its correlation with contractile force. J Mol Biol. 1986 Jul 5;190(1):69–82. doi: 10.1016/0022-2836(86)90076-8. [DOI] [PubMed] [Google Scholar]
- White S., Martin A. F., Periasamy M. Identification of a novel smooth muscle myosin heavy chain cDNA: isoform diversity in the S1 head region. Am J Physiol. 1993 May;264(5 Pt 1):C1252–C1258. doi: 10.1152/ajpcell.1993.264.5.C1252. [DOI] [PubMed] [Google Scholar]