Abstract
Cardiomyocytes derived from human induced pluripotent stem cells (iPSC-CMs) hold great promise for modeling human heart diseases. However, iPSC-CMs studied to date resemble immature embryonic myocytes and therefore do not adequately recapitulate native adult cardiomyocyte phenotypes. Since extracellular matrix plays an essential role in heart development and maturation in vivo, we sought to develop a synthetic culture matrix that could enhance functional maturation of iPSC-CMs in vitro. In this study, we employed a library of combinatorial polymers comprising of three functional subunits - poly-ε-caprolacton (PCL), polyethylene glycol (PEG), and carboxylated PCL (cPCL) - as synthetic substrates for culturing human iPSC-CMs. Of these, iPSC-CMs cultured on 4%PEG-96%PCL (each % indicates the corresponding molar ratio) exhibit the greatest contractility and mitochondrial function. These functional enhancements are associated with increased expression of cardiac myosin light chain-2v, cardiac troponin I and integrin alpha-7. Importantly, iPSC-CMs cultured on 4%PEG-95%PCL demonstrate troponin I (TnI) isoform switch from the fetal slow skeletal TnI (ssTnI) to the postnatal cardiac TnI (cTnI), the first report of such transition in vitro. Finally, culturing iPSC-CMs on 4%PEG-96%PCL also significantly increased expression of genes encoding intermediate filaments known to transduce integrin-mediated mechanical signals to the myofilaments. In summary, our study demonstrates that synthetic culture matrices engineered from combinatorial polymers can be utilized to promote in vitro maturation of human iPSC-CMs through the engagement of critical matrix-integrin interactions.
Keywords: iPSC, cardiomyocytes, maturation, combinatorial polymer, myosin light chain-2v, troponin I
1. Introduction
Heart disease is the leading cause of death in developed countries, accounting for over 36% of all deaths in the United States [1], yet the mechanistic study of human heart disease beginning at the cellular level has been limited by the lack of suitable human cardiomyocyte models. However, thanks to recent revolutionary advances in cellular reprogramming, and directed differentiation of human induced pluripotent stem cells (iPSCs) into cardiomyocytes (iPSC-CMs), a number of in vitro models of healthy and diseased human cardiac tissues have been developed.[2, 3] Despite these advances, an important concern regarding the use of iPSC-CMs is their functional immaturity relative to primary cardiomyocytes. For instance, early studies indicated that cardiomyocytes generated in vitro from pluripotent stem cells exhibit fetal phenotypes with respect to their physiological performance.[4, 5] Several recent studies have addressed maturation of iPSC-CMs or embryonic stem cell-derived cardiomyocytes (ESC-CMs),[6–10] but the majority were limited to calcium handling and electrophysiological evaluation. Thus, considerable unmet needs remain for the adequate study of in vitro maturation of iPSC-CMs, particularly at the cell and molecular levels, and factors that modulate it. One such factor influencing cell maturation, including cardiomyocytes, is the tissue microenvironment. In particular, cell-substratum interaction is essential for proper development and maintenance of tissue architecture and function. In many complex organisms, the extracellular matrix (ECM) plays a critical role in cardiomyocyte development, but the full mechanism of its impact remains unknown due to the ECM’s heterogeneity in both composition and structural orientation. Yet, despite considerable progress being made to engineer niches that control cellular responses through purpose-specific biomaterial designs (e.g., surface patterning, biomolecule addition) that would encompass some of the native ECM properties, the direct effects of characteristic biochemical and biophysical properties of unmodified materials alone have largely been underexplored. To address the need, we employed a library of copolymer scaffolds with varying physicochemical properties as culture substrates.[11] The copolymer library contained different mole percentages of three components: hydrophilic poly(ethylene glycol) (PEG), hydrophobic poly(ε-caprolacton) (PCL), and negatively-charged carboxylated-PCL (cPCL). Each copolymer subunit was selected for the specific properties it contributed to the resulting copolymer: PCL is a semi-crystalline, biodegradable, and hydrophobic, as well as being FDA-approved in medical devices[12]; PEG is a biocompatible, hydrophilic, and repellent polymer that reduces protein adsorption and cell attachment through steric exclusion[13, 14]; and cPCL facilitates cell attachment to the scaffold surface by providing a negative charge, effectively counteracting the PEG’s repellant effects.[14] These combinatorial polymers were electrospun to make fiber mesh scaffolds that mimic ECM fiber structure and orientation, and subsequently used as test culture substrates.
Human iPSCs were differentiated into human iPSC-CMs through a directed differentiation protocol.[15] After 15 to 30 days of culture on each copolymer scaffold, we examined the effects of the copolymer composition on iPSC-CM phenotype by evaluating beating behavior, mitochondrial function and gene expression profiles. Our results indicate that certain combinatorial polymer scaffolds, especially a 4%PEG-96%PCL copolymer, promote the acquisition of several phenotypic features of mature ventricular myocytes including organized sarcomeres, abundant mitochondria, increased contractility and higher expression of cardiac myosin light chain-2v, cardiac troponin I and integrin alpha-7, each of which have been associated with cardiac/ventricular maturation.[16–18] Moreover, 4%PEG-96%PCL was associated with enhanced expression of intermediate filament-associated proteins involved in transducing integrin-mediated mechanical signals to the myofilaments. These results suggest the synthetic biomaterial promoted cardiac maturation by mimicking some features of basement membrane-integrin/sarcolemma interactions seen in normal development. In summary, our study suggests that specific chemical compositions of synthetic extracellular substrates can exert profound influence on in vitro maturation of iPSC-CMs.
2. Materials and Methods
2.1 Reprogramming of human dermal fibroblasts and maintenance of human iPSCs
A human iPSC line (CC2) was generated from a healthy control subject using an episomal approach and validated, as we have previously described, following the work of Dr. Shinya Yamanaka.[19–21] Maintenance and culture of human iPSCs followed our established methods.[19–22] Pluripotency was validated by PluriTest, a bioinformatics assay,[23] using a teratoma-validated line as a positive control, and normal chromosomal karyotype was confirmed (Genetic Associates, Nashville TN) as previously described (Fig. S1a and S1b).[19, 23] The absence of episomal vector genomic integration was confirmed by PCR (data not shown). Immunostaining for pluripotency markers used the following antibodies; OCT4 (mouse monoclonal, Millipore), NANOG (affinity purified anti-goat IgG), and SSEA4 (rabbit monoclonal, Millipore) (Fig. S2a).
2.2 Differentiation of human iPSCs to cardiomyocytes, and isolation of rabbit ventricular myocytes
iPSCs were washed with DMEM/F12 (1:1, Invitrogen) and PBS, followed by incubation with 1 mL/well Versene (Invitrogen) for 10 minutes at 37°C. iPSCs were seeded on growth factor reduced Matrigel (BD Biosciences) coated plates at a density of 1 million cells per well in mTeSR1 medium supplemented with 10 μM ROCK inhibitor (Y-27632, CalBiochem). iPSCs were overlaid with mTeSR1 supplemented Matrigel (70–150 μg). After 24 hours, medium was changed to RPMI 1640 medium plus B-27 and Activin A (R&D Systems) without insulin supplement (Invitrogen). Fresh Matrigel (83 μg Matrigel/well) mixed with RPMI 1640 basal medium was then overlaid for 30 minutes on the cells at 24 hours post-Activin A treatment at day 0 of cardiac induction. After 20 minutes of Matrigel gelation, RPMI 1640 basal medium with B-27, BMP4 (5ng/mL, R&D Systems) and bFGF (5ng/mL, Invitrogen) without insulin supplement was replaced on the cells (day 1), and then incubated for 4 days without changing medium. At day 5, the medium was changed with RPMI 1640 with B27 complete supplement (Invitrogen) (Fig. S2b) and replaced every 2–3 days. After 15 days of cardiac differentiation, the medium was changed to DMEM/F12 including 2% fetal bovine serum (FBS), 2mM L-glutamine, 0.1mM nonessential amino acids, 0.1mM β-mercaptoethanol, and 50u/mL penicillin/streptomycin for maintenance of iPSC-CMs.
Ventricular myocytes from adult rabbits were isolated by a modified collagenase/protease method, as previously described.[24]
2.3 Traction force microscopy
Traction force maps of spontaneously beating iPSC-CMs were generated at room temperature using the traction force microscopy (N=15 cells from four experimental replicates). Briefly, iPSC-CMs, grown on various culture matrices, were dissociated and replated on Matrigel containing 0.75 μm fluorescence beads (Polysciences) diluted at 1:50. Contraction motion of individual iPSC-CMs was video-captured by phase-contrast and fluorescent imaging, and the particle image velocimetry (PIV) was applied to assess displacement deformation of Matrigel substrate. Fourier transform traction cytometry (FTTC) was used to quantify contractile force using an ImageJ plugin. [25, 26]
2.4 Flow cytometry
iPSC-CMs maintained on each matrix for 30 days were detached with Triplexpress (Invitrogen). Cells were lightly vortexed to break up large cell aggregates and quenched with RMPI1640 with complete B27 supplement. Cells were fixed in 1% paraformaldehyde at room temperature for 10 minutes, permeabilized by FACS buffer (PBS without Ca/Mg2+, 1% FBS, 0.1% NaN3) with 0.1% saponin. Cells were washed once in FACS buffer and centrifuged. The supernatant was then discarded leaving about 50 μl of cell suspension. Troponin T (mouse monoclonal, Santa Cruz Biotechnology), RYR (rabbit polyclonal, Santa Cruz Biotechnology), and a-actinin (mouse monoclonal, Sigma) were diluted in 50 μl /sample FACS buffer plus 0.1% Triton X-100. Cells were incubated with the primary antibodies for 1 hour at 4°C; washed once in 3 ml FACS buffer plus 0.1% Triton X-100; centrifuged; and supernatant discarded leaving about 50 μl of cell suspension. Secondary antibodies specific to the primary antibody IgG isotype were diluted in FACS buffer plus 0.1% Triton X-100 for a final sample volume of 100 μl at 1:1000 dilution. Cells were incubated for 30 minutes in the dark at 4°C; washed in FACS buffer plus 0.1% Triton X-100; and resuspended in 500 μl FACS buffer for analysis (N=4). Resulting data were analyzed using FlowJo v8.5.2 and data reported are based on 10,000 gated events.
2.5 Western Blotting and Immunostaining
For western blotting, iPSC-CMs were lysed in RIPA buffer containing protease and phosphatase inhibitors (Thermo Scientific), followed by boiling the samples in Lamelli Buffer (Bio-Rad) Extracted proteins were run on 12% (wt/vol) Trisglycine acrylamide gels (Bio-Rad) for separation and transferred onto a nitrocellulose membrane. The membrane was incubated with primary antibody overnight at 4°C after blocking, washed, and incubated with the appropriate secondary antibody conjugated with an infrared fluorescent dye for 1 hour at 25°C. Antibodies to cardiac troponin I (cTnI; rabbit polyclonal, Abcam) and slow skeletal troponin I (ssTnI; rabbit polyclonal, Sigma) were used as primary antibodies along with α-tubulin antibody (rabbit monoclonal, Abcam) as a loading protein control. Odyssey (LiCor) was used for scanning, and analyses of western blot images were performed using Image J software (N=4). Each value of cTnI and ssTnI protein expression was normalized to the α-tubulin intensity.
For immunofluorescent imaging, iPSC-CMs were harvested from the matrix sandwich culture or polymer matrices using Triplexpress (Invitrogen). Cells were washed fixed in 4% paraformaldehyde for 15 minutes at room temperature and permeabilized in 0.1% Triton X-100 (Sigma) for 1 hour at room temperature. Samples were blocked with 5% non-fat dry milk (Bio-Rad) in PBS with 0.1% Triton X-100 and incubated for 1 hour at room temperature. Troponin T (mouse monoclonal, Santa Cruz Biotehnology), a-actinin (mouse monoclonal, Sigma), MLC-2a (mouse polyclonal, Santa Cruiz Biotehnology), MLC-2v (rabbit polyclonal Abcam), cardiac troponin I (rabbit polyclonal, Abcam), slow skeletal troponin I (rabbit polyclonal, Sigma) and Mitofusin 2 (mouse monoclonal, Abcam) were added in 0.1% Triton X-100, 1% BSA in PBS solution and incubated for 1 hour at 37°C. After multiple washing with 0.2% Tween 20 in PBS and 1X PBS twice, secondary antibodies specific to the primary antibody IgG isotype were diluted (1:1000) in the same solution as the primary antibodies and incubated at room temperature for 1 hour. Samples were washed with 0.2% Tween 20 in PBS twice and 1X PBS twice and treated with Gold Anti-fade Reagent (Invitrogen). Slides were examined under a confocal microscope (Olympus FV-1000) and analyzed with NIS-Elements BR3.0 (9–10 images per test group from four experimental replicates).
2.6 Synthesis of combinatorial polymer library
Test copolymers were synthesized as previously reported.[11] Briefly, caprolactone (100×10−6 mol, 11.4 g, 10.96 mL), Sn(Oct)2 (100×10−6 mol, 40 mg), and monomethoxy-PEG (100×10−6 mol, 0.10 g, 0.10 ml) were reacted and degassed for 30 min with three freeze-pump-thaw cycles. The ampoule was immersed in an oil bath at 140°C. Cooling after 4 hours stopped polymerization. For synthesizing carboxylated PCL (cPCL), PCL (10g, 0.067 mol) in 500 mL of anhydrous THF was added under dry nitrogen. After the solution was placed in a dry ice/acetone bath and degassed, a solution of LDA, 2 M in THF/n-heptane (33.5 mL, 0.067 mol: 1 equivalent per monomeric unit), was added drop wise into the reaction. Dry CO2 gas was then generated by addition of concentrated H2SO4 to dry Na2CO3, and allowed to bubble through the solution for 30 minutes. A solution of NH4Cl was added to the flask to quench the reaction mixture. The resulting solution was acidified with a solution of concentrated HCl to pH 2–3. The combined organic phases were washed twice with 50 mL of distilled water and dried over anhydrous Na2SO4. The polymer was precipitated from the resulting concentrated solution by addition of diethyl ether, producing x%PEG-b-y%PCL-co-z%cPCL (x, y, z: molar %). The resulting polymers were confirmed by nuclear magnetic resonance (NMR); For 4%PEG-96%PCL: 1H NMR (CDCl3) =d 4.06 (t, 3H, -OCH2), 2.31(t, 2H, -CH2), 1.66 (m, 2H, -CH2), 1.37 (m, 4H, -CH2) ppm. For 4%PEG-86%PCL-10%cPCL: 1H NMR (CDCl3) = δ 4.06 (t, 3H, -OCH2), 3.4 (m, 1H, - CH-COOH), 2.31(t, 2H, -CH2), 1.66 (m, 2H, -CH2), 1.37 (m, 2H, -CH2) ppm. For 90%PCL-10%cPCL: 1H NMR (CDCl3) = δ 4.06 (t, 3H, -OCH2), 3.4 (m, 1H, -CH-COOH), 2.31(t, 2H, -CH2), 1.66 (m, 2H, -CH2), 1.37 (m, 2H, -CH2) ppm.
2.7 Characterization of x%PEG-b-y%PCL-co-z%cPCL
Polymer molecular weight was determined by gel permeation chromatography (GPC) [27] on a Tosoh Biosciences TSKGel SuperHZ-M mixed bed column incubated at 40°C, with a Shimadzu SPD-10A UV detector and RID-10A refractive index detector (Shimadzu Scientific Instruments, Columbia, MD, USA). Molecular weight (Mn) was calculated against monodisperse poly(methyl methacrylate) standards (PMMA; Varian Inc., Palo Alto, CA, USA). 1H NMR spectra were recorded on a Brüker 400 MHz spectrometer with CDCl3 as solvent. At least three samples per each polymer composition were used for these studies (N≥3).
2.8 Electrospinning and coating
Polymer solutions (10% w/v) in a mixture of chloroform and methanol (4:1 by volume) were loaded into a glass syringe with a needle connected to a high-voltage power supply (15 kV). The solution was continuously supplied using a syringe pump at a rate of 0.5 ml/hour for 20 minutes. The resulting fibers were collected over glass cover slips placed on a rotating mandrel (10 cm from the needle) at 1300 rpm. The scaffolds were dried under vacuum for 72 hours and sterilized by UV irradiation for 30 minutes. Substrates were coated with vitronectin (Life Technology, 50ng/ml) to maintain the same level of cell adhesion among test polymers. This procedure has been shown to remove toxic organic solvents and sterilize polymer substrates effectively in previous studies [11].
2.9 Contact Angle and Dynamic Mechanical Analysis (DMA)
Contact angles of spin coated polymers were measured by sessile drop method using an in-house goniometer [28]. Data points were collected at 10 seconds intervals for a total of 10 min for each polymer. Dry elastic moduli of electrospun polymer scaffolds were measured by DMA (Q800 DMA, TA Instruments, New Castle, DE, USA). Matrices were prepared in a uniform rectangular form with the dimension of 15.0 (l)×6.6 (w) mm2. A stress-strain curve was obtained using a tension clamp at 37°C. A preload force of 0.1 N was applied to each sample, and force was increased at a rate of 0.1 N/min until failure. At least three samples per each polymer composition were used for these studies (N≥3).
2.10 Mitochondria staining with TMRM
TMRM, 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide (Invitrogen) was used to monitor mitochondrial membrane potentials on live iPSC-CMs. Briefly, iPSC-CMs were incubated for 4 hours in RPMI 1640 with B27 complete supplement containing 2 nM TMRM, [29] followed by washes with the above media without TMRM. Cells were imaged using a fluorescent microscope (Olympus FV-1000, 20X objective), and the fluorescence intensity was quantified using Image J software (9–10 images per test group from four experimental replicates), calibrated as described in [30], and normalized to TMRM intensity of iPSC-CMs on Matrigel. Also, adult rabbit cardiomyocytes were used for TMRM experiment as a positive control.
2.11 Sample preparation and RNA sequencing for molecular profiles of iPSC-CMs
An Illumina TruSeq Stranded mRNA Sample Preparation Kit was used to convert the total mRNA (100 ng) into a library of template molecules suitable for subsequent cluster generation and sequencing on the Illumina HiSeq 2500. A quality check of the input total RNA was done by running an aliquot on the Agilent Bioanalyzer to confirm integrity. The Qubit RNA fluorometry assay was then conducted to measure the RNA concentration. The input to library prep was 48 ul of 100 ng of total RNA (2 ng/ul) and 2 ul of a 1:1000 dilution of the ERCC RNA Spike-In Control Mix 1 to provide a set of external RNA controls for gene expression analysis. The poly-A containing mRNA molecules were selected using poly-T oligo-attached magnetic beads, and the eluted poly-A RNA molecules were cleaved into small fragments of 120–210 bp using divalent cations under elevated temperature. The cleaved RNA fragments were copied into first strand cDNA using SuperScript II reverse transcriptase and random primers, followed by second strand cDNA synthesis using DNA polymerase I and RNase H. The cDNA fragments then went through an end repair process, the addition of a single ‘A’ base, and then ligation of the Illumina multiplexing adapters. The products were purified and enriched by PCR to create the final cDNA sequencing library. The quality of the cDNA library was controlled using the Agilent Bioanalyzer HS DNA assay to confirm the final library size. An Agilent Mx3005P qPCR machine was also used with the KAPA Illumina library quantification kit to determine the product concentration. From the pool, 12 pM was loaded into each well of the flow cell for the Illumina cBot. The flow cell was then loaded onto the Illumina HiSeq 2500 utilizing v3 chemistry and HTA 1.8. The raw sequencing readouts in BCL format were processed through CASAVA-1.8.2 for FASTQ conversion. The RTA chastity filter was used, and only the PF (passfilter) reads were retained for further analysis. HiSeq 2500 readouts were aligned by Cufflink (http://http://cufflinks.cbcb.umd.edu/).
2.12 Evaluation of gene candidates by qRT-PCR
Genes obtained from RNA seq were evaluated by qRT-PCR using TaqMan technology in the ViiA7 Fast Real-Time RT-PCR System according to the manufacturer’s instructions (Applied Biosystems). We designed all TaqMan probes and obtained them from Applied Biosystems. Information of designed TaqMan probes is shown in Table S1. GAPDH was used to normalize all qRT-PCR products. To determine transcript quantity, ddCt-based fold-change calculations were used.[31] All qPCR experiments were done with triplicates, each set from at least three biological replicates.
2.13 Statistical analysis
All experiments were replicated at least three times (N≥3). For image analysis of cell staining (TMRM or immunostaining) and traction force microscopy, 9–10 images and 15 isolated cells were analyzed, respectively, from four replicate experiments (N≥9). Comparisons between two groups were performed with a Student’s unpaired t test. Comparisons between multiple groups were performed with a one-way analysis of variance (ANOVA) with a Tukey post hoc test to adjust p-values for multiple comparisons. In all cases, p < 0.05 is considered statistically significant. Mean ± standard deviation is reported, unless otherwise noted.
3. Results
3.1. Generation of human iPSC-CMs
Validated human iPSCs from a healthy control subject, generated via an episomal-based reprogramming method developed by Shinya Yamanaka and colleagues, were used for all studies (Fig. S2a).[19–21] Direct differentiation of these iPSCs cultured on Matrigel, outlined in Fig. S2b, successfully yielded iPSC-CMs. Visual inspection of the iPSC-CMs 7–8 days after differentiation showed individual beating cells, and 10–13 days after differentiation the iPSC-CMs formed beating sheets of cells (Fig. S2c, Movie S1). Using this cardiac induction method, more than 95% of the cells were positive for troponin T at 15 days post-differentiation (Fig. S2d). Immunostaining of iPSC-CMs for cardiac α-actinin and troponin T revealed distinct Z lines and well-organized cardiac myofilaments (Fig. S2e). Additionally, surface plot analysis assessed the movement of spontaneously beating sheets of iPSC-CMs (Fig. S2f). Collectively, the cardiac induction processes resulted in an actively beating and uniform iPSC-CM population for subsequent use on the copolymer matrices.
3.2. Synthesis and characterization of copolymer matrices
To determine the impact of synthetic culture substrates on human iPSC-CMs maturation, we used a library of copolymers, which had previously been used to enhance cardiac differentiation of mouse embryonic stem cells.[11] Copolymers were generated by ring opening polymerization of monomeric ε-caprolactone with methoxy-PEG at desired molar ratios, followed by production of carboxylated PCL (cPCL) at random positions of the copolymers. All resulting copolymers were formulated with x%PEG-y%PCL-z%cPCL (x, y, z: molar ratio) (Fig. 1a) and electrospun on glass coverslips to form fiber-mesh substrates for cell culture.[32] All copolymer fiber matrices exhibited consistent random mesh structures with an average fiber diameter of 0.6 μm (data not shown).
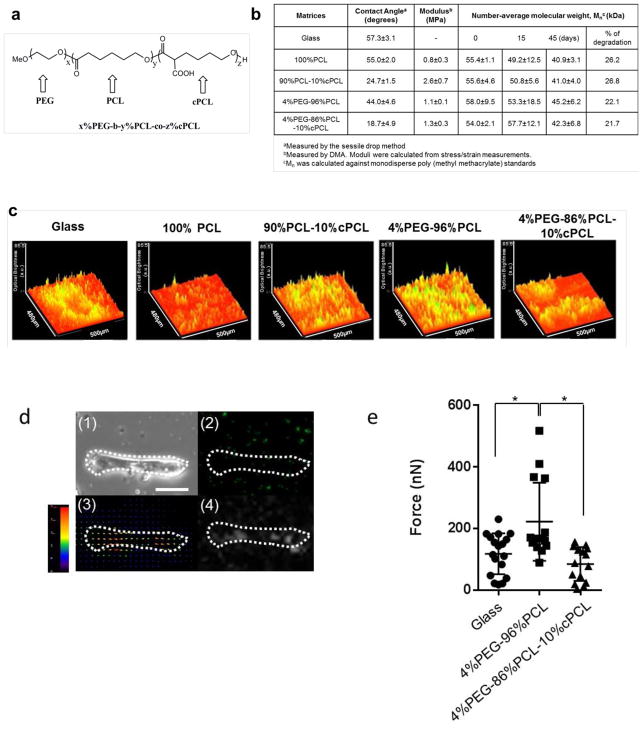
Fig. 1. Characterization of combinatorial copolymer matrices, and their effect on iPSC-CM contractility.
(a) Copolymer structure represented as x%PEG-y%PCL-z%cPCL where x, y and z represent the respective molar ratios of the subunits (PEG, PCL and cPCL). (b) Surface hydrophilicity, modulus and degradation of combinatorial polymers with different molar ratios of subunits. (c) Surface plot image on each polymer matrix represents the contractile activity of iPSC-CMs at the 15-day point after seeding. iPSC-CMs maintained on 90%PCL-10%cPCL and 4%PEG-96%PCL exhibited stronger contractile movement, compared to two controls (Glass and 100% PCL). (d) Parts (1) and (2) show the phase-contrast and fluorescent images (single cell size) from fluorescence beads (0.75um) mixed with Matrigel after culturing an iPSC-CM on glass for 15 days. Scale bar indicates 20μm. Part (3) is an example of a displacement field image of fluorescence beads calculated by the particle image velocimetry (PIV) algorithm. Part (4) illustrates an example of a traction force field image calculated using the PIV result from part (3) by the Fourier transform traction cytometry (FTTC) method. (e) Traction force generated from individual iPSC-CMs on Matrigel after 15-day culture on glass, 4%PEG-96%PCL, and 4%PEG-86%PCL-10%cPCL. *p<0.05.
Copolymers were then characterized for hydrophilicity, stiffness and degradation (Fig. 1b). Water contact angles of each copolymer and glass substrate were measured using the sessile drop method. The average contact angles of glass, 100%PCL, 90%PCL-10%cPCL, 4%PEG-96%PCL, and 4%PEG-86%PCL-10%cPCL were 57.3°, 55°, 24.7°, 44°, and 18.7°, respectively. Thus, incorporation of the hydrophobic PEG subunit into the copolymer had the net effect of decreasing the contact angle in comparison to 100%PCL and glass. Incorporation of 10%cPCL further decreased the contact angle, with the contact angle for 4%PEG-86%PCL-10%cPCL demonstrating the additive hydrophillic effect of PEG and cPCL. With respect to intrinsic mechanical properties, elastic moduli of the test polymers were similar, except for 90%PCL-10%cPCL, which had a slightly higher elastic modulus (stiffness) than the others (Fig. 1b). The elastic moduli were measured under dry conditions. Under wet condition, we previously showed that the elastic moduli of these copolymers decreased approximately 10 to 20-fold [11]. Furthermore, hydrolytic degradation of the synthetic materials is predicted to makes them even more mechanically compliant. Considering that the modulus of native rat myocardium has been shown to range from about 10 to 150 kPa [33–35], our results indicate that elastic moduli of the polymers, which do not differ significantly from one another, probably fall within the range for the mechanical properties of intact heart tissue in vivo. Analysis by gel permeation chromatography (GPC) also revealed that the number average molecular weight (Mn) of all copolymers ranged from 54kDa to 58kDa. After a 45-day degradation period, the new Mn range spanned from 38kDa to 45kDa (Fig. 1b), indicating 21–26% degradation of the test copolymers during 45 days of incubation.
3.3. Beating behavior of iPSC-CMs on copolymer matrices, and traction force generated by them
Human iPSC-CMs at day 15 of cardiac induction were plated onto vitronectin-coated electrospun copolymer matrices and cultured for 15 days. Since the degree of contractility is an important indicator of cardiomyocyte maturity,[36–38] the contractile behavior of spontaneously beating iPSC-CM sheets was analyzed with surface plots generated by ImageJ. Of the substrates tested, iPSC-CMs on glass coverslips and 100%PCL displayed relatively weak, heterogeneous beating, while iPSC-CMs on the copolymer matrices exhibited significantly higher movement during each beat. Of these, iPSC-CMs sheets maintain on the 4%PEG-96%PCL displayed the greatest motion (Fig. 1c), suggesting this particular substrate enhanced iPSC-CM contractility.
In addition, contractile force exerted by iPSC-CMs cultured on glass, 4%PEG-96%PCL, and 4%PEG-86%PCL-10%cPCL were quantified by traction force microscopy after re-plating the cells on Matrigel.[25, 26] Representative phase-contrast and fluorescence images of an iPSC-CM transferred from a copolymer substrate onto Matrigel are shown in Fig. 1d. The traction force maps revealed that the average force generated by individual iPSC-CMs previously maintained on glass, 4%PEG-96%PCL, and 4%PEG-86%PCL-10%cPCL were 117.6, 221.9, and 84.5 nN, respectively (Fig. 1e). Thus, analyses of iPSC-CM sheet movement and traction force performance of individual iPSC-CMs indicate that, of the substrates tested, iPSC-CMs maintained on 4%PEG-96%PCL exhibited the highest contractility.
3.4. Mitochondrial function of iPSC-CMs on copolymer matrices
We next examined whether the differences in contractility was associated with changes in mitochondrial function, as mitochondrial energy output is closely tied to cardiomyocyte contractility.[38, 39] We assessed the mitochondrial function of iPSC-CMs using tetramethyl rhodamine methylester (TMRM), an in vitro fluorescent probe serving as a reporter of the inner mitochondrial membrane potential (Δψm), which is critical for maintaining adequate cellular ATP levels.[40] Previous studies have shown that the enhancement of mitochondrial function resulting from cardiomyocyte maturation can be assessed by an increased TMRM florescence intensity.[29, 41, 42] Significantly higher TMRM fluorescence intensity was observed in iPSC-CMs cultured on 4%PEG-96%PCL than on other substrates (Fig. 2a and 2b). Although the TMRM fluorescence intensity for iPSC-CMs was not equivalent to that of adult rabbit myocytes, maintaining iPSC-CMs on 4%PEG-96%PCL enhances contractility and mitochondrial function in comparison to other culture substrates, including the standard substrate Matrigel.
Fig. 2. Effects of combinatorial polymer matrix on mitochondria functions of iPSC-CMs.
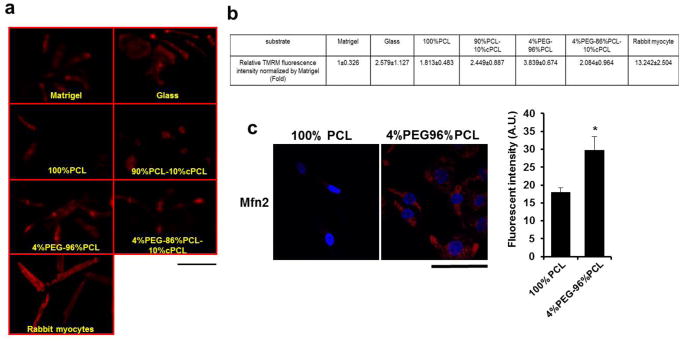
(a) Mitochondrial function of iPSC-CMs maintained on each polymer matrix was assessed by tetramethyl rhodamine methylester (TMRM), a fluorescent probe of inner mitochondrial membrane potential (red). Scale bar is 100 μm. (b) A higher intensity of TMRM staining was seen on iPSC-CMs maintained on 4%PEG-96%PCL compared to glass and other matrices. *p<0.05: compared to other substrates. (c) Immunostaining of iPSC-CMs for mitofusin-2 (Mfn2), which is involved in the mitochondrial maturation, revealed higher level of Mfn2 expression in iPSC-CMs maintained on 4%PEG-96%PCL compared to 100%PCL. *p<0.05: compared to PCL as a polymer control. Scale bar is 100 μm.
We also examined Mitofusin-2 (Mfn2) protein expression as a cell biological correlate for mitochondrial network maturation. Mfn2 is a mitochondrial membrane protein required for mitochondrial fusion and maintenance of functional mitochondrial network.[43] Consistent with the TMRM results, immunostained Mfn2 showed considerably greater mitochondrial network expression in iPSC-CMs on 4%PEG-96%PCL than on 100%PCL (Fig. 2c). Taken together, our results indicate that 4%PEG-96%PCL substantially enhances both contractility and mitochondrial maturation in iPSC-CMs compared to other matrices.
3.5. MLC-2v and cardiac troponin I (cTnI) expression in iPSC-CMs on copolymer matrices
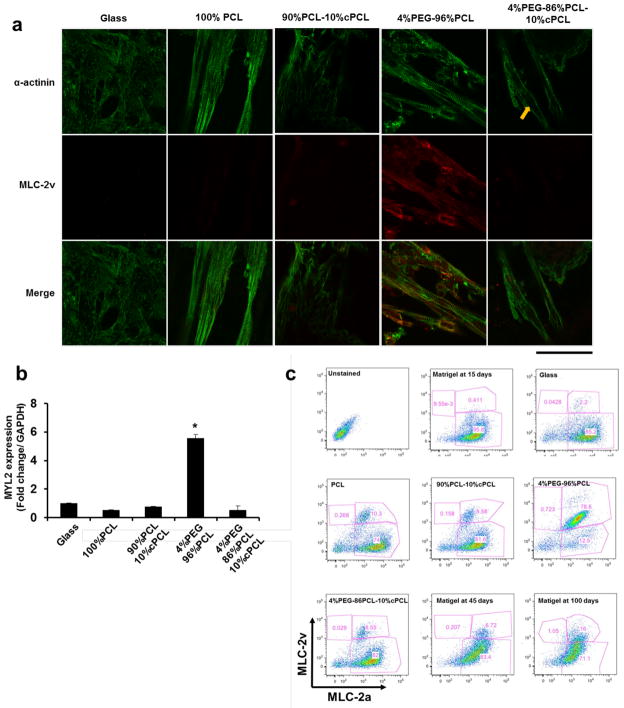
When immunostained for cardiac α-actinin, iPSC-CMs maintained on each substrate displayed varying degrees of sarcomere-like pattern (Fig. 3a). Interestingly, when immunostained for myosin light chain-2v (MLC-2v, encoded by MYL2), a commonly accepted marker of ventricular cardiomyocyte maturation,[15] only iPSC-CMs cultured on 4%PEG-96%PCL exhibited consistently detectable MLC-2v expression. Quantitative RT-PCR mRNA expression analysis confirmed that 4%PEG-96%PCL dramatically enhanced MYL2 gene expression compared to other matrices (Fig. 3b). To quantify the MLC-2v expressing cells, FACS analysis was performed with iPSC-CMs cultured for 15 days on each matrix. Expression levels of myosin light chain-2a (MLC-2a, encoded by MYL7) were simultaneously evaluated as a marker of immature cardiomyocyte phenotype.[15] 78.6% of iPSC-CMs on 4%PEG-96%PCL were MLC-2v-positive whereas only about 10% of cells grown on other matrices were MLC-2v-positive (Fig. 3c). By comparison, only 6.7% and 16% iPSC-CMs cultured on Matrigel for 45 days and 100 days respectively were MLC-2v-positive. FACS analysis also revealed that the majority of iPSC-CMs maintained on each matrix were MLC-2a–positive including cells on 4%PEG-96%PCL despite the latter’s high-frequency of MLC-2v expression. In adult human hearts, MLC-2a expression is detected in both atria and ventricles whereas MLC-2v expression is restricted to ventricles [15, 44–46]; thus, our results suggest that 4%PEG-96%PCL specifically promotes human iPSC-CM maturation toward the ventricular myocyte phenotype.
Fig. 3. Effects of polymer matrices on maturation of iPSC-CMs.
(a) Immunofluorescence staining for α-actinin and myosin light chain-2v (MLC-2v; encoded by MYL2). The iPSC-CMs maintained on 4%PEG-96%PCL exhibited clear sarcomere structures with distinct z-lines and myofilament structures. Arrow indicates an undefined sarcomere structures. Scale bar is 100 μm. (b) MYL2 expression is dramatically higher in iPSC-CMs on 4%PEG-96%PCL than other test matrices at 30 days after re-plating. *p<0.001 compared to other matrices. (c) FACS analysis showed the expression pattern of myosin light chain 2 isoforms, myosin light chain-2a (atrial) and -2v (ventricular). It determined higher expression of MLC-2v, a reported marker of ventricular maturation in iPSC-CMs maintained on 4%PEG-96%PCL for 30 days, compared to the other test substrates.
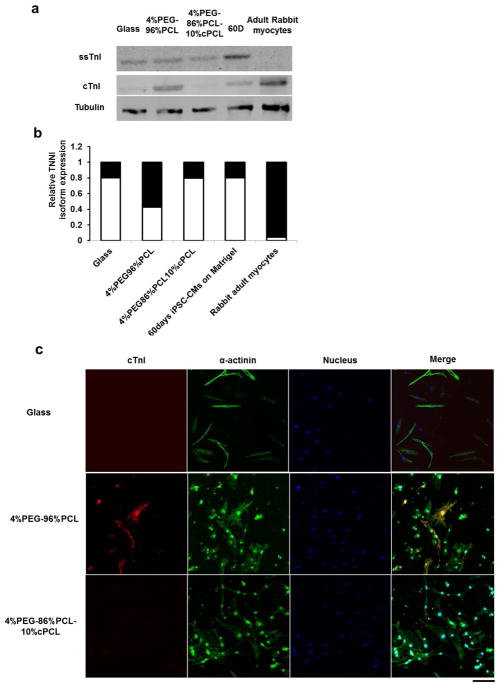
During the fetal to postnatal transition, the mammalian heart undergoes a troponin I isoform switch from the slow skeletal troponin I (ssTnI, encoded by the TNNI1 gene) to the cardiac troponin I (cTnI, encoded by TNNI3), and recent studies indicate that this switch, involving stoichiometric replacement of ssTnI in the troponin complex with cTnI represents a key hallmark of human cardiomyocyte maturation in vivo.[18, 47, 48] Importantly, Metzger and colleagues have shown that the troponin complex in human iPSC-CMs are comprised largely of ssTnI even after 9 months of culture.[49]. RNA-seq and RT-PCR analyses indicated that culturing iPSC-CMs on 4%PEG-96%PCL increased TNNI3 mRNA expression versus other substrates (Fig. 5a and 6a). Consistent with the TNNI3 mRNA induction, Western blot indicates a robust induction in cTnI protein levels in iPSC-CMs maintained on 4%PEG-96%PCL for 30 days, compared to other culture substrates (Fig. 4a). Although the ssTnI expression in iPSC-CMs on 4%PEG-96%PCL is not completely inhibited as it is in adult rabbit cardiomyocytes (Fig. 4a), our Western blot analysis indicates there is a dramatic increase in the cTnI:ssTnI ratio following culturing on 4%PEG-96%PCL even when compared to 60 day culture on standard Matrigel (Fig 4b). Interestingly, on immunostaining cTnI expression is found in a subset of iPSC-CMs maintained on 4%PEG-96%PCL (Fig. 4c). Taken together, these results indicate that culturing iPSC-CMs on 4%PEG-96%PCL promotes their maturation in vitro, resulting in the troponin I isoform switch in subset of cells.
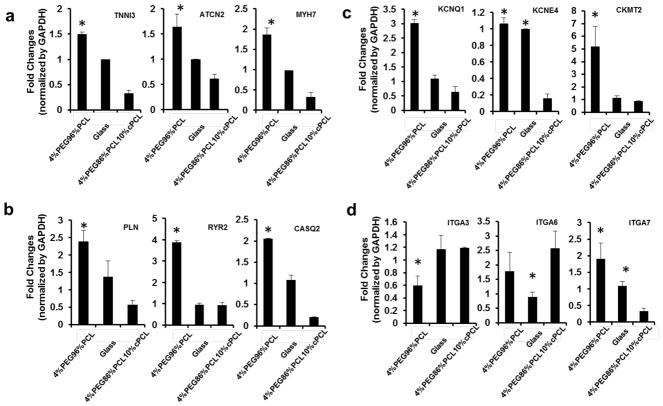
Fig. 5. RNA-sequencing expression profiles of iPSC-CMs maintained on 4%PEG-96%PCL versus 4%PEG-86%PCL-10%cPCL.
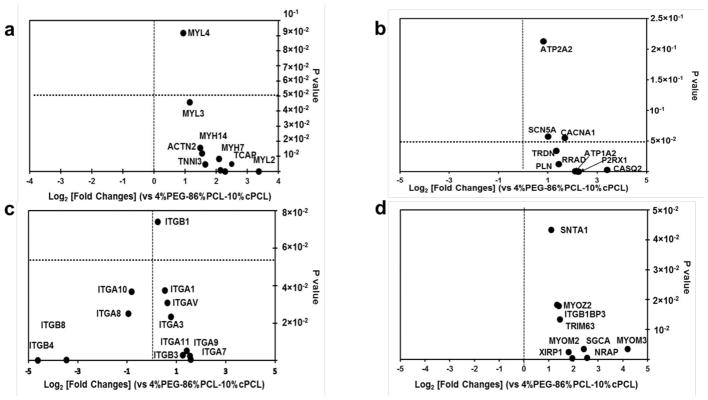
(a) iPSC-CMs maintained on 4%PEG-96%PCL expressed higher levels of sarcomere genes (MYH7, TCAP, MYH14, TNNI3, ACTN2, MYL3, MYL4, and MYL2) compared to cells maintained on 4%PEG-86%PCL-10%cPCL. Of note, MYL2 (encoding MLC-2v) expression was increased by over 8-fold in 4%PEG-96%PCL iPSC-CMs (p-value of 5×10−5). (b) iPSC-CMs maintained on 4%PEG-96%PCL expressed higher levels of genes associated with calcium handling and ion fluxes (ATP2A1, SCN5A, CACNA1, PLN, TRDN, RRAD, P2RX1, and CASQ2). Notably, expression of CASQ2, a marker of mature sarcoplasmic reticulum, was increased by over 8-fold in 4%PEG-96%PCL iPSC-CMs. (c) iPSC-CMs maintained on either polymer matrix exhibited dramatically different expression profiles of integrin subunits which mediate cell-matrix interaction. For instance, maintaining cells on 4%PEG-96%PCL significantly enhanced the expression of ITGA7 (p-value of 0.005) whereas 4%PEG-86%PCL-10%cPCL increased the expression of ITGB8 and ITGB4 (p-value of 0.002 and 5×10−5, respectively). (d) iPSC-CMs maintained on 4%PEG-96%PCL expressed much higher levels of the intermediate filament genes (MYOM3, SGCA, NRAP, TRIM63, XIRP1, SNTA1, MYOZ2, ITGBIBP3, and MYOM2).
Fig. 6. Quantitative PCR analyses of the impact of different substrates on gene expression of iPSC-CMs.
Relative expression of the representative genes associated with the (a) sarcomere, (b) calcium handling, (c) voltage-gated potassium channel and the sarcomeric mitochondrial creatine kinase2, and (d) integrins were determined by qRT-PCR. Distinct matrices, notably the 4%PEG-96%PCL copolymer, exert differential effects on iPSC-CM maturation, and that these matrix-dependent influences involve distinct subset of integrins and intermediate filaments that serve as mechanical link to the myofibrils. *p<0.05 compared to 4%PEG-86%PCL-10%cPCL
Fig. 4. Troponin I isoform transition from ssTnI to cTnI.
(a) Western blotting for ssTnI and cTnI. “60D” indicates iPSC-CMs maintained on traditional substrate (Matrigel) for 60 days after cardiac differentiation. (b) Ratio of cTnI/ssTnI protein expressed under each condition. Black bar represents percentage of cTnI with respect to the total Troponin I protein amount. White bar represents percentage of ssTnI with respect to the total Troponin I protein amount. (c) Representative iPSC-CM images of cTnI immunofluorescence in iPSC-CMs on glass, 4%PEG-96%PCL, and 4%PEG-86%PCL-10%cPCL. Scale bar indicates 100um.
3.6. RNA sequencing of iPSC-CMs on 4%PEG-96%PCL versus 4%PEG-86%PCL-10%cPCL
Next, we screened the global gene expression profiles of iPSC-CMs maintained on 4%PEG-96%PCL and 4%PEG-86%PCL-10%cPCL by RNA-Seq. This provided the opportunity to compare the impact of slightly repellent (4%PEG-96%PCL) versus slightly adhesive (4%PEG-86%PCL-10%cPCL) surface conditions while keeping mechanical properties relatively constant (Fig. 1b). Analysis of the RNA-seq data revealed that 464 genes (fold change > 1.5, p<0.05) were upregulated and 2,361 genes (fold change > 1.5, p<0.05) were downregulated in iPSC-CMs on 4%PEG-96%PCL compared to cells on 4%PEG-86%PCL-10%cPCL. Of particular interest were expression changes in the genes within four distinct categories: sarcomere structural components, calcium handling components, integrins, and intermediate filament-associated proteins.
Representative sarcomere structural genes (MYH7, TCAP, MYH14, TNNI3, ACTN2, MYL3, MYL4, and MYL2) were all upregulated over 2-fold in iPSC-CMs on 4%PEG-96%PCL (Fig. 5a). Interestingly, the myosin heavy chain 7 (MYH7), which is predominantly expressed in the ventricles, was dramatically increased by more than 4-fold (p<0.001). Telethonin (TCAP), which regulates sarcomere assembly, was also upregulated over 4-fold (p=5×10−5), implying 4%PEG-96%PCL promotes a more effective formation of the sarcomere.
In addition, the expression of genes related to excitation-contraction coupling - ATP1A2 (Na+/K+ ATPase), SCN5A (Na+ gated voltage channel), CACNA1 (Ca2+ gated voltage channel), PLN (Phospholamban), TRDN (Triadin), P2RX1 (ATP gated ion channel) and CASQ2 (Calsequestrin 2) - were analyzed (Fig. 5b). The expression levels of key calcium handling genes, PLN (p=0.035), CASQ2 (p=0.0026), and TRDN (p=0.012) were increased multiple-fold in iPSC-CMs maintained on 4%PEG-96%PCL over those on 4%PEG-86%PCL-10%cPCL. Additionally, SCN5A (p=0.05), CACNA1 (p=0.05), and P2RX1 (p=0.00025) were also significantly up-regulated, suggestive of the enhanced maturation of calcium handling in cells maintained on 4%PEG-96%PCL.
Integrins, key transmembrane receptors of ECM components, serve as major signal transducers responsive to chemical and mechanical status of ECM. Since the copolymer matrices were designed as a synthetic surrogate for ECM, we investigated whether gene expression of the integrin family members was altered when iPSC-CMs were cultured on 4%PEG-96%PCL versus 4%PEG-86%PCL-10%cPCL. On genome-wide RNA-seq analysis, the expression of only a few integrin subunits were found to be significantly altered. The ITGA7 gene, encoding the primary receptor for laminin in cardiomyocytes, was expressed at significantly higher levels in iPSC-CMs maintained on 4%PEG-96%PCL than cells on 4%PEG-86%PCL-10%cPCL (p=0.005). Conversely, two beta integrin genes ITGB8 (p=0.0022) and ITGB4 (p<0.0001), respectively encoding distinct laminin and fibronectin receptor subdomains, were expressed at significantly higher levels on 4%PEG-86%PCL-10%cPCL. Despite the limited number of altered integrin genes between the two selected copolymer substrates, this data suggests the effects of 4%PEG-96%PCL on iPSC-CM maturation might involve the selective engagement of distinct integrin-mediated cell signaling cascades (Fig. 5c).
Finally, genes associated with intermediate filaments – such as MYOM2 (Myomesin 2), MYOM3 (Myomesin3), SGCA (sarcoglycan), NRAP (nebulin-related anchoring protein), TRIM63 (Tripartite motif containing 63), XIRP1 (E3 ubiquitin protein ligase, Xin actin binding repeated protein), SNTA1 (Syntrophin1), and MYOZ2 (Myozenin2) – were expressed at significantly higher levels in iPSC-CMs maintained on 4%PEG-96%PCL than those on 4%PEG-86%PCL-10%cPCL (p<0.05) (Fig. 5d). Thus, enhanced maturation of iPSC-CMs on 4%PEG-96%PCL is associated with substantial changes to the intermediate filaments, important mediators of mechanotransduction in cells.[50–52]
3.7. Confirmation of RNA sequencing results by qRT-PCR
To confirm some of the RNA-seq data, qRT-PCR analysis was performed. All results were normalized to iPSC-CMs cultured on glass. Among the genes encoding myofilament components, TNNI3, ATCN2 and MYH7 were selected. Consistent with the RNA-seq data, qRT-PCR analysis revealed significantly increased expression of TNNI3 (~3 fold, p<0.05), ACTN2 (~2 fold, p<0.05), and MYH7 (≥3 fold, p<0.05) in iPSC-CMs cultured on 4%PEG-96%PCL compared to 4%PEG-86%PCL-10%cPCL (Fig. 6a). Since both TNNI3 and MYH7 are markers of a mature ventricular myocytes, the results further suggest that 4%PEG-96%PCL enhances cardiac maturation of iPSC-CMs in vitro.
Among genes that encode for calcium handling, the expression of Phospholamban (PLN), Ryanodine receptor 2 (RYR2), Calsequestrin 2 (CASQ2, and ATP2A2 (SERCA2) were further interrogated by qRT-PCR. In addition, given their role in regulating myocyte action potential, the expression of KCNQ1 and KCNE4 (K+ channels) genes were analyzed. Compared to those cultured on 4%PEG-86%PCL-10%cPCL, iPSC-CMs on 4%PEG-96%PCL expressed markedly higher expression of PLN (~ 3 fold, p<0.05), RYR2 (≥ 3 fold, p<0.05), CASQ2 (≥ 4 fold, p<0.001), KCNQ1 (≥ 3 fold, p<0.05) and KCNE4 (~ 5 folds, p<0.05). By comparison, no significant difference was seen in ATP2A2 expression (Fig. 6b, 6c, and Fig. S3a).
Among the genes involved in cell-matrix interactions, RT-PCR analysis of integrin subunit genes revealed a substantially higher expression of ITGA7 (≥ 4-fold, p<0.05) and decreased expression of ITGA3, ITGA6, ITGA8, and ITGB4 in iPSC-CMs maintained on 4%PEG-96%PCL versus 4%PEG-86%PCL-10%cPCL (Fig. 6d, Fig. S3b). RT-PCR results of additional integrin subunit genes are presented in Fig. S3b. Since ITGA3 and ITGA6 are highly expressed in the fetal hearts whereas ITGA7 is expressed predominatly in the postnatal hearts [17, 53, 54], our results provide additional evidence that 4%PEG-96%PCL enhances iPSC-CM maturation in vitro.
Finally, qRT-PCR analysis again confirmed that culturing iPSC-CMs in 4%PEG-96%PCL led to significant increases in the expression of many intermediate filament genes, including SGCA (≥ 10-fold, p<0.05), NRAP (≥ 10-fold, p<0.05), NMRK2 (3-fold, p<0.05), DES (≥ 5-fold, p<0.05), and MYOZ2 (3-fold, p<0.05) (Fig. S3a).
4. Discussion
New breakthroughs in cellular reprogramming to generate human iPSCs, which are capable of differentiating into cardiomyocytes, offer extraordinary opportunities to study human heart diseases at the cellular level. However, despite their vast potential, iPSC-CMs generated with existing methods appear to be immature, resembling embryonic or fetal myocytes; thus, iPSC-CMs do not yet fully recapitulate characteristics of adult human myocytes. In this study, we succeeded in enhancing the maturation of human iPSC-CMs by culturing them on a synthetic polymer matrix.
The ECM plays a central role in controlling cell-cell signaling and cell-matrix interactions critical for cardiomyocyte development and maturation in vivo [55–57] and undoubtedly exerts influence on iPSC-CMs in vitro as well. However, given the complexity of tissue ECM and matrix-substitutes like Matrigel, very little is known about the specific mechanisms by which these substrates influence maturation of iPSC-CMs. In particular, how biochemical functionalities and biophysical properties of matrices modulate the signaling cascades involved in CM maturation has been underexplored. In order to better define the functionally relevant matrix characteristics in vitro, we have employed combinatorial polymer matrices as a synthetic, physicochemically-defined model.[58] A copolymerization technique[11] was used for synthesizing copolymers of different mole percentages of three components known to alter the physicochemical properties which can affect cardiac maturation: PCL was used as the primary component due to its biocompatibility, hydrophophilicity, and slow degradation rate[12]; PEG was used to promote hydrophilicity and water adsorption and to repel proteins and cells[1, 13]; and cPCL was used for increased hydrophilicity and to expose a negative surface charge that was found to reduce the repellent effect of PEG.[14] Since each of these three subunits exhibits distinct material properties, the resulting copolymer can be tailored to modulate cellular responses (Fig. 1a) when copolymerized at different molar ratios.
As evidenced by changes in contact angles among test materials, the addition of PEG or cPCL significantly augmented surface hydrophilicity (Fig. 2b). However, hydrolytic degradation and modulus did not vary significantly between test matrix materials. These results are consistent with our previous studies [11, 13] and suggest a dominant role of surface chemistry in modulating iPSC-CM phenotype.
Of the test matrices, 4%PEG-96%PCL conferred highest level of contractility in iPSC-CMs (Fig. 1c–e). Given the repellent nature of PEG, small amounts (4–8%) of PEG present in cell substrates are known to enhance cell-cell interactions.[59] Such repellent effects are minimal in hydrophobic materials (i.e., glass and 100%PCL), and the repellent effect of PEG is diminished when 10%cPCL is added to the 4%PEG-96%PCL [14]. Indeed, compared to 4%PEG-96%PCL, iPSC-CM contractility is significantly lower when cultured on 100%PCL or copolymers containing cPCL such as 4%PEG-86%PCL-10%cPCL. Thus, our results suggest that the repellent effects of PEG in 4%PEG-96%PCL enhance myocyte contractility by increasing cell-cell interactions critical for mechanical and electrical coupling of cardiomyocytes in vitro.
A substantial enhancement of myocyte contractility on 4%PEG-96%PCL is associated with enhanced mitochondrial function, as assessed by inner mitochondrial membrane potential (Δψm).[29, 60–63] Our results are consistent with previously published reports that show a direct correlation between mitochondrial energy production and sarcomere contractility.[38, 39] Moreover, iPSC-CMs cultured on 4%PEG-96%PCL exhibited greater expression of genes involved in calcium handling, which is also associated with functional cardiomyocyte maturation.[64–67] Notably, two key genes involved in calcium handling CASQ2 and RYR2, which are generally not expressed in embryonic stem cell (ESC)-CMs or iPSC-CMs maintained by traditional methods [4, 8, 68–70], were significantly upregulated by 4%PEG-96%PCL. While the iPSC-CMs on 4%PEG-96%PCL are not functionally equivalent to adult rabbit cardiomyocyte, our results indicate that adjusting the chemical composition of combinatorial polymer matrices, specifically 4%PEG-96%PCL, enhances functional maturation of iPSC-CM in vitro.
Culturing iPSC-CMs on 4%PEG-96%PCL led to significant increases in the expression of ITGA7, a marker of postnatal heart, and simultaneous decreases in the expression of ITGA3 and ITGA6, both found predominantly in fetal hearts (Fig. 6d).[17, 53, 54] Moreover, compared to other culture substrates, 4%PEG-96%PCL significantly increased the expression of MYL2, encoding MLC-2v, a marker of mature ventricular myocytes [15, 36, 44–46, 71].
At just 30 days of cardiac induction, almost 80% of cells maintained on 4%PEG-96%PCL were double positive for MLC-2a and MLC-2v compared to ~10% or less for cells on other matrices. Whereas MLC-2a is considered an atrial-specific marker in mice [72], MLC-2a expression in the absence of MCL-2v expression is also considered a marker of immature human CM [15, 71, 73]. Moreover, because MLC-2a expression persists in both atria and ventricle of adult human hearts [44, 73], co-expression of MLC-2a and MLC-2v in iPSC-CMs cultured on 4%PEG-96%PCL suggests that 4%PEG-96%PCL promotes iPSC-CM maturation specifically toward the ventricular myocyte phenotype [15, 44, 71].
Another key feature of cardiomyocyte maturation is the troponin I isoform switch, which was not previously achieved in human iPSC-CMs.[18, 47] According to our Western analysis, culturing iPSC-CMs on 4%PEG-96%PCL for 30 days dramatically increased cTnI:ssTnI ratio to approximately 70:30, which was not achieved in human iPSC-CMs even after 9 months of culture.[18, 47] Immunostaining with cTnI suggests that the increase in cTni:ssTnI ratio may reflect a ssTnI to cTnI switch in a subset of iPSC_CMs maintained on 4%PEG-96%PCL. Finally, the fraction of cells that expressed MLC-2v was about 5-fold higher for 30-day old iPSC-CMs maintain on 4%PEG-96%PCL than for 100-day old iPSC-CMs maintained on traditional substrates like Matrigel. Thus, synthetic copolymer matrices like 4%PEG-96%PCL may prove valuable for greatly accelerating the pace, and hence reducing the cost, of iPSC-based approaches to model postnatal human heart diseases.
Our gene expression analysis implicates involvement of cell-matrix components and intermediate filaments in 4%PEG-96%PCL-facilitated iPSC-CM maturation. For instance, the expression of ITGA7, the primary laminin receptor in cardiomyocytes, was dramatically increased on 4%PEG-96%PCL compared to other matrices (Fig. 6d). This is associated with striking decreases in the expression of IGTA8 and IGTB4 (Fig. S3), suggesting different copolymer substrates engage different integrins on iPSC-CM surfaces, perhaps altering the pattern of integrin-transduced cell signaling. Consistent with the involvement of an integrin-mediated mechanosensory transduction, 4%PEG-96%PCL significantly induced expression of a number of intermediate filament genes, which serve as critical mechanosensory links between the plasma membrane and underlying cell structures such as myofilaments and the nuclear envelope.[74] Our results collectively suggest that the synthetic copolymer matrices, specifically 4%PEG-96%PCL, engage distinct subsets of integrins, leading to the induction of mechanotransductory pathways by which intermediate filaments activate the iPSC-CM maturation program.
Future work will involve additional functional characterization of iPSC-CMs (e.g., calcium handling and electrophysiological studies) to verify cardiac maturation induced by the polymer matrix. For instance, we anticipate that iPSC-CM maturation is accompanied by a reduced resting membrane potential and increased upstroke velocities [75] along with a larger calcium transient amplitude and faster decay, as shown in our previous study. [11]
5. Conclusion
Synthetic combinatorial polymer substrates were found to have distinct effects on in vitro maturation of human iPSC-CMs. Specifically, the 4%PEG-96%PCL scaffold enhanced iPSC-CM contractility and mitochondrial activity. This functional improvement was accompanied by the increased expression of MLC-2v, a marker of mature ventricular myocytes, and cTnI, a cardiac adult marker. While the precise mechanism is unknown, our results suggest that the chemical composition of a synthetic substrate greatly influences iPSC-CMs maturation. At the cellular level, certain synthetic substrates such as 4%PEG-96%PCL selectively engage a distinct subset of integrins, resulting in the activation of a mechanosensory transduction pathway involving intermediate filaments that promote iPSC-CM maturation. Thus, by altering chemical composition, we have identified a synthetic culture substrate that dramatically enhances and accelerates in vitro cardiomyocyte maturation by recapitulating some of the critical cell-matrix interactions found in in vivo tissue environment. In summary, our work explored the effects of synthetic biomaterials on human pluripotent stem cell differentiation, and our results could pave the way for a successful translation of ongoing advances in tissue engineering and regenerative biology into new diagnostic and therapeutic paradigms for heart diseases.
Supplementary Material
Fig. S1. Karyotype and PluriTest. (a) Karyotype analysis was performed by Genetics Associates (Nashville, TN), only cells with normal euploid karyotype (higher metaphase nuclei without abnormal karyotype) were included. (b) PluriTest analysis of CC2 (Green circle) used in this study and a representative human fibroblast RNA sample (black square). CC2, except for the human fibroblasts, showed “Pluripotency” Scores within the acceptable range of the testing criteria and scored similar to our teratoma previously validated human iPSCs (red triangle), validating their pluripotency.
Fig. S2. Cardiomyocytes generated from human induced pluripotent stem cells (iPSCs). (a) Human iPSCs reprogramed from donor adult epidermal fibroblasts express pluripotency markers (NANOG, Oct4, and SSEA4). (b) Schematic diagram of a robust, efficient cardiac induction protocol. (c) Representative microscopy image of cardiomyocytes derived from iPSCs (iPSC-CMs) at day 15 of cardiac induction. (d) FACS analysis determined that the cardiac differentiation method produced over 97% cardiac troponin T-positive cells. (e) Sarcomere structures of iPSC-CMs immunostained for cardiac troponin T and a-actinin. (f) The surface plot shows the active movement of a beating iPSC-CMs at 15-day post cardiac induction. (a, c, and e) Scale bars are 100μm.
Fig. S3. Quantitative PCR analyses of gene expression of iPSC-CMs maintained on different matrices. (a) Gene expression related to sarcomere, calcium handling and intermediate filaments, and (b) integrins were investigated by qRT-PCR. *p< 0.05 compared to 4%PEG-86%PCL-10%cPCL
Table S1. The primers used for qRT-PCR
Movie S1. Beating sheets of 15 days old iPSC-CMs through cardiac differentiation
Acknowledgments
Authors thank Dr. Bjorn Knollman’s laboratory for serving healthy rabbit adult myocytes to us for mitochondrial functional assay. This research work was funded and supported by NIH R01HL104040, NIH UH2 TR000491, NIH R01ES010563, NIH R01ES016931, 1R01 NS078289, VA Merit 101BX000771, NSF CAREER CBET 1056046 and Clinical Translational Science Award No. UL1TR000445 from the National Center for Advancing Translational Sciences. Polymer characterization was conducted through the use of the core facilities of the Vanderbilt Institute of Nanoscale Sciences and Engineering (VINSE), which was supported by NSF EPS 1004083. We thank Drs. Asad Aboud and Kevin Kumar for technical assistance in generation and validation of the CC2 human iPSCs.
Appendix
Supplementary data related to this article can be found at Elsevier website
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lloyd-Jones D, Adams R, Carnethon M, De Simone G, Ferguson TB, Flegal K, et al. Heart disease and stroke statistics--2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;119:e21–181. doi: 10.1161/CIRCULATIONAHA.108.191261. [DOI] [PubMed] [Google Scholar]
- 2.Ma Z, Koo S, Finnegan MA, Loskill P, Huebsch N, Marks NC, et al. Three-dimensional filamentous human diseased cardiac tissue model. Biomaterials. 2014;35:1367–77. doi: 10.1016/j.biomaterials.2013.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nature medicine. 2014;20:616–23. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dolnikov K, Shilkrut M, Zeevi-Levin N, Gerecht-Nir S, Amit M, Danon A, et al. Functional properties of human embryonic stem cell-derived cardiomyocytes: Intracellular Ca2+ handling and the role of sarcoplasmic reticulum in the contraction. Stem cells. 2006;24:236–45. doi: 10.1634/stemcells.2005-0036. [DOI] [PubMed] [Google Scholar]
- 5.Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–8. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 6.Foldes G, Mioulane M, Wright JS, Liu AQ, Novak P, Merkely B, et al. Modulation of human embryonic stem cell-derived cardiomyocyte growth: a testbed for studying human cardiac hypertrophy? Journal of molecular and cellular cardiology. 2011;50:367–76. doi: 10.1016/j.yjmcc.2010.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim C, Majdi M, Xia P, Wei KA, Talantova M, Spiering S, et al. Non-cardiomyocytes influence the electrophysiological maturation of human embryonic stem cell-derived cardiomyocytes during differentiation. Stem cells and development. 2010;19:783–95. doi: 10.1089/scd.2009.0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Lieu DK, Siu CW, Fu JD, Tse HF, Li RA. Facilitated maturation of Ca2+ handling properties of human embryonic stem cell-derived cardiomyocytes by calsequestrin expression. American journal of physiology Cell physiology. 2009;297:C152–9. doi: 10.1152/ajpcell.00060.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sartiani L, Bettiol E, Stillitano F, Mugelli A, Cerbai E, Jaconi ME. Developmental changes in cardiomyocytes differentiated from human embryonic stem cells: a molecular and electrophysiological approach. Stem cells. 2007;25:1136–44. doi: 10.1634/stemcells.2006-0466. [DOI] [PubMed] [Google Scholar]
- 10.Hwang HS, Kryshtal DO, Feaster TK, Sanchez-Freire V, Zhang J, Kamp TJ, et al. Comparable calcium handling of human iPSC-derived cardiomyocytes generated by multiple laboratories. Journal of molecular and cellular cardiology. 2015;85:79–88. doi: 10.1016/j.yjmcc.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta MK, Walthall JM, Venkataraman R, Crowder SW, Jung DK, Yu SS, et al. Combinatorial Polymer Electrospun Matrices Promote Physiologically-Relevant Cardiomyogenic Stem Cell Differentiation. Plos One. 2011;6 doi: 10.1371/journal.pone.0028935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinha VR, Bansal K, Kaushik R, Kumria R, Trehan A. Poly-epsilon-caprolactone microspheres and nanospheres: an overview. International journal of pharmaceutics. 2004;278:1–23. doi: 10.1016/j.ijpharm.2004.01.044. [DOI] [PubMed] [Google Scholar]
- 13.Crowder SW, Gupta MK, Hofmeister LH, Zachman AL, Sung HJ. Modular polymer design to regulate phenotype and oxidative response of human coronary artery cells for potential stent coating applications. Acta biomaterialia. 2012;8:559–69. doi: 10.1016/j.actbio.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sung HJ, Luk A, Murthy NS, Liu E, Jois M, Joy A, et al. Poly(ethylene glycol) as a sensitive regulator of cell survival fate on polymeric biomaterials: the interplay of cell adhesion and pro-oxidant signaling mechanisms. Soft Matter. 2010;6:5196–205. [Google Scholar]
- 15.Zhang J, Klos M, Wilson GF, Herman AM, Lian X, Raval KK, et al. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circulation research. 2012;111:1125–36. doi: 10.1161/CIRCRESAHA.112.273144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belkin AM, Zhidkova NI, Balzac F, Altruda F, Tomatis D, Maier A, et al. beta 1D integrin displaces the beta 1A isoform in striated muscles: Localization at junctional structures and signaling potential in nonmuscle cells. J Cell Biol. 1996;132:211–26. doi: 10.1083/jcb.132.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flintoff-Dye NL, Welser J, Rooney J, Scowen P, Tamowski S, Hatton W, et al. Role for the alpha 7 beta 1 integrin in vascular development and integrity. Dev Dynam. 2005;234:11–21. doi: 10.1002/dvdy.20462. [DOI] [PubMed] [Google Scholar]
- 18.Bedada FB, Chan SS, Metzger SK, Zhang L, Zhang J, Garry DJ, et al. Acquisition of a quantitative, stoichiometrically conserved ratiometric marker of maturation status in stem cell-derived cardiac myocytes. Stem Cell Reports. 2014;3:594–605. doi: 10.1016/j.stemcr.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aboud AA, Tidball AM, Kumar KK, Diana Neely M, Han B, Ess KC, et al. PARK2 patient neuroprogenitors show increased mitochondrial sensitivity to copper. Neurobiology of disease. 2014 doi: 10.1016/j.nbd.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aboud AA, Tidball AM, Kumar KK, Neely MD, Ess KC, Erikson KM, et al. Genetic risk for Parkinson’s disease correlates with alterations in neuronal manganese sensitivity between two human subjects. Neurotoxicology. 2012;33:1443–9. doi: 10.1016/j.neuro.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, et al. A more efficient method to generate integration-free human iPS cells. Nature methods. 2011;8:409–12. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 22.Neely MD, Litt MJ, Tidball AM, Li GG, Aboud AA, Hopkins CR, et al. DMH1, a highly selective small molecule BMP inhibitor promotes neurogenesis of hiPSCs: comparison of PAX6 and SOX1 expression during neural induction. ACS Chem Neurosci. 2012;3:482–91. doi: 10.1021/cn300029t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller FJ, Schuldt BM, Williams R, Mason D, Altun G, Papapetrou EP, et al. A bioinformatic assay for pluripotency in human cells. Nature methods. 2011;8:315–7. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knollmann BC, Chopra N, Hlaing T, Akin B, Yang T, Ettensohn K, et al. Casq2 deletion causes sarcoplasmic reticulum volume increase, premature Ca2+ release, and catecholaminergic polymorphic ventricular tachycardia. The Journal of clinical investigation. 2006;116:2510–20. doi: 10.1172/JCI29128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayakawa T, Kunihiro T, Ando T, Kobayashi S, Matsui E, Yada H, et al. Image-based evaluation of contraction-relaxation kinetics of human-induced pluripotent stem cell-derived cardiomyocytes: Correlation and complementarity with extracellular electrophysiology. Journal of molecular and cellular cardiology. 2014;77:178–91. doi: 10.1016/j.yjmcc.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Hazeltine LB, Simmons CS, Salick MR, Lian X, Badur MG, Han W, et al. Effects of substrate mechanics on contractility of cardiomyocytes generated from human pluripotent stem cells. International journal of cell biology. 2012;2012:508294. doi: 10.1155/2012/508294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sung HJ, Meredith C, Johnson C, Galis ZS. The effect of scaffold degradation rate on three-dimensional cell growth and angiogenesis. Biomaterials. 2004;25:5735–42. doi: 10.1016/j.biomaterials.2004.01.066. [DOI] [PubMed] [Google Scholar]
- 28.Kwok DY, Gietzelt T, Grundke K, Jacobasch HJ, Neumann AW. Contact angle measurements and contact angle interpretation.1. Contact angle measurements by axisymmetric drop shape analysis and a goniometer sessile drop technique. Langmuir. 1997;13:2880–94. [Google Scholar]
- 29.Hattori F, Chen H, Yamashita H, Tohyama S, Satoh YS, Yuasa S, et al. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nature methods. 2010;7:61–6. doi: 10.1038/nmeth.1403. [DOI] [PubMed] [Google Scholar]
- 30.Waters JC. Accuracy and precision in quantitative fluorescence microscopy. J Cell Biol. 2009;185:1135–48. doi: 10.1083/jcb.200903097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Greiner A, Wendorff JH. Electrospinning: A fascinating method for the preparation of ultrathin fibres. Angew Chem Int Edit. 2007;46:5670–703. doi: 10.1002/anie.200604646. [DOI] [PubMed] [Google Scholar]
- 33.Jawad H, Lyon AR, Harding SE, Ali NN, Boccaccini AR. Myocardial tissue engineering. British medical bulletin. 2008;87:31–47. doi: 10.1093/bmb/ldn026. [DOI] [PubMed] [Google Scholar]
- 34.Engelmayr GC, Jr, Cheng M, Bettinger CJ, Borenstein JT, Langer R, Freed LE. Accordion-like honeycombs for tissue engineering of cardiac anisotropy. Nature materials. 2008;7:1003–10. doi: 10.1038/nmat2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhana B, Iyer RK, Chen WL, Zhao R, Sider KL, Likhitpanichkul M, et al. Influence of substrate stiffness on the phenotype of heart cells. Biotechnology and bioengineering. 2010;105:1148–60. doi: 10.1002/bit.22647. [DOI] [PubMed] [Google Scholar]
- 36.Lee MY, Sun BN, Schliffke S, Yue ZC, Ye MY, Paavola J, et al. Derivation of functional ventricular cardiomyocytes using endogenous promoter sequence from murine embryonic stem cells. Stem Cell Res. 2012;8:49–57. doi: 10.1016/j.scr.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lundy SD, Zhu WZ, Regnier M, Laflamme MA. Structural and Functional Maturation of Cardiomyocytes Derived from Human Pluripotent Stem Cells. Stem cells and development. 2013;22:1991–2002. doi: 10.1089/scd.2012.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marchini T, Magnani N, D’Annunzio V, Tasat D, Gelpi RJ, Alvarez S, et al. Impaired cardiac mitochondrial function and contractile reserve following an acute exposure to environmental particulate matter. Biochimica et biophysica acta. 2013;1830:2545–52. doi: 10.1016/j.bbagen.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 39.Kaasik A, Joubert F, Ventura-Clapier R, Veksler V. A novel mechanism of regulation of cardiac contractility by mitochondrial functional state. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2004;18:1219–27. doi: 10.1096/fj.04-1508com. [DOI] [PubMed] [Google Scholar]
- 40.Joshi DC, Bakowska JC. Determination of mitochondrial membrane potential and reactive oxygen species in live rat cortical neurons. Journal of visualized experiments : JoVE. 2011 doi: 10.3791/2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loew LM, Tuft RA, Carrington W, Fay FS. Imaging in 5 Dimensions - Time-Dependent Membrane-Potentials in Individual Mitochondria. Biophys J. 1993;65:2396–407. doi: 10.1016/S0006-3495(93)81318-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scaduto RC, Grotyohann LW. Measurement of mitochondrial membrane potential using fluorescent rhodamine derivatives. Biophys J. 1999;76:469–77. doi: 10.1016/S0006-3495(99)77214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Archer SL. Mitochondrial dynamics--mitochondrial fission and fusion in human diseases. The New England journal of medicine. 2013;369:2236–51. doi: 10.1056/NEJMra1215233. [DOI] [PubMed] [Google Scholar]
- 44.Chuva de Sousa Lopes SMC, Hassink RJ, Feijen A, van Rooijen MA, Doevendans PA, Tertoolen L, et al. Patterning the heart, a template for human cardiomyocyte development. Dev Dynam. 2006;235:1994–2002. doi: 10.1002/dvdy.20830. [DOI] [PubMed] [Google Scholar]
- 45.Kubalak SW, Millerhance WC, Obrien TX, Dyson E, Chien KR. Chamber Specification of Atrial Myosin Light Chain-2 Expression Precedes Septation during Murine Cardiogenesis. Journal of Biological Chemistry. 1994;269:16961–70. [PubMed] [Google Scholar]
- 46.Obrien TX, Lee KJ, Chien KR. Positional Specification of Ventricular Myosin Light Chain-2 Expression in the Primitive Murine Heart Tube. P Natl Acad Sci USA. 1993;90:5157–61. doi: 10.1073/pnas.90.11.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhavsar PK, Dhoot GK, Cumming DV, Butler-Browne GS, Yacoub MH, Barton PJ. Developmental expression of troponin I isoforms in fetal human heart. FEBS letters. 1991;292:5–8. doi: 10.1016/0014-5793(91)80820-s. [DOI] [PubMed] [Google Scholar]
- 48.Siedner S, Kruger M, Schroeter M, Metzler D, Roell W, Fleischmann BK, et al. Developmental changes in contractility and sarcomeric proteins from the early embryonic to the adult stage in the mouse heart. The Journal of physiology. 2003;548:493–505. doi: 10.1113/jphysiol.2002.036509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bedada Fikru B, Chan Sunny S-K, Metzger Stefania K, Zhang L, Zhang J, Garry Daniel J, et al. Acquisition of a Quantitative, Stoichiometrically Conserved Ratiometric Marker of Maturation Status in Stem Cell-Derived Cardiac Myocytes. Stem Cell Reports. doi: 10.1016/j.stemcr.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Al-Jassar C, Bikker H, Overduin M, Chidgey M. Mechanistic basis of desmosome-targeted diseases. Journal of molecular biology. 2013;425:4006–22. doi: 10.1016/j.jmb.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker LK, Gillis DC, Sharma S, Ambrus A, Herrmann H, Conover GM. Nebulin binding impedes mutant desmin filament assembly. Molecular biology of the cell. 2013;24:1918–32. doi: 10.1091/mbc.E12-11-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hnia K, Ramspacher C, Vermot J, Laporte J. Desmin in muscle and associated diseases: beyond the structural function. Cell and tissue research. 2014 doi: 10.1007/s00441-014-2016-4. [DOI] [PubMed] [Google Scholar]
- 53.Maitra N, Flink IL, Bahl JJ, Morkin E. Expression of alpha and beta integrins during terminal differentiation of cardiomyocytes. Cardiovasc Res. 2000;47:715–25. doi: 10.1016/s0008-6363(00)00140-1. [DOI] [PubMed] [Google Scholar]
- 54.Ross RS. Molecular and mechanical synergy: cross-talk between integrins and growth factor receptors. Cardiovasc Res. 2004;63:381–90. doi: 10.1016/j.cardiores.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 55.Giancotti FG, Ruoslahti E. Transduction - Integrin signaling. Science. 1999;285:1028–32. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 56.Hynes RO. Cell adhesion: old and new questions. Trends Biochem Sci. 1999;24:M33–M7. [PubMed] [Google Scholar]
- 57.Pham CG, Harpf AE, Keller RS, Vu HT, Shai SY, Loftus JC, et al. Striated muscle-specific beta(1D)-integrin and FAK are involved in cardiac myocyte hypertrophic response pathway. Am J Physiol-Heart C. 2000;279:H2916–H26. doi: 10.1152/ajpheart.2000.279.6.H2916. [DOI] [PubMed] [Google Scholar]
- 58.Yu SS, Koblin RL, Zachman AL, Perrien DS, Hofmeister LH, Giorgio TD, et al. Physiologically Relevant Oxidative Degradation of Oligo(proline) Cross-Linked Polymeric Scaffolds. Biomacromolecules. 2011;12:4357–66. doi: 10.1021/bm201328k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crowder SW, Horton LW, Lee SH, McClain CM, Hawkins OE, Palmer AM, et al. Passage-dependent cancerous transformation of human mesenchymal stem cells under carcinogenic hypoxia. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27:2788–98. doi: 10.1096/fj.13-228288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gandra PG, Nogueira L, Hogan MC. Mitochondrial activation at the onset of contractions in isolated myofibres during successive contractile periods. The Journal of physiology. 2012;590:3597–609. doi: 10.1113/jphysiol.2012.232405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gerencser AA, Chinopoulos C, Birket MJ, Jastroch M, Vitelli C, Nicholls DG, et al. Quantitative measurement of mitochondrial membrane potential in cultured cells: calcium-induced de- and hyperpolarization of neuronal mitochondria. The Journal of physiology. 2012;590:2845–71. doi: 10.1113/jphysiol.2012.228387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sobolewski P, Kandel J, Eckmann DM. Air bubble contact with endothelial cells causes a calcium-independent loss in mitochondrial membrane potential. Plos One. 2012;7:e47254. doi: 10.1371/journal.pone.0047254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Venable PW, Taylor TG, Sciuto KJ, Zhao J, Shibayama J, Warren M, et al. Detection of mitochondrial depolarization/recovery during ischaemia--reperfusion using spectral properties of confocally recorded TMRM fluorescence. The Journal of physiology. 2013;591:2781–94. doi: 10.1113/jphysiol.2012.248153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gyorke I, Hester N, Jones LR, Gyorke S. The role of calsequestrin, triadin, and junctin in conferring cardiac ryanodine receptor responsiveness to luminal calcium. Biophys J. 2004;86:2121–8. doi: 10.1016/S0006-3495(04)74271-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu J, Lieu DK, Siu CW, Fu JD, Tse HF, Li RA. Facilitated maturation of Ca2+ handling properties of human embryonic stem cell-derived cardiomyocytes by calsequestrin expression. Am J Physiol-Cell Ph. 2009;297:C152–C9. doi: 10.1152/ajpcell.00060.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.MacLennan DH, Kranias EG. Phospholamban: A crucial regulator of cardiac contractility. Nat Rev Mol Cell Bio. 2003;4:566–77. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- 67.Robertson C, Tran DD, George SC. Concise review: maturation phases of human pluripotent stem cell-derived cardiomyocytes. Stem cells. 2013;31:829–37. doi: 10.1002/stem.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kehat I, Snir M, Gepstein A, Gepstein L. Assessment of the ultrastructural and proliferative properties of human embryonic stem cell-derived cardiomyocytes. Circulation. 2003;108:244. doi: 10.1152/ajpheart.00020.2003. [DOI] [PubMed] [Google Scholar]
- 69.Lee YK, Ng KM, Lai WH, Chan YC, Lau YM, Lian QZ, et al. Calcium Homeostasis in Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Stem Cell Rev Rep. 2011;7:976–86. doi: 10.1007/s12015-011-9273-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu J, Fu JD, Siu CW, Li RA. Functional sarcoplasmic reticulum for calcium handling of human embryonic stem cell-derived cardiomyocytes: Insights for driven maturation. Stem cells. 2007;25:3038–44. doi: 10.1634/stemcells.2007-0549. [DOI] [PubMed] [Google Scholar]
- 71.Segev H, Kenyagin-Karsenti D, Fishman B, Gerecht-Nir S, Ziskind A, Amit M, et al. Molecular analysis of cardiomyocytes derived from human embryonic stem cells. Dev Growth Differ. 2005;47:295–306. doi: 10.1111/j.1440-169X.2005.00803.x. [DOI] [PubMed] [Google Scholar]
- 72.Huang C, Sheikh F, Hollander M, Cai C, Becker D, Chu PH, et al. Embryonic atrial function is essential for mouse embryogenesis, cardiac morphogenesis and angiogenesis. Development. 2003;130:6111–9. doi: 10.1242/dev.00831. [DOI] [PubMed] [Google Scholar]
- 73.Bizy A, Guerrero-Serna G, Hu B, Ponce-Balbuena D, Willis BC, Zarzoso M, et al. Myosin light chain 2-based selection of human iPSC-derived early ventricular cardiac myocytes. Stem Cell Res. 2013;11:1335–47. doi: 10.1016/j.scr.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lund LM, Kerr JP, Lupinetti J, Zhang Y, Russell MA, Bloch RJ, et al. Synemin isoforms differentially organize cell junctions and desmin filaments in neonatal cardiomyocytes. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012;26:137–48. doi: 10.1096/fj.10-179408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sallam K, Li Y, Sager PT, Houser SR, Wu JC. Finding the rhythm of sudden cardiac death: new opportunities using induced pluripotent stem cell-derived cardiomyocytes. Circulation research. 2015;116:1989–2004. doi: 10.1161/CIRCRESAHA.116.304494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Karyotype and PluriTest. (a) Karyotype analysis was performed by Genetics Associates (Nashville, TN), only cells with normal euploid karyotype (higher metaphase nuclei without abnormal karyotype) were included. (b) PluriTest analysis of CC2 (Green circle) used in this study and a representative human fibroblast RNA sample (black square). CC2, except for the human fibroblasts, showed “Pluripotency” Scores within the acceptable range of the testing criteria and scored similar to our teratoma previously validated human iPSCs (red triangle), validating their pluripotency.
Fig. S2. Cardiomyocytes generated from human induced pluripotent stem cells (iPSCs). (a) Human iPSCs reprogramed from donor adult epidermal fibroblasts express pluripotency markers (NANOG, Oct4, and SSEA4). (b) Schematic diagram of a robust, efficient cardiac induction protocol. (c) Representative microscopy image of cardiomyocytes derived from iPSCs (iPSC-CMs) at day 15 of cardiac induction. (d) FACS analysis determined that the cardiac differentiation method produced over 97% cardiac troponin T-positive cells. (e) Sarcomere structures of iPSC-CMs immunostained for cardiac troponin T and a-actinin. (f) The surface plot shows the active movement of a beating iPSC-CMs at 15-day post cardiac induction. (a, c, and e) Scale bars are 100μm.
Fig. S3. Quantitative PCR analyses of gene expression of iPSC-CMs maintained on different matrices. (a) Gene expression related to sarcomere, calcium handling and intermediate filaments, and (b) integrins were investigated by qRT-PCR. *p< 0.05 compared to 4%PEG-86%PCL-10%cPCL
Table S1. The primers used for qRT-PCR
Movie S1. Beating sheets of 15 days old iPSC-CMs through cardiac differentiation