Abstract
The establishment of a productive symbiosis between Euprymna scolopes, the Hawaiian bobtail squid, and its luminous bacterial symbiont, Vibrio fischeri, is mediated by transcriptional changes in both partners. A key challenge to unraveling the steps required to successfully initiate this and many other symbiotic associations is characterization of the timing and location of these changes. We report on the adaptation of hybridization chain reaction-fluorescent in situ hybridization (HCR-FISH) to simultaneously probe the spatiotemporal regulation of targeted genes in both E. scolopes and V. fischeri. This method revealed localized, transcriptionally coregulated epithelial cells within the light organ that responded directly to the presence of bacterial cells while, at the same time, provided a sensitive means to directly show regulated gene expression within the symbiont population. Thus, HCR-FISH provides a new approach for characterizing habitat transition in bacteria and for discovering host tissue responses to colonization.
INTRODUCTION
Juveniles of the squid Euprymna scolopes hatch as aposymbiotic squid, i.e., squid without bacterial symbionts, but are rapidly and efficiently colonized by a single species of bacteria, Vibrio fischeri. In coastal regions with established populations of these squid, V. fischeri constitutes only 0.01 to 0.15% of the ∼106 bacterial cells present per ml of seawater (1). Thus, as it initiates the association, V. fischeri transitions from constituting just a small fraction of the total bacterioplankton to being the exclusive symbiont within the host's light-emitting organ (Fig. 1A and B). Previous studies of the symbiosis indicate that in this highly selective process, both the host and the bacterium have evolved mechanisms to ensure the specificity of the symbiotic relationship (2). Furthermore, the presence of the bacterial symbiont triggers a specific developmental program in the host (reviewed by McFall-Ngai [3]). Not surprisingly, transcriptional responses in both the host and the bacteria play a role in establishing the symbiosis as well as in mediating light organ development (4, 5). However, as in most other host-microbe interactions, a formidable challenge has been to identify exactly where within the target tissues and at what stages in the colonization process these transcriptional changes take place in each partner.
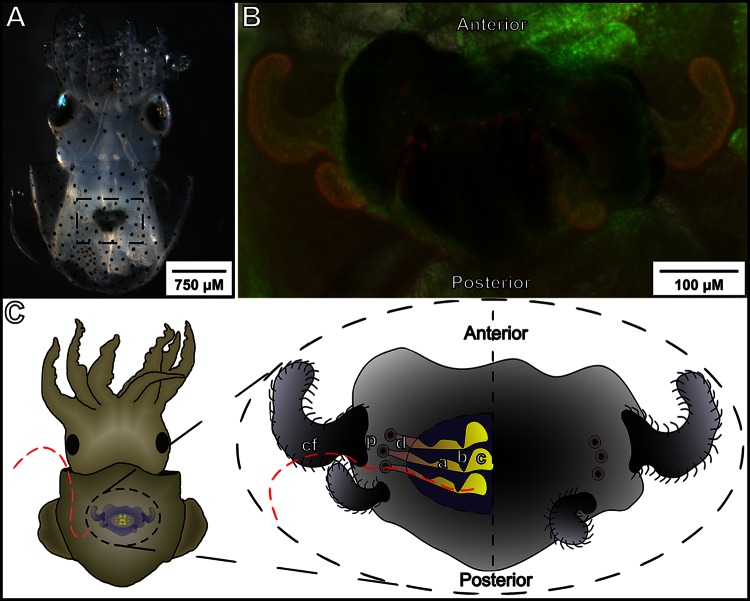
FIG 1.
Anatomy of the E. scolopes light organ. (A) An anesthetized animal was imaged using a dissection scope, revealing the light organ and ink sac (dashed box) through the translucent mantle tissue. (B) The host light organ counterstained using acridine orange as a live stain. The light organ is visible above the ink sac, and two pairs of ciliated appendages splayed out to each side can be seen. The light organ is bilobed, with a pair of appendages, three pores, and three crypts being present on each side. (C) Model of the colonization area and process. The red dashed line indicates the general path that V. fischeri cells must follow as they pass from the ambient seawater to the interior of the host's light organ. Once they are inside the mantle cavity, the bacteria attach to the ciliated field (cf) before migrating to the pores (p) at the base of the light organ appendages. They then pass into the ducts (d) and through the antechamber (a) before pausing at the bottleneck (b). Only one or a few bacteria pass the bottleneck before it constricts, and once they are past the bottleneck, they enter the deep crypts (c), where they are able to multiply and begin to luminesce.
During their first 12 h of initiating colonization, the bacteria traverse several distinct regions of the host light organ (Fig. 1C). Briefly, planktonic bacteria in ambient seawater are drawn into the body cavity of the squid, where they associate with epithelial extensions of a ciliated surface on each side of the light organ. These ciliated appendages help direct bacteria to three pores at their base. Once they pass through the pores, the bacteria navigate along ducts into an antechamber, and from there they navigate into deep crypts. The medial end of the antechamber serves as a bottleneck (6), and only a small number of bacteria are able to migrate into the crypts before the bottleneck restricts (7). After reaching the crypts, the symbionts multiply, eventually achieving a density sufficient to induce their lux genes and, thus, bioluminescence, the bacterial product used by the host in its nocturnal behavior (8). During this first 24 h of symbiotic development, the host tissues also respond to the presence of bacteria, changing the expression of a number of genes (4), including the induction of some, like the peptidoglycan-receptor E. scolopes proteins PGRP1 (EsPGRP1) and PGRP2 (EsPGRP2), that are likely derived from those involved in host immunity (9, 10). Other protein transcripts not associated with immunity, such as the E. scolopes protocadherin-like CadDP1 (EsCadDP1), were found to be upregulated ∼7-fold at 24 h postcolonization in an independent transcriptome sequencing study of whole-organ extracts (S. Moriano-Gutierrez, personal communication). Protocadherin is an adhesion molecule that is expected to be present in the brush border of the epithelium leading to the crypts (11) but whose regulation and localization remain undescribed.
To investigate the spatiotemporal regulation of gene expression in both the host and the symbiont, we have adapted hybridization chain reaction-fluorescent in situ hybridization (HCR-FISH) (12–14) to the squid-vibrio system. This newly established technique was developed for the probing of zebrafish embryos and has recently been used to investigate the patterns of expression of the microbial community of the termite gut (15) and mRNA expression patterns in mouse embryos (16) and for the sensitive detection of environmental organisms (17). In this study, we show that HCR-FISH allows multiple genes to be probed simultaneously in whole mounts of both a host and its symbiont in order to study the transcriptional events surrounding microbial colonization. In addition, HCR-FISH provides a means to visualize the regulation of rare transcripts unapproachable by traditional in situ hybridization (12). Specifically, with HCR-FISH, the amplified fluorescent signal can be detected throughout an intact sample like the light organ, and a suite of unique hairpin sequences allows multiple transcripts to be probed simultaneously. The timing and localization of gene expression by bacterial symbionts within this tissue have previously been accomplished using promoter fusion constructs (18, 19). However, visualization of such fluorescent protein reporters can be problematic because individual bacterial cells are not easily resolved, the signal is often too weak, and its accumulation and maturation lag behind the induction of gene expression. In contrast, by using multiple probes per transcript, the HCR-FISH protocol permits the signal to be easily increased in a roughly linear fashion, permitting direct detection of the transcripts in single cells at the time of their synthesis.
Here we demonstrate the strength of the HCR-FISH method by simultaneously probing several transcripts in both the bacterial symbiont and the host, thereby revealing (i) the time during colonization of lux expression by the symbiont and (ii) newly localized patterns of gene expression in the host. Successful adaptation of this technique to the squid-vibrio system and, by extension, other host-microbe interactions greatly increases the sensitivity with which transcriptional changes can be observed and allows a degree of spatial and temporal precision not previously possible in studies of host-symbiont engagement.
MATERIALS AND METHODS
General methods.
Adult E. scolopes squid were gathered off the coast of Oahu, HI, and maintained and bred as previously described (20). Newly hatched juvenile squid were placed in filter-sterilized Instant Ocean (IO; Aquarium Systems, Mentor, OH, USA) from the University of Wisconsin facility's aquaria. To inoculate the squid, V. fischeri strains were grown overnight as previously described (21) at 28°C with shaking in Luria-Bertani salt (LBS) medium (22), containing antibiotics where appropriate. On the next morning, cultures were diluted into a seawater-based tryptone (SWT) medium (23) and allowed to regrow to mid-log phase. The bacteria were then added at a concentration of ∼5,000 CFU ml−1 of IO containing the newly hatched juveniles. After 3 h, the squid were moved to sterile IO to ensure relatively synchronized colonization. The inoculum concentration was verified by plating a small volume on LBS agar medium to determine the number of CFU of bacteria present milliliter−1.
HCR-FISH.
The nucleic acid probes and hairpin sequences used in this study were obtained from Molecular Instruments (www.molecularinstruments.org), which also assisted with the probe design. Juvenile squid were collected prior to or 3, 6, or 24 h after an initial exposure to V. fischeri. Animals were immediately anesthetized in artificial seawater containing 2% ethanol and fixed overnight at 4°C in 4% paraformaldehyde in marine phosphate-buffered saline (mPBS; 50 mM phosphate buffer, pH 7.4, 0.45 M NaCl). The light organs were removed by dissection and permeabilized overnight at room temperature in mPBS containing 1% Tween 20 detergent. For additional permeabilization, samples were treated with 0.01 mg proteinase K (catalog number AM2546; Ambion) per ml of permeabilization buffer for 15 min at room temperature; the reaction was halted by two washes of glycine (2 mg ml−1) in permeabilization buffer. The sample was then postfixed for 1 h in 4% paraformaldehyde (PFA) in mPBS at room temperature and washed five times for 5 min each time in permeabilization buffer.
The solutions used for HCR-FISH were identical to those described previously (12), except that the probe hybridization buffer was diluted 1:1 with 2× mPBS to prevent the lysis of the V. fischeri cells. Samples were equilibrated in 500 μl mPBS-supplemented probe hybridization buffer at 65°C for 30 min, followed by 2.5 h of incubation in fresh solution. The samples were then incubated overnight at 45°C in mPBS-supplemented probe hybridization buffer, to which up to 5 probes per gene of interest were added to a final concentration of 2 μmol per probe. To remove nonspecifically bound probe, samples were sequentially incubated for 15 min at 45°C in 500 μl probe wash buffer to which 5× SSC (750 mM NaCl, 75 mM sodium citrate, pH 7) had been added to final concentrations (vol/vol) of 0%, 25%, 50%, and then 75%. This wash sequence was followed by two 15-min washes and then two 30-min washes at 45°C in 500 μl 5× SSC. After the probe washes, 6 pmol of each hairpin sequence was separately heated to 95°C for 90 s and allowed to cool at room temperature in the dark for 30 min. During this period, samples were equilibrated twice with 500 μl DNA amplification buffer at room temperature for 30 min. Hairpin sequences were then added to 100 μl DNA amplification buffer, in which samples were then incubated overnight at room temperature in the dark. To remove unbound hairpin sequences, samples were washed four times in 500 μl 5× SSC containing 0.05% Tween 20 for 5 min, followed by two 30-min washes. Samples were either imaged immediately or stored for up to 3 days in permeabilization buffer; all samples comparing the same gene were imaged by confocal microscopy on the same day. When only bacteria within squid tissue were being probed, samples were counterstained overnight in 1:25 Alexa Fluor 633-phalloidin (catalog number A22284; Molecular Probes) in permeabilization buffer. All probe sequences are available in Table S1 in the supplemental material. A more complete protocol is available in the supplemental material.
Imaging.
Samples were imaged using a Zeiss LSM 510 confocal microscope. Figure panels were arranged and labeled using the Inkscape program. The brightness of the final figures was adjusted for visual clarity using Adobe Photoshop software.
RESULTS
Simultaneous labeling of multiple host transcripts.
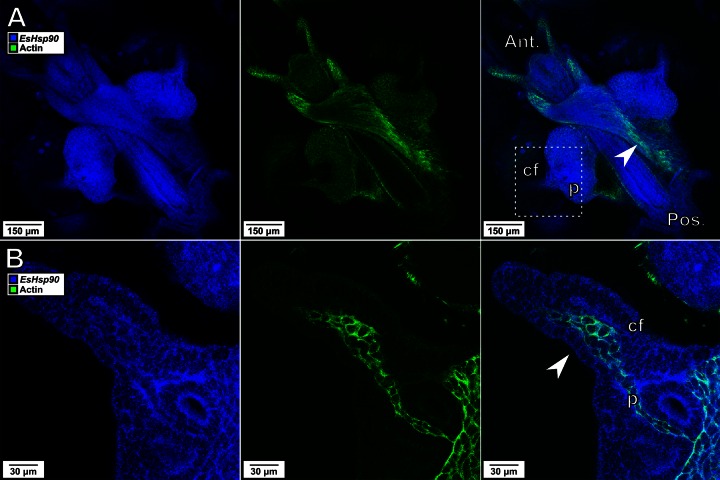
To determine the efficacy of HCR-FISH in colonized light organ tissue and to identify broadly distributed, normalizing transcripts, we probed whole-mount Euprymna light organs for transcripts of two widely distributed host genes: those encoding the structural protein actin and the chaperone Hsp90 (Fig. 2). We found that the Hsp90 transcript was distributed evenly throughout the tissues, allowing it to serve both as a control for uniform probe permeation and as a normalization factor to identify areas of increased expression of other genes. Interestingly, while the actin transcript appeared to be abundant in striated muscle cells, it was conspicuously less visible in the organ's ciliated surface and appendages. Moreover, these low actin transcript levels were unlikely due to poor permeation, as demonstrated by both the clear presence of the probe signal for Hsp90 and the strong staining of the vascular tissue in the interior of the appendage. These results demonstrate the ability of HCR-FISH to label and detect transcripts from multiple genes in host tissue and the uniformity of the distribution of the Hsp90 transcript, making its use as an internal control preferable to that of actin for HCR-FISH experiments in the squid light organ.
FIG 2.
Visualization of multiple transcripts within animal tissues. Representative images at ×10 (A) and ×40 (B) magnifications of the EsHsp90 transcript (blue) and the actin transcript (green) are shown. (A) The actin transcript is present throughout much of the light organ and strongly expressed in striated muscle (arrowhead). (B) In the enlarged area encompassed by the dashed box, the actin transcript is not detectable either in the ciliated field (cf) around the pore (p) or in the epithelium of the appendages, although it is easily observed in the vascular tissue within the appendage (arrowhead). In contrast to the actin transcript, the probe for the EsHsp90 transcript was detected strongly and uniformly throughout the appendages and ciliated field. Ant., anterior; Pos., posterior.
Bacterial transcript labeling in the squid.
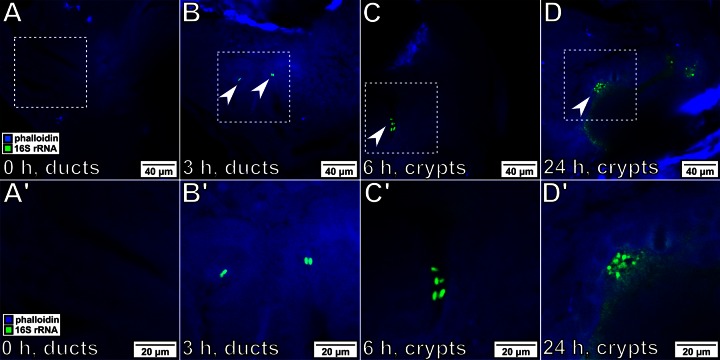
To localize the symbionts during colonization, probes specific for the bacterial 16S rRNA were designed. Because V. fischeri is the only bacterium present within the light organ, the species specificity of the probes was not a concern. Labeled bacteria were easily localized within the host tissue at different time points during the initiation of symbiosis (Fig. 3). Due to the high copy number of the 16S rRNA transcript, the intensity of the signal was sufficient to distinguish individual cells within the host, even when a low-magnification (i.e., ×10) lens was used (see Fig. S1 in the supplemental material). Bacteria were observed at almost all stages of their migration into the host, and no signal from the host tissue was observed; therefore, rRNA provides an effective marker for following bacterial migration. In addition, these samples were stained with Alexa Fluor 633-phalloidin after the amplification stage of the HCR-FISH protocol, illustrating the compatibility of this method with alternate fluorescent staining methods.
FIG 3.
Tracking the bacterial position during initiation of colonization. For all images, bacteria were labeled with probes specific for 16S rRNA (green), and squid tissue was stained with Alex Fluor 633-phalloidin (blue). (A′ to D′) Enlargements of the boxed regions in panels A to D, respectively. Magnifications, ×40. (A, A′) Before exposure, no bacteria were visible within the light organ of the host. (B, B′) After 3 h of exposure, bacteria (arrowheads) have associated with the host and begin to be visible within the light organ ducts, located immediately in the interior of the pores (Fig. 1C). (C, C′) By 6 h postexposure, bacteria have migrated into the light organ and have begun to colonize the crypt space. (D, D′) After 24 h of exposure, the host is bioluminescent and the symbionts are visible throughout the crypts.
HCR-FISH visualization of the spatiotemporal regulation of gene expression in the host and symbiont.
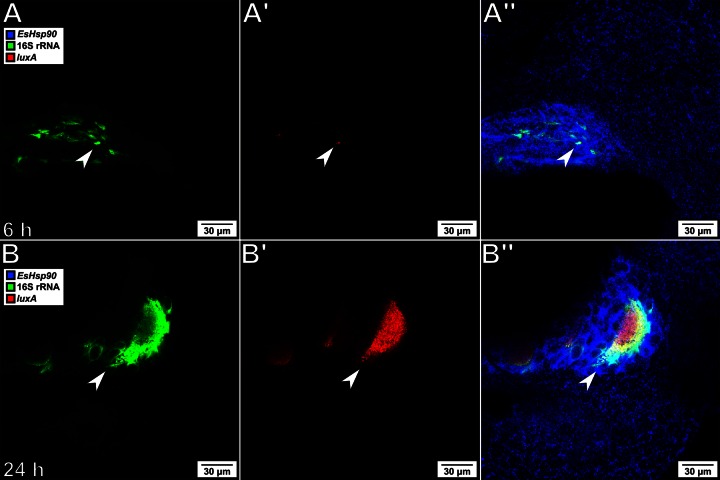
To determine the ability to visualize transcriptional changes in the symbiont, probes specific for the luxA gene were designed. The lux locus has been well studied in V. fischeri, where it is transcriptionally regulated via quorum sensing (24). As the bacteria grow in the host crypts, they reach a density sufficient to induce transcription of the lux operon, leading to the production of bioluminescence. To track the pattern of lux expression during colonization, bacteria were labeled with probes specific for both the 16S rRNA and the luxA transcript, and the host tissue was labeled with probes specific for EsHsp90 (Fig. 4). This combination of probes allows localization of symbionts within the host tissue through the visual identification of the crypt structure and further demonstrates that mixed host and symbiont probe/hairpin sequence sets can simultaneously be used on the same tissue sample. The bacteria in samples fixed at 6 h postcolonization showed only a weak, isolated signal from the luxA transcript, while those in samples fixed at 24 h revealed a clear and robust signal throughout the population, consistent with the onset of the animal's luminescence when recorded at the same time points. The possibility of nonspecific amplification of off-target bacterial mRNA was addressed by repeating the experiment with a Δlux mutant strain (25). As predicted, no signal was observed at 24 h in animals colonized by this mutant strain, despite the presence of large numbers of bacteria within the host crypts (see Fig. S2 in the supplemental material).
FIG 4.
Tracking expression of luxA during host colonization. (A and B) Visualization of V. fischeri by labeling of the 16S rRNA (green); (A′ and B′) expression of the luxA gene (red); (A″ and B″) visualization of host tissue by labeling the EsHsp90 transcript (blue), in which the label is overlaid with the two bacterial labels; (A, A′, and A″) at 6 h after inoculation, V. fischeri (arrows) has migrated into the crypts but has not yet grown to a sufficient density to induce strong expression of the luxA gene; (B, B′, and B″) after 24 h, when the symbionts (arrows) are densely packed and brightly bioluminescent, V. fischeri shows strong expression of the luxA gene.
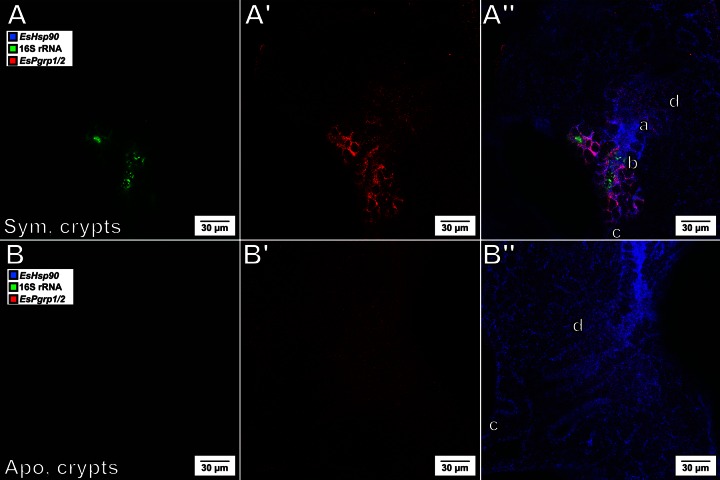
EsPGRP2 is an N-acetyl-muramyl-l-alanine amidase that localizes to the apical cytoplasm of the host epithelial tissue (10). Persistent colonization by V. fischeri induces the release of EsPGRP2 into the lumen of the deep crypts, resulting in the degradation of the peptidoglycan derivative tracheal cytotoxin (TCT), a powerful bacterial inducer of host development (26). While the EsPgrp2 transcript level was previously shown to increase in the light organ at 18 h postcolonization (4), it was unknown whether this increase was uniform throughout the organ or localized to a specific region of cells. We probed for EsPGRP1/2 expression at 24 h postcolonization and found that the upregulation of this transcript in response to colonization by V. fischeri was confined essentially to the deep crypt epithelium (Fig. 5), seemingly in the same cells that released the protein into the crypts.
FIG 5.
The EsPgrp1/2 transcript level is elevated predominantly in the crypts by 24 h postcolonization. (A and B) Visualization of V. fischeri by labeling of the 16S rRNA (green); (A′ and B′) expression of EsPgrp1/2 (red); (A″ and B″) visualization of host tissue by labeling the EsHsp90 transcript (blue), in which the label is overlaid with the two other labels; (A, A′, and A″) EsPgrp1/2 is expressed in the symbiotic (Sym.) crypt epithelium at 24 h postcolonization; (B, B′, and B″) in contrast, its expression is undetectable in the crypts of aposymbiotic (Apo.) animals at the same stage of development. d, duct; a, antechamber; b, bottleneck; c, crypt (as shown in Fig. 1).
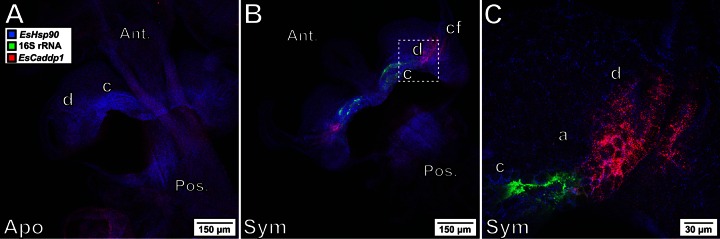
To further test the ability of HCR-FISH to localize host gene expression, we probed for another transcript (EsCadDP1) encoding a protocadherin-like protein. Whereas we observed only diffuse expression of this transcript in the light organ tissues of aposymbiotic animals (Fig. 6A), symbiotic animals expressed EsCadDP1 in the epithelial cells lining the ducts and antechambers at detectably higher levels (Fig. 6B). Intriguingly, the epithelium of the crypt cells, which is in direct contact with the bacterial population, showed no change in this gene's expression compared with that of the epithelium of the crypt cells of aposymbiotic animals (Fig. 6C). This visualization of the transcript encoding EsCadDP1 exclusively in the light organ ducts and that encoding EsPGRP1/2 solely within the crypts demonstrated the induction of specific genes in specific locations along the 100-μm path traversed by V. fischeri during its migration through host tissues.
FIG 6.
Tracking of a host transcriptional response to the bacterial symbiont. The light organs from aposymbiotic (A) and symbiotic (B) animals at 24 h postcolonization were labeled with three probes. EsHsp90 (blue) labels the light organ throughout, but EsCadDP1 (red) is found only in the ducts (d) and antechamber (a) adjacent to the V. fischeri (whose 16S rRNA is labeled green)-colonized crypts (c). (C) The EsCadDP1 signal is localized to the ducts and not the epithelium in contact with the bacteria. Ant., anterior; Pos., posterior; cf, ciliated field (as in Fig. 1). Magnifications, ×10 (A and B) and ×40 (C).
DISCUSSION
The natural colonization of an animal host by either beneficial or pathogenic bacteria is a complex process whose trajectory and genetic underpinnings are still poorly understood. Nevertheless, it is likely that, during establishment of such associations, reciprocally induced transcriptional changes occur in both the bacteria and the host cells with which they interact. Because host tissues are often complex and inaccessible and small numbers of bacteria typically initiate a colonization, it is challenging to define either the particular cells that undergo these changes or the precise stage of the colonization process during which symbiosis-induced regulation of gene expression occurs. Application of HCR-FISH permits the visualization of transcriptional responses by multiple, specific genes that occur in individual cells, providing a detailed temporally and spatially defined picture of the program of gene expression (12). As such, this capability, when applied to symbiotic systems, promises to reveal the dynamics of gene expression underlying the host-microbe interaction with a temporal and spatial precision and sensitivity not previously possible.
In adapting this approach for use with the Euprymna-Vibrio system and perhaps others, there are several caveats and concerns to keep in mind. As with other in situ hybridization techniques, it is important to perform probe validation experiments (see Fig. S2 in the supplemental material) with gene deletion mutants when possible. Whereas the gene knockout technology is not yet available for the squid host, such tools have been developed for its symbiont. It is also advisable to perform autofluorescence and nonspecific amplification controls for each experiment. In both the host and the symbiont under study here, these controls revealed that no observable fluorescence was present at the signal amplification levels used. Finally, significant lysis of V. fischeri was observed in the probe hybridization buffer originally reported by Choi et al. (12). V. fischeri cells suspended in this buffer lysed when heated to 65°C, the temperature used for probe prehybridization; such a problem may be encountered when applying this method to other marine or structurally sensitive bacteria. It was resolved by diluting the probe hybridization buffer in which V. fischeri was suspended 1:1 with mPBS. Surprisingly, it was determined that buffer osmolarity was not the sole reason for the observed bacterial lysis.
Our application of HCR-FISH revealed several interesting patterns of gene expression in both the squid and its symbiont. Constitutively expressed transcripts in both partners (i.e., E. scolopes Hsp90 [EsHsp90] in the host and 16S rRNA in the bacteria) allowed analysis of their colocalization as well as provided controls for probe penetration. In the bacteria, the observed pattern of luxA expression correlated well with that observed in previous studies done with lux promoter fusions and observations of squid bioluminescence (18, 27). The host EsPgrp1/2 transcript is upregulated in cells in close proximity to the symbionts, similar to the cells that secrete EsPGRP2 itself into the crypts (10). These data provide evidence that the provision of this host protein into the crypts is a process controlled at the level of gene transcription. We also observed a highly localized response of the transcript for a cadherin domain-containing protein, EsCadDP1, in the ducts and antechambers (Fig. 6C). The spatial and temporal expression patterns of these two genes suggest that the onset of symbiosis sets up biochemically discrete regions of the light organ that reflect the restriction of the symbionts to the crypt spaces. The family of cadherin-like proteins comprises cadherins, protocadherins, and atypical cadherins (28). While many proteins in this family are involved in mechanical cell-cell adhesion, others function in cell signaling pathways or are of unknown function. Bioinformatic analysis of EsCadDP1 revealed only one cadherin domain; unlike other cadherins, the domain is not repeated, nor does it possess a cytoplasmic cadherin domain. The lack of repeating cadherin domains calls into question the degree to which the protein encoded by this transcript is capable of adhering to other cadherin domain-containing proteins. Thus, while its function remains unknown, EsCadDP1 is an excellent candidate for further study due to its strong and cell-specific responses to the presence of the bacterial symbionts.
HCR-FISH is a powerful technique for investigating gene expression within a model of symbiosis. By adapting the protocol developed by Choi et al. (12) to the squid-vibrio model system, we have demonstrated that this technique uniquely addresses specific questions of host-symbiont responses. HCR-FISH provides several advantages over previously available techniques. First, the use by HCR-FISH of multiple probe/hairpin sequence combinations within the same sample allows an investigation of the expression patterns of several genes simultaneously both in the host and in the symbiont population. As such, the number of transcripts analyzed becomes limited only by the number of wavelengths that can be detected on a particular microscope system. Second, as illustrated in Fig. 5 and 6, HCR-FISH greatly increases the resolution at which transcriptional responses can be localized within host tissue. Similarly, while promoter fusion constructs have allowed the observation of symbiont gene activation (18, 19), they rely on the presence of a strong native promoter to express a fluorescent reporter and, thus, are unlikely to detect genes expressed at moderate or low levels. Our observations of small amounts of a fluorescent signal from luxA transcripts in a subpopulation of cells at 6 h after exposure (Fig. 4A) show that lux transcription occurs at very low levels even at low cell densities. Previous promoter fusion studies of lux expression first showed a detectable signal starting at 8 h after exposure (18). These results are consistent with the higher sensitivity of HCR-FISH transcript labeling. Furthermore, in promoter fusion experiments, the time of appearance of the fluorescent signal lags behind the time of transcriptional initiation, complicating the proper recognition of regulatory cascades or development programs. Overall, HCR-FISH provides researchers in the area of host-symbiont interactions, both beneficial and pathogenic, with a tool that is a significant improvement over previously available techniques.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Moriano-Gutierrez and B. Krasity for advice concerning host gene expression, N. Pierce and his lab for sharing HCR-FISH protocols before publication, and T. Truong and V. Trivedi for assistance with initial protocol development.
This work was funded by the Gordon and Betty Moore Foundation, Marine Microbiology Initiative 3396, and NIH grants OD11024 (to E.G.R. and M.J.M.-N.) and AI050661 (to M.J.M.-N.). K.N. was supported by a Ruth L. Kirschstein National Research Service Award from the NIGMS (F32GM112214).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00890-15.
REFERENCES
- 1.Ruby EG, Lee KH. 1998. The Vibrio fischeri-Euprymna scolopes light organ association: current ecological paradigms. Appl Environ Microbiol 64:805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nyholm SV, McFall-Ngai MJ. 2004. The winnowing: establishing the squid-vibrio symbiosis. Nat Rev Microbiol 2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 3.McFall-Ngai MJ. 2014. The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Annu Rev Microbiol 68:177–194. doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chun CK, Troll JV, Koroleva I, Brown B, Manzella L, Snir E, Almabrazi H, Scheetz TE, de Fatima Bonaldo M, Casavant TL, Soares MB, Ruby EG, McFall-Ngai MJ. 2008. Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-vibrio association. Proc Natl Acad Sci U S A 105:11323–11328. doi: 10.1073/pnas.0802369105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kremer N, Philipp EE, Carpentier MC, Brennan CA, Kraemer L, Altura MA, Augustin R, Hasler R, Heath-Heckman EA, Peyer SM, Schwartzman J, Rader BA, Ruby EG, Rosenstiel P, McFall-Ngai MJ. 2013. Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe 14:183–194. doi: 10.1016/j.chom.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sycuro LK, Ruby EG, McFall-Ngai M. 2006. Confocal microscopy of the light organ crypts in juvenile Euprymna scolopes reveals their morphological complexity and dynamic function in symbiosis. J Morphol 267:555–568. doi: 10.1002/jmor.10422. [DOI] [PubMed] [Google Scholar]
- 7.Wollenberg MS, Ruby EG. 2009. Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from two Oahu (Hawaii) populations. Appl Environ Microbiol 75:193–202. doi: 10.1128/AEM.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Visick KL, Ruby EG. 2006. Vibrio fischeri and its host: it takes two to tango. Curr Opin Microbiol 9:632–638. doi: 10.1016/j.mib.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Troll JV, Adin DM, Wier AM, Paquette N, Silverman N, Goldman WE, Stadermann FJ, Stabb EV, McFall-Ngai MJ. 2009. Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis. Cell Microbiol 11:1114–1127. doi: 10.1111/j.1462-5822.2009.01315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Troll JV, Bent EH, Pacquette N, Wier AM, Goldman WE, Silverman N, McFall-Ngai MJ. 2010. Taming the symbiont for coexistence: a host PGRP neutralizes a bacterial symbiont toxin. Environ Microbiol 12:2190–2203. doi: 10.1111/j.1462-2920.2009.02121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McConnell RE, Benesh AE, Mao S, Tabb DL, Tyska MJ. 2011. Proteomic analysis of the enterocyte brush border. Am J Physiol Gastrointest Liver Physiol 300:G914–G926. doi: 10.1152/ajpgi.00005.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi HM, Beck VA, Pierce NA. 2014. Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability. ACS Nano 8:4284–4294. doi: 10.1021/nn405717p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi HM, Chang JY, Trinh LA, Padilla JE, Fraser SE, Pierce NA. 2010. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat Biotechnol 28:1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dirks RM, Pierce NA. 2004. Triggered amplification by hybridization chain reaction. Proc Natl Acad Sci U S A 101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenthal AZ, Zhang X, Lucey KS, Ottesen EA, Trivedi V, Choi HM, Pierce NA, Leadbetter JR. 2013. Localizing transcripts to single cells suggests an important role of uncultured deltaproteobacteria in the termite gut hydrogen economy. Proc Natl Acad Sci U S A 110:16163–16168. doi: 10.1073/pnas.1307876110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huss D, Choi HM, Readhead C, Fraser SE, Pierce NA, Lansford R. 2015. Combinatorial analysis of mRNA expression patterns in mouse embryos using hybridization chain reaction. Cold Spring Harb Protoc 2015:259–268. doi: 10.1101/pdb.prot083832. [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi T, Kawakami S, Hatamoto M, Imachi H, Takahashi M, Araki N, Yamaguchi T, Kubota K. 19 December 2014. In situ DNA-hybridization chain reaction (HCR): a facilitated in situ HCR system for the detection of environmental microorganisms. Environ Microbiol doi: 10.1111/1462-2920.12745. [DOI] [PubMed] [Google Scholar]
- 18.Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol 72:802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Dunn AK, Wilneff J, McFall-Ngai MJ, Spiro S, Ruby EG. 2010. Vibrio fischeri flavohaemoglobin protects against nitric oxide during initiation of the squid-vibrio symbiosis. Mol Microbiol 78:903–915. doi: 10.1111/j.1365-2958.2010.07376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montgomery MK, McFall-Ngai M. 1994. Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development 120:1719–1729. [DOI] [PubMed] [Google Scholar]
- 21.Naughton LM, Mandel MJ. 2012. Colonization of Euprymna scolopes squid by Vibrio fischeri. J Vis Exp 2012:e3758. doi: 10.3791/3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunlap PV. 1989. Regulation of luminescence by cyclic AMP in cya-like and crp-like mutants of Vibrio fischeri. J Bacteriol 171:1199–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boettcher KJ, Ruby EG. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol 172:3701–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupp C, Urbanowski M, Greenberg EP, Ruby EG. 2003. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol Microbiol 50:319–331. doi: 10.1046/j.1365-2958.2003.t01-1-03585.x. [DOI] [PubMed] [Google Scholar]
- 25.Bose JL, Rosenberg CS, Stabb EV. 2008. Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch Microbiol 190:169–183. doi: 10.1007/s00203-008-0387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McFall-Ngai M, Heath-Heckman EA, Gillette AA, Peyer SM, Harvie EA. 2012. The secret languages of coevolved symbioses: insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Semin Immunol 24:3–8. doi: 10.1016/j.smim.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruby EG, Asato LM. 1993. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol 159:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- 28.Halbleib JM, Nelson WJ. 2006. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev 20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.