Abstract
The scyphozoan Aurelia aurita is recognized as a key player in marine ecosystems and a driver of ecosystem change. It is thus intensely studied to address ecological questions, although its associations with microorganisms remain so far undescribed. In the present study, the microbiota associated with A. aurita was visualized with fluorescence in situ hybridization (FISH) analysis, and community structure was analyzed with respect to different life stages, compartments, and populations of A. aurita by 16S rRNA gene amplicon sequencing. We demonstrate that the composition of the A. aurita microbiota is generally highly distinct from the composition of communities present in ambient water. Comparison of microbial communities from different developmental stages reveals evidence for life stage-specific community patterns. Significant restructuring of the microbiota during strobilation from benthic polyp to planktonic life stages is present, arguing for a restructuring during the course of metamorphosis. Furthermore, the microbiota present in different compartments of the adult medusa (exumbrella mucus and gastric cavity) display significant differences, indicating body part-specific colonization. A novel Mycoplasma strain was identified in both compartment-specific microbiota and is most likely present inside the epithelium as indicated by FISH analysis of polyps, indicating potential endosymbiosis. Finally, comparison of polyps of different populations kept under the same controlled laboratory conditions in the same ambient water showed population-specific community patterns, most likely due the genetic background of the host. In conclusion, the presented data indicate that the associated microbiota of A. aurita may play important functional roles, e.g., during the life cycle.
INTRODUCTION
Multicellular eukaryotes in marine systems provide diverse surfaces for attachment of and colonization by microbes, e.g., mucosa-covered epithelia. Bacteria, often present in high numbers in the surrounding marine environment, tend to colonize these surfaces or even invade the surface epithelium. Multicellular organisms nowadays are largely recognized as “metaorganisms” comprising the macroscopic host and its synergistic interdependence with bacteria, archaea, and fungi specifically associated with the host (1, 2). Depending on the variety of different niches provided by the host, which can change according to developmental stage, diet, or other environmental factors, diverse and specific microbial communities can establish themselves with hosts (3). Previous investigations demonstrated that marine invertebrates harbor bacteria as stable associates recruited from the ocean's pool of potential colonizers (4–6). A well-studied example is the symbiosis between the squid Euprymna scolopes and Vibrio fischeri, which is crucial for certain developmental steps of the squid (7, 8). Additionally, endosymbionts living inside host cells or in the intracellular space of the host's tissue are well documented for marine invertebrates, e.g., in hydrothermal vent systems, including the giant tubeworm Riftia pachyptila and associated chemolithoautotrophic bacteria capable of oxidizing hydrogen sulfide (9). Furthermore, members of the class Mollicutes are known to be (endo)symbionts of various invertebrates (10–12).
Scyphozoa (jellyfish) represent a class of Cnidaria, which is at the base of the metazoan tree. They play an important role in the marine ecosystem by substantially affecting the structure of the planktonic food web (13–15). They release large amounts of nutrients and dissolved organic matter (16, 17) resulting from their metabolic activities, presumably directly stimulating bacterial growth and potentially influencing bacterial community composition (18–20). The cosmopolitan moon jelly Aurelia aurita, likely representing a collection of species as recently shown by molecular markers (21), is one of the most widely distributed scyphozoans and is found in almost all warm and temperate waters of coastal zones worldwide (22–24). Due to its ability to tolerate a wide range of environmental conditions (14, 25), especially temperature and salinity, and its highly diverse food spectrum, it successfully colonizes different environments (26) and often causes jellyfish blooms around the world. Such blooms not only impact food web structures by predation but also play an important role in the dynamics of nutrients and oxygen in planktonic food webs (14). The diphasic life cycle of A. aurita alternates between free-living pelagic sexual (medusa) and sessile benthic asexual (polyp) phases. After release from sexual reproduction, larvae settle on suitable seafloor substrata, followed by metamorphosis into the sessile benthic stage, the polyp. Transition from the benthic phase to the pelagic phase occurs when the juvenile pelagic stage, the ephyra, is formed and released by the polyp through the process of strobilation (see Fig. S1 in the supplemental material). The medusa has a very simple morphology, with two tissue layers: the epidermis, covering the outer body surface, and the gastrodermis, forming the boundary of the gastric cavity. The mesoglea between these tissue layers, consisting mainly of collagen, provides stability for the animal. The prominent oral arms of the medusa harbor brood sacs, where the developing planula larvae are sheltered until release. The polyp exhibits an even simpler anatomy, lacking tissues for sexual reproduction. Based on strobilation frequency and the temperatures required to induce strobilation, different populations of A. aurita can be distinguished, e.g., the arctic boreal lineage (Baltic and North Seas) and the temperate Mediterranean lineage (e.g., Eastern Atlantic-Roscoff). In addition, Schroth et al. showed that limited gene flow among populations around the world causes cryptic speciation and proposed at least seven different species of the genus Aurelia based on mitochondrial and nuclear DNA analyses (16S and internal transcribed spacer 1 [ITS-1]/5.8S rRNA gene) (27). In summary, different A. aurita subpopulations can be correlated with their geographic origin; for example, the BOR lineage includes Baltic Sea and North Sea specimens, whereas the UBI lineage, among others, includes Eastern Atlantic-Roscoff specimens. To date, most studies have addressed ecological questions (22, 27, 28) as well as the physiology (29–31) and development (32–34) of A. aurita; however, no studies on associated microbes are available. Consequently, due to its simple anatomy, evolutionary age, alteration of different life stages, wide distribution, and important role in the marine ecosystem, A. aurita is an attractive model organism to study the metaorganism concept (2).
The establishment of next-generation sequencing techniques has revolutionized the study of microorganisms in natural environments (35) and profoundly altered our awareness of microbial ecosystems (36, 37). Thus, the present study aimed to analyze the microbiota associated with different life stages, compartments, and populations of A. aurita by using 16S rRNA gene-based amplicon sequencing to gain insight into transiently and specifically associated bacteria of A. aurita and, ultimately, into interactions between the host and its microbiota. Moreover, in the long term, detailed knowledge of the microbial consortia of scyphozoa such as A. aurita may provide critical insights for understanding a microbe-dependent life history and its evolutionary consequences.
MATERIALS AND METHODS
Medusa sampling.
Individual A. aurita (Linnaeus) medusae (mean umbrella diameter, 26 cm) were sampled from one location in the Kiel Bight, Baltic Sea (54°19.4′N, 10°08.5′E) in June 2013 by using a dip net. The animals were transported immediately to the laboratory, washed thoroughly with sterile filtered artificial seawater (ASW) to remove nonassociated microbes, and processed for further analysis. Processing of animals started with sampling of mucus from ex- and subumbrellas by scraping and sampling the slime. Furthermore, samples of the gastric cavity were taken with a pipette by penetrating the mouth. Water from the same site was collected by bucket sampling, from which reference water samples were taken. Water samples were filtered through 0.22-μm polyethersulfone membrane filters (Millipore Corporation, Bedford, MA), and DNA was extracted from the respective filters as described below.
Aurelia aurita husbandry.
Polyp populations originating from the Southern English Channel (Roscoff, France) and the North Sea (Helgoland, Germany) were provided by Thomas Bosch (CAU Kiel) and were kept in the laboratory under artificial conditions for 10 years. Baltic Sea specimens were raised from planula larvae originating from medusae caught in the Kiel Bight in September 2009 and transferred to the laboratory for rearing under artificial conditions. The polyps were kept in 2-liter vessels at 20°C in aerated artificial seawater (Reef Crystals; Aquarium Systems, Sarrebourg, France) with a salinity of 30 practical salinity units (PSU). Twice a week, animals were fed freshly hatched Artemia salina, and the water was changed completely. Feeding of animals was ceased at least 4 days prior to the analysis of microbial consortia to exclude contamination by food. Strobilation of Roscoff polyps for the production of ephyrae was induced in the laboratory after reducing the temperature to 8°C for at least 1 month. A subset of obtained ephyrae continued to develop into juvenile medusae.
Generation of sterile A. aurita polyps and ephyrae.
In order to obtain sterile/germfree polyps, polyps were treated with an antibiotic mixture present in the water (Provasoli's antibiotic [AB] mixture with final concentrations of 360,000 U/liter penicillin G, 1.5 mg/liter chloramphenicol, 1.8 mg/liter neomycin, and 9,000 U/liter polymyxin B [all components from Carl Roth, Karlsruhe, Germany]), kept under the above-mentioned conditions in sterilized (by filtration through 0.22-μm filters) artificial seawater, and fed antibiotic-treated A. salina. Antibiotic-treated ephyrae originated from AB-treated polyps. The absence of bacteria was confirmed by plating of homogenized polyps/ephyrae onto R2A agar plates (Roth, Karlsruhe, Germany). After incubation at 19°C for 5 days, the CFU were determined. An absence of CFU indicated successful antibiotic treatment. For culture-independent analysis, total DNA was extracted, and the 16S rRNA genes were amplified as described below. The respective impacts of AB treatment are depicted in Fig. 3 and 4D.
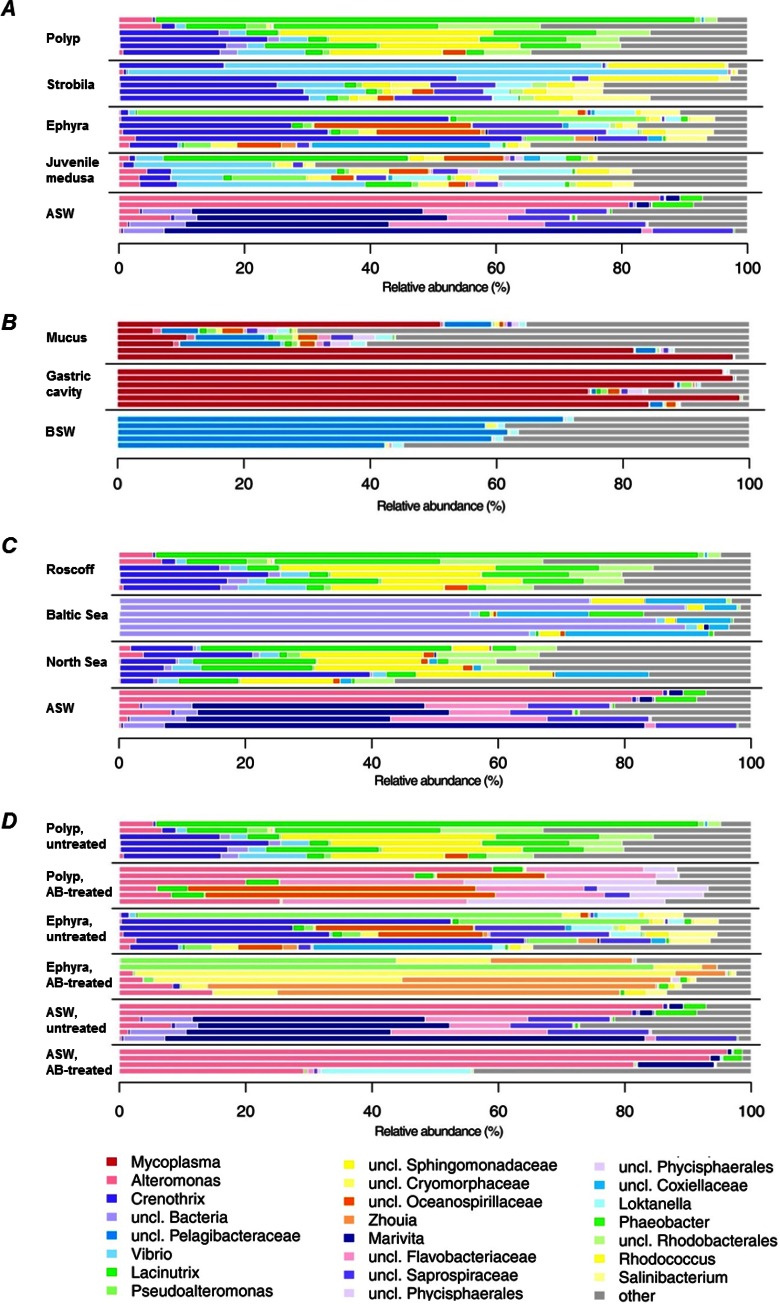
FIG 3.
Composition of microbiota associated with individual A. aurita samples. Microbial communities were analyzed by sequencing of the V1-V2 region of 16S bacterial rRNA genes from different life stages (A), compartments (B), populations of A. aurita living under the same environmental conditions (C), and induced sterile conditions (D). Taxonomic assemblages were inferred from nonchimeric operational taxonomic units using mothur at a 97% pairwise similarity cutoff; distribution patterns were calculated by using R. OTU abundances were summarized at the genus level and normalized by the total number of reads per sample. Bar plots are grouped according to sample type, each including 4 to 6 replicates. Taxa with a mean relative abundance of <1% across all samples are subsumed under “other.” ASW, artificial seawater; BSW, Baltic Sea water.
FIG 4.
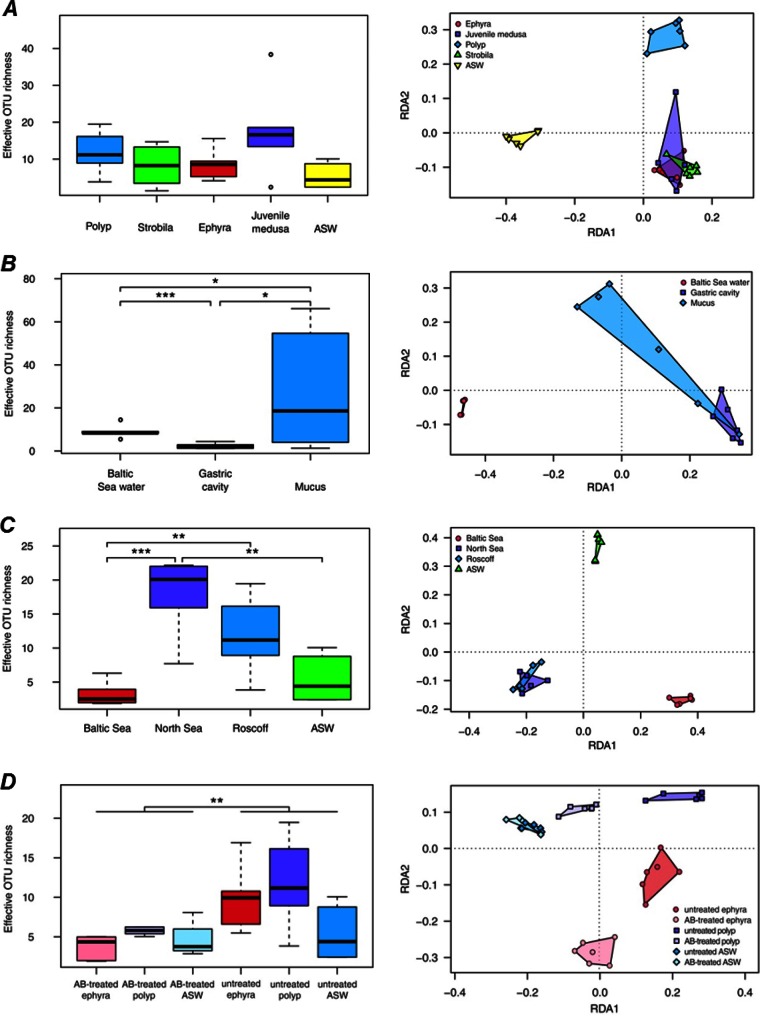
Distribution of effective OTU richnesses of bacterial communities associated with A. aurita and RDA models of Hellinger-transformed OTU abundances. (Left) Distribution of effective OTU richnesses of bacterial communities associated with A. aurita life stages (A), A. aurita medusa compartments (B), populations (C), and induced sterile conditions (D). Box plots mark the medians (bold lines), interquartile ranges (colored boxes), outliers ≤1.5 times the interquartile range from the box (whiskers), and outliers beyond (circles). Square brackets denote pairwise comparisons with P values of between 5% and 1% (*), between 1% and 0.1% (**), and between 0.1% and 0.01% (***). (Right) Graphical representation (distance plots) of the RDA model of Hellinger-transformed OTU abundances. Each point represents the whole microbial community of an individual sample, as mentioned above for panels A to C. Groups of related sample points are framed by polygons filled with the corresponding color to elucidate the distribution and variability of sample groups in the ordination space.
Phylogenetic classification of A. aurita polyps.
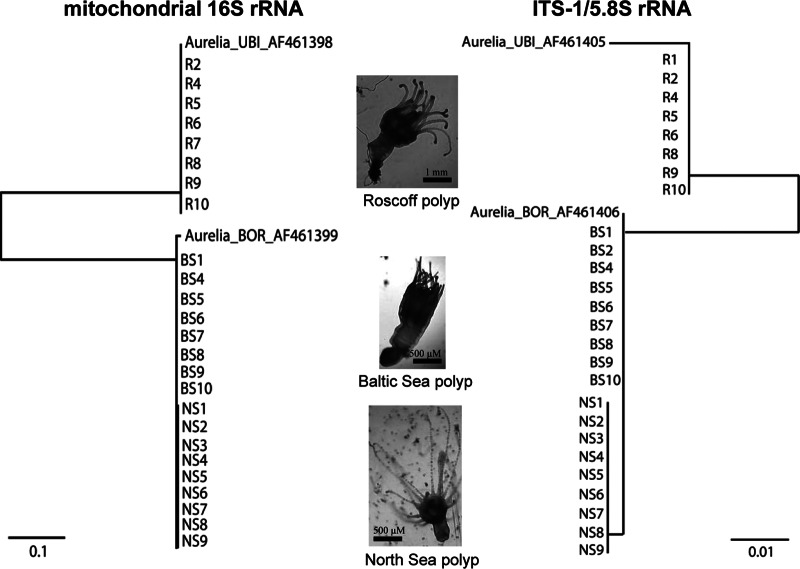
DNA was extracted from 10 single polyps from each respective Roscoff, Baltic Sea, and North Sea population kept in the laboratory under the same artificial conditions by using the Wizard genomic purification kit (Promega, Madison, WI, USA). Polyps were homogenized in 480 μl 50 mM EDTA with a pestle and incubated at 37°C for 30 min after the addition of proteinase K (Life Technologies, Darmstadt, Germany). The remaining preparation steps were performed according to the manufacturer's protocol. DNA templates (10 ng) were subjected to PCR with a 25-μl total volume by using a GoTaq DNA polymerase kit (Promega, Madison, WI, USA). Cycling conditions were 30 cycles of 30 s at 95°C, 30 s at 50°C, and 60 s at 72°C. Aurelia-specific mitochondrial 16S rRNA primers (L5′-CTCTTGTAAGGTGAAGCC and H5′-CATAATTCAACATCGAGG) and specific primers encompassing the ITS-1/5.8S rRNA region of nuclear 18S rRNA (F5′-TAACAAGGTTTCCGTAGG and R5′-CTC AGA CAG ACA TGC TCC) were used as described previously by Schroth et al. (27). PCRs were used to determine the DNA sequences, performed by the sequencing facility at the Institute of Clinical Molecular Biology (IKMB), University of Kiel, Kiel, Germany. Tree reconstruction was performed with both mitochondrial 16S rRNA and nuclear 18S rRNA sequences by using Phylogeny.fr (http://www.phylogeny.fr/) (38, 39) with ClustalW for multiple alignment, Gblocks for alignment curation, and the maximum likelihood/PhyML method for construction of the phylogenetic tree. Previously reported sequence data from Aurelia populations deposited in GenBank (accession numbers AF461398 and AF461399 for 16S rRNA and AF461405 and AF461406 for ITS-1/5.8S rRNA) were used for classification of polyps into known subpopulations.
Fluorescence in situ hybridization and microscopic analysis.
Polyps (Roscoff population, 6 to 8 replicates) were washed three times with filter-sterilized artificial seawater and relaxed by incubation with 2% urethane (Sigma-Aldrich, Schnelldorf, Germany) for 5 min. Polyps were incubated in Carnoy fixative (60% [vol/vol] ethanol, 30% [vol/vol] chloroform, 10% [vol/vol] glacial acetic acid) for 2 h at 4°C. Fixed samples were dehydrated three times with 100% ethanol for 5 min each. Semithin sections (0.5 μm) were prepared after several dehydration steps (5 min each, with 50%, 75%, and 100% ethanol), embedding in LR White (Sigma-Aldrich), and polymerization of the samples in LR White at high temperature. Whole polyps and semithin sections were further incubated with 10 μg/ml proteinase K (Fermentas, St. Leon-Rot, Germany) and 10 mg/ml lysozyme (Roth, Karlsruhe, Germany) in PBT (phosphate-buffered saline [PBS], 1% Tween 80 [pH 7.4]) for 1 h at 37°C. This was followed by two washing steps with Milli-Q water and further dehydration. Hybridization was performed in hybridization buffer (900 mM NaCl, 20 mM Tris-HCl [pH 7.4], various concentrations of formamide [see Table S2 in the supplemental material], 0.01% SDS) with the respective probes (working solution of 5 ng/μl; Firmicutes probes LGC0354a, -b, and -c were mixed in equimolar proportions) at the specified temperature for 3 h (see Table S2 in the supplemental material). Finally, samples were incubated in washing buffer (80 mM NaCl with 35% formamide or 225 mM NaCl with 20% formamide in hybridization buffer, 20 mM Tris-HCl [pH 7.4], 20 mM EDTA, and 0.01% SDS) for 15 min at a temperature 2°C below the specified temperature and washed with Milli-Q water. Probes with the corresponding formamide concentrations in the buffers and incubation temperatures are listed in Table S2 in the supplemental material. For counterstaining, samples were incubated with SYTO9 (Invitrogen, Darmstadt, Germany) or 4′,-6-diamidino-2-phenylindole (DAPI) (Roth, Karlsruhe, Germany) at room temperature for 20 min and washed with Milli-Q water. Light microscopy and fluorescence microscopy were performed with an Axio Scope microscope and Axio Vision software (Zeiss, Jena, Germany). The entire three-dimensional structure of the polyp and its associated microbiota was recorded by scanning along the depth by using a TCS SP confocal laser scanning microscope (Leica, Wetzlar, Germany) and recording the stacks of cross sections simultaneously at the corresponding excitation wavelengths. For each field of view, an appropriate number of optical slices were acquired with a Z-step of 1 μm. Digital image acquisition, postprocessing, analysis of the confocal laser scanning microscopy (CLSM) optical thin sections, three-dimensional reconstructions, and calculation of bacterial coverage were performed with the corresponding Leica software (provided for the TCS SP confocal laser scanning microscope). For detection of each bacterial phylum, three independent fluorescence in situ hybridization (FISH) experiments were performed, and 10 confocal stacks were compared for each independent experiment. Six polyps for each treatment were used for calculation of bacterial coverage of polyps.
Nucleic acid isolation and 16S rRNA gene amplicon sequencing.
Bacterial genomic DNA of the A. aurita microbiota was isolated with the AquaPure genomic DNA kit (Bio-Rad, Munich, Germany) from 10 mg mucus; 5 mg of the gastric cavity sample; pools of 10 polyps, 20 ephyrae, 1 strobila, and 1 juvenile medusa per sample type (all samples representing approximately the same wet weight); and 1 liter of reference water filtered through a 0.22-μm-pore-size membrane filters. DNA isolation was performed with 6 replicates for each sample type. PCR amplification of the V1-V2 hypervariable region of the 16S rRNA gene was performed with primers listed in Table S3 in the supplemental material. Amplicons were size checked and purified by using the MinElute gel extraction kit (Qiagen, Hilden, Germany). Purified amplicons were quantified by using the Quant-iT PicoGreen kit (Invitrogen, Darmstadt, Germany), and equal amounts thereof were pooled for subsequent pyrosequencing. Pyrosequencing was carried out according to the manufacturer's instructions, using the GS FLX Titanium series kit (sequencing kit XLR70, Pico Titer plate kit 70x75, SV emulsion PCR [emPCR] kit/Lib-A, and Maintenance Wash kit; Roche, Mannheim, Germany). The libraries were sequenced by using the 454 GS-FLX Titanium sequencer (Roche) at the Max Planck Institute for Evolutionary Biology (Plön, Germany).
Study design.
In order to reduce practical and computational workloads, an incomplete block design was conceived, corresponding to four experimental setups, which in part made use of the same samples. These setups comprised examination of differences in epibacterial community compositions according to the following factors (Table 1): (i) four life stages (polyp, strobila, ephyra, and juvenile medusa) with individuals from the Roscoff population, (ii) compartment types of medusae from the Baltic Sea with microbial samples from the mucus and gastric cavity, (iii) individuals from different populations (Roscoff, Baltic Sea, and North Sea), and (iv) AB-treated versus untreated control polyps from the Roscoff population. Each factor level encompassed six replicates, four to six of which yielded read libraries sufficient for analysis.
TABLE 1.
Study design
| Study type | Sample | Population | Keeping condition(s) | Origin |
|---|---|---|---|---|
| Life stage | Polyp | Roscoff | 10 yr in the laboratory, artificial seawater, temp of 20°C | Laboratory stock |
| Strobila | Roscoff | Artificial seawater, temp of 10°C | Polyp | |
| Ephyra | Roscoff | Artificial seawater, temp of 10°C | Strobila | |
| Juvenile medusa | Roscoff | Artificial seawater, temp of 20°C | Ephyra | |
| Compartment | Medusa mucus | Baltic Sea | Freshly hatched | Baltic Sea |
| Medusa gastric cavity | Baltic Sea | Freshly hatched | Baltic Sea | |
| Population | Polyp | Roscoff | 10 yr in the laboratory, artificial seawater, temp of 20°C | Laboratory stock |
| Polyp | Baltic Sea | 10 yr in the laboratory, artificial seawater, temp of 20°C | Laboratory stock | |
| Polyp | North Sea | 10 yr in the laboratory, artificial seawater, temp of 20°C | Laboratory stock | |
| AB treatment | Polyp | Roscoff | 10 yr in the laboratory, artificial seawater, temp of 20°C | Laboratory stock |
| Ephyra | Roscoff | Artificial seawater, temp of 10°C | Polyp | |
| AB-treated polyp | Roscoff | 10 yr in the laboratory, artificial seawater, temp of 20°C | Laboratory stock | |
| AB-treated ephyra | Roscoff | Artificial seawater, temp of 10°C | AB-treated polyp |
Sequence processing.
Sequence processing was conducted with mothur v1.33.3 (40), as recently described (41). The mothur greengenes reference taxonomy (http://www.mothur.org/w/images/9/9d/Gg_13_5_99.taxonomy.tgz) built from the May 2013 release of the greengenes database (42) was used for sequence and operational taxonomic unit (OTU) classification. This database was complemented by selected sequences of Labrenzia spp. (National Center for Biotechnology Information [NCBI] GenBank accession numbers AJ582083, AY628423, and AJ878875). Reference sequences were trimmed to the V1-V2 primer region to improve the accuracy of the classification (43). A random subset of 3,034 sequences per sample (corresponding to the smallest number of reads across samples) was generated to eliminate bias due to unequal sampling effort. A total of 1,865 OTUs were identified at the 97% similarity level.
Sequence abundance plots.
Downstream statistical analysis was performed in R v3.1.1 (44) with custom scripts (available upon request). Initial characterization of bacterial community composition was performed by summarizing OTU abundances at the genus level, normalized by the total number of reads per sample. Normalized genus abundances were visualized as stacked bar plots.
Alpha diversity analysis.
Effective OTU richness (Shannon number equivalent [1D]) (45, 46) was calculated from the sample-by-OTU table with the R package vegan (47). Effects of A. aurita life stages, provenance, compartments, and AB treatment in connection with sample type on 1D were assessed. For this purpose, 1D was fitted to these experimental factors in generalized linear models by using the glm function of the R stats package. In the case of the factor “compartment,” samples of A. aurita were paired (i.e., for each mucus sample, there was a corresponding gastric cavity sample from the same medusa), while reference water samples were unpaired. In order to analyze both sample types in the same statistical model, all samples had to be treated as unpaired. The stochastic part of all models was described either by a gamma or an inverse Gaussian distribution. Models were compared by means of the Akaike information criterion (AIC) and validated according to recommendations described previously (48). As a measure of model performance, the ratio between explained and residual deviances (so-called pseudo-R2) was calculated and adjusted by sample size and the number of explanatory variables using Ezekiel's formula (49). Since the different experimental setups were used for some of the same samples, model significances were corrected for multiple testing with the Bonferroni-Holm method (“q values”) (50). Upon significance of the overall model corresponding to a certain experiment, pairwise factor-level combinations of the respective descriptor(s) were tested for significance where applicable. Pairwise tests were corrected for multiple testing by the Benjamini-Hochberg procedure (false discovery rate [FDR]) (51).
Beta diversity analysis.
The extent of change in OTU composition explained by the above-mentioned experimental factors was explored by redundancy analysis (RDA) with Hellinger-transformed OTU abundances (41, 52, 53). In general, the RDA model was formulated as “OTU abundances ∼ F,” with factor F being either “life stage,” “provenance,” or “compartment.” The factors “antibiotic” and “type” were tested for interaction as “OTU abundances ∼ antibiotic × type.”
Models were evaluated with the vegan function rda, followed by anova.cca, using 10,000 random permutations. Homoscedasticity was checked by using the vegan betadisper function, followed by anova.betadisper to assess significance. Upon determination of the significance of the overall model, pairwise factor-level combinations were tested for significance. Corrections for multiple testing across and within models were conducted as described above for alpha diversity analysis. OTUs significantly correlated with any axis in a significant RDA model were determined by using the envfit function with 105 permutations, followed by Benjamini-Hochberg correction. In order to reduce the number of tests in this procedure, OTUs were prefiltered according to their vector lengths calculated from the corresponding RDA scores (scaling, 1) by profile likelihood selection (54). OTUs that were significant at an FDR of 10% were further subjected to indicator analysis with the multipatt function of the R package indicspecies v1.7.1 (55) with 105 permutations. Indicator OTUs, in analogy to indicator species in the sense described in reference 55, are OTUs that prevail in a certain sample type while being found only infrequently and irregularly in other sample types compared to the former. Thus, an indicator OTU can be considered “characteristic” for its sample type.
Phylogenetic tree construction for Mycoplasma associated with A. aurita.
DNA sequences were taxonomically classified via a BLAST search using the 16S rRNA database of the NCBI. Tree reconstruction was performed with full-length 16S rRNA sequences by using Phylogeny.fr (http://www.phylogeny.fr/) (38, 39) with the maximum likelihood method.
Nucleotide sequence accession numbers.
Sequences were submitted to the NCBI GenBank database under accession number KP299289. Sequence data were submitted to the NCBI Sequence Read Archive under accession number SRP049658.
RESULTS
Detection of bacteria accumulated in the mucus of A. aurita polyps by fluorescence in situ hybridization.
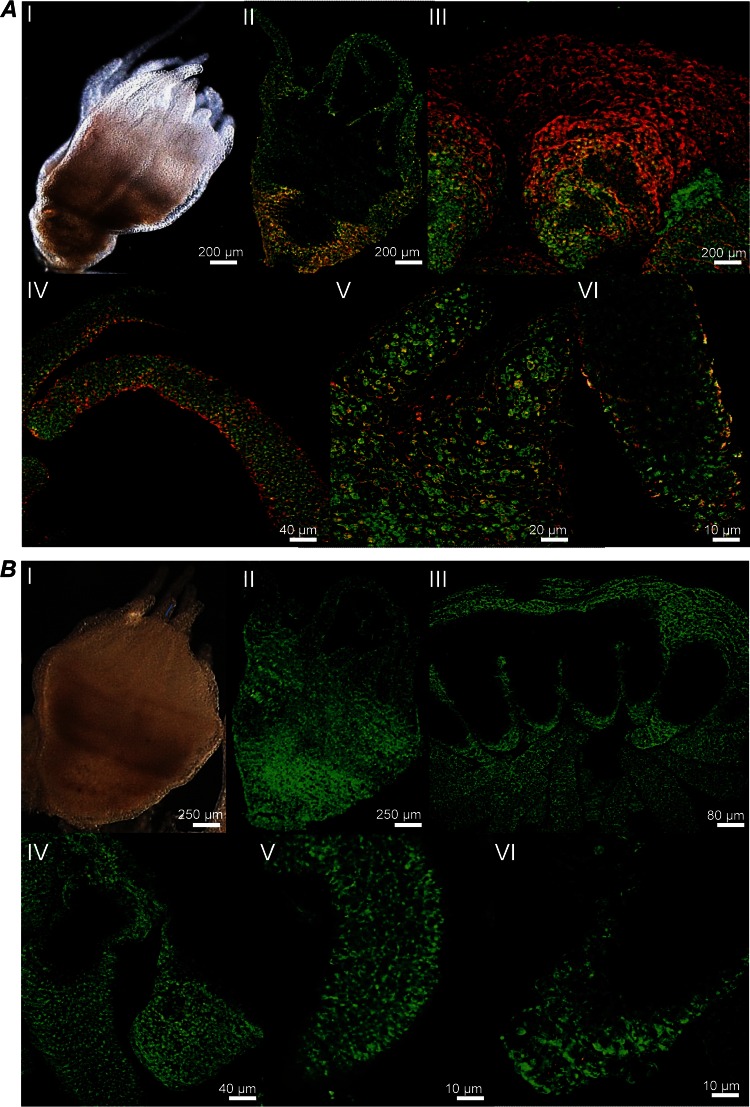
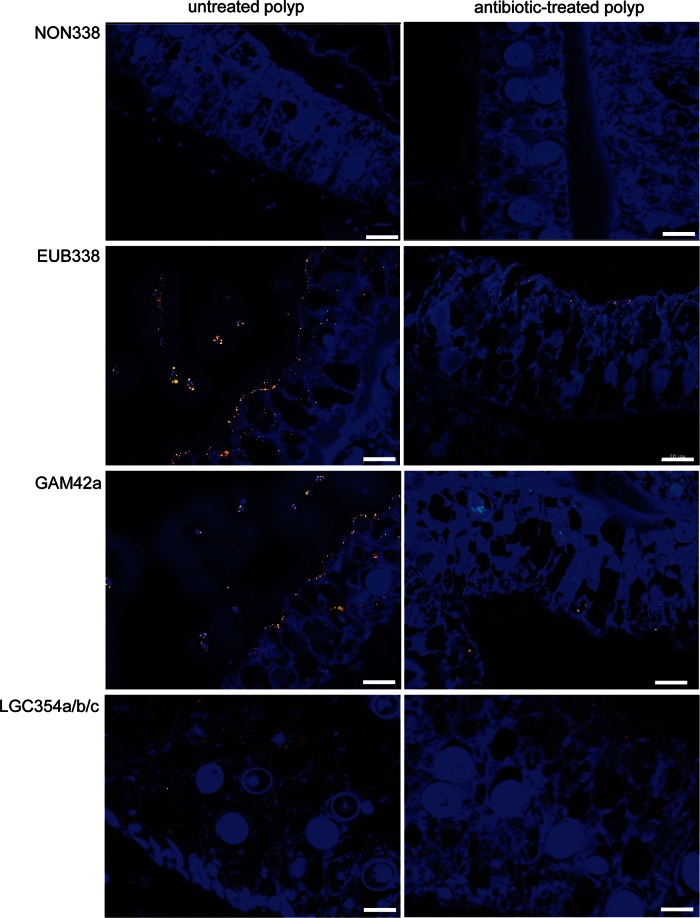
To detect and localize potential associated microbial colonizers, the protocol for fluorescence in situ hybridization (FISH) analysis was successfully modified and established for A. aurita polyps (see Materials and Methods). This is, to our knowledge, the first successful attempt, as extensive autofluorescence of tissues previously limited the application of standard fluorescence in situ hybridization techniques to detect and identify the associated bacterial communities on jellyfish surfaces. The protocol used here overcomes these limitations by combining FISH analysis and spectral imaging, enabling discrimination between autofluorescence and FISH probe-fluorochrome emissions (56). Sterile control polyps were generated from the Roscoff population by using a broad-spectrum antibiotic (AB) mixture (see Materials and Methods). Analysis of untreated and AB-treated whole polyps using the bacterial domain-specific probe EUB338 confirmed our hypothesis that the complete epithelial surface of untreated A. aurita polyps is covered by bacteria, which attach to and accumulate in the mucus (Fig. 1). Calculation of the surface coverage of bacteria using Leica software (see Materials and Methods) resulted in significantly different amounts of colonizing bacteria on polyps with and those without antibiotic treatment. The surface of untreated polyps was up to 45% covered, whereas the surface of AB-treated polyps was covered up to only 4%. Epifluorescence micrographs of semithin sections, as shown in Fig. 2, further demonstrate that bacteria (monitored with probe EUB338-Cy3) are located mainly on the outer surface of the coating mucus. Initial sequence data for cloned 16S rRNA genes of microbes associated with A. aurita polyps from previous Sanger sequencing data demonstrate the presence of highly abundant Gammaproteobacteria and a potentially novel Mycoplasma sp. (phylum Firmicutes) (GenBank accession number KP299289) (data not shown). Consequently, FISH analysis of semithin sections was performed by using the GAM42a probe specific for Gammaproteobacteria (57), confirming that the majority of all detected bacteria in the mucus represent Gammaproteobacteria. Moreover, by using a mixture of Firmicutes-specific probes (mixture of probes LGC354a, -b, and -c [51]) (see Materials and Methods), bacteria were detected inside and between cells of the host tissue, which were still detectable after AB treatment (Fig. 2, bottom). Based on these findings, we hypothesize that A. aurita establishes endosymbiosis with a bacterium, which is most likely the putatively novel Mycoplasma strain identified by the above-described sequencing approach. However, further experiments using probes specific for the novel Mycoplasma sp. strain are required to confirm this hypothesis.
FIG 1.
Bacterial colonization of untreated and antibiotic-treated A. aurita polyps. Confocal laser scanning microscope images show FISH probe hybridization (EUB338 probe) on untreated (A) and antibiotic-treated (B) A. aurita polyps (Roscoff). (I) Light microscope images. (II to V) Fluorescently labeled bacteria are shown in red, and the fluorescence due to SYTO9 in the epithelial tissue is shown in green. (II) Whole polyp; (III) head region with tentacles; (IV) tentacles; (V) zoom view of tentacle; (VI) tentacle tip.
FIG 2.
Bacterial communities associated with A. aurita polyps identified by FISH analysis. Shown are epifluorescence micrographs of bacteria on semithin sections (0.5 μm) of untreated (left) and antibiotic-treated (right) A. aurita polyps (Roscoff). Images are overlays of DAPI signals of polyp cell nuclei (blue) and hybridization signals of the probes NONEUB338 (Cy3) (yellow), EUB338 (Cy3) (yellow), GAM42a (ATTO550) (orange), and LGC345a/b/c (ATTO647N) (red). Bars, 10 μm.
Phylogenetic composition of A. aurita-associated microbiota determined by amplicon sequencing reveals sample type-specific colonization.
The microbiota associated with different life stages, compartments, and populations of A. aurita were analyzed by sequencing of the V1-V2 region of the 16S rRNA gene. DNA isolation, PCR amplification, and sequencing were successful with all but a few samples; thus, all sample types had sufficient replication of 4 to mostly 6 samples. Fig. 3 depicts abundances of OTUs (97% sequence similarity) between individual samples with regard to life stages (panel A), compartments (panel B), populations (panel C), and induced sterile conditions (panel D). While the variability of genus abundances between samples within the same sample type was generally high, some types could be clearly distinguished from others solely by dominant taxa: Alteromonas in the majority of artificial seawater samples, an unclassified Pelagibacteraceae bacterium in Baltic Sea water, Mycoplasma in gastric cavity and mucus samples of medusae, one unclassified bacterium in polyps from the Baltic Sea population, and Phaeobacter in polyps from the Roscoff population (see Table S1 and Fig. S3 in the supplemental material). A more detailed differentiation between sample types was achieved by examining the distributions of OTUs. Tables 2 and 3 summarize the results of the statistical models of alpha diversity (distribution of effective OTU richness associated with a certain sample type) and beta diversity (distribution of individual OTUs across samples), respectively. All models were significant and explained between 19 and 67% of the variance in the data. Table 4 lists the results for the corresponding pairwise factor-level combinations. The main results of these analyses are presented below.
TABLE 2.
Overview of statistical models in alpha diversity analysisa
| Factor | Model | df | Pseudo-R2adj | P value | q value |
|---|---|---|---|---|---|
| Life stage | GLM, gamma; link, log | 4 | 0.19 | 0.010 | 0.010 |
| Compartment | GLM, inverse Gaussian; link, μ−2 | 2 | 0.50 | 0.000 | |
| Provenance (population) | GLM, gamma; link, log | 3 | 0.60 | 0.000 | 0.000 |
| Antibiotic | GLM, gamma; link, log | 1 | 0.27 | 0.000 | 0.001 |
1D was fitted to the respective experimental factor(s) given in by a generalized linear model (GLM). Model indicates the generalized linear model based on the stated distribution (gamma/inverse Gaussian) of the response variable with the stated link function between the stochastic (response variable) and systematic model parts. df, degree of freedom; pseudo-R2adj, measure of model performance (ratio between explained and residual deviances in the generalized linear model), adjusted by using Ezekiel's formula (49) (see Materials and Methods); P value, type I error probability of the respective model; q value, Bonferroni-Holm-corrected P value. Significant results are in boldface type.
TABLE 3.
Overview of statistical models in beta diversity analysisa
| Factor | df | R2adj | Pmhv value | P value | q value |
|---|---|---|---|---|---|
| Life stage | 4 | 0.39 | 0.96 | 0.000 | 0.000 |
| Compartment | 2 | 0.23 | 0.00 | 0.000 | |
| Provenance (population) | 3 | 0.46 | 0.18 | 0.000 | 0.000 |
| Antibiotic × type | 5 | 0.58 | 0.98 | 0.000 | 0.000 |
Hellinger-transformed OTU abundances were fitted to the respective experimental factor(s) by RDA. df, degree of freedom; R2adj, measure of model performance (variance explained by the RDA model, adjusted to sample size and degree of freedom); Pmhv, type I error probability of the test for multivariate homogeneity of variance (homoscedasticity); P value, type I error probability of the respective model; q value, Bonferroni-Holm-corrected P value. Significant results are in boldface type.
TABLE 4.
Results of pairwise tests in alpha and beta diversity analysesa
| Factor | Comparison | Alpha diversity |
Beta diversity |
||||
|---|---|---|---|---|---|---|---|
| Pseudo-R2adj | P value | FDR | R2adj | P value | FDR | ||
| Life stage | Ephyra-juvenile medusa | 0.16 | 0.089 | 0.177 | 0.17 | 0.002 | 0.003 |
| Ephyra-polyp | −0.06 | 0.514 | 0.570 | 0.34 | 0.003 | 0.004 | |
| Ephyra-strobila | −0.06 | 0.570 | 0.570 | 0.20 | 0.002 | 0.003 | |
| Ephyra-ASW | 0.20 | 0.074 | 0.177 | 0.37 | 0.002 | 0.003 | |
| Juvenile medusa-polyp | 0.03 | 0.214 | 0.356 | 0.29 | 0.003 | 0.004 | |
| Juvenile medusa-strobila | 0.14 | 0.085 | 0.177 | 0.12 | 0.024 | 0.024 | |
| Juvenile medusa-ASW | 0.41 | 0.012 | 0.120 | 0.34 | 0.001 | 0.003 | |
| Polyp-strobila | 0.02 | 0.305 | 0.418 | 0.35 | 0.002 | 0.003 | |
| Polyp-ASW | 0.27 | 0.035 | 0.173 | 0.43 | 0.002 | 0.003 | |
| Strobila-ASW | −0.03 | 0.334 | 0.418 | 0.41 | 0.000 | 0.001 | |
| Compartment | Baltic Sea water-gastric cavity | 0.69 | 0.001 | 0.003 | 0.88 | 0.000 | 0.000 |
| Baltic Sea water-mucus | 0.17 | 0.045 | 0.045 | 0.46 | 0.004 | 0.005 | |
| Gastric cavity-mucus | 0.52 | 0.020 | 0.030 | 0.19 | 0.029 | 0.029 | |
| Provenance | Baltic Sea-North Sea | 0.79 | 0.000 | 0.000 | 0.61 | 0.000 | 0.000 |
| Baltic Sea-Roscoff | 0.61 | 0.001 | 0.002 | 0.61 | 0.002 | 0.003 | |
| Baltic Sea-ASW | 0.11 | 0.182 | 0.182 | 0.60 | 0.002 | 0.003 | |
| North Sea-Roscoff | 0.20 | 0.082 | 0.099 | 0.08 | 0.031 | 0.031 | |
| North Sea-ASW | 0.66 | 0.001 | 0.002 | 0.44 | 0.000 | 0.000 | |
| Roscoff-ASW | 0.27 | 0.035 | 0.052 | 0.43 | 0.002 | 0.003 | |
| Antibiotic × type | Ephyra (AB)-ephyra (no AB) | − | − | − | 0.35 | 0.002 | 0.004 |
| Polyp (AB)-polyp (no AB) | − | − | − | 0.46 | 0.000 | 0.000 | |
| ASW (AB)-ASW (no AB) | − | − | − | 0.24 | 0.018 | 0.018 | |
| Ephyra (AB)-polyp (no AB) | − | − | − | 0.45 | 0.000 | 0.000 | |
| Ephyra (AB)-ASW (no AB) | − | − | − | 0.42 | 0.004 | 0.006 | |
| Polyp (AB)-ASW (no AB) | − | − | − | 0.40 | 0.005 | 0.006 | |
Tests were conducted for the pairs of factor levels or factor-level combinations stated in the “Comparison” column, as shown in Tables 1 and 2 for alpha and beta diversity analyses, respectively. FDR, Benjamini-Hochberg-corrected P value (false discovery rate). “−” denotes values that were not determined. Results significant at the 5% level (both P values and FDRs) are in boldface type.
Life stages: significant restructuring of the microbiota during transition from benthic polyp to planktonic life stages.
First, microbial community patterns of different A. aurita life stages (polyp, strobila, ephyra, and juvenile medusa) of the Roscoff population were analyzed. Polyps were kept in artificial seawater as described in Materials and Methods. Strobilation of polyps for the production of ephyrae was induced by lowering the temperature, and a subset of the resulting ephyrae continued to develop into juvenile medusae (see Materials and Methods). Considering the main representatives of associated bacteria, as shown on the genus level in Fig. 3A, the dominating colonizers on polyps were Lacinutrix, Phaeobacter, and Crenothrix; those on strobilating polyps were Crenothrix, Vibrio, and Rhodococcus; those on released ephyrae were Crenothrix, Pseudoalteromonas, Loktanella, and Salinibacterium; and those on juvenile medusae were Lacinutrix, Labrenzia, Loktanella, and Vibrio, whereas ambient water (artificial seawater) harbored mainly Alteromonas and Marivita. The effective numbers of OTUs (OTU richness [1D]) were approximately the same in all examined life stages, as depicted in Fig. 4A (left), with the only notable difference in 1D being that between juvenile medusae and ambient water, which, however, is not significant after correcting for multiple testing (Table 4). In contrast, beta diversity shows a marked difference in community composition between benthic polyps and planktonic life stages (Table 4 and Fig. 4A, right), despite A. aurita jellyfish at all life stages being kept under the same controlled laboratory conditions with the same pool of potential bacterial colonizers in ambient water. Pairwise comparisons show that the group effect (expressed as explained variance [R2adj] in Table 4) between the polyp group and the other planktonic life stages (strobila, ephyra, and juvenile medusa) is comparable to that between these life stages and ambient water. Differences among the three planktonic life stages, while also significant, are less pronounced (Table 4 and Fig. 4A). In conclusion, the different life stages are characterized by a specific microbiota distinct from that of ambient water. Moreover, the stages of the strobilation event—strobila, ephyra, and juvenile medusa—harbor a similar microbiota, which significantly differs from the microbiota of the polyp. In addition, untreated and AB-treated polyps and ephyrae were analyzed, illustrating that the polyp-associated microbiota is strongly affected by AB treatment, in agreement with data from the FISH analysis (Fig. 1, panels II to VI). Figure 4D illustrates a significant trend of higher effective OTU richness in untreated samples (polyps and ephyrae) than in AB-treated samples.
Compartment-specific colonization of medusae.
Amplified bacterial 16S rRNA gene sequences were retrieved from six adult medusae freshly hatched from the Kiel Bight in the Baltic Sea and studied with respect to potential differences in microbial composition between the gastric cavity and exumbrellar mucus. As depicted in Fig. 3B, the microbial composition of ambient water (Baltic Sea) is distinct from the microbiota detected in the gastric cavity and mucus. The main representative species in ambient water is an unclassified Pelagibacteraceae species. The microbial consortia associated with the medusa compartments are both dominated by one unclassified Mycoplasma sequence. This is in agreement with the above-mentioned detection of a Mycoplasma sp. by Sanger sequencing and FISH analysis, most likely indicating the presence of a possible Mycoplasma endosymbiont (Fig. 2). A small number of sequences in the gastric cavity were identified as the alphaproteobacterium Labrenzia, and a small number of sequences in the mucus were identified as the alphaproteobacterium Roseovarius. Alpha diversity analysis showed significant differences in 1D values, both between the two compartments and between both compartments and Baltic Sea water, with mucus revealing a substantially larger variability in OTU richness than the other two samples (Table 4 and Fig. 4B, left). Beta diversity analysis revealed conspicuous compositional differences among the three groups at the OTU level, with the largest difference being between Baltic Sea water and the gastric cavity, followed by that between Baltic Sea water and mucus (Table 4 and Fig. 4B, right). In contrast, bacterial communities of the gastric cavity and mucus differed considerably less from each other. Mucus-associated microbes were more variable in composition than were those from the gastric cavity and Baltic Sea water. In summary, adult medusae from the Kiel Bight harbor specific bacterial consortia that are distinct from those in seawater communities, especially in their gastric cavity.
Population-specific community patterns most likely due the genetic background of the host.
Next, microbial community patterns of A. aurita polyps from the Roscoff, Baltic Sea, and North Sea populations were analyzed. In order to use comparable cultivation conditions, polyps were kept under identical artificial conditions (temperature, salinity, and bacterial colonizer pool in ASW) in the laboratory for >10 years. Figure 3C illustrates that the major microbes are Lacinutrix, Phaeobacter, and Crenothrix for the Roscoff population; unclassified Bacteria, unclassified Coxiellaceae, and unclassified Sphingomonadaceae for the Baltic Sea population; and Lacinutrix, unclassified Sphingomonadaceae, and Crenothrix for the North Sea population. The Baltic Sea A. aurita polyps exhibited a markedly lower alpha diversity (∼3 effective OTUs) than that of their North Sea and Roscoff counterparts (∼20 and ∼11 effective OTUs, respectively), while the latter two populations did not differ measurably in this respect (Table 4 and Fig. 4C, left). This constellation was mirrored in beta diversity, with the strongest group effects being observed with samples from the Baltic Sea population (R2adj Baltic-North Sea = 61%; R2adj Baltic-Roscoff = 61%) (Table 4 and Fig. 4C, right). With the exception of North Sea polyps, the alpha diversity was not significantly higher in polyps than in ASW, whereas the difference in OTU composition between polyps and ASW was significant in all cases (Table 4). While OTU composition also differed significantly between polyps from the North Sea and Roscoff populations, the respective group effect was much lower (R2adj = 8%) (Table 4). The discrepancy between the Baltic Sea population and other polyp populations could be traced back to two indicator OTUs specific to the former: OTU 3 (unclassified Bacteria), showing 96% similarity to an uncultured marine bacterial clone, “B1G5” (GenBank accession number GU317735), from the mesopelagic Northeast Pacific Ocean (58), and OTU 19 (unclassified Coxiellaceae). The latter belongs to the order Legionellales, which is commonly known for its many pathogenic species. Given that the three populations were kept under identical conditions in the same artificial seawater for several years, the observed discrepancies are remarkable. The findings indicate that specific microbiota are most likely due to the genetic background (subpopulation) of the host population, irrespective of the ambient water (and the respective microbial colonizer pool).
In order to characterize the genetic relationship between host populations, we sequenced mitochondrial and nuclear markers (16S and ITS-1/5.8S rRNA) and classified the samples as described previously by Schroth et al. (27). This analysis revealed that the 10 replicate Roscoff polyps belong to the UBI lineage. Baltic Sea and North Sea polyps, on the other hand, were classified as belonging to the BOR lineage (Fig. 5). The latter clustered into two different clades (populations) within the BOR lineage. Based on these data, we conclude that the three different polyp laboratory stocks represent three different subpopulations of A. aurita, all harboring a distinct microbiota according to their genetic background and independent of the ambient water.
FIG 5.
Phylogenetic classification of A. aurita polyp laboratory stocks. DNA was extracted from 10 single polyps of each respective Roscoff, Baltic Sea, and North Sea population, kept under the same artificial conditions. Aurelia-specific mitochondrial 16S rRNA (left) and ITS-1/5.8S rRNA (right) fragments were amplified by PCR and sequenced. Phylogenetic reconstruction was performed by using Phylogeny.fr (http://www.phylogeny.fr/) (38, 39) with the maximum likelihood method. Previously reported sequences from Aurelia populations deposited in GenBank (accession numbers AF461398 and AF461399 for 16S rRNA and AF461405 and AF461406 for ITS-1/5.8S rRNA) were used for classification of polyps into known subpopulations. Phenotypic differences between populations are visualized in the middle.
DISCUSSION
Our results clearly demonstrate that A. aurita is colonized by specific bacterial communities that significantly differ in diversity and composition from those of ambient water. Below, we discuss the main conclusions, association of microbiota, restructuring of microbiota during the course of transformation from polyp to planktonic life stages, and host population-specific colonization.
Association of microbes with A. aurita tissues.
FISH analysis confirms that the whole epithelial surface of A. aurita polyps is covered by bacteria (Fig. 1, panel II). As reported previously for other animals, most interactions between the potential colonizers and A. aurita appear to occur at the mucosal surface, forming a barrier against ambient seawater (59–61). The majority of bacteria identified on polyps by FISH analysis are Gammaproteobacteria, which is one of the dominant bacterial classes associated with different surfaces of multicellular eukaryotic organisms in the ocean (62–65) and appears to outcompete other bacterial classes due its effective attachment and colonization abilities. Moreover, data from the FISH analysis reported here and previous 16S rRNA gene analyses (GenBank accession number KP299289) (R. Kraemer and R. A. Schmitz, unpublished data) argue for the presence of a potential endosymbiont in A. aurita tissues, possibly belonging to the genus Mycoplasma (class Mollicutes). The identified Mycoplasma sequences obtained from both medusa compartments show 84% similarity to an uncultured bacterial clone, “CFI73” (GenBank accession number AB703186), derived from the digestive tract of Octopus mimus (66), which is closely related to members of the orders Entomoplasmatales and Spiroplasmatales (see Fig. S2 in the supplemental material). The A. aurita-associated Mycoplasma species may thus represent a hitherto-unrecognized novel Mycoplasma species, most likely an endosymbiont. Surprisingly, the Mycoplasma species was not detected in the 454 sequencing libraries from polyps, which suggests that this particular animal batch may have lost the Mollicutes, e.g., due to undetected changes in husbandry conditions, or that they were in very low abundance in the 454 library. These explanations are further supported by very recent analyses using the Illumina MiSeq platform, which clearly detected the presence of this Mycoplasma species at all life stages of A. aurita (data not shown).
In general, the class Mollicutes represents a unique category of bacteria that have in common a small cell size, the absence of a cell wall, a reduced genome, and simplified metabolic pathways. Mollicutes are widespread commensals or pathogens of humans, mammals, reptiles, fish, plants, and other arthropods (reviewed in reference 67) and have also been reported to be associated with different invertebrates, e.g., earthworms (12), isopods (11), oysters (68, 69), crustaceans (10), bryozoans (70), ctenophores (63, 71), and also cnidarians (72–74). Parasitic, commensal, and mutualistic endosymbioses are widely distributed among invertebrate taxa and have arguably played a major role in the evolution of several invertebrate families, classes, and phyla (75). Sometimes accounting for as much as 50% of the invertebrate volume or biomass, endosymbionts can profoundly affect the ecology, physiology, development, and behavior of invertebrate hosts (76). Our modified FISH protocol now allows precise localization of A. aurita-associated bacteria and potential endosymbionts. Thus, in future studies, the proposed endosymbiont Mycoplasma sp. will be further analyzed with genus-specific 16S rRNA probes to identify its cellular localization. Furthermore, genome analysis employing, e.g., a single-cell sequencing approach will allow prediction of its potential functions.
Microbial community patterns change according to developmental stages. (i) Microbial colonization of A. aurita kept under artificial conditions at different life stages.
Current knowledge of the early steps of bacterial colonization of Scyphozoa larvae, the establishment of its microbiota, and stability at different life stages, especially after strobilation, is limited. Thus, the finding that community patterns of different developmental stages of A. aurita differ raises the question of the functional role of associated microbiota in general and concerning development. The fact that A. aurita jellyfish at stages of strobilation harbor similar microbiota but differ significantly in their community patterns compared to those of polyps argues that the A. aurita-associated microbiota undergoes significant restructuring during the transition from the benthic and immortal to the planktonic mortal life phases of their host. Moreover, polyps also show the highest number of indicator OTUs compared to those at other life stages. This further supports the idea that polyps harbor a specific bacterial community of high diversity, potentially essential for their sessile lifestyle (e.g., providing vitamins, amino acids, or secondary metabolites) and possibly important for the initiation of later developmental stages (e.g., strobilation initiation) but less important after the transition to planktonic stages. Thus, restructuring of microbial communities during strobilation may represent a life cycle strategy of A. aurita, where certain bacterial communities include members with specific (metabolic) functions important for specific developmental stages of the host. On the other hand, it is also possible that animal development shapes a new microbiota due to changes in the surface architecture or the production of host compounds (e.g., differential expression patterns of antimicrobial peptides during different life stages). In the long term, detailed knowledge on the composition of bacteria associated with different life stages of A. aurita might provide insight into a microbiota-dependent life history and its evolutionary consequences. Moreover, the role of the associated microbiota might also be important to understand and control reproductive mechanisms to restrict jellyfish blooms. In this respect, it was recently demonstrated that a water-soluble membrane-diffusible molecule (an indole derivative) is significantly involved in the induction of A. aurita proliferation (77). Since it is currently not clear whether the host exclusively produces the effector molecule, it is attractive to speculate that similar or antagonistic molecules might be synthesized by the associated microbiota. Identification of such potential strobilation antagonists might facilitate the control of jellyfish blooms.
In regard to proposed functions of the microbiota during a life cycle, it was previously reported that bacteria are important for the larval settlement processes of most marine invertebrates (78), such as sponges (79), cnidarians (80), ascidians (81), and bryozoans (82). In agreement with these findings, no settlement of planulae of the scyphozoans A. aurita (80) and Cassiopea andromeda (83) or the hydrozoan Hydractinia echinata (84) occurred in the absence of microbes. However, the presence of bacteria induced the settlement and metamorphosis of each of these animals (85).
(ii) Microbial colonization of compartments of medusae caught in the Baltic Sea.
Living medusae were caught in the Baltic Sea to analyze the community patterns of two compartments, the exumbrella mucus and gastric cavity. Significant differences among the two studied compartments and Baltic Sea water were identified. Furthermore, the composition of the mucus-associated microbiota was more variable than those of the gastric cavity and Baltic Sea water. Overall, adult medusae from the Kiel Bight apparently harbor bacterial consortia that are distinct from seawater communities, especially in their gastric cavity. Bacterial consortia associated with the animal's outer surface are most likely more influenced by the surrounding microbial community of ambient water and external disturbance. This is likely the reason for the high similarity of the exumbrella mucus samples to the seawater consortia and their high compositional variability compared to that of gastric cavity-associated bacteria.
Provenance matters: population-specific community patterns.
In this study, polyps of the Roscoff, North Sea, and Baltic Sea populations originating from geographically distinct regions were kept under the same controlled artificial conditions in the laboratory for >10 years (common garden experiment). Phylogenetic analyses revealed a specific microbiota for each population distinct from and irrespective of that of the ambient water (Fig. 3C and 4C), arguing for a significant influence of genetic background. The molecular phylogeny of A. aurita has been studied extensively, based on nuclear internal transcribed spacer 1 (ITS-1) and mitochondrial cytochrome c subunit I (COI), which led to taxonomic revisions. For example, Schroth et al. (27) suggested historical speciation events and reconstructed the taxonomic classification of the genus Aurelia to at least seven different species. Dawson and colleagues (21, 86) proposed a taxonomic system combining macromorphology and ITS-1, in addition to partial COI gene sequences. Altogether, the global phylogeny of the genus Aurelia reveals at least 16 phylogenetic branches, including seven populations of A. aurita, and most of them appear to be regionally restricted.
The largest differences in microbial composition among the polyp populations studied in this report were found between the North Sea/Roscoff and Baltic Sea populations. Phylogenetic classification based on mitochondrial and nuclear DNA data revealed three different subpopulations corresponding to the geographic occurrence of natural A. aurita populations. North Sea and Baltic Sea polyps were more similar and thus clustered. This cluster (BOR lineage) deeply branched from the Roscoff polyps (UBI lineage) (Fig. 5). Of note, the differential subpopulations do not necessarily match the phenotypic grouping of the populations. The two populations of the North Sea and Baltic Sea are genetically unified by one lineage (BOR), but the phenotypic differences are significant (Fig. 5). Baltic Sea specimens have much larger polyps. North Sea and Roscoff specimens are substantially smaller than those from the Baltic Sea and phenotypically more similar, although they differ genetically. Overall, our findings indicate that a correlation between the composition of the microbiota, genetic background, and phenotype of the host should be taken into account. In nature, both host genetics as well as environmental factors are likely important in controlling the acquisition and stability of a healthy microbiota. Indeed, several animal-bacterium studies show that the three components environment, host genetics, and microbiome are important to maintain homeostasis in the host (87–89).
In conclusion, it appears that A. aurita-associated microbial communities are indeed unique and sample type specific (compartment, life stage, and population). Taking the metaorganism concept into account, we hypothesize that these specific microbiota serve numerous important functions, including contributing to the development of different life stages.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the Federal Ministry of Education and Research (BMBF) (ChemBiofilm Cluster of the GenoMik-Transfer network).
Polyp populations were provided by the work group of T. C. G. Bosch (Institute of Zoology, Christian Albrecht University Kiel). We thank the staff of the Centre of Microscopy at Christian Albrecht University Kiel for their assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01601-15.
REFERENCES
- 1.Bosch TC, McFall-Ngai MJ. 2011. Metaorganisms as the new frontier. Zoology (Jena) 114:185–190. doi: 10.1016/j.zool.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Loso T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A 110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zilber-Rosenberg I, Rosenberg E. 2008. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32:723–735. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
- 4.Dinasquet J, Granhag L, Riemann L. 2012. Stimulated bacterioplankton growth and selection for certain bacterial taxa in the vicinity of the ctenophore Mnemiopsis leidyi. Front Microbiol 3:302. doi: 10.3389/fmicb.2012.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Matos AE, Rosado W, Govind NS. 2007. Bacterial diversity associated with the Caribbean tunicate Ecteinascidia turbinata. Antonie Van Leeuwenhoek 92:155–164. doi: 10.1007/s10482-007-9143-9. [DOI] [PubMed] [Google Scholar]
- 6.Piskorska M, Smith G, Weil E. 2007. Bacteria associated with the coral Echinopora lamellosa (Esper 1795) in the Indian Ocean-Zanzibar region. Afr J Environ Sci Technol 1:93–98. [Google Scholar]
- 7.Koch EJ, Miyashiro T, McFall-Ngai MJ, Ruby EG. 2014. Features governing symbiont persistence in the squid-vibrio association. Mol Ecol 23:1624–1634. doi: 10.1111/mec.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McFall-Ngai MJ. 2014. The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Annu Rev Microbiol 68:177–194. doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cary SC, Cottrell MT, Stein JL, Camacho F, Desbruyeres D. 1997. Molecular identification and localization of filamentous symbiotic bacteria associated with the hydrothermal vent annelid Alvinella pompejana. Appl Environ Microbiol 63:1124–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi K, Huang H, Gu W, Wang J, Wang W. 2008. Phylogenetic analysis of spiroplasmas from three freshwater crustaceans (Eriocheir sinensis, Procambarus clarkia and Panaeus vannamei) in China. J Invertebr Pathol 99:57–65. doi: 10.1016/j.jip.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Fraune S, Zimmer M. 2008. Host-specificity of environmentally transmitted Mycoplasma-like isopod symbionts. Environ Microbiol 10:2497–2504. doi: 10.1111/j.1462-2920.2008.01672.x. [DOI] [PubMed] [Google Scholar]
- 12.Nechitaylo TY, Timmis KN, Golyshin PN. 2009. ‘Candidatus Lumbricincola’, a novel lineage of uncultured Mollicutes from earthworms of family Lumbricidae. Environ Microbiol 11:1016–1026. doi: 10.1111/j.1462-2920.2008.01837.x. [DOI] [PubMed] [Google Scholar]
- 13.Brodeur RD, Sugisaki H, Hunt GL Jr. 2002 Increases in jellyfish biomass in the Bering Sea: implications for the ecosystem. Mar Ecol Prog Ser 233:89–103. doi: 10.3354/meps233089. [DOI] [Google Scholar]
- 14.Purcell JE. 2005. Climate effects on formation of jellyfish and ctenophore blooms: a review. J Mar Biol Assoc UK 85:461–476. doi: 10.1017/S0025315405011409. [DOI] [Google Scholar]
- 15.Sommer U, Lengfellner K. 2008. Climate change and the timing, magnitude, and composition of the phytoplankton spring bloom. Glob Chang Biol 14:1199–1208. doi: 10.1111/j.1365-2486.2008.01571.x. [DOI] [Google Scholar]
- 16.Nemazie DA, Purcell JE, Glibert PM. 1993. Ammonium excretion by gelationous zooplankton and their contribution to the ammonium requirements of microplankton in Chesapeake Bay. Mar Biol 116:451–458. doi: 10.1007/BF00350062. [DOI] [Google Scholar]
- 17.Schneider G, Behrends G. 1998. Top-down control in a neritic plankton system by Aurelia aurita medusae—a summary. Ophelia 48:71–82. doi: 10.1080/00785236.1998.10428677. [DOI] [Google Scholar]
- 18.Blanchet M, Pringault O, Bouvy M, Catala P, Oriol L, Caparros J, Ortega-Retuerta E, Intertaglia L, West N, Agis M. 20 November 2014. Changes in bacterial community metabolism and composition during the degradation of dissolved organic matter from the jellyfish Aurelia aurita in a Mediterranean coastal lagoon. Environ Sci Pollut Res Int doi: 10.1007/s11356-014-3848-x. [DOI] [PubMed] [Google Scholar]
- 19.Tinta T, Kogovsek T, Malej A, Turk V, Kirchman DL. 2012. Jellyfish modulate bacterial dynamic and community structure. PLoS One 7:e39274. doi: 10.1371/journal.pone.0039274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tinta T, Malej A, Kos M, Turk V. 2010. Degradation of the Adriatic medusa Aurelia sp. by ambient bacteria. Hydrobiologia 645:179–191. doi: 10.1007/s10750-010-0223-x. [DOI] [Google Scholar]
- 21.Dawson MN, Sen Gupta A, England MH. 2005. Coupled biophysical global ocean model and molecular genetic analyses identify multiple introductions of cryptogenic species. Proc Natl Acad Sci U S A 102:11968–11973. doi: 10.1073/pnas.0503811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dawson MN, Jacobs DK. 2001. Molecular evidence for cryptic species of Aurelia aurita (Cnidaria, Scyphozoa). Biol Bull 200:92–96. doi: 10.2307/1543089. [DOI] [PubMed] [Google Scholar]
- 23.Gröndahl F. 1989. Evidence of gregarious settlement of planula larvae of the scyphozoan Aurelia aurita: an experimental study. Mar Ecol Prog Ser 56:119–125. doi: 10.3354/meps056119. [DOI] [Google Scholar]
- 24.Mutlu E. 2001. Distribution and abundance of moon jellyfish (Aurelia aurita) and its zooplankton food in the Black Sea. Mar Biol 138:329–339. doi: 10.1007/s002270000459. [DOI] [Google Scholar]
- 25.Lucas CH. 2001. Reproduction and life history strategies of the common jellyfish, Aurelia aurita, in relation to its ambient environment. Hydrobiologia 451:229–246. doi: 10.1023/A:1011836326717. [DOI] [Google Scholar]
- 26.Greenberg N, Garthwaite RL, Potts DC. 1996. Allozyme and morphological evidence for a newly introduced species of Aurelia in San Francisco Bay, California. Mar Biol 125:401–410. doi: 10.1007/BF00346320. [DOI] [Google Scholar]
- 27.Schroth W, Jarms G, Streit B, Schierwater B. 2002. Speciation and phylogeography in the cosmopolitan marine moon jelly, Aurelia sp. BMC Evol Biol 2:1. doi: 10.1186/1471-2148-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe T, Ishii H. 2001. In situ estimation of ephyrae liberated from polyps of Aurelia aurita using settling plates in Tokyo Bay, Japan. Hydrobiologia 451:247–258. doi: 10.1023/A:1011856929443. [DOI] [Google Scholar]
- 29.Berking S, Czech N, Gerharz M, Herrmann K, Hoffmann U, Raifer H, Sekul G, Siefker B, Sommerei A, Vedder F. 2005. A newly discovered oxidant defence system and its involvement in the development of Aurelia aurita (Scyphozoa, Cnidaria): reactive oxygen species and elemental iodine control medusa formation. Int J Dev Biol 49:969–976. doi: 10.1387/ijdb.052024sb. [DOI] [PubMed] [Google Scholar]
- 30.Hansson LJ. 1997. Effect of temperature on growth rate of Aurelia aurita (Cnidaria, Scyphozoa) from Gullmarsfjorden, Sweden. Mar Ecol Prog Ser 161:145–153. doi: 10.3354/meps161145. [DOI] [Google Scholar]
- 31.Lucas CH. 1994. Biochemical composition of Aurelia aurita in relation to age and sexual maturity. J Exp Mar Biol Ecol 183:179–192. doi: 10.1016/0022-0981(94)90086-8. [DOI] [Google Scholar]
- 32.Berking S, Herrmann K. 2007. Compartments in Scyphozoa. Int J Dev Biol 51:221–228. doi: 10.1387/ijdb.062215sb. [DOI] [PubMed] [Google Scholar]
- 33.Ishii H, Takagi AI. 2003. Development time of planula larvae on the oral arms of the scyphomedusa Aurelia aurita. J Plankton Res 25:1447–1450. doi: 10.1093/plankt/fbg094. [DOI] [Google Scholar]
- 34.Yuan D, Nakanishi N, Jacobs DK, Hartenstein V. 2008. Embryonic development and metamorphosis of the scyphozoan Aurelia. Dev Genes Evol 218:525–539. doi: 10.1007/s00427-008-0254-8. [DOI] [PubMed] [Google Scholar]
- 35.Metzker ML. 2010. Sequencing technologies—the next generation. Nat Rev Genet 11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 36.Lee OO, Wang Y, Yang J, Lafi FF, Al-Suwailem A, Qian PY. 2011. Pyrosequencing reveals highly diverse and species-specific microbial communities in sponges from the Red Sea. ISME J 5:650–664. doi: 10.1038/ismej.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Webster NS, Taylor MW, Behnam F, Lucker S, Rattei T, Whalan S, Horn M, Wagner M. 2010. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ Microbiol 12:2070–2082. doi: 10.1111/j.1462-2920.2009.02065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dereeper A, Audic S, Claverie J-M, Blanc G. 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol 10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langfeldt D, Neulinger SC, Heuer W, Staufenbiel I, Kunzel S, Baines JF, Eberhard J, Schmitz RA. 2014. Composition of microbial oral biofilms during maturation in young healthy adults. PLoS One 9:e87449. doi: 10.1371/journal.pone.0087449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Werner JJ, Koren O, Hugenholtz P, DeSantis TZ, Walters WA, Caporaso JG, Angenent LT, Knight R, Ley RE. 2012. Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J 6:94–103. doi: 10.1038/ismej.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.R Core Team. 2014. R package v3.1.1. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 45.Jost L. 2006. Entropy and diversity. Oikos 113:363–375. doi: 10.1111/j.2006.0030-1299.14714.x. [DOI] [Google Scholar]
- 46.Jost L. 2007. Partitioning diversity into independent alpha and beta components. Ecology 88:2427–2439. doi: 10.1890/06-1736.1. [DOI] [PubMed] [Google Scholar]
- 47.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O′Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2013. Package ‘vegan’. R Package version 254, p 20–28. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 48.Zuur AF, Ieno EN, Walker NJ, Saveliev AA, Smith GM. 2009. Mixed effects models and extensions in ecology with R. Springer, New York, NY. [Google Scholar]
- 49.Ezekiel M. 1930. Methods of correlational analysis. Wiley, New York, NY. [Google Scholar]
- 50.Holm S. 1979. A simple sequentially rejective multiple test procedure. Scand J Stat 6:65–70. [Google Scholar]
- 51.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. [Google Scholar]
- 52.Stratil SB, Neulinger SC, Knecht H, Friedrichs AK, Wahl M. 2014. Salinity affects compositional traits of epibacterial communities on the brown macroalga Fucus vesiculosus. FEMS Microbiol Ecol 88:272–279. doi: 10.1111/1574-6941.12292. [DOI] [PubMed] [Google Scholar]
- 53.Stratil SB, Neulinger SC, Knecht H, Friedrichs AK, Wahl M. 2013. Temperature-driven shifts in the epibiotic bacterial community composition of the brown macroalga Fucus vesiculosus. Microbiologyopen 2:338–349. doi: 10.1002/mbo3.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu M, Ghodsi A. 2006. Automatic dimensionality selection from the scree plot via the use of profile likelihood. Comput Stat Data Anal 51:918–930. doi: 10.1016/j.csda.2005.09.010. [DOI] [Google Scholar]
- 55.De Cáceres M, Legendre P. 2009. Associations between species and groups of sites: indices and statistical inference. Ecology 90:3566–3574. doi: 10.1890/08-1823.1. [DOI] [PubMed] [Google Scholar]
- 56.Ainsworth TD, Fine M, Blackall LL, Hoegh-Guldberg O. 2006. Fluorescence in situ hybridization and spectral imaging of coral-associated bacterial communities. Appl Environ Microbiol 72:3016–3020. doi: 10.1128/AEM.72.4.3016-3020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagner M, Amann R, Lemmer H, Schleifer KH. 1993. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol 59:1520–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao W, Shi X, Wu J, Jin Y, Zhang W, Meldrum DR. 2011. Phylogenetic and gene expression analysis of cyanobacteria and diatoms in the twilight waters of the temperate northeast Pacific Ocean. Microb Ecol 62:765–775. doi: 10.1007/s00248-011-9891-y. [DOI] [PubMed] [Google Scholar]
- 59.Daniels CA, Zeifman A, Heym K, Ritchie KB, Watson CA, Berzins I, Breitbart M. 2011. Spatial heterogeneity of bacterial communities in the mucus of Montastraea annularis. Mar Ecol Prog Ser 426:29–40. doi: 10.3354/meps09024. [DOI] [Google Scholar]
- 60.Mullen KM, Peters EC, Harvell CD. 2004. Coral resistance to disease, p 377–399. In Rosenberg E, Loya Y (ed), Coral health and disease. Springer, New York, NY. [Google Scholar]
- 61.Ritchie KB. 2006. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser 322:1–14. doi: 10.3354/meps322001. [DOI] [Google Scholar]
- 62.Franzenburg S, Fraune S, Künzel S, Baines JF, Domazet-Loso T, Bosch TCG. 2012. MyD88-deficient Hydra reveal an ancient function of TLR signaling in sensing bacterial colonizers. Proc Natl Acad Sci U S A 109:19374–19379. doi: 10.1073/pnas.1213110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hao W, Gerdts G, Peplies J, Wichels A. 2015. Bacterial communities associated with four ctenophore genera from the German Bight (North Sea). FEMS Microbiol Ecol 91:1–11. doi: 10.1093/femsec/fiu006. [DOI] [PubMed] [Google Scholar]
- 64.Schmitt S, Tsai P, Bell J, Fromont J, Ilan M, Lindquist N, Perez T, Rodrigo A, Schupp PJ, Vacelet J, Webster N, Hentschel U, Taylor MW. 2012. Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges. ISME J 6:564–576. doi: 10.1038/ismej.2011.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sfanos K, Harmody D, Dang P, Ledger A, Pomponi S, McCarthy P, Lopez J. 2005. A molecular systematic survey of cultured microbial associates of deep-water marine invertebrates. Syst Appl Microbiol 28:242–264. doi: 10.1016/j.syapm.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 66.Iehata S, Fernando V, Esteban M, Carlos R. 2012. Phylogenetic analysis of the bacterial community associated with digestive tract of wild Chilean octopus (Octopus mimus Gould, 1852). University of Antofagasta, Bioinnovation Center, Faculty of Marine Resources, Antofagasta, Chile. [Google Scholar]
- 67.Razin S. 1985. Molecular biology and genetics of mycoplasmas (Mollicutes). Microbiol Rev 49:419–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harshbarger JC, Chang SC, Otto SV. 1977. Chlamydiae (with phages), mycoplasmas, and rickettsiae in Chesapeake Bay bivalves. Science 196:666–668. doi: 10.1126/science.193184. [DOI] [PubMed] [Google Scholar]
- 69.McCoy RE. 1984. Mycoplasma-like organisms of plants and invertebrates, p 792–793. In Bergey DH, Krieg NR, Holt JG (ed), Bergey's manual of systematic bacteriology, vol 1 Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 70.Zimmer RL, Woollacott RM. 1983. Mycoplasma-like organisms: occurrence with the larvae and adults of a marine bryozoan. Science 220:208–210. doi: 10.1126/science.220.4593.208. [DOI] [PubMed] [Google Scholar]
- 71.Daniels C, Breitbart M. 2012. Bacterial communities associated with the ctenophores Mnemiopsis leidyi and Beroe ovata. FEMS Microbiol Ecol 82:90–101. doi: 10.1111/j.1574-6941.2012.01409.x. [DOI] [PubMed] [Google Scholar]
- 72.Gray MW, Burger G, Lang BF. 1999. Mitochondrial evolution. Science 283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 73.Kellogg CA, Lisle JT, Galkiewicz JP. 2009. Culture-independent characterization of bacterial communities associated with the cold-water coral Lophelia pertusa in the northeastern Gulf of Mexico. Appl Environ Microbiol 75:2294–2303. doi: 10.1128/AEM.02357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Neulinger SC, Gärtner A, Järnegren J, Ludvigsen M, Lochte K, Dullo W-C. 2009. Tissue-associated “Candidatus Mycoplasma corallicola” and filamentous bacteria on the cold-water coral Lophelia pertusa (Scleractinia). Appl Environ Microbiol 75:1437–1444. doi: 10.1128/AEM.01781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wernegreen JJ. 2012. Endosymbiosis. Curr Biol 22:R555–R561. doi: 10.1016/j.cub.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 76.Saffo MB. 1992. Invertebrates in endosymbiotic associations. Am Zool 32:557–565. [Google Scholar]
- 77.Fuchs B, Wang W, Graspeuntner S, Li Y, Insua S, Herbst E-M, Dirksen P, Böhm A-M, Hemmrich G, Sommer F. 2014. Regulation of polyp-to-jellyfish transition in Aurelia aurita. Curr Biol 24:263–273. doi: 10.1016/j.cub.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 78.Wieczorek SK, Murray AW, Todd CD. 1996. Seasonal variation in the effects of hard substratum biofilming on settlement of marine invertebrate larvae. Biofouling 10:309–330. doi: 10.1080/08927019609386289. [DOI] [PubMed] [Google Scholar]
- 79.Woollacott RM, Hadfield MG. 1996. Induction of metamorphosis in larvae of a sponge. Invertebr Biol 115:257–262. [Google Scholar]
- 80.Schmahl G. 1985. Induction of stolon settlement in the scyphopolyps of Aurelia aurita (Cnidaria, Scyphozoa, Semaeostomeae) by glycolipids of marine bacteria. Helgoländer Meeresuntersuchungen 39:117–127. [Google Scholar]
- 81.Schuett C, Doepke H, Groepler W, Wichels A. 2005. Diversity of intratunical bacteria in the tunic matrix of the colonial ascidian Diplosoma migrans. Helgoland Mar Res 59:136–140. doi: 10.1007/s10152-004-0212-4. [DOI] [Google Scholar]
- 82.Kittelmann S, Harder T. 2005. Species- and site-specific bacterial communities associated with four encrusting bryozoans from the North Sea, Germany. J Exp Mar Biol Ecol 327:201–209. doi: 10.1016/j.jembe.2005.06.020. [DOI] [Google Scholar]
- 83.Hofmann DK, Fitt WK, Fleck J. 1996. Checkpoints in the life-cycle of Cassiopea spp.: control of metagenesis and metamorphosis in a tropical jellyfish. Int J Dev Biol 40:331–338. [PubMed] [Google Scholar]
- 84.Leitz T, Wagner T. 1993. The marine bacterium Alteromonas espejiana induces metamorphosis of the hydroid Hydractinia echinata. Mar Biol 115:173–178. doi: 10.1007/BF00346332. [DOI] [Google Scholar]
- 85.Müller WA, Leitz T. 2002. Metamorphosis in the Cnidaria. Can J Zool 80:1755–1771. doi: 10.1139/z02-130. [DOI] [Google Scholar]
- 86.Dawson MN, Martin LE. 2001. Geographic variation and ecological adaptation in Aurelia (Scyphozoa, Semaeostomeae): some implications from molecular phylogenetics. Hydrobiologia 451:259–273. doi: 10.1023/A:1011869215330. [DOI] [Google Scholar]
- 87.Campbell JH, Foster CM, Vishnivetskaya T, Campbell AG, Yang ZK, Wymore A, Palumbo AV, Chesler EJ, Podar M. 2012. Host genetic and environmental effects on mouse intestinal microbiota. ISME J 6:2033–2044. doi: 10.1038/ismej.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koch H, Schmid-Hempel P. 2012. Gut microbiota instead of host genotype drive the specificity in the interaction of a natural host-parasite system. Ecol Lett 15:1095–1103. doi: 10.1111/j.1461-0248.2012.01831.x. [DOI] [PubMed] [Google Scholar]
- 89.Kostic AD, Howitt MR, Garrett WS. 2013. Exploring host-microbiota interactions in animal models and humans. Genes Dev 27:701–718. doi: 10.1101/gad.212522.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.