Abstract
While rain and irrigation events have been associated with an increased prevalence of foodborne pathogens in produce production environments, quantitative data are needed to determine the effects of various spatial and temporal factors on the risk of produce contamination following these events. This study was performed to quantify these effects and to determine the impact of rain and irrigation events on the detection frequency and diversity of Listeria species (including L. monocytogenes) and L. monocytogenes in produce fields. Two spinach fields, with high and low predicted risks of L. monocytogenes isolation, were sampled 24, 48, 72, and 144 to 192 h following irrigation and rain events. Predicted risk was a function of the field's proximity to water and roads. Factors were evaluated for their association with Listeria species and L. monocytogenes isolation by using generalized linear mixed models (GLMMs). In total, 1,492 (1,092 soil, 334 leaf, 14 fecal, and 52 water) samples were collected. According to the GLMM, the likelihood of Listeria species and L. monocytogenes isolation from soil samples was highest during the 24 h immediately following an event (odds ratios [ORs] of 7.7 and 25, respectively). Additionally, Listeria species and L. monocytogenes isolates associated with irrigation events showed significantly lower sigB allele type diversity than did isolates associated with precipitation events (P = <0.001), suggesting that irrigation water may be a point source of L. monocytogenes contamination. Small changes in management practices (e.g., not irrigating fields before harvest) may therefore reduce the risk of L. monocytogenes contamination of fresh produce.
INTRODUCTION
Foodborne outbreaks have been increasingly linked to fresh produce in the United States (1–6). In fact, the proportion of foodborne outbreaks that were attributed to produce between 1998 and 2008, 46%, was over twice that attributed to meat, 22% (6). Similarly, between 2002 and 2011, produce-associated outbreaks caused, on average, more illnesses per outbreak than any other food (1). As a result, the safety of fresh produce has come into question, negatively affecting produce growers, the food industry, and local economies (7, 8). For example, as a consequence of a 2011 listeriosis outbreak linked to fresh cantaloupes in the United States (9), cantaloupe consumption dropped nationwide by 53% (10). The instability of the cantaloupe market following the 2011 outbreak is indicative of a larger trend of wide-scale consumer avoidance of products associated with outbreaks, even when the outbreak is associated with point source events (7, 8). Thus, prevention of produce-associated outbreaks is a key concern of the produce industry. Although most listeriosis outbreaks associated with fresh produce are traced back to processing environments, the prevention of produce contamination in production environments is crucial. In fact, previous studies have shown that low-level sporadic contamination of produce in production environments can result in pathogen proliferation and widespread contamination throughout the supply chain (11–13). In order to minimize preharvest produce contamination, it is necessary to understand how different spatial (e.g., proximity to water and roads) and temporal (e.g., time since irrigation) factors affect the likelihood of a contamination event in production environments.
Numerous studies have examined the relationship between environmental factors and the prevalence of Listeria monocytogenes (14–18) and L. monocytogenes surrogates (e.g., Listeria spp.) (19, 20) in produce production environments. Many of these studies (14–16) determined that water-related factors were significantly associated with the isolation of L. monocytogenes from environmental samples. Similar studies conducted in nonagricultural environments also found similar results (16, 21, 22). For example, Ivanek et al. (21) found that the isolation of Listeria spp. from samples collected in forested environments was positively associated with rainfall. Additionally, Strawn et al. (15) developed a geospatial algorithm, which included several water-related factors (e.g., available water storage [AWS] and proximity to water), to predict L. monocytogenes prevalence in New York State preharvest environments. The findings of Strawn et al. (15) also suggest that not all fields are at equal risk of pathogen contamination. Therefore, to identify and develop effective mitigation strategies to reduce the risk of on-farm produce contamination, it is essential to understand how contamination risk differs within and between fields due to variation in spatial (e.g., proximity to water) and temporal (e.g., time since a rain event) factors.
Previous studies have also found that management practices affect the risk of contamination by L. monocytogenes (14, 23–28) and L. monocytogenes surrogates (19, 20). For example, irrigation has repeatedly been associated with an increased risk of preharvest produce contamination by L. monocytogenes (14, 15, 26) and other foodborne pathogens (29–31). In fact, two studies (14, 25) found that irrigation was one of the most important risk factors associated with L. monocytogenes isolation from samples collected in preharvest environments; both studies collected samples from multiple farms growing a variety of crops. Moreover, contaminated irrigation water has been identified or suspected as the source of contamination in several produce-associated Escherichia coli and Salmonella outbreaks (32–36). Despite the repeated identification of irrigation as a risk factor for preharvest produce contamination, no study, to our knowledge, has reported, quantitatively, the impact of irrigation over time (i.e., over subsequent 24-h periods following an irrigation event) on the risk of produce contamination in production environments.
Therefore, the purpose of this study was (i) to quantify the effects of various spatial (e.g., proximity to water) and temporal (e.g., time since an irrigation event) factors on the risk of produce contamination after rain and irrigation events and (ii) to determine the impact of rain and irrigation events on Listeria species and L. monocytogenes diversity in spinach fields. The ultimate goal of this research was to identify potential mitigation strategies that can reduce the risk of produce contamination at the preharvest level.
MATERIALS AND METHODS
Study design.
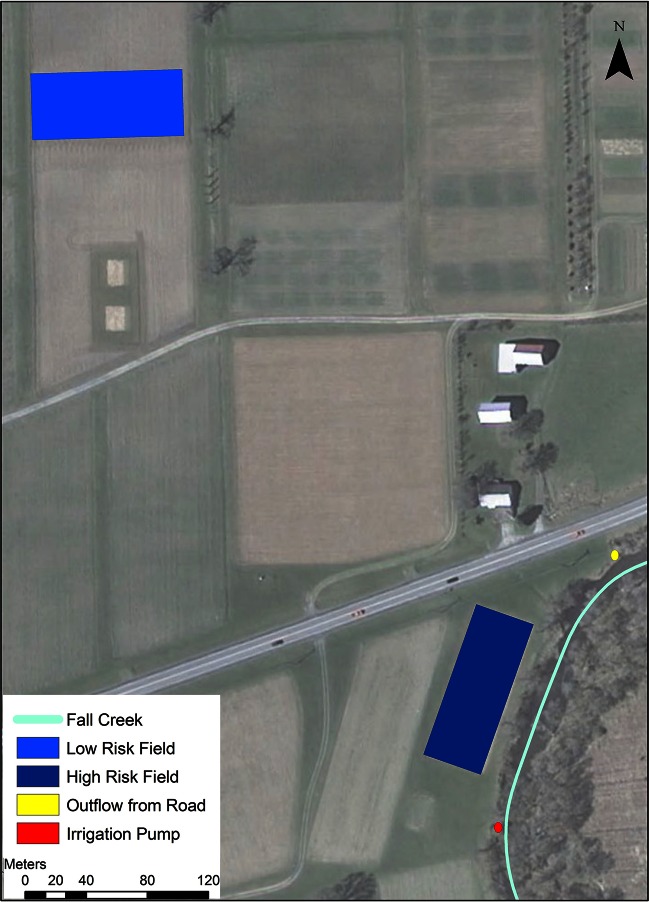
A longitudinal study was conducted in two spinach (Unipack 15-F1; Harris Seeds) fields at the Homer C. Thompson Vegetable Research Farm over a 7-week period in May, June, and July 2014. Two 0.2-ha fields (Fig. 1) were selected based on their respective predicted prevalence of L. monocytogenes (i.e., one high-risk and one low-risk field), which was a function of the fields' proximity to water and roads (see “Landscape data and determining predicted field risk,” below). Fields were prepared for planting by harrowing and treatment with a 13-13-13 fertilizer at a rate of 789 kg per ha. The herbicide metolachlor (DuPont, Wilmington, DE) was applied at a rate of 0.58 liters per ha immediately following seeding. Irrigation water was drawn from Fall Creek (Fig. 1).
FIG 1.
Locations of the low- and high-risk fields and the surface water sampling sites included in this study.
Each field was divided into 21 13- by 13-m plots. Soil sample sites were randomly selected from each plot by using the “Create Random Points” function in ArcGIS (version 10.2.2; Environmental Systems Research Institute, Redlands, CA) for each sampling trip (i.e., the same location within each plot was sampled only once during the course of the study). New sampling sites were selected for each sampling trip to ensure that (i) a representative sample of Listeria spp. and L. monocytogenes was collected from each plot during the course of the study and (ii) there was sufficient variation in sample location to statistically determine the effect of spatially specific factors (e.g., distance of a sampling site to water) on the likelihood of Listeria species and L. monocytogenes isolation. Soil samples were collected on the day of planting and 24, 48, 72, and 144 to 192 h after an “irrigation” or “rain” event. An irrigation event was defined as any time that irrigation water was applied to the field. An irrigation event was defined as any time irrigation water was applied to the field (via overhead irrigation using untreated surface water). A rain event was defined as >6 mm of rain over a 24-h time period (i.e., 9 a.m. to 9 a.m.). If multiple subsequent 24-h periods each received >6 mm of rain, the first sample collection (i.e., at 24 h) was performed 24 h after the last 24-h time period with >6 mm of rain (i.e., if it rained >6 mm on Tuesday and Wednesday, the 24-h samples were collected on Thursday). Two multiday rain events lasting 48 h occurred during the course of the study. To account for the effect of a multiday rain event on our results, the amounts of rainfall 0, 1, 2, 3, 0 to 1, 0 to 2, and 0 to 3 days preceding sample collection were included as risk factors in the statistical analyses (see “Statistical analysis,” below, for more information). If a rain or irrigation event did not occur between 144 and 192 h after a rain event, “dry”-event sampling was performed. Sampling at a later time point (e.g., 192 h versus 144 h) was given preference, if forecasts clearly indicated an absence of rainfall for >144 h. Each set of samples (i.e., 24, 48, 72, and 144 to 192 h, if collected) was defined as representing either an irrigation or a rain event depending on which “event type” initiated sample collection. Overall, seven sets of samples were collected: (i) five sets that represented rain events, including three sets where samples were collected 144 to 192 h after the event, and (ii) two sets that represented irrigation events, including one set where samples were collected 144 to 192 h after the event. Additionally, a set of samples was collected prior to seeding on the day of planting. Overall, each plot was sampled 26 times.
Water, leaf, and fecal samples were also collected. Water samples were collected from Fall Creek (Fig. 1), the water source used for irrigation. Fecal samples were collected when they were detected within 5 m of the sampled fields or Fall Creek. Fecal samples were not identified to the species level due to the high misclassification rate associated with visual identification of wildlife scat (37, 38). Composite leaf samples were collected for each plot once the spinach plants were large enough to survive harvesting (i.e., 36 days after planting). Composite leaf samples were hand collected by gathering leaves from 6 to 12 spinach plants growing along the perimeter and diagonals of each plot. Global positioning system (GPS) coordinates were recorded for each soil and water sample. In total, 1,092 soil, 52 water, 334 leaf, and 14 fecal samples were collected (n = 1,492 total).
Sample collection and preparation.
Samples were collected and tested as previously described by Strawn et al. (15). Briefly, latex gloves (Nasco, Fort Atkinson, WI) were worn and changed for each sample collected. For each plot, a soil sample was collected ∼4 in. (10.16 cm) below the soil surface by using 5-ml sterile scoops (Fisher Scientific, Hampton, NH) and placed into a sterile Whirl-Pak bag (Nasco, Fort Atkinson, WI). Twenty-five grams of soil was then weighed in a separate sterile filter Whirl-Pak bag. Water samples were collected directly into sterile jars by using a sampling pole (Nasco) and processed according to Environmental Protection Agency (EPA) standard methods (39). Briefly, a 250-ml water sample was passed through a 0.45-μm filter (Nalgene, Rochester, NY), and the filter was aseptically transferred to a sterile Whirl-Pak bag. Additionally, 10 g of each fecal sample and 25 g of each composite leaf sample were weighed out and aseptically transferred to separate sterile filter Whirl-Pak bags. All samples were transported on ice and processed within 3 h of collection.
Bacterial enrichment and isolation.
To enrich and isolate Listeria spp. and L. monocytogenes, samples were prepared as previously described by Strawn et al. (15). Briefly, each sample was diluted 1:10 with buffered Listeria enrichment broth (Becton Dickinson, Franklin Lakes, NJ) and incubated at 30°C for 24 h. After 4 h, Listeria selective enrichment supplement was added to each sample enrichment bag. At 24 and 48 h, 50 μl of each sample enrichment was plated onto Listeria monocytogenes plating medium (LMPM) agar (Biosynth International, Itasca, IL) and modified Oxford agar (MOX; Becton Dickinson). After incubation for 48 h at 35°C (LMPM) and 30°C (MOX), up to four presumptive Listeria colonies were substreaked from LMPM and MOX onto brain heart infusion (BHI) agar plates (Becton Dickinson). The BHI plates were then incubated at 37°C for 24 h. Presumptive Listeria colonies were confirmed by PCR amplification and sequencing of the partial sigB gene as previously described (40–42). Isolates were identified to the allelic type (AT) by comparison of partial sigB sequences to an internal reference database (Food Safety Laboratory, Cornell University, Ithaca, NY) (40–42). We acknowledge that there are more discriminatory subtyping methods that are more translatable to other subtyping schemes commonly used (e.g., multilocus sequence typing and multilocus genotyping); however, a previous study (43) showed that DNA-based subtyping methods, such as sigB AT identification, can efficiently differentiate between species of Listeria. More advanced subtyping schemes, such as whole-genome sequencing, should be used in future studies to assess the relatedness of isolates across time and space.
Positive and negative controls were processed in parallel for each sample. L. monocytogenes FSL R3-001 (44) inoculated in BHI broth was used as the positive control, and uninoculated enrichment medium was used as the negative control. All isolates were preserved at −80°C. Isolate information can be found at the Food Microbe Tracker website (http://www.FoodMicrobeTracker.com/) and in Table S1 in the supplemental material.
Landscape data and determining predicted field risk.
Landscape data (see Table S2 in the supplemental material) were derived by using ArcGIS as described previously by Weller et al. (14). Predicted risk was based on a geospatial algorithm described previously by Strawn et al. (15). Briefly, the GPS coordinates for each field and soil sampling site were imported into ArcMap by using the Universal Transverse Mercator, North American Datum, 1983. Road and hydrologic data were downloaded from the Cornell University Geospatial Information Repository (http://cugir.mannlib.cornell.edu/). Soil data were obtained from the Natural Resources Conservation Service Web Soil Survey (http://websoilsurvey.sc.egov.usda.gov/) Shape files for field edge and irrigation lines were created by using the “Create Features (Editor)” function. Data on the proximity of each sample collection point to field edge, irrigation lines, roads, and surface water were derived by using the “Near (Analysis)” function.
Based on the data and models described previously by Strawn et al. (15), a field was considered to be at high risk for L. monocytogenes contamination if it was ≤37.5 m from water and ≤9.5 m from a road (15). A field was considered to be at low risk for L. monocytogenes contamination if it was >37.5 m from water and >9.5 m from a road (15). The high-risk field also had, on average, more AWS (i.e., AWS 0 to 100 cm below the soil surface of >4.2 cm) than did the low-risk field. Soil in the high-risk field was Eel silt loam, and soil 0 to 100 cm below the soil surface was, on average, composed of 60% sand, 30% silt, 11% clay, and 2% organic matter; these values are based on representative values for several soil layers and a large area (30 m2) and therefore do not add up to 100%. Soil in the low-risk field was Howard gravely loam, and soil 0 to 100 cm below the soil surface was, on average, composed of 47% silt, 39% sand, 13% clay, and 1% organic matter content. Both fields were level (i.e., slope of <5%). Finally, spinach and a clover-rye cover crop were planted in the high-risk field in 2013 and 2012, respectively, while cucurbits and broccoli were planted in the low-risk field in 2013 and 2012, respectively.
Meteorological data.
Meteorological data (see Table S2 in the supplemental material) were obtained from the Cornell University weather station located at the Homer C. Thompson Vegetable Research Farm (Rainwise Inc., Trenton, NJ). Data on leaf wetness were obtained from the Cornell University Network for the Environment and Weather Applications (Ithaca, NY). Data were downloaded for each sample collection date and the three preceding 24-h periods (i.e., 9 a.m. to 9 a.m.). Average values for each factor 0 to 1, 0 to 2, and 0 to 3 days before sample collection were also calculated. For a description of all meteorological factors included in this study, see Table S2 in the supplemental material.
Statistical analysis.
All statistical analyses were performed in R (version 3.1; R Core Team, Vienna, Austria). Prevalence was calculated for each field (high or low risk), time period (24, 48, 72, and 144 to 192 h), event type (rain versus irrigation event), and sample type (leaf, soil, and water). The total number of ATs (i.e., allele type richness) for Listeria spp. and L. monocytogenes was determined, and the Shannon-Wiener index was calculated. A Hutcheson t test (45) was performed to compare the Shannon-Wiener indices for the high-risk and low-risk fields and for irrigation and rain events.
Univariable analyses were performed to determine the effect of spatial and meteorological factors, time since event, predicted field risk, and event type (i.e., irrigation versus rain event) on the odds of Listeria species and L. monocytogenes isolation. Correlation between significant factors (at a P value of ≤0.20) was assessed by using the corrplot package (version 0.73 [http://cran.r-project.org/web/packages/corrplot]). Principal component analysis (PCA) was performed on each set of meteorological factors (e.g., all humidity factors), with the exception of rainfall. PCA was performed only if the factors were significant by univariable analysis and correlated and if the combination was biologically plausible. The first eigenvector from each PCA was added to the data set as a potential covariate for inclusion in the final model. Factors that were identified as significant by univariable analysis but not included in a PCA were included as potential covariates in the final model as well.
Generalized linear mixed models (GLMMs) (79) were developed by using the logit link function. The outcome was the presence or absence of Listeria spp. or L. monocytogenes. Event type, hours, and either predicted field risk or proximity to water and road were included as fixed effects. Set and plot were included as random effects. The model was built by using a backwards selection method (i.e., factors were removed from the model until only factors significant at a P value of ≤0.05 remained).
Spatial analysis.
Model residuals were obtained for each GLMM, and a residual variogram was created to determine if there were spatial dependencies in the data that were not accounted for by the multivariable model (46).
RESULTS
Prevalence and diversity of L. monocytogenes and Listeria spp. in produce production environments.
The overall prevalence of Listeria spp. was 14% (204/1,492). The prevalence of Listeria spp. was higher in water samples (90%; 47/52) and in fecal samples (79%; 11/14) than in soil samples (12%; 126/1,092) and leaf samples (6%; 19/334) (Table 1). The prevalence of Listeria spp. was higher in soil samples collected from the high-risk field (15%; 84/546) than in those from the low-risk field (8%; 42/546) (Table 2 and Fig. 2). The prevalence of Listeria spp. was higher in soil samples collected 24 h after irrigation and rain events (23%; 68/294) than in soil samples collected at 48 h (10%; 28/294), 72 h (5%; 14/294), and 144 to 192 h (3%; 5/168) after irrigation and rain events (Table 2). The prevalence of Listeria spp. was higher in soil samples collected after irrigation events (14%; 40/294) than in those collected after rain events (10%; 75/756) (Table 2).
TABLE 1.
Effect of sample type on frequency and prevalence of Listeria spp. and L. monocytogenes isolates from soil samples collected from spinach fields previously identified as being at high or low risk for L. monocytogenes isolation
| Sampling site and sample type (no. of samples) | No. (%) of samples positive for: |
|
|---|---|---|
| Listeria spp.a | L. monocytogenes | |
| High-risk field (726) | 109 (15) | 73 (10) |
| Fecal (13) | 11 (85) | 9 (69) |
| Leaf (167) | 14 (8) | 2 (1) |
| Soil (546) | 84 (15) | 62 (11) |
| Low-risk field (714) | 48 (7) | 24 (3) |
| Fecal (1) | 0 (0) | 0 (0) |
| Leaf (167) | 5 (3) | 0 (0) |
| Soil (546) | 43 (8) | 24 (4) |
| Surface waterb (52) | 47 (90) | 33 (63) |
Listeria spp. including L. monocytogenes.
Surface water used for irrigation.
TABLE 2.
Frequency and prevalence of Listeria spp. and L monocytogenes in soil samples collected 24, 48, 72, and 144 to 192 h after irrigation and rain events from two spinach fields previously identified as being at high or low risk for L. monocytogenes isolation
| Event typea | Time (h)b (no. of samples) | No. (%) of samples positive for: |
|
|---|---|---|---|
| Listeria spp.c | L. monocytogenes | ||
| Low-risk field | |||
| Presample | NA (21) | 1 (5) | 1 (5) |
| Irrigation | 24 (42) | 8 (19) | 7 (17) |
| 48 (42) | 3 (7) | 2 (5) | |
| 72 (42) | 2 (5) | 2 (5) | |
| 144–192 (21) | 0 (0) | 0 (0) | |
| Rain | 24 (105) | 16 (15) | 7 (7) |
| 48 (105) | 7 (7) | 2 (3) | |
| 72 (105) | 4 (4) | 2 (3) | |
| 144–192 (63) | 2 (3) | 1 (1) | |
| High-risk field | |||
| Presample | NA (21) | 4 (19) | 2 (10) |
| Irrigation | 24 (42) | 11 (26) | 10 (24) |
| 48 (42) | 11 (26) | 11 (26) | |
| 72 (42) | 5 (12) | 2 (5) | |
| 144–192 (21) | 0 (0) | 0 (0) | |
| Rain | 24 (105) | 33 (31) | 28 (27) |
| 48 (105) | 7 (7) | 3 (3) | |
| 72 (105) | 3 (3) | 5 (12) | |
| 144–192 (63) | 3 (5) | 1 (16) | |
Event type (i.e., irrigation or rain event) that initiated sample collection.
Time in hours (i.e., 24, 48, 72, or 144 to 192 h) since the event. NA (not applicable) indicates that samples were collected before study initiation.
Listeria spp. including L. monocytogenes.
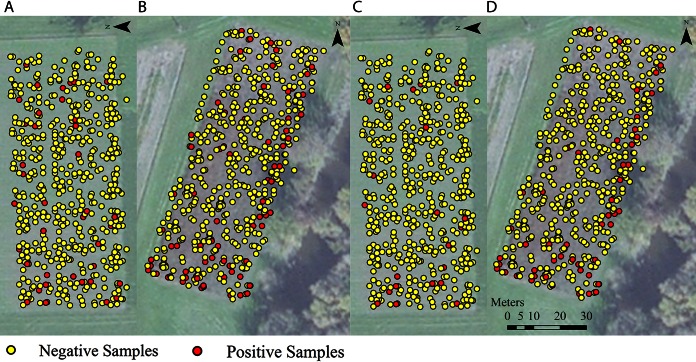
FIG 2.
Distribution of Listeria species (including L. monocytogenes)-positive and -negative samples in the low-risk (A) and high-risk (B) fields and of L. monocytogenes-positive and -negative samples in the low-risk (C) and high-risk (D) fields. Fall Creek, the source of irrigation water in this study, is visible in the bottom right-hand corner of panels B and D.
Twenty-seven different Listeria species allelic types were isolated from the Listeria species-positive soil samples collected in this study (see Table S3 in the supplemental material). While there was a greater diversity of ATs in soil samples collected from the low-risk field than in those from the high-risk field, the difference was not statistically significant according to a T-Hutcheson test (P = 0.08) (Table 3 and Fig. 3). The diversity of Listeria species ATs isolated from soil samples following rain events was significantly greater (P < 0.001) than the diversity of allelic types isolated from soil samples following irrigation events (Table 3). The diversity of Listeria species ATs isolated from water samples was not significantly different from the diversity of Listeria species ATs isolated from soil samples following irrigation events (P = 0.36). However, the diversity of Listeria species ATs isolated from water samples was significantly lower than the diversity of the Listeria species ATs isolated from soil samples following rain events (P < 0.001).
TABLE 3.
Diversity of Listeria species and L. monocytogenes allelic types isolated from soil and water samples collected from spinach fields previously identified as being at high or low risk for L. monocytogenes isolation
| Sampling site | Event typea |
Listeria spp.b |
L. monocytogenes |
||
|---|---|---|---|---|---|
| No. of allelic types | Shannon-Weiner index | No. of allelic types | Shannon-Weiner index | ||
| Low-risk field | — | 18 | 2.4 | 7 | 1.2 |
| Irrigation | 4 | 0.84 | 2 | 0.33 | |
| Rain | 16 | 2.5 | 6 | 1.4 | |
| High-risk field | — | 21 | 2.0 | 7 | 1.1 |
| Irrigation | 8 | 1.1 | 4 | 0.53 | |
| Rain | 18 | 2.2 | 6 | 1.2 | |
| Surface water | — | 14 | 0.85 | 6 | 0.99 |
| Irrigation | 4 | 0.67 | 3 | 0.39 | |
| Rain | 12 | 1.1 | 5 | 0.60 | |
Event type (i.e., irrigation or rain event) that initiated sample collection. — indicates information for all samples collected from the high-risk field, the low-risk field, or surface water regardless of the event type that initiated collection. The number of allelic types is not a simple summation of the numbers of ATs found following irrigation and rain events, as some ATs may have been found following events of both types.
Listeria spp. including L. monocytogenes.
FIG 3.
Distribution of L. monocytogenes allelic types in the low-risk (A) and high-risk (B) fields. Fall Creek, the source of irrigation water in this study, is visible in the bottom right-hand corner of panel B.
The overall prevalence of L. monocytogenes was 9% (130/1,492). The prevalence of L. monocytogenes was higher in fecal samples (64%; 9/14) and water samples (63%; 33/52) than in soil samples (8%; 86/1,092) and leaf samples (0.6%; 2/334) (Table 1). The prevalence of L. monocytogenes was higher in soil samples collected from the high-risk field (11%; 62/546) than in those collected from the low-risk field (4%; 24/546) (Table 1 and Fig. 2). The prevalence of L. monocytogenes was higher in soil samples collected 24 h after irrigation and rain events (18%; 52/294) than in soil samples collected 48 h (6%; 18/294), 72 h (4%; 11/294), and 144 to 192 h (1%; 2/168) after irrigation and rain events (Table 2). Finally, the prevalence of L. monocytogenes was higher in soil samples collected after irrigation events (12%; 34/294) than in those collected after rain events (6%; 49/756) (Table 2).
Nine different L. monocytogenes ATs were isolated from L. monocytogenes-positive soil samples (see Table S3 in the supplemental material); all isolates were of lineage I or II. While there was a greater diversity of ATs in soil samples collected from the low-risk field than in soil samples collected from the high-risk field (Fig. 3), the difference was not statistically significant according to a T-Hutcheson test (P = 0.39) (Table 3). The diversity of L. monocytogenes ATs isolated from soil samples following rain events was significantly greater (P < 0.001) than the diversity of L. monocytogenes ATs isolated from soil samples following irrigation events (Table 3). The diversity of L. monocytogenes ATs isolated from water samples was not significantly different from the diversity of L. monocytogenes ATs isolated from soil samples following irrigation events (P = 0.45). However, the diversity of L. monocytogenes ATs isolated from water samples was significantly lower than the diversity of L. monocytogenes ATs isolated from soil samples following rain events (P = 0.03).
Risk factors associated with Listeria species isolation from soil samples.
Of the 107 factors that were evaluated, 39 factors were significantly associated with Listeria species-positive soil samples by univariable analysis, including 2 study parameters, 2 spatial factors, 1 dew point factor, 6 humidity factors, 3 irrigation factors, 3 leaf wetness factors, 15 temperature factors, 3 precipitation factors, and 4 wind speed factors (see Table S4 in the supplemental material). PCA was performed for the leaf wetness factors as a group, the temperature factors as a group, and the wind speed factors as a group (see Table S5 in the supplemental material).
In the multivariable analysis, four factors (hours since the event occurred, amount of irrigation water applied to the fields 2 days before sampling, amount of rain water that precipitated 2 days before sampling, and predicted field risk) were retained (Table 4). Although event type was not found to be significant by multivariable analysis, it was retained in the final model so that the effect of irrigation events compared to rain events could be quantified, as this was of interest to the study. All factors retained in the final model were also retained when proximity to water and proximity to road were substituted for predicted field risk (see Table S6 in the supplemental material). No significant interactions between any factors were identified for either of the models. The model containing predicted field risk was selected as the final model because it had a lower Akaike information criterion (AIC) value than did the model containing proximity to water and road (AIC values of 654.7 and 658.0, respectively). The odds of Listeria species isolation in soil samples were 8 times greater (odds ratio [OR] = 7.7; 95% confidence interval [CI] = 2.9, 20) for samples collected 24 h after an event than for soil samples collected 144 to 192 h after any event. The odds of Listeria species isolation in soil samples were 2 and 3 times greater for samples collected 48 and 72 h after an event (OR = 2.1 and 95% CI = 0.74 to 6.2, and OR = 2.5 and 95% CI = 0.94 to 6.9, respectively) than for soil samples collected 144 to 192 h after an event. The odds of Listeria species isolation were 2 times greater (OR = 2.3; 95% CI = 1.5, 3.5) for soil samples collected from the high-risk field than for soil samples collected from the low-risk field. Finally, for each 1-mm increase in the amount of irrigation water applied to the field, the odds of Listeria species isolation increased (OR = 1.1; 95% CI = 1.0, 1.2), and for each 1-mm increase in the amount of rain that fell on the field, the odds of Listeria species isolation also increased (OR = 1.4; 95% CI = 1.1, 1.8). The residual variogram for the Listeria species final model (see Fig. S1 in the supplemental material) suggests that the final model effectively accounts for all spatial dependencies within the data.
TABLE 4.
Final multivariable model for the likelihood of isolating Listeria spp. and L. monocytogenes from spinach fields based on testing of soil samples and given a P value cutoff of 0.05
| Factor | ORa | 95% CIb | P value |
|---|---|---|---|
| Factors significant for Listeria spp.c | |||
| Amt of irrigation water (mm) applied to fields 2 days before sample collection | 1.1 | 1.0, 1.2 | 0.04 |
| Event type that initiated sample collection | |||
| Irrigation | 0.71 | 0.40, 1.2 | 0.22 |
| Rain | 1.0 | ||
| Time since event occurred (h) | |||
| 24 | 7.7 | 2.9, 20 | <0.01 |
| 48 | 2.1 | 0.74, 6.2 | 0.16 |
| 72 | 2.5 | 0.94, 6.9 | 0.07 |
| 144–192 | 1.0 | ||
| Predicted field risk | |||
| Low | 1.0 | ||
| High | 2.3 | 1.5, 3.5 | <0.01 |
| Total amt of rain (mm) on day 2 before sample collection | 1.4 | 1.1, 1.8 | <0.01 |
| Factors significant for L. monocytogenes | |||
| Amt of irrigation water applied to fields 2 days before sample collection | 1.2 | 1.1, 1.3 | <0.01 |
| Event type that initiated sample collection | |||
| Irrigation | 0.74 | 0.41, 1.3 | 0.33 |
| Rain | 1.0 | ||
| Time (h) since event occurred | |||
| 24 | 25 | 5.7, 99 | <0.01 |
| 48 | 2.5 | 0.49, 12 | 0.27 |
| 72 | 3.4 | 0.74, 15 | 0.11 |
| 144–192 | 1.0 | ||
| Predicted field risk | |||
| Low | 1.0 | ||
| High | 3.5 | 2.0, 6.0 | <0.01 |
For continuous factors, OR refers to the change in the odds of isolating Listeria spp. or L. monocytogenes associated with a one-unit increase in the factor (e.g., a 1-mm increase in the amount of irrigation water applied).
CI, confidence interval for the odds ratio.
Listeria spp. including L. monocytogenes.
Risk factors associated with L. monocytogenes isolation from soil samples.
Of the 107 factors that were evaluated (see Table S2 in the supplemental material), 46 were significantly associated with L. monocytogenes-positive soil samples by univariable analysis, including 2 study parameters, 2 spatial factors, 5 dew point factors, 10 humidity factors, 3 irrigation factors, 2 leaf wetness factors, 20 temperature factors, 1 precipitation factor, and 1 wind direction factor (see Table S4 in the supplemental material). PCA was performed for the dew point factors as a group, the humidity factors as a group, the leaf wetness factors as a group, and the temperature factors as a group (see Table S5 in the supplemental material).
In the multivariable analysis, three factors (hours since the event occurred, amount of irrigation water applied to the fields 2 days before sampling, and predicted field risk) were retained (Table 4). Although event type was not significant, it was retained in the final model. All factors retained in the final model were also retained when proximity to water and proximity to road were substituted for predicted field risk (see Table S6 in the supplemental material). No significant interactions between any factors were identified for either of the models. The model containing predicted field risk was selected as the final model because it had a lower AIC than did the model containing proximity to water and road (AIC values of 461.5 and 465.9, respectively). The odds of L. monocytogenes isolation in soil samples were 25 times greater (OR = 25; 95% CI = 5.7, 99) for samples collected 24 h after an event than for soil samples collected 144 to 192 h after any event. The odds of L. monocytogenes isolation in soil samples were about 3 times greater for samples collected 48 h (OR = 2.5; 95% CI = 0.49, 12) and 72 h (OR = 3.4; 95% CI = 0.74, 15) after an event than for soil samples collected 144 to 192 h after an event. While, statistically, the odds of isolating L. monocytogenes were greatest during the 24 h immediately following an irrigation or rain event, for the high-risk field, the observed prevalence of L. monocytogenes was higher 48 h than 24 h after irrigation (Table 2). The odds of L. monocytogenes isolation were 3.5 times greater (OR = 3.5; 95% CI = 2.0, 6.0) for soil samples collected from the high-risk field than for soil samples collected from the low-risk field. Finally, for each 1-mm increase in the amount of irrigation water applied to a field, the odds of L. monocytogenes isolation increased (OR = 1.2; 95% CI = 1.1, 1.3). The residual variogram (see Fig. S1 in the supplemental material) for the final model also suggests that the final model effectively accounted for all spatial dependencies within the data.
DISCUSSION
The objectives of this study were (i) to quantify the effects of different spatial and temporal factors associated with the isolation of Listeria spp. and L. monocytogenes from produce fields following rain and irrigation events and (ii) to determine how rain and irrigation events affect the detection frequency and diversity of Listeria spp. and L. monocytogenes in produce fields. Our study showed that the likelihood of isolating Listeria spp. and L. monocytogenes was greatest during the 24 h immediately following rain or irrigation events and that the diversity of Listeria species and L. monocytogenes subtypes (ATs) was lower after irrigation events than after rain events. Additionally, we show that proximity to water and roads was associated with an increased likelihood of isolating L. monocytogenes from soil samples collected in produce fields. These findings are consistent with data from previous research performed in New York State produce production environments (14–16, 25, 47), supporting a robust relationship between these factors and Listeria species and L. monocytogenes isolation. Our study is unique as it quantified changes in Listeria species and L. monocytogenes prevalence over subsequent 24-h periods following rain and irrigation events. It is important to note that these findings are based on a single study conducted on two fields over the course of one growing season and that additional studies are needed to determine if our findings are translatable to other farms. However, the results support conclusions from previous studies (14, 25, 48–50) that management practice-based interventions may reduce the risk of preharvest produce contamination.
Irrigation can be a point source of Listeria contamination, while rain appears to increase the prevalence of Listeria detection through non-point-source mechanisms.
Analysis of sigB AT diversity showed that in our study here, the diversity of Listeria species and L. monocytogenes isolates was significantly lower in soil samples collected after irrigation events than in those collected after rain events. The lower diversity following irrigation events suggests that irrigation water served as a homogenous point source of Listeria species and L. monocytogenes contamination in the produce fields studied here. This is supported by the fact that the diversity of ATs in soil samples collected after irrigation events was not statistically different from the diversity of ATs in water samples collected from Fall Creek, the source of irrigation water used in this study. These findings are consistent with findings of previous studies (14, 25, 26, 47, 51, 52) that identified irrigation water as a potential preharvest source of bacterial contamination of produce. Multiple studies have also reported significant associations between irrigation water and fresh produce contamination (12, 13, 19, 20). The relationship between irrigation and L. monocytogenes isolation in this study may be explained by the fact that surface water is a known reservoir of foodborne pathogens in produce production environments (22, 52–56). As our study and other studies (14, 21, 25, 47, 51, 52) have demonstrated, irrigation is an important risk factor for preharvest produce contamination, particularly if the irrigation water is drawn from a surface water source. Therefore, intervention at the irrigation level may decrease the risk of L. monocytogenes contamination of produce. For example, in a review of pre- and postharvest measures to reduce microbial contamination of fresh produce, Gil et al. (48) identified selection of proper irrigation methods, protection of surface water sources, and periodic testing of irrigation water as critical interventions for preventing microbial contamination.
The higher diversity of Listeria species and L. monocytogenes ATs associated with rain events suggests that rain increases the likelihood of Listeria species and L. monocytogenes detection. For example, rain may facilitate the movement of diverse Listeria strains into field environments or may facilitate the growth and detection of Listeria strains already present in the field. This is supported by previous studies (15, 57) that found that pathogens transmitted by runoff and splash associated with rain events can bypass physical barriers to movement into and within fields. The findings of these previous studies may also explain why all of the positive leaf samples in our study were associated with rain events. Additionally, rain events may create favorable conditions for foodborne pathogen growth (e.g., higher soil moisture [55, 58–60]), amplifying existing Listeria populations within the field and increasing the likelihood of detection during sampling. Similarly, higher nutrient loads associated with runoff (61–64) could facilitate microbial growth in fields (62, 65–67). As rain and irrigation events can affect the diversity of Listeria spp. and L. monocytogenes in produce production environments differently, interventions to reduce the risk of pathogen contamination in fields may need to take into account the water source (i.e., surface water versus rain).
Meteorological factors are significantly associated with isolation of Listeria monocytogenes from produce production environments.
In previous studies, temperature-related (e.g., heat index and maximum temperature) (15, 19–21, 60, 68, 69) and water-related (e.g., humidity and leaf wetness) (20, 21, 69) meteorological factors were significantly associated with pathogen isolation from produce production environments. For example, in a study conducted on Spanish lettuce fields, Oliveira et al. (20) found that humidity and temperature influence Listeria innocua survival following irrigation. Interestingly, in the study reported here, multiple meteorological factors (except rain) were significantly associated with L. monocytogenes isolation by univariable analysis, but no meteorological factors were retained in the final multivariable model for L. monocytogenes; this is consistent with the findings of Weller et al. (14). Moreover, in the model developed by Strawn et al. (15) to predict the risk of L. monocytogenes isolation from produce production environments, temperature was ranked below proximity to water, suggesting that spatial factors (e.g., proximity to water) have a greater influence on L. monocytogenes isolation than do meteorological factors (e.g., temperature). Combined, the findings reported here and in other studies (14, 15) may indicate that although meteorological factors are associated with L. monocytogenes isolation, they are not the most important risk factors for L. monocytogenes isolation. Thus, meteorological factors (other than rain) should not be the primary focus of risk management strategies for L. monocytogenes in produce production environments; rather, risk management strategies that focus on landscape factors or management practices may be more beneficial long term. However, due to the above-mentioned weaknesses of this study, further research is needed to determine if these conclusions are translatable to other farms both within and outside New York State.
Risk of produce contamination is highest within 24 h of irrigation and rain events.
In our study, the odds of isolating L. monocytogenes in soil samples were greatest during the 24 h immediately following rain or irrigation events, compared to 48, 72, or 144 to 192 h following rain or irrigation events. Overall, our findings suggest that L. monocytogenes levels spike after an initial inoculation event, such as irrigation, and then decrease over subsequent 24-h periods. While no other study, to our knowledge, has investigated L. monocytogenes survival in produce production environments over subsequent 24-h periods, previous studies (17, 19, 60, 68, 70) that investigated the persistence and survival of L. monocytogenes in non-produce-production environments found similar patterns. For example, McLaughlin et al. (60) found that L. monocytogenes populations in soil samples collected from urban and forest environments declined after inoculation and were undetectable in 8 to 10 days. Similarly, both Castro-Ibáñez et al. (68) and Taylor et al. (70) showed that levels of fecal indicator bacteria peaked immediately following flooding events and then declined over subsequent sampling events. In the context of these studies, our findings suggest that waiting 24 h after irrigation and rain events to harvest crops may significantly reduce the risk of L. monocytogenes contamination. This time frame offers a tangible solution to growers that can be implemented with limited economic impact.
Landscape factors accurately predict the risk of L. monocytogenes contamination.
In our study, the odds of isolating L. monocytogenes were significantly higher for samples collected from the high-risk field than for samples collected from the low-risk field, suggesting that landscape factors (e.g., proximity to road and water) may be useful for accurately predicting the likelihood of L. monocytogenes isolation from produce production environments. This is not surprising, since previous studies have repeatedly associated landscape factors with foodborne pathogen isolation from produce production environments (14–16, 22, 71). However, it is important to note that the model that included predicted risk fit the data better than did the model containing proximity to water and roads. This may suggest that for the data set discussed here, the model containing predicted risk accounted for additional differences between the two fields, such as soil type and field history. As mentioned above, differences in soil properties are known to affect the likelihood of isolating L. monocytogenes from soil samples, and the high-risk field had, on average, more AWS than did the low-risk field. Overall, the findings reported here and in other studies (14–16, 22, 60) support the conclusion that not all cropland is at equal risk of foodborne pathogen contamination. Clearly, preharvest contamination of fresh produce is the result of complex interactions between factors, including factors that were not included in this study (e.g., seasonal effects and worker activity). However, our findings suggest that the use of landscape factors to predict risk and to tailor cropping schemes to reduce risk (e.g., planting high-risk crops in low-risk areas) may be useful for developing targeted on-farm food safety risk management plans.
The association between L. monocytogenes prevalence and proximity to water and road found in this study is consistent with data in the existing literature (14–16, 71). For example, Strawn et al. (15) and Sauders et al. (71) found that the closer a field or location was to a road, the greater the likelihood of L. monocytogenes isolation. Roadside ditches, like surface water, may act as a reservoir and transmission pathway for foodborne pathogens in produce production environments (72). Heavy rain, melting snow, wind, flooding, and human activity may also act as mechanisms for the spread of foodborne pathogens from ditches and waterways to produce fields. Additionally, roads, roadside ditches, and riparian areas may act as corridors for animal movement. Therefore, fields that are closer to roads and water may be at greater risk for wildlife intrusion, which was previously associated with produce contamination by foodborne pathogens (73–75). Since previous studies have found that buffer zones (25, 76, 77) and wetlands (78) reduce the risk of microbial contamination in produce production and other environments, the construction of buffer zones and the conservation of wetlands around fields may reduce the risk of L. monocytogenes contamination of produce. However, more research is needed to quantify the impact of buffer zones and wetlands on the risk of produce contamination and to determine how buffer zones and wetlands can be most effectively used to reduce produce contamination risks.
Overall, our findings suggest that small changes in management practices may have a significant effect on the risk of L. monocytogenes contamination in produce production environments. For example, growers may reduce L. monocytogenes contamination risk by waiting 24 h to harvest crops following rain events or by not performing irrigation within 24 h of harvest. Additionally, interventions at the irrigation level, such as treatment of irrigation water (e.g., by chlorine tabs), may reduce the risk of preharvest contamination. Other potential intervention strategies may include constructing buffer zones or conserving wetlands around fields near water or roads, altering cropping schemes (e.g., planting high-risk crops in low-risk fields), and monitoring pathogen levels in irrigation water.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Center for Produce Safety.
We are grateful for the technical assistance of Maureen Gunderson, Sherry Roof, Jeff Tokman, and Silin Tang as well as the statistical support of Yrjo Grohn, Saurabh Mehta, Julia Finkelstein, Erika Mudrak, Francoise Vermeylen, and David Kent.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01286-15.
REFERENCES
- 1.DeWaal CS, Glassman M. 2014. Outbreak alert 2014: a review of foodborne illness in America from 2002–2011. Center for Science in the Public Interest, Washington, DC. [Google Scholar]
- 2.Sivapalasingam S, Friedman CR, Cohen L, Tauxe RV. 2004. Fresh produce: a growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J Food Prot 67:2342–2353. [DOI] [PubMed] [Google Scholar]
- 3.Bean NH, Griffin PM. 1990. Foodborne disease outbreaks in the United States, 1973–1987: pathogens, vehicles, and trends. J Food Prot 53:804–817. [DOI] [PubMed] [Google Scholar]
- 4.Olsen S, MacKinnon L, Goulding J, Bean N, Slutsker L. 2000. Surveillance for foodborne disease outbreaks—United States, 1993-1997. MMWR CDC Surveill Summ 49:1–62. http://www.cdc.gov/foodsafety/outbreaks/multistate-outbreaks/reports.html. [PubMed] [Google Scholar]
- 5.Bean N, Griffin P, Goulding J, Ivey C. 1990. Foodborne disease outbreaks, 5-year summary, 1983-1987. MMWR CDC Surveill Summ 39:15–57. http://www.cdc.gov/MMWR/preview/mmwrhtml/00001597.htm. [PubMed] [Google Scholar]
- 6.Painter JA, Hoekstra RM, Ayers T, Tauxe RV, Braden CR, Angulo FJ, Griffin PM. 2013. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998-2008. Emerg Infect Dis 19:407–415. doi: 10.3201/eid1903.111866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dillaway R, Messer KD, Bernard JC, Kaiser HM. 2011. Do consumer responses to media food safety information last? Appl Econ Perspect Policy 33:363–383. doi: 10.1093/aepp/ppr019. [DOI] [Google Scholar]
- 8.Peake WO, Detre JD, Carlson CC. 2014. One bad apple spoils the bunch? An exploration of broad consumption changes in response to food recalls. Food Policy 49:13–22. [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2012. Multistate outbreak of listeriosis linked to whole cantaloupes from Jensen Farms, Colorado. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/listeria/outbreaks/cantaloupes-jensen-farms/index.html. [Google Scholar]
- 10.Bailin D. 2013. Killer cantaloupes: ignoring the science behind food safety. Union of Concerned Scientists, Cambridge, MA: http://www.ucsusa.org/center-for-science-and-democracy/case-studies/killer-cantaloupes-case-study.html. [Google Scholar]
- 11.US Food and Drug Administration. 2011. Environmental assessment: factors potentially contributing to the contamination of fresh whole cantaloupe implicated in a multi-state outbreak of listeriosis. US Food and Drug Administration, Washington, DC: http://www.fda.gov/Food/RecallsOutbreaksEmergencies/Outbreaks/ucm276247.htm. [Google Scholar]
- 12.Materon LA, Martinez-Garcia M, McDonald V. 2007. Identification of sources of microbial pathogens on cantaloupe rinds from pre-harvest to post-harvest operations. World J Microbiol Biotechnol 23:1281–1287. doi: 10.1007/s11274-007-9362-2. [DOI] [Google Scholar]
- 13.Gagliardi JV, Millner PD, Lester G, Ingram D. 2003. On-farm and postharvest processing sources of bacterial contamination to melon rinds. J Food Prot 66:82–87. [DOI] [PubMed] [Google Scholar]
- 14.Weller D, Wiedmann M, Strawn LK. 2015. Irrigation is significantly associated with an increased prevalence of Listeria monocytogenes in produce production environments in New York State. J Food Prot 78:1132–1141. doi: 10.4315/0362-028X.JFP-14-584. [DOI] [PubMed] [Google Scholar]
- 15.Strawn LK, Fortes ED, Bihn EA, Nightingale KK, Gröhn YT, Worobo RW, Wiedmann M, Bergholz PW. 2013. Landscape and meteorological factors affecting prevalence of three food-borne pathogens in fruit and vegetable farms. Appl Environ Microbiol 79:588–600. doi: 10.1128/AEM.02491-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapin TK, Nightingale KK, Worobo RW, Wiedmann M, Strawn LK. 2014. Geographical and meteorological factors associated with isolation of Listeria species in New York State produce production and natural environments. J Food Prot 77:1919–1928. doi: 10.4315/0362-028X.JFP-14-132. [DOI] [PubMed] [Google Scholar]
- 17.Dowe MJ, Jackson ED, Mori JG, Bell CR. 1997. Listeria monocytogenes survival in soil and incidence in agricultural soils. J Food Prot 1158–1286. [DOI] [PubMed] [Google Scholar]
- 18.Castro-Ibáñez I, Gil MI, Tudela JA, Ivanek R, Allende A. 2015. Assessment of microbial risk factors and impact of meteorological conditions during production of baby spinach in the Southeast of Spain. Food Microbiol 49:173–181. doi: 10.1016/j.fm.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Girardin H, Morris CE, Albagnac C, Dreux N, Glaux C, Nguyen-The C. 2005. Behaviour of the pathogen surrogates Listeria innocua and Clostridium sporogenes during production of parsley in fields fertilized with contaminated amendments. FEMS Microbiol Ecol 54:287–295. doi: 10.1016/j.femsec.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Oliveira M, Usall J, Viñas I, Solsona C, Abadias M. 2011. Transfer of Listeria innocua from contaminated compost and irrigation water to lettuce leaves. Food Microbiol 28:590–596. doi: 10.1016/j.fm.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Ivanek R, Gröhn YT, Wells MT, Lembo A, Sauders JBD, Wiedmann M. 2009. Modeling of spatially referenced environmental and meteorological factors influencing the probability of Listeria species isolation from natural environments. Appl Environ Microbiol 75:5893–5909. doi: 10.1128/AEM.02757-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linke K, Rückerl I, Brugger K, Karpiskova R, Walland J, Muri-Klinger S, Tichy A, Wagner M, Stessl B. 2014. Reservoirs of Listeria species in three environmental ecosystems. Appl Environ Microbiol 80:5583–5592. doi: 10.1128/AEM.01018-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliveira M, Usall J, Viñas I, Anguera M, Gatius F, Abadias M. 2010. Microbiological quality of fresh lettuce from organic and conventional production. Food Microbiol 27:679–684. doi: 10.1016/j.fm.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Selma MV, Allende A, López-Gálvez F, Elizaquível P, Aznar R, Gil MI. 2007. Potential microbial risk factors related to soil amendments and irrigation water of potato crops. J Appl Microbiol 103:2542–2549. doi: 10.1111/j.1365-2672.2007.03504.x. [DOI] [PubMed] [Google Scholar]
- 25.Strawn LK, Gröhn YT, Warchocki S, Worobo RW, Bihn EA, Wiedmann M. 2013. Risk factors associated with Salmonella and Listeria monocytogenes contamination of produce fields. Appl Environ Microbiol 79:7618–7627. doi: 10.1128/AEM.02831-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ijabadeniyi OA, Debusho LK, Vanderlinde M, Buys EM. 2011. Irrigation water as a potential pre-harvest source of bacterial contamination of vegetables. J Food Saf 31:452–461. doi: 10.1111/j.1745-4565.2011.00321.x. [DOI] [Google Scholar]
- 27.Jiang X, Islam M, Morgan J, Doyle MP. 2004. Fate of Listeria monocytogenes in bovine manure-amended soil. J Food Prot 67:1676–1681. [DOI] [PubMed] [Google Scholar]
- 28.Islam M, Morgan J, Doyle MP, Phatak SC, Millner P, Jiang X. 2004. Fate of Salmonella enterica serovar Typhimurium on carrots and radishes grown in fields treated with contaminated manure composts or irrigation water. Appl Environ Microbiol 70:2497–2502. doi: 10.1128/AEM.70.4.2497-2502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pagadala S, Marine SC, Micallef SA, Wang F, Pahl DM, Melendez MV, Kline WL, Oni RA, Walsh CS, Everts KL, Buchanan RL. 2015. Assessment of region, farming system, irrigation source and sampling time as food safety risk factors for tomatoes. Int J Food Microbiol 196:98–108. doi: 10.1016/j.ijfoodmicro.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Mañas P, Castro E, de Las Heras J. 2009. Irrigation with treated wastewater: effects on soil, lettuce (Lactuca sativa L.) crop and dynamics of microorganisms. J Environ Sci Health A Tox Hazard Subst Environ Eng 44:1261–1273. doi: 10.1080/10934520903140033. [DOI] [PubMed] [Google Scholar]
- 31.Ibekwe AM, Watt PM, Shouse PJ, Grieve CM. 2004. Fate of Escherichia coli O157:H7 in irrigation water on soils and plants as validated by culture method and real-time PCR. Can J Microbiol 50:1007–1014. doi: 10.1139/w04-097. [DOI] [PubMed] [Google Scholar]
- 32.Gelting RJ, Baloch MA, Zarate-Bermudez MA, Selman C. 2011. Irrigation water issues potentially related to the 2006 multistate E. coli O157:H7 outbreak associated with spinach. Agric Water Manag 98:1395–1402. doi: 10.1016/j.agwat.2011.04.004. [DOI] [Google Scholar]
- 33.Ackers M, Mahon BE, Leahy E, Goode B, Damrow T, Hayes PS, Bibb WF, Rice DH, Barrett TJ, Hutwagner L, Griffin PM, Slutsker L. 1998. An outbreak of Escherichia coli O157:H7 infections associated with leaf lettuce consumption. J Infect Dis 177:1588–1593. doi: 10.1086/515323. [DOI] [PubMed] [Google Scholar]
- 34.Nygård K, Lassen J, Vold L, Andersson Y, Fisher I, Löfdahl S, Threlfall J, Luzzi I, Peters T, Hampton M, Torpdahl M, Kapperud G, Aavitsland P. 2008. Outbreak of Salmonella Thompson infections linked to imported rucola lettuce. Foodborne Pathog Dis 5:165–173. doi: 10.1089/fpd.2007.0053. [DOI] [PubMed] [Google Scholar]
- 35.Crawford W, Baloch MA, Gerrity K. 2010. Environmental assessment: non-O157 Shiga toxin-producing E. coli (STEC). US Food and Drug Administration, Washington, DC. http://www.fda.gov/Food/RecallsOutbreaksEmergencies/Outbreaks/ucm235477.htm.
- 36.California Food Emergency Response Team. 2008. Investigation of the Taco John's Escherichia coli O157:H1 outbreak associated with iceberg lettuce. California Food Emergency Response Team, Sacramento, CA: http://www.cdph.ca.gov/pubsforms/Documents/fdb%20eru%20IceLet%20TacoJohn022008.pdf. [Google Scholar]
- 37.Monterroso P, Castro D, Silva TL, Ferreras P, Godinho R, Alves PC. 2013. Factors affecting the (in)accuracy of mammalian mesocarnivore scat identification in South-western Europe. J Zool 289:243–250. doi: 10.1111/jzo.12000. [DOI] [Google Scholar]
- 38.Harrington LA, Harrington AL, Hughes J, Stirling D, Macdonald DW. 2009. The accuracy of scat identification in distribution surveys: American mink, Neovison vison, in the northern highlands of Scotland. Eur J Wildl Res 56:377–384. [Google Scholar]
- 39.Environmental Protection Agency. 2002. Method 1603: Escherichia coli (E. coli) in water by membrane filtration using modified membrane-thermotolerant Escherichia coli agar (modified mTEC). Office of Science and Technology, Environmental Protection Agency, Washington, DC: http://www.epa.gov/nerlcwww/documents/1603sp02.pdf. [Google Scholar]
- 40.Nightingale KK, Windham K, Wiedmann M. 2005. Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J Bacteriol 187:5537–5551. doi: 10.1128/JB.187.16.5537-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.den Bakker HC, Bundrant BN, Fortes ED, Orsi RH, Wiedmann M. 2010. A population genetics-based and phylogenetic approach to understanding the evolution of virulence in the genus Listeria. Appl Environ Microbiol 76:6085–6100. doi: 10.1128/AEM.00447-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bundrant BN, Hutchins T, den Bakker HC, Fortes E, Wiedmann M. 2011. Listeriosis outbreak in dairy cattle caused by an unusual Listeria monocytogenes serotype 4b strain. J Vet Diagn Invest 23:155–158. doi: 10.1177/104063871102300130. [DOI] [PubMed] [Google Scholar]
- 43.Cai S, Kabuki DY, Kuaye AY, Cargioli TG, Chung MS, Nielsen R, Wiedmann M. 2002. Rational design of DNA sequence-based strategies for subtyping Listeria monocytogenes. J Clin Microbiol 40:3319–3325. doi: 10.1128/JCM.40.9.3319-3325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts AJ, Wiedmann M. 2006. Allelic exchange and site-directed mutagenesis probe the contribution of ActA amino-acid variability to phosphorylation and virulence-associated phenotypes among Listeria monocytogenes strains. FEMS Microbiol Lett 254:300–307. doi: 10.1111/j.1574-6968.2005.00041.x. [DOI] [PubMed] [Google Scholar]
- 45.Hutcheson K. 1970. A test for comparing diversities based on the Shannon formula. J Theor Biol 29:151–154. doi: 10.1016/0022-5193(70)90124-4. [DOI] [PubMed] [Google Scholar]
- 46.Bivand R, Pebesma E, Gomez-Rubio V. 2013. Applied spatial data analysis with R, 2nd ed Springer Science and Business Media, New York, NY. [Google Scholar]
- 47.Jones LA, Worobo RW, Smart CD. 2014. Plant-pathogenic oomycetes, Escherichia coli strains, and Salmonella spp. frequently found in surface water used for irrigation of fruit and vegetable crops in New York State. Appl Environ Microbiol 80:4814–4820. doi: 10.1128/AEM.01012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gil MI, Selma MV, Suslow T, Jacxsens L, Uyttendaele M, Allende A. 2015. Pre- and postharvest preventive measures and intervention strategies to control microbial food safety hazards of fresh leafy vegetables. Crit Rev Food Sci Nutr 55:453–468. doi: 10.1080/10408398.2012.657808. [DOI] [PubMed] [Google Scholar]
- 49.Izumi H, Tsukada Y, Poubol J, Hisa K. 2008. On-farm sources of microbial contamination of persimmon fruit in Japan. J Food Prot 71:52–59. [DOI] [PubMed] [Google Scholar]
- 50.Izumi H, Poubol J, Hisa K, Sera K. 2008. Potential sources of microbial contamination of Satsuma mandarin fruit in Japan, from production through packing shed. J Food Prot 71:530–538. [DOI] [PubMed] [Google Scholar]
- 51.Park S, Navratil S, Gregory A, Bauer A, Srinath I, Jun M, Szonyi B, Nightingale K, Anciso J, Ivanek R. 2013. Generic Escherichia coli contamination of spinach at the preharvest stage: effects of farm management and environmental factors. Appl Environ Microbiol 79:4347–4358. doi: 10.1128/AEM.00474-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pachepsky Y, Shelton D, McLain J, Patel JET, Mandrell R. 2011. Irrigation waters as a source of pathogenic microorganisms in produce: a review. Adv Agron 113:73–138. [Google Scholar]
- 53.Lyautey E, Lapen DR, Wilkes G, McCleary K, Pagotto F, Tyler K, Hartmann A, Piveteau P, Rieu A, Robertson WJ, Medeiros DT, Edge TA, Gannon V, Topp E. 2007. Distribution and characteristics of Listeria monocytogenes isolates from surface waters of the South Nation River watershed, Ontario, Canada. Appl Environ Microbiol 73:5401–5410. doi: 10.1128/AEM.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cooley MB, Quiñones B, Oryang D, Mandrell RE, Gorski L. 2014. Prevalence of Shiga toxin producing Escherichia coli, Salmonella enterica, and Listeria monocytogenes at public access watershed sites in a California Central Coast agricultural region. Front Cell Infect Microbiol 4:30. doi: 10.3389/fcimb.2014.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haley BJ, Cole DJ, Lipp EK. 2009. Distribution, diversity, and seasonality of waterborne salmonellae in a rural watershed. Appl Environ Microbiol 75:1248–1255. doi: 10.1128/AEM.01648-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilkes G, Edge TA, Gannon VPJ, Jokinen C, Lyautey E, Neumann NF, Ruecker N, Scott A, Sunohara M, Topp E, Lapen DR. 2011. Associations among pathogenic bacteria, parasites, and environmental and land use factors in multiple mixed-use watersheds. Water Res 45:5807–5825. doi: 10.1016/j.watres.2011.06.021. [DOI] [PubMed] [Google Scholar]
- 57.Boyer DG. 2008. Fecal coliform dispersal by rain splash on slopes. Agric For Meteorol 148:1395–1400. doi: 10.1016/j.agrformet.2008.04.001. [DOI] [Google Scholar]
- 58.Bergholz PW, Noar JD, Buckley DH. 2011. Environmental patterns are imposed on the population structure of Escherichia coli after fecal deposition. Appl Environ Microbiol 77:211–219. doi: 10.1128/AEM.01880-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winfield MDM, Groisman EA. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl Environ Microbiol 69:3687–3694. doi: 10.1128/AEM.69.7.3687-3694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McLaughlin HP, Casey PG, Cotter J, Gahan CGM, Hill C. 2011. Factors affecting survival of Listeria monocytogenes and Listeria innocua in soil samples. Arch Microbiol 193:775–785. doi: 10.1007/s00203-011-0716-7. [DOI] [PubMed] [Google Scholar]
- 61.Brezonik PL, Stadelmann TH. 2002. Analysis and predictive models of stormwater runoff volumes, loads, and pollutant concentrations from watersheds in the Twin Cities metropolitan area, Minnesota, USA. Water Res 36:1743–1757. doi: 10.1016/S0043-1354(01)00375-X. [DOI] [PubMed] [Google Scholar]
- 62.Jeppesen E, Kronvang B, Meerhoff M, Søndergaard M, Hansen KM, Andersen HE, Lauridsen TL, Liboriussen L, Beklioglu M, Ozen A, Olesen JE. 2009. Climate change effects on runoff, catchment phosphorus loading and lake ecological state, and potential adaptations. J Environ Qual 38:1930–1941. doi: 10.2134/jeq2008.0113. [DOI] [PubMed] [Google Scholar]
- 63.Sonzogni WC, Chesters G, Coote DR, Jeffs DN, Konrad JC, Ostry RC, Robinson JB. 1980. Pollution from land runoff. Environ Sci Technol 14:148–153. doi: 10.1021/es60162a003. [DOI] [Google Scholar]
- 64.Jordan TE, Correll DL, Weller DE. 1997. Relating nutrient discharges from watersheds to land use and streamflow variability. Water Resour Res 33:2579–2590. doi: 10.1029/97WR02005. [DOI] [Google Scholar]
- 65.Correll DL. 1998. The role of phosphorus in the eutrophication of receiving waters: a review. J Environ Qual 27:261–266. doi: 10.2134/jeq1998.00472425002700020004x. [DOI] [Google Scholar]
- 66.Rose JB, Epstein PR, Lipp EK, Sherman BH, Bernard SM, Patz JA. 2001. Climate variability and change in the United States: potential impacts on water- and foodborne diseases caused by microbiologic agents. Environ Health Perspect 109(Suppl):211–221. doi: 10.2307/3435011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Michael Beman J, Arrigo KR, Matson PA. 2005. Agricultural runoff fuels large phytoplankton blooms in vulnerable areas of the ocean. Nature 434:211–214. doi: 10.1038/nature03370. [DOI] [PubMed] [Google Scholar]
- 68.Castro-Ibáñez I, Gil MI, Tudela JA, Allende A. 2015. Microbial safety considerations of flooding in primary production of leafy greens: a case study. Food Res Int 68:62–69. doi: 10.1016/j.foodres.2014.05.065. [DOI] [Google Scholar]
- 69.Park S, Navratil S, Gregory A, Bauer A, Srinath I, Szonyi B, Nightingale K, Anciso J, Jun M, Han D, Lawhon S, Ivanek R. 2015. Multifactorial effects of ambient temperature, precipitation, farm management, and environmental factors determine the level of generic Escherichia coli contamination on preharvested spinach. Appl Environ Microbiol 81:2635–2650. doi: 10.1128/AEM.03793-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taylor J, Davies M, Canales M, Lai KM. 2013. The persistence of flood-borne pathogens on building surfaces under drying conditions. Int J Hyg Environ Health 216:91–99. doi: 10.1016/j.ijheh.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 71.Sauders BD, Overdevest J, Fortes E, Windham K, Schukken Y, Lembo A, Wiedmann M. 2012. Diversity of Listeria species in urban and natural environments. Appl Environ Microbiol 78:4420–4433. doi: 10.1128/AEM.00282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Falbo K, Schneider RL, Buckley DH, Walter MT, Bergholz PW, Buchanan BP. 2013. Roadside ditches as conduits of fecal indicator organisms and sediment: implications for water quality management. J Environ Manage 128:1050–1059. doi: 10.1016/j.jenvman.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 73.Kilonzo C, Li X, Vivas EJ, Jay-Russell MT, Fernandez KL, Atwill ER. 2013. Fecal shedding of zoonotic food-borne pathogens by wild rodents in a major agricultural region of the central California coast. Appl Environ Microbiol 79:6337–6344. doi: 10.1128/AEM.01503-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jay M, Cooley M. 2007. Escherichia coli O157:H7 in feral swine near spinach fields and cattle, central California coast. Emerg Infect Dis 13:1908–1911. doi: 10.3201/eid1312.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Atwill ER. 2008. Implications of wildlife in E. coli outbreaks associated with leafy green produce, p 5–6. In Timm R, Madon M (ed), Proceedings of the 23rd Vertebrate Pest Conference University of California, Davis, Davis, CA. [Google Scholar]
- 76.Tate KW, Atwill ER, Bartolome JW, Nader G. 2006. Significant Escherichia coli attenuation by vegetative buffers on annual grasslands. J Environ Qual 35:795–805. doi: 10.2134/jeq2005.0141. [DOI] [PubMed] [Google Scholar]
- 77.Koelsch RK, Lorimer JC, Mankin KR. 2006. Vegetative treatment systems for management of open lot runoff: review of literature. Appl Eng Agric 22:141–153. doi: 10.13031/2013.20190. [DOI] [Google Scholar]
- 78.El-Khateeb MA, Al-Herrawy AZ, Kamel MM, El-Gohary FA. 2009. Use of wetlands as post-treatment of anaerobically treated effluent. Desalination 245:50–59. doi: 10.1016/j.desal.2008.01.071. [DOI] [Google Scholar]
- 79.Bates D, Mächler M, Bolker BM, Walker SC. 2014. Fitting linear mixed-effects models using lme4. arXiv arXiv:1406.5823v1 [stat.CO]. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.