Abstract
An outstanding challenge toward efficient production of biofuels and value-added chemicals from plant biomass is the impact that lignocellulose-derived inhibitors have on microbial fermentations. Elucidating the mechanisms that underlie their toxicity is critical for developing strategies to overcome them. Here, using Escherichia coli as a model system, we investigated the metabolic effects and toxicity mechanisms of feruloyl amide and coumaroyl amide, the predominant phenolic compounds in ammonia-pretreated biomass hydrolysates. Using metabolomics, isotope tracers, and biochemical assays, we showed that these two phenolic amides act as potent and fast-acting inhibitors of purine and pyrimidine biosynthetic pathways. Feruloyl or coumaroyl amide exposure leads to (i) a rapid buildup of 5-phosphoribosyl-1-pyrophosphate (PRPP), a key precursor in nucleotide biosynthesis, (ii) a rapid decrease in the levels of pyrimidine biosynthetic intermediates, and (iii) a long-term generalized decrease in nucleotide and deoxynucleotide levels. Tracer experiments using 13C-labeled sugars and [15N]ammonia demonstrated that carbon and nitrogen fluxes into nucleotides and deoxynucleotides are inhibited by these phenolic amides. We found that these effects are mediated via direct inhibition of glutamine amidotransferases that participate in nucleotide biosynthetic pathways. In particular, feruloyl amide is a competitive inhibitor of glutamine PRPP amidotransferase (PurF), which catalyzes the first committed step in de novo purine biosynthesis. Finally, external nucleoside supplementation prevents phenolic amide-mediated growth inhibition by allowing nucleotide biosynthesis via salvage pathways. The results presented here will help in the development of strategies to overcome toxicity of phenolic compounds and facilitate engineering of more efficient microbial producers of biofuels and chemicals.
INTRODUCTION
Lignocellulosic biomass constitutes a renewable substrate for the sustainable production of biofuels and other added-value chemicals (1). However, the sugars in lignocellulosic biomass are not easily accessible to most microbial fermenters, as they exist as sugar polymers (cellulose and hemicellulose) tightly bound by lignin. Biomass pretreatment processes coupled to enzymatic hydrolysis are typically required to break down this lignin barrier and transform sugar polymers into easily fermentable monosaccharides such as glucose and xylose (2–4).
Unfortunately, biomass pretreatment processes are often accompanied by the generation of a variety of lignocellulose-derived compounds that are detrimental to microbial fermentations and lead to inefficient conversion of sugars into biofuels (5–8). Elucidating the mechanisms underlying the toxicity of this diverse set of microbial inhibitors, and finding ways to overcome them, continues to be an area of intense research (9–12).
The most commonly used biomass pretreatment processes are acid based, which generate toxic sugar-derived inhibitors such as furfural and 5-hydroxymethyl-furfural (HMF) (13–19). Microbes such as Saccharomyces cerevisiae and Escherichia coli are capable of detoxifying these compounds via energy-consuming, NADPH-dependent processes (15, 16, 20–23). However, these detoxification pathways are thought to drain cellular resources and result in depletion of key intracellular metabolites and redox cofactors (17, 18, 24, 25). For instance, when exposed to furfural, E. coli increases expression of cysteine and methionine biosynthetic genes as a response to decreased levels of sulfur-containing amino acids. It was proposed that the reductive detoxification of furfural leads to NADPH depletion, which in turn limits sulfur assimilation into amino acids and leads to growth inhibition (11). Supporting this hypothesis, it was shown that overexpression of a NADH-dependent furfural reductase prevents NADPH depletion and leads to increased furfural tolerance in E. coli (14). Studies in other biofuel producers, such as Clostridium thermocellum (13), Clostridium acetobutylicum (26), and Clostridium beijerinckii (27), also support the idea that furfural detoxification leads to NADPH depletion, which could hinder sulfur assimilation and other important cellular processes.
Alkaline pretreatments such as ammonia fiber expansion (AFEX) are a favorable alternative to acid-based pretreatments since they produce smaller amounts of HMF and furfural and are better at preserving xylose and other essential nutrients present in plant biomass (28). Nonetheless, ammonia-based pretreatments generate a variety of lignocellulose-derived phenolic inhibitors (LDPIs), including phenolic amides, carboxylates, and aldehydes (29). The toxicity mechanisms of these aromatic inhibitors, especially phenolic amides, remain largely unexplored. LDPIs affect microbial growth on glucose and xylose, although their inhibitory effects are considerably stronger for xylose utilization (9). Most LDPIs (e.g., feruloyl amide, coumaroyl amide, and their carboxylate counterparts) cannot be metabolized by biofuel producers such as E. coli, which arguably makes them a more persistent problem during fermentations than other known inhibitors (10). Although the inhibitory mechanisms of individual LDPIs have never been reported, a recent study in E. coli explored the transcriptional regulatory responses to the set of inhibitors present in AFEX-pretreated corn stover hydrolysates (ACSHs), which are characterized by high concentrations of phenolic amides and phenolic carboxylates (30). Aldehyde detoxification and aromatic carboxylate efflux pumps were shown to be transcriptionally upregulated in response to this set of inhibitors. This upregulation was accompanied by a buildup of pyruvate, depletion of ATP and NAD(P)H, and a strong inhibition of xylose utilization. It was suggested that inhibitor efflux and detoxification exhaust cellular energy, thereby inhibiting growth and biofuel production (30).
Despite these recent advances, much remains to be learned about the toxicity of LDPIs. In this study, we used liquid chromatography-mass spectrometry (LC-MS)-based metabolomics, isotopic tracers, and biochemical assays to investigate the metabolic effects and underlying toxicity mechanisms of feruloyl amide and coumaroyl amide, the predominant phenolic inhibitors found in ACSH. Using E. coli fermentations as a model system, we explored the hypothesis that these phenolic amides might be direct inhibitors of bacterial metabolism. We report that both feruloyl amide and coumaroyl amide act as potent and fast-acting inhibitors of purine and pyrimidine de novo biosynthesis and that these deleterious effects are at least partially mediated via direct inhibition of the glutamine amidotransferases that participate in these biosynthetic pathways.
MATERIALS AND METHODS
Media, culture conditions, and metabolite extractions.
Escherichia coli RL3000 (E. coli K-12 substrain MG1655 rph+ ilvG+; a gift from Robert Landick, University of Wisconsin—Madison) was maintained on Luria-Bertani (LB) agar plates. Fermentations and metabolite extractions were performed under anaerobic conditions (<300 ppm O2) inside an environmental chamber (Coy Laboratory) with an atmosphere of 90% nitrogen, 5% hydrogen, and 5% carbon dioxide. All experiments were done in M9 liquid minimal medium (1.28% [wt/vol] Na2HPO4·7H2O, 0.3% [wt/vol] KH2PO4, 0.05% [wt/vol] NaCl, 0.1% [wt/vol] NH4Cl, 1 mM MgSO4, 0.1 mM CaCl2, 7.2 μM FeCl3, 1% [wt/vol] xylose or glucose, pH 7.0).
For the preparation of anaerobic fermentations, an overnight anaerobic culture was used to inoculate (at at least a 1:100 dilution) a liquid minimal medium culture to an initial optical density at 600 nm (OD600) of 0.05. When the culture reached mid-exponential phase (OD600 of ∼0.4), it was divided into separate flasks containing medium with different concentrations of lignin-derived phenolic inhibitors (LDPIs) (see Table S1 in the supplemental material) and/or nucleoside cocktail. At 0, 10, 30, 60, 120, 180, and 240 min after the addition of LDPIs, intracellular metabolite samples were collected by rapidly filtering 5 ml of culture through 47-mm-diameter round hydrophilic nylon filters (Millipore catalog no. HNWP04700). Filters containing the cells were then immediately submerged into 1.5 ml of −20°C acetonitrile-methanol-water (40:40:20), which quenches metabolism and extracts metabolites. Metabolites and cell debris were thoroughly washed from filter before the solvent was subject to centrifugation at 20,817 × g for 5 min at 4°C. The supernatant was stored at −20°C until analysis.
To determine xylose consumption rates, 1-ml bacterial culture samples were collected by centrifugation, and the supernatant was analyzed by high-pressure liquid chromatography (HPLC)-MS using [13C]xylose as an internal standard.
The feruloyl amide and coumaroyl amide used in this study were synthesized as previously described (30). Synthesized amides exceeding ∼90% purity were used for experiments. Feruloyl amide contained ∼10% (wt/wt) ammonium hydroxide, while coumaroyl amide was ammonia free. All other LDPIs were purchased from Sigma-Aldrich. The nucleoside cocktail used in nucleoside supplementation experiments contained adenosine, guanosine, cytidine, thymidine, and uridine in equimolar concentrations, either 0.25 or 0.75 mM. Cellular growth was monitored using OD600.
Metabolite measurements.
Metabolites in extraction solvent were dried with N2 and resuspended in LC-MS-grade water (Sigma-Aldrich). Samples were analyzed using an HPLC-tandem MS (HPLC-MS/MS) system consisting of a Dionex UHPLC coupled by electrospray ionization (ESI) (negative mode) to a hybrid quadrupole–high-resolution mass spectrometer (Q Exactive Orbitrap; Thermo Scientific) operated in full-scan mode for detection of targeted compounds based on their accurate masses. Liquid chromatography (LC) separation was achieved using either a Synergi Fusion-RP 100A column (100 by 2 mm, 2.5-μm particle size; Phenomenex, Torrance, CA) or an Acquity UPLC BEH C18 column (2.1- by 100-mm column, 1.7-μm particle size). Solvent A was 97:3 water-methanol with 10 mM tributylamine (TBA) and 10 mM acetic acid, pH ∼8.2; solvent B was 100% methanol. The total run time was 25 min with the following gradient: 0 min, 5% B; 2.5 min, 5% B; 5 min, 20% B; 7.5 min, 20% B; 13 min, 55% B; 15.5 min, 95% B; 18.5 min, 95% B; 19 min, 5% B; and 25 min, 5% B. Metabolite peaks were identified using the Metabolomics Analysis and Visualization Engine (MAVEN) (31, 32). Measurement of xylose concentrations was achieved similarly using a 15-min chromatographic run. Xylose uptake rates (millimolar/hour/OD unit) were obtained from nonlinear least squares (curve fitting).
Isotope tracer experiments using 13C and 15N.
Using LC-MS, we followed the dynamic incorporation of 13C from [1,2-13C]xylose into downstream metabolites after exposure to phenolic amides. Exponential-phase E. coli cells growing anaerobically on [12C]xylose were pretreated with feruloyl or coumaroyl amide for 5 min; cells were then rapidly switched to medium containing [1,2-13C]xylose (0.3%, wt/vol) and sampled for intracellular metabolite analysis at 0.5, 1, 2, 4, 7, 10, and 15 min. Rapid interchange of nonlabeled to 13C-labeled medium was performed by rapid filtration using syringe filters (nylon membrane filter, 0.45 μm, 25 mm; Fisher Scientific, Pittsburgh, PA). Briefly, a 10-ml culture was filtered to remove the nonlabeled medium and immediately switched to medium containing [1,2-13C]xylose (and the appropriate phenolic inhibitor concentration) by passing 3 ml of the labeled medium through the syringe filter. Metabolite extractions were then done by passing 1.5 ml of −20°C acetonitrile-methanol-water (40:40:20) and collecting the eluent. Metabolite measurements were performed as described above. The isotopic tracer experiments using universally labeled [13C]glucose and 15NH4Cl were performed similarly. All isotopic tracers were purchased from Cambridge Isotope Laboratories (Andover, MA).
For obtaining positional labeling information of nucleoside triphosphates (ATP and UTP), cell extracts were subjected to MS/MS fragmentation. Instrument parameters for MS/MS analysis are shown in Table S2 in the supplemental material.
Enzymatic assays.
E. coli AG1 overexpressing glutamine-dependent amidotransferases, glutamine 5-phosphoribosyl-1-pyrophosphate (PRPP) amidotransferase (PurF), CTP synthetase (PyrG), GMP synthetase (GuaA), and carbamoyl phosphate synthase (CarAB), were obtained from the ASKA collection (National BioResource Project, Japan) (33) and maintained on LB with 30 μg/ml chloramphenicol. The structure of open reading frame (ORF) clones was confirmed as described in reference 33. Affinity chromatography was performed for enzyme purification as per the manufacturer's instructions (Ni-nitrilotriacetic acid [NTA] fast-start kit; Qiagen). Guanylate kinase (Gmk) was purified from Sinorhizobium meliloti (34), and pyruvate kinase was purchased from Sigma-Aldrich.
Enzymatic reactions for PurF, PyrG, GuaA, and CarAB were performed in Tris-HCl buffer (50 mM Tris-HCl, 10 mM MgCl2, pH 8.0). Tested substrates and their concentrations were 1 mM glutamine and 0.25 mM PRPP for PurF activity, 1 mM glutamine, 0.1 mM ATP, and 0.1 mM UTP for PyrG activity, 1 mM glutamine, 0.1 mM XMP, and 0.1 mM ATP for GuaA activity, 0.12 mM glutamine, 1 mM ATP, and 20 mM bicarbonate (HCO3−) for CarA/CarB activity, 1 mM GMP and 1 mM ATP for Gmk activity, and 1 mM phosphoenolpyruvate (PEP) and 1 mM ADP for Pyk activity. Enzymatic reactions were monitored by measuring substrate consumption and/or product formation using LC-MS; samples were taken at various time intervals and rapidly quenched using cold organic solvent (40:40:20 methanol-acetonitrile-water) before analysis. Enzyme activity was determined as initial velocity (V0) obtained from the slope of product concentration versus time; V0 under each condition was then normalized to that of the no-inhibitor control (100% activity). Enzymatic reactions for Gmk and Pyk were carried out at 30°C in Tris-HCl buffer (100 mM Tris-HCl [pH 7.5], 100 mM KCl, 10 mM MgCl2). Substrate concentrations were 1 mM ATP and 1 mM GMP for Gmk and 1 mM ADP and 1 mM phosphoenolpyruvate for Pyk.
To determine the mode of PurF inhibition by feruloyl amide, cell lysates from E. coli RL3000 aerobically grown in M9 minimal medium with 1% glucose were prepared in Tris-HCl buffer. Glutamine transfer reactions were carried out at 37°C in Tris-HCl buffer containing 4 mM PRPP and 2.5, 5, 10, 15, or 20 mM glutamine. Reactions were quenched at 2, 5, 10, 15, 20, 30, and 40 min after initiation. Glutamate concentrations measured by LC-MS were used for calculation of v0. Data were fitted into a competitive inhibition equation: V0 = Vmax · [S]/(Km · (1 + [I]/Ki) + [S]) using GraphPad Prism version 6.0f for Mac OSX (GraphPad Software, San Diego, CA, USA). Data points were transformed into the Hanes-Woolf equation [S]/V0 = [S]/Vmax + Km/Vmax.
RESULTS
Effects of lignocellulose-derived phenolic amides on the anaerobic growth of E. coli.
Ammonia fiber expansion (AFEX) is a biomass pretreatment technology that can be applied to agricultural residues such as corn stover, straw, or switchgrass. It allows near-complete conversion of cellulose and hemicellulose to fermentable, monomeric sugars and produces a liquid hydrolysate that contains approximately 6% glucose and 3% xylose (35). As shown in Table S1 in the supplemental material, the recently characterized components of AFEX-pretreated corn stover hydrolysate (ACSH) include phenolic amides, phenolic carboxylates, and phenolic aldehydes (30). Here, using E. coli xylose fermentations as our primary experimental system, we focused on investigating the metabolic effects and possible inhibitory mechanisms of feruloyl amide and coumaroyl amide, the two most abundant lignocellulose-derived phenolic inhibitors (LDPIs) in ACSH.
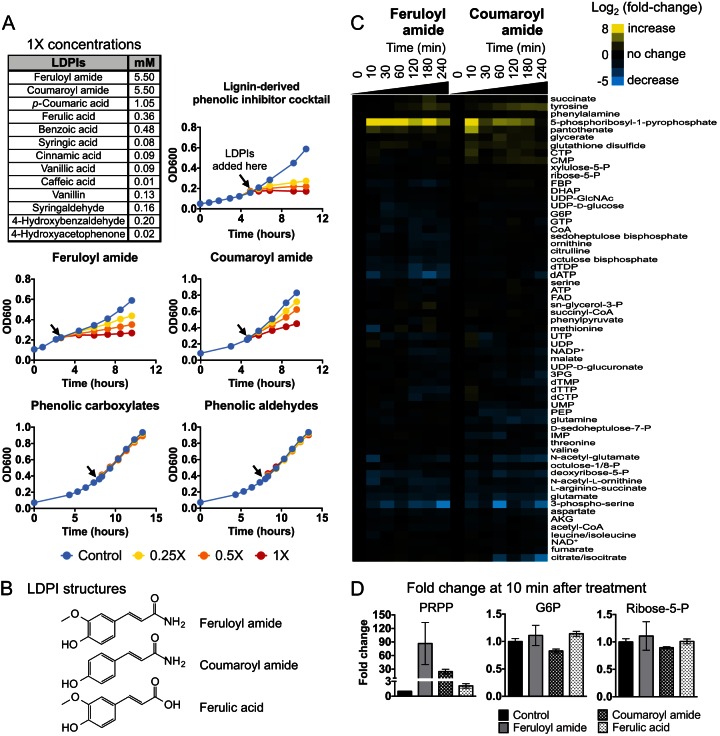
To test the inhibitory effects of LDPIs on anaerobic growth on xylose, we treated exponential-phase E. coli cultures with either the full set of, a subset of, or individual LDPIs present in ACSH (Fig. 1A). The complete LDPI cocktail, composed of 13 different phenolic compounds, rapidly and effectively inhibited growth in a dose-dependent manner. However, using individual LDPIs, strong growth inhibition was observed only with feruloyl amide and coumaroyl amide; phenolic carboxylates and phenolic aldehydes showed no inhibitory effects at the concentrations present in ACSH (Fig. 1A). Growth inhibition by feruloyl amide and coumaroyl amide correlated with an inhibition of xylose consumption (see Fig. S1 in the supplemental material), and their combined effects nearly recapitulated the growth inhibition of the complete LDPI cocktail (see Fig. S2 in the supplemental material). Therefore, these two phenolic amides likely represent the major contributors to anaerobic growth inhibition in this system.
FIG 1.
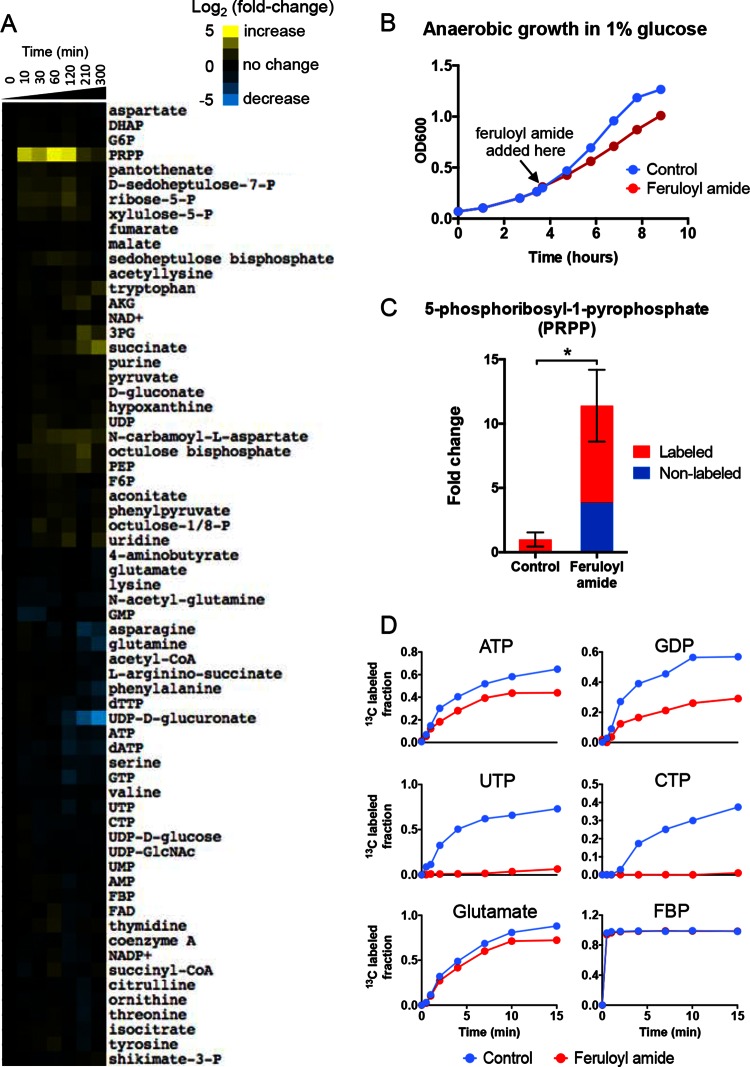
Phenolic amides hinder anaerobic growth and alter intracellular metabolite levels. (A) E. coli cultures were grown anaerobically in M9 minimal medium containing 1% (wt/vol) xylose as the single carbon source. At the time indicated by the arrows (early/mid-exponential phase), lignocellulose-derived phenolic inhibitors (LDPIs) were added at various concentrations (1×, 0.5×, and 0.25×). The 1× concentrations are shown in the top-left table and refer to those present in AFEX-pretreated corn stover hydrolysate (29). Feruloyl amide and coumaroyl amide were the two major contributors to growth inhibition among this set of LDPIs. Phenolic carboxylates and aldehydes showed no growth-inhibitory effects at the concentrations shown here. The phenolic carboxylate mix contained ferulic acid, p-coumaric acid, benzoic acid, syringic acid, cinnamic acid, vanillic acid, and caffeic acid. The phenolic aldehyde mix contained vanillin, syringaldehyde, 4-hydroxybenzaldehyde, and 4-hydroxyacetophenone. The data shown are representative of three biological replicates. (B) Molecular structures of feruloyl amide, coumaroyl amide, and ferulic acid. (C) Exponential-phase E. coli cultures grown anaerobically on xylose were treated with 5.5 mM feruloyl amide or coumaroyl amide. Intracellular metabolites were extracted at 10, 30, 60, 120, 180, and 240 min after exposure to phenolic amides and measured by LC-MS. Metabolite levels in treated samples were normalized against those in controls at each corresponding time point. Metabolites whose level increases upon phenolic amide treatment are shown in shades of yellow, and those that decrease are shown in shades of blue based on their log2 fold change over nontreated controls. The data represent the averages of two biological replicates. (D) Average fold changes in 5-phosphoribosyl-1-pyrophosphate (PRPP), glucose-6-phosphate (G6P), and ribose-5-phosphate at 10 min after treatment with 5.5 mM feruloyl amide, coumaroyl amide, or ferulic acid. Bars represent the mean of four biological replicates ± standard error of the mean (SEM). Abbreviations: P, phosphate; FBP, fructose-1,6-bisphosphate; DHAP, dihydroxyacetone phosphate; G6P, glucose-6-phosphate; PEP, phosphoenolpyruvate; AKG, α-ketoglutarate.
Exposure to phenolic amides results in rapid accumulation of the biosynthetic intermediate PRPP.
Using liquid chromatography coupled with mass spectrometry (LC-MS), we looked for alterations in intracellular metabolite levels resulting from the addition of inhibitory concentrations (5.5 mM) of feruloyl or coumaroyl amide to exponential-phase E. coli cultures growing anaerobically on xylose. At various time points after addition of each phenolic amide, samples were collected for intracellular metabolite extraction and analyzed by a set of LC-MS methods which together enable the analysis of ∼200 known metabolites in central catabolic and biosynthetic pathways. Quantitative data encompassing both treatments and the entire time course were obtained for 62 metabolites. The dynamics of these metabolites after phenolic amide treatment (normalized to the levels of nontreated controls) are shown in heat map format in Fig. 1C.
Starting with feruloyl amide, our analysis revealed that addition of this compound led to drastic changes in the intracellular levels of a large number of metabolites. In many instances, these alterations were seen within 10 min after treatment. There was also a general trend of long-term decreases in the levels of a large number of metabolites, including nucleotides, amino acids, and intermediates in glycolysis and the tricarboxylic acid (TCA) cycle. Interestingly, the levels of intermediates in the pentose phosphate pathway (PPP) (i.e., xylulose-5-phosphate, ribose-5-phosphate, and d-sedoheptulose-7-phosphate) were not strongly affected. Among all of the alterations elicited by feruloyl amide treatment, the most striking one was a rapid and sustained increase (>50-fold at 10 min after treatment [Fig. 1D]) in the level of 5-phosphoribosyl-1-pyrophosphate (PRPP). PRPP is an intermediate produced from ribose-5-phosphate; it serves as the phospho-ribose donor in all de novo nucleotide biosynthetic pathways and in a subset of nucleotide salvage pathways. PRPP is also used for the biosynthesis of NAD both through de novo biosynthesis and via salvage pathways (36).
We observed similar results when E. coli cells were exposed to coumaroyl amide. As with feruloyl amide treatment, one of the strongest effects was the large and rapid increase in the levels of PRPP (over 20-fold at 10 min after treatment [Fig. 1D]). Here, we also found a generalized, albeit less strong in some cases, long-term decrease in the levels of several glycolytic and TCA cycle intermediates, amino acids, and nucleotides. PPP intermediates still remained minimally affected.
As a comparison to the effects of phenolic amides, we also tested for alterations in intracellular metabolite levels resulting from the addition of ferulic acid (5.5 mM), the acid counterpart of feruloyl amide. Treatment with ferulic acid displayed many similarities to feruloyl amide and coumaroyl amide treatments, particularly with respect to the decrease in deoxyribonucleotide levels (see Fig. S3 in the supplemental material). However, it resulted in only a comparatively small increase in the levels of PRPP (∼2-fold) (Fig. 1D), which suggests that this effect is enhanced by the presence of the amide group in feruloyl amide.
The strong similarity between the effects on metabolite levels of feruloyl amide and coumaroyl amide suggests that these two compounds likely share common mechanisms of action, which is reasonable given their structural similarities (Fig. 1B). Most notably, the rapid accumulation of PRPP and the concomitant long-term decrease in nucleotide levels that occurred in response to feruloyl amide and coumaroyl amide treatment, pointed to metabolic blockages downstream of PRPP and suggested that these two compounds might be inhibitors of nucleotide biosynthetic pathways.
Metabolic flux into purine and pyrimidine biosynthesis is inhibited by phenolic amides. (i) 13C-labeling experiments.
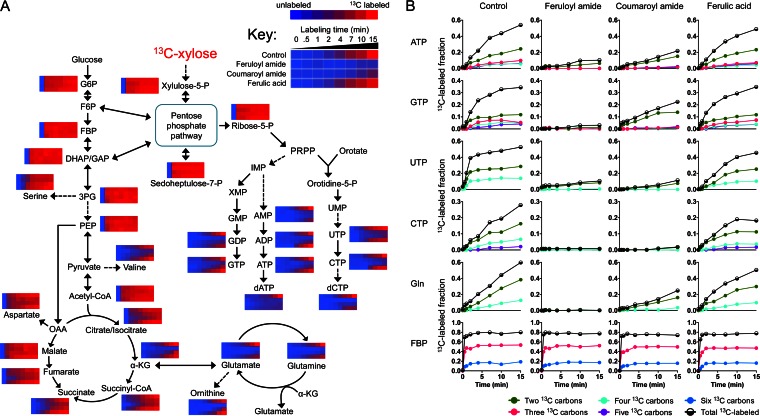
To test the hypothesis that feruloyl amide and coumaroyl amide act as inhibitors of nucleotide biosynthetic pathways, we performed isotopic tracer analyses with [13C]xylose. Using LC-MS, we followed the dynamic incorporation of 13C from [1,2-13C]xylose into downstream metabolites after exposure to feruloyl or coumaroyl amide. In these experiments, exponential-phase E. coli cells growing anaerobically on nonlabeled xylose were pretreated with feruloyl or coumaroyl amide (5.5 mM) for 5 min; cells were then rapidly switched to medium containing [1,2-13C]xylose (maintaining the inhibitor concentration at 5.5 mM) and sampled for intracellular metabolite analysis at 0.5, 1, 2, 4, 7, 10, and 15 min (see Materials and Methods).
As shown in Fig. 2, the incorporation of 13C into metabolites in the PPP, glycolysis, or reductive part of the TCA cycle was not greatly affected in cells exposed to feruloyl or coumaroyl amide. Specifically, the 13C-labeling dynamics of the PPP intermediates xylulose-5-phosphate, ribose-5-phosphate, and d-sedoheptulose-7-phosphate were comparable between controls and feruloyl or coumaroyl amide-treated samples. This pattern was also true for all measured glycolytic intermediates, including glucose-6-phosphate (G6P), fructose-1,6-bisphosphate (FBP), phosphoenolpyruvate (PEP), and the downstream metabolite acetyl coenzyme A (acetyl-CoA). Similarly, 13C-labeling dynamics of metabolites in the reductive TCA cycle (i.e., malate, fumarate, and succinate) were only minimally affected.
FIG 2.
Carbon flux into purine and pyrimidine biosynthetic pathways is inhibited by phenolic amides. (A) Using LC-MS, we followed the dynamic incorporation of 13C from [1,2-13C]xylose into downstream metabolites after exposure to phenolic amides or ferulic acid. Exponential-phase E. coli cultures growing anaerobically on nonlabeled xylose were pretreated with 5.5 mM feruloyl amide, coumaroyl amide, or ferulic acid for 5 min before switching them from nonlabeled xylose to [1,2-13C]xylose. Samples for intracellular metabolite analysis were then taken at 0.5, 1, 2, 4, 7, 10, and 15 min thereafter. The blue-to-red gradient represents the amount of 13C label incorporation (sum of all 13C-labeled forms) into each metabolite as a fraction of its total pool of the control sample; for each metabolite, the lowest value (nonlabeled) was set to blue, while the highest 13C-label fraction value in control samples was set to red. The data represent the average of two biological replicates. (B) E. coli cells were pretreated with 5.5 mM feruloyl amide, coumaroyl amide, or ferulic acid for 5 min before switching to [1,2-13C]xylose. The x axis represents minutes after the switch to [1,2-13C]xylose, and the y axis represents the fraction of the observed compound of the indicated isotopic form (containing different number of 13C atoms). The data shown are representative of two biological replicates.
In contrast, the incorporation of 13C into intermediates and downstream products of the purine and pyrimidine biosynthetic pathways was strongly impaired by phenolic amides (Fig. 2). Compared to controls, the 13C-labeling dynamics of all measured nucleoside monophosphates, diphosphates, and triphosphates were slowed in the presence of both feruloyl amide and coumaroyl amide. This negative effect on 13C flux was further reflected in downstream deoxyribonucleotides (e.g., dATP and dCTP). Interestingly, we also observed that 13C incorporation into intermediates in nitrogen/ammonia assimilation (i.e., α-ketoglutarate, glutamate, and glutamine) was also delayed. The effects of both phenolic amides on 13C-labeling dynamics were conserved, although there was an overall trend for these effects to be somewhat stronger with feruloyl amide than with coumaroyl amide.
With few exceptions, the observed effects on 13C-labeling dynamics discussed above were generally not reproduced in cultures treated with ferulic acid. As shown in Fig. 2, feruloyl amide and coumaroyl amide elicited a stronger inhibitory effect on nucleotides and nitrogen assimilation metabolites than ferulic acid. However, the 13C incorporation into UTP was slightly hindered, and that into dCTP and dATP was considerably impeded.
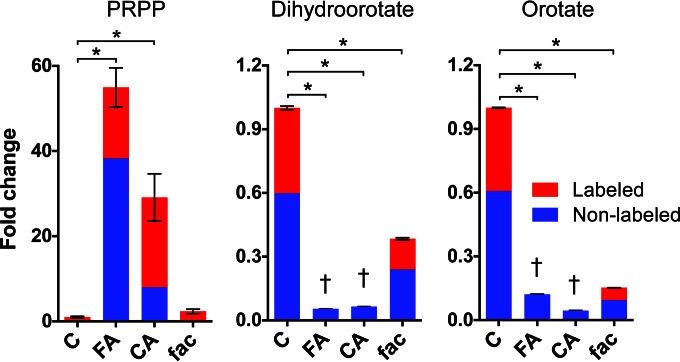
Figure 3 presents a closer examination of the 13C-labeling patterns in the purine biosynthetic precursor PRPP and the pyrimidine biosynthetic intermediates dihydroorotate and orotate at 10 min after the switch to [1,2-13C]xylose (i.e., 10 min after the 5-min pretreatment with inhibitors). As we saw before, PRPP levels increased dramatically after treatment with feruloyl amide and coumaroyl amide. The accumulation of both nonlabeled and [13C]PRPP denotes a severe metabolic block downstream of PRPP biosynthesis, which is consistent with the inhibition of 13C incorporation into downstream purine nucleotides just described. Since we did not detect the accumulation of any other purine biosynthetic intermediate, this suggested that feruloyl amide and coumaroyl amide might be inhibiting the glutamine-dependent conversion of PRPP into 5-phosphoribosylamine. This reaction represents the first committed step in purine biosynthesis and is catalyzed by glutamine PRPP amidotransferase (PurF in E. coli, also known as amidophosphoribosyl transferase). In contrast to the case for PRPP, the intracellular levels and 13C incorporation into the pyrimidine biosynthetic intermediates dihydroorotate and orotate decreased drastically after feruloyl amide and coumaroyl amide treatment (Fig. 3), which suggested inhibition of a biosynthetic step upstream of dihydroorotate. Interestingly, the first committed step in pyrimidine biosynthesis is also catalyzed by a glutamine amidotransferase, carbamoyl phosphate synthetase (CarAB), which transfers the side chain amino group of glutamine into bicarbonate to form carbamoyl-phosphate. It is therefore possible that feruloyl amide and coumaroyl amide might also be inhibitors of this glutamine amidotransferase enzyme. An alternative hypothesis to explain the low levels of dihydroorotate and orotate after phenolic amide treatment is that the large increase in PRPP levels shifts the equilibrium of the reaction catalyzed by orotate phosphoribosyltransferase, which combines orotate and PRPP to produce orotidine-5-phosphate, toward product formation. However, this hypothesis seems less likely given the strong inhibition of 13C incorporation into downstream metabolites such as UTP and CTP. Finally, although ferulic acid treatment had only a small effect on PRPP levels, it resulted in a decrease in dihydroorotate and orotate levels (Fig. 3). Nonetheless, we still observed 13C incorporation into orotate and dihydroorotate in the presence of ferulic acid. These partial and relatively small inhibitory effects on purine and pyrimidine biosynthesis by ferulic acid might help explain the comparatively small but still visible inhibitory effects on 13C incorporation into downstream dATP, UTP, and dCTP that we described above.
FIG 3.
Alterations in 13C-labeling patterns and intracellular levels of purine and pyrimidine precursors in response to phenolic amides. Anaerobic, exponential-phase E. coli cultures were exposed to 5.5.mM feruloyl amide, coumaroyl amide, or ferulic acid for 5 min before switching them to [1,2-13C]xylose. The graphs show the average fold change (versus nontreated controls) in intracellular levels and 13C-labeled fractions at 10 to 15 min after the addition of [13C]xylose. The large accumulation of both nonlabeled and [13C]PRPP in the presence of phenolic amides denotes a severe metabolic block downstream of PRPP biosynthesis. The decrease in pyrimidine biosynthetic intermediates dihydroorotate and orotate suggests inhibition of a biosynthetic step upstream of dihydroorotate. Bars represent averages for four biological samples ± SEM. t-test statistical significance: *, P < 0.001. 13C-labeled forms of these metabolites were below background noise levels. C, control; FA, feruloyl amide; CA, coumaroyl amide; fac, ferulic acid.
(ii) 15N-labeling experiments.
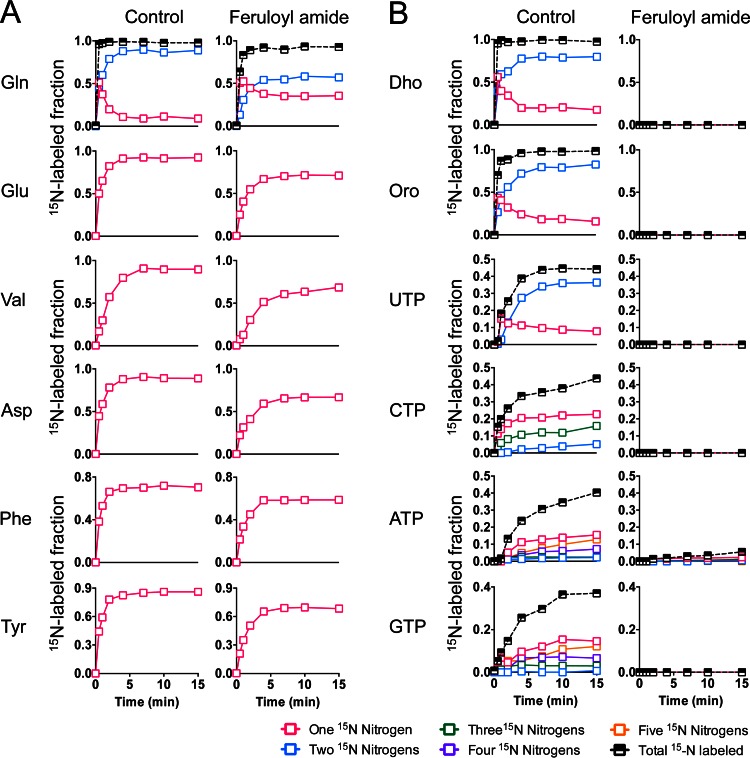
The strong inhibition of 13C incorporation into glutamate and glutamine suggested that coumaroyl amide and feruloyl amide might be interfering with ammonia assimilation pathways, either glutamate dehydrogenase (GDH) or the glutamine synthetase/glutamate synthase cycle (GS/GOGAT). This potential inhibition of nitrogen assimilation might in turn hinder glutamine-dependent de novo nucleotide biosynthesis. To investigate whether the inhibition of nucleotide synthesis in the presence of phenolic amides might be a downstream effect of the inhibition of ammonia assimilation pathways, we performed isotopic tracer experiments using 15N-labeled ammonia (15NH4Cl). Using the same experimental setup that was used for 13C-labeling experiments, exponential-phase E. coli cells growing anaerobically on xylose were pretreated with feruloyl amide for 5 min before nonlabeled ammonia was replaced with 15NH4Cl. We then followed the dynamic incorporation of 15N into amino acids, nucleotides, and the pyrimidine biosynthetic intermediates dihydroorotate and orotate. As shown in Fig. 4A, the 15N-labeling dynamics of glutamine, glutamate, and most other amino acids were only minimally affected in the presence of feruloyl amide (the small effect on final 15N-labeled fractions was due to small amounts of nonlabeled ammonia present in feruloyl amide [see Materials and Methods]). In contrast, 15N incorporation into dihydroorotate, orotate, and nucleotides was blocked almost completely in most instances. These data independently support the findings from our 13C-labeling experiments and indicate that the strong inhibition of de novo nucleotide biosynthetic pathways is not driven primarily by an inhibition of ammonia assimilation pathways.
FIG 4.
15N-labeling dynamics of nucleotides, amino acids, and biosynthetic intermediates following feruloyl amide treatment. Nitrogen incorporation into purine and pyrimidine biosynthetic pathways is inhibited by phenolic amides. Exponential-phase E. coli cultures exposed to 5.5 mM feruloyl amide for 5 min were switched into 15NH4Cl. The data represent the average of two biological replicates. Incorporation of 15N into glutamine, glutamate, and other amino acids was minimally affected (A), while that into nucleotides and intermediates in pyrimidine biosynthesis was strongly inhibited (B), by feruloyl amide. Abbreviations: FBP, fructose-1,6-bisphosphate; Gln, glutamine; Glu, glutamate; Val, valine; Asp, aspartate; Phe, phenylalanine; Tyr, tyrosine; Dho, dihydroorotate; Oro, orotate.
Interestingly, although 15N incorporation into nucleotides such as UTP or ATP was almost completely blocked in the presence of feruloyl amide (Fig. 4B), we still observed some incorporation of 13C into these metabolites (Fig. 2B). However, while in nontreated controls we observed the appearance of different labeled forms of UTP (containing 2 or 4 13C atoms) and ATP (containing 2, 3, 4, or 5 13C atoms), in the presence of feruloyl amide we observed only the appearance of UTP and GTP containing two 13C atoms. We suspected that in the presence of feruloyl amide, 13C incorporation into these nucleotides occurs exclusively into the ribose sugar moiety and not into the pyrimidine or purine. To test this hypothesis we used tandem mass spectrometry (MS/MS) to obtain positional 13C-labeling information. We observed that while 13C atoms were present in both the ribose sugars and nucleobases of UTP and GTP in the controls, in feruloyl amide-treated cells the two 13C atoms were found exclusively in the ribose sugar moiety (see Fig. S5 in the supplemental material). This observation indicated that the small amount of 13C incorporation into nucleotides in the presence of phenolic amides might be taking place via nucleobase recycling using salvage pathways.
(iii) The inhibitory effects of feruloyl amide are conserved in glucose fermentations.
Growth inhibition by feruloyl amide was considerably stronger in xylose fermentations than in glucose fermentations (see Fig. S2 in the supplemental material). Nonetheless, we found that the inhibitory effects of feruloyl amide on nucleotide biosynthetic pathways were well conserved across both carbon sources (Fig. 5). Specifically, the rapid and large increase in PRPP levels after feruloyl amide treatment was also observed in glucose fermentations (Fig. 5C). In addition, isotopic tracers experiments using [13C]glucose also showed a strong inhibition of 13C incorporation into purine and pyrimidine nucleotides. However, while the effects on pyrimidine biosynthesis were almost equally strong, those on purine biosynthesis were less pronounced (Fig. 5D).
FIG 5.
The inhibitory effects of feruloyl amide are conserved in glucose fermentations. All experiments were performed similarly to xylose fermentations. (A) Anaerobic E. coli culture were grown on 1% glucose and treated with 5.5 mM feruloyl amide at mid-exponential phase (OD600 of ∼0.4). Intracellular metabolites were collected at 0, 10, 30, 60, 120, 210, and 300 min after exposure to feruloyl amide and analyzed by LC-MS. The data represent the average of two biological replicates. (B) Anaerobic growth inhibition by 5.5 mM feruloyl amide. (C) Rapid PRPP accumulation after the feruloyl amide treatment. Similar to the observation in cells grown in xylose, PRPP rapidly increased upon exposure to 5.5 mM feruloyl amide. The accumulated PRPP was 13C labeled, indicating a metabolic bottleneck downstream of PRPP synthesis. Bars represent means of four biological replicates ± SEM. The fold change shown is nontreated control cells. (D) Isotopic tracers experiments using [13C]glucose also showed inhibition of 13C incorporation into purine and pyrimidine nucleotides.
Nucleoside supplementation relieves growth inhibition by phenolic amides.
Our results have shown that feruloyl amide and coumaroyl amide strongly inhibit de novo purine and pyrimidine biosynthetic pathways. The inhibition of these biosynthetic pathways is likely a primary contributor to the observed growth inhibition. Since the results of isotopic tracer experiments also suggested that salvage pathways remain active in the presence of phenolic amides, we hypothesized that external nucleoside supplementation should allow the production of nucleotides via salvage pathways and relieve this growth inhibition.
We found that nucleoside supplementation rescues growth in the presence of feruloyl amide and coumaroyl amide to levels that are comparable to those in unexposed cells (see Fig. S6 in the supplemental material). The external addition of nucleosides (a mix of adenosine, guanosine, uridine, thymidine, and cytidine) resulted in a larger percent increase in growth rate in the presence of feruloyl amide or coumaroyl amide than in cells unexposed to these phenolic amides (Table 1). For example, the addition of 0.75 mM nucleosides improved growth in the presence of feruloyl or coumaroyl amide (2.75 mM) by 88% and 82%, respectively, but improved that in unexposed cells by only 32%. These observations indicate that nucleotide biosynthesis becomes a specific growth-limiting factor in the presence of phenolic amides. Interestingly, nucleoside supplementation rescued growth to a lesser degree in cells exposed to ferulic acid. This is consistent with our previous observation that ferulic acid does not inhibit nucleotide biosynthesis as strongly as phenolic amides.
TABLE 1.
Nucleoside supplementation relieves growth inhibition by phenolic amides
| Condition | Normalized growth rate (%) with nucleoside supplementationa at: |
|
|---|---|---|
| 0.25 mM | 0.75 mM | |
| Control | 115.90 ± 1.16 | 132.44 ± 6.21 |
| Feruloyl amide | 164.44 ± 6.91 | 188.06 ± 8.15 |
| Coumaroyl amide | 151.83 ± 14.14 | 182.44 ± 5.40 |
| Ferulic acid | 126.91 ± 2.05 | 139.74 ± 1.82 |
The nucleoside cocktail contained equimolar concentrations of adenosine, guanosine, cytidine, thymidine, and uridine. The growth rate under each condition was normalized against that of the corresponding nonsupplemented control (shown as a percentage). Data represent means of two at least two biological replicates ± standard deviations (n = 5 for controls, n = 3 for feruloyl amide treatment, and n = 2 for coumaroyl amide and ferulic acid treatments). Differences between nucleoside-supplemented and nonsupplemented samples were significant in all cases (P < 0.005 by linear regression).
Inhibition of glutamine amidotransferase activity by phenolic amides.
Our isotopic labeling data predicted that inhibition of nucleotide de novo biosynthesis by phenolic amides might be localized to early steps in these pathways, specifically glutamine PRPP amidotransferase (PurF) and carbamoyl phosphate synthetase (CarAB), which, respectively, catalyze the first committed steps in purine and pyrimidine biosynthesis (Fig. 6D). We therefore tested whether feruloyl amide was a direct inhibitor of these two glutamine amidotransferases.
FIG 6.
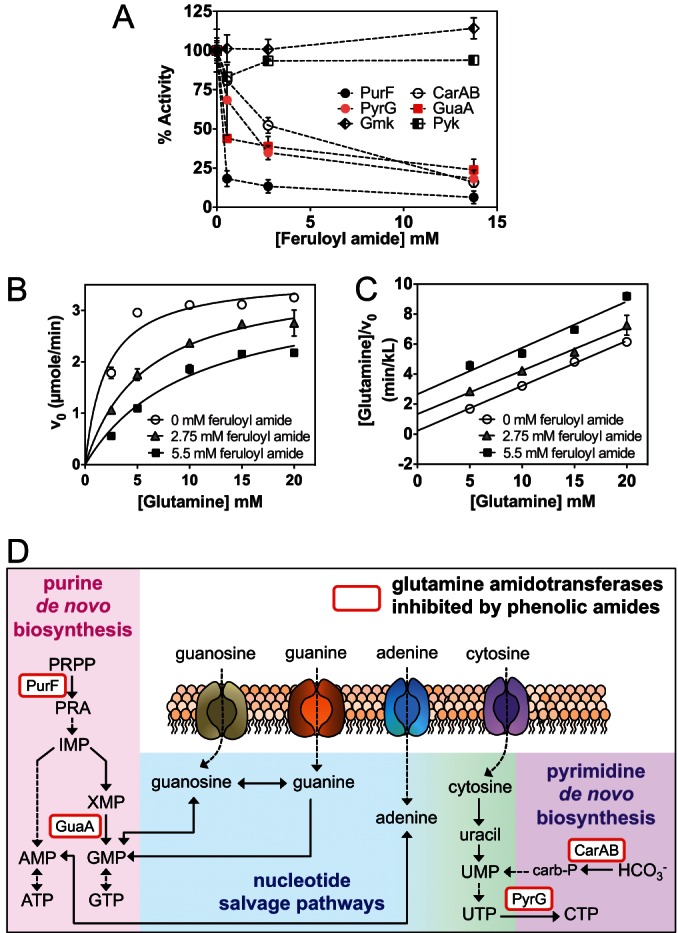
Phenolic amides inhibit glutamine amidotransferases in vitro. (A) Feruloyl amide inhibits glutamine amidotransferases in vitro. Enzymatic assay showed that feruloyl amide directly inhibits glutamine-PRPP-amidotransferase (PurF), GMP synthetase (GuaA), CTP synthetase (PyrG), and carbamoyl phosphate synthetase (CarA/CarB) but not guanylate kinase (Gmk) and pyruvate kinase (Pyk). The activity of each enzyme (measured as initial velocity) was normalized against that of the control. Data shown are an average of at least two technical replicates ± standard deviation (SD). (B and C) Feruloyl amide competitively inhibits PurF activity. (B) Averages of two biological replicates ± SD are fitted into a competitive inhibition equation, V0 = Vmax · [S]/(Km · (1 + [I]/Ki) + [S]). Data points in panel B were transformed into the Hanes-Woolf equation [S]/V0 = [S]/Vmax + Km/Vmax. (C) Parallel slopes in the Hanes-Woolf plot suggest competitive inhibitory mechanism. (D) Glutamine amidotransferases (rounded rectangles) directly inhibited by feruloyl amide and coumaroyl amide are involved in de novo nucleotide biosynthesis. Abbreviations: PRA, 5-phosphoribosylamine; IMP, inosine-5-monophosphate; XMP, xanthosine-5-monophosphate; carb-P, carbamoyl phosphate. Enzyme names: PurF, glutamine-PRPP-amidotransferase; GuaB, IMP dehydrogenase; GuaA, GMP synthetase; PyrG, CTP synthetase; CarAB, carbamoyl phosphate synthase.
Using purified enzymes, we found that the activities of both glutamine PRPP amidotransferase and carbamoyl phosphate synthetase were inhibited by feruloyl amide in a dose-dependent manner (Fig. 6A). The inhibition of these two enzymes in vitro is consistent with the observed large and rapid increase in PRPP levels and the decrease in dihydroorotate and orotate levels observed upon exposure of whole cells to phenolic amides. In addition, we found that the activities of other glutamine amidotransferases that participate in nucleotide biosynthesis were also inhibited by feruloyl amide. Specifically, we found that GMP synthase, which catalyzes the synthesis of GMP from XMP, and CTP synthase, which catalyzes UTP-to-CTP conversion, were both inhibited by feruloyl amide (Fig. 6A).
Nucleotide monophosphate kinases are another important class of enzymes involved in nucleotide metabolism. Thus, we also tested for inhibitory effects of feruloyl amide on guanylate kinase (Gmk), which plays an important role in balancing nucleotide pools by reversibly transferring the γ-phosphoryl group of ATP to GMP or dGMP to produce GDP or dGDP and ADP (37). Feruloyl amide did not have an inhibitory effect on the activity of this enzyme (Fig. 6B). We also tested the effects of feruloyl amide on the activity of pyruvate kinase (Pyk), a key enzyme in lower glycolysis responsible for the production of pyruvate and ATP from PEP and ADP. Data from 13C tracer experiments showed no major effects of phenolic amides on carbon flux into lower glycolysis (Fig. 3), and, as predicted from this finding, Pyk activity was not affected by feruloyl amide (Fig. 6A).
We next investigated the mode of inhibition of glutamine PRPP amidotransferase (PurF) by feruloyl amide using kinetic assays. The effects of various levels of feruloyl amide on the initial velocities of the PurF-catalyzed reaction were measured as a function of glutamine concentration (Fig. 6B). The kinetic data were examined graphically using the Hanes-Woolf analysis, in which competitive inhibition yields parallel lines with y intercepts that depend on inhibitor concentration. As shown in Fig. 6C, parallel lines in the Hanes-Woolf transformation were observed, which indicates that feruloyl amide is a competitive inhibitor of glutamine PRPP amidotransferase. Collectively, these observations indicate that feruloyl amide acts as a direct inhibitor of glutamine amidotransferase enzymes that participate in nucleotide biosynthesis pathways (Fig. 6D).
DISCUSSION
The inhibition of microbial processes by plant-derived inhibitors is currently a major challenge for the efficient production of biofuels and added-value chemicals from plant biomass. The adverse effects of lignocellulose-derived inhibitors on growth and sugar conversion have been well documented in biofuel producers such as yeast, Zymomonas mobilis, and E. coli (9, 38). Although there have been several recent efforts at elucidating the mechanisms underlying the toxicity of these microbial inhibitors, most of them have been focused on the sugar-derived furfural and aldehyde inhibitors predominantly found in acid-pretreated biomass hydrolysates (7, 8, 11, 38–41). In contrast, the molecular basis for toxicity of phenolic amides and carboxylates derived from biomass processing remains largely unknown.
Here, we focused on investigating the metabolic effects of feruloyl amide and coumaroyl amide, the predominant lignocellulose-derived inhibitors present in AFEX-pretreated corn stover and possibly other ammonia-based biomass hydrolysates. Using metabolomics and isotopic tracers, we have shown that these two phenolic amides are potent inhibitors of de novo purine and pyrimidine biosynthesis. Our results also indicate that these inhibitory effects are at least partially mediated via direct inhibition of the glutamine amidotransferases involved in these biosynthetic pathways. We also found that exogenous nucleosides rescue growth inhibition by phenolic amides. These observations provide both a mechanistic basis for the observed growth inhibition and a target for future strain improvement.
Phenolic amides act as fast-acting inhibitors of nucleotide metabolism; within 10 min, we observed a large accumulation of the biosynthetic precursor PRPP, decreased levels of pyrimidine intermediates, and decreased carbon and nitrogen flux into nucleotides. Although these fast inhibitory effects are unlikely to be mediated via transcriptional responses, it is possible that these phenolic amides might also elicit additional, direct or indirect, long-term effects on transcription or other cellular processes (30). Regardless of this, the inhibition of nucleotide metabolism by phenolic amides is a persisting effect from which E. coli is unable to recover. As shown in Fig. S4 in the supplemental material, nucleotide de novo biosynthesis remains blocked after 2 h of a continuous feruloyl amide treatment, although there is some attenuation on the inhibition of carbon flux into α-ketoglutarate, glutamate, and glutamine.
Our results suggest that the inhibition of sugar catabolism by phenolic amides is an effect secondary to nucleotide biosynthesis inhibition. Interestingly, the reduction in sugar consumption was not fully proportional to growth inhibition (see Fig. S1 in the supplemental material), and sugar catabolism continued, albeit very slowly, even in instances where cell proliferation had almost completely stalled. The regulatory connections that coordinate nucleotide biosynthesis and sugar catabolism remain poorly understood. If these were known, it might be possible to engineer biofuel producers in which sugar catabolic pathways are uncoupled from nucleotide biosynthesis. Such a system would be more resistant to phenolic inhibitors and allow fermentation to continue even when nucleotide biosynthesis has been inhibited.
Tracer experiments using [13C]xylose revealed a strong inhibition of 13C incorporation into α-ketoglutarate, glutamate, and glutamine in the presence of phenolic amides (Fig. 2). However, [15N]ammonium labeling experiments showed that nitrogen assimilation was only slightly affected (Fig. 4A). While these observations indicated that the strong inhibition of nucleotide biosynthesis did not stem primarily from obstructed ammonium assimilation, they also suggested that phenolic amides produce an additional metabolic bottleneck on the right half of TCA cycle. Interestingly, though, this inhibition was attenuated 2 h after phenolic amide treatment (see Fig. S4 in the supplemental material). Further studies are required to understand how E. coli is capable of partially relieving this inhibition.
Although ferulic acid treatment did not have a strong direct effect on purine and pyrimidine biosynthetic pathways, it still resulted in a long-term decrease in the levels of downstream deoxyribonucleotides (see Fig. S3 in the supplemental material). This observation was consistent with impaired 13C incorporation into deoxynucleotides (Fig. 2; see Fig. S3 in the supplemental material). It is therefore possible that ferulic acid might specifically inhibit deoxynucleotide production. This view helps explain why external nucleoside supplementation is less effective at rescuing growth inhibition by ferulic acid than by feruloyl or coumaroyl amide (Fig. 6A; see Fig. S6 in the supplemental material).
Growth inhibition by phenolic amides was considerably stronger in xylose fermentations than in glucose fermentations. This agrees well with previous observations in Z. mobilis and yeast that show a greater toxicity of lignocellulosic inhibitors in xylose fermentations (9). We found that the inhibitory effects of phenolic amides on metabolism were well conserved across both carbon sources, although the inhibitory effects on purine biosynthesis were somewhat less strong in glucose fermentations (Fig. 5). The mechanisms underlying the greater susceptibility of cells grown on xylose versus glucose remain to be elucidated.
We found that feruloyl amide is a direct inhibitor of glutamine amidotransferase enzymes that participate in de novo nucleotide biosynthesis. In particular, feruloyl amide is a competitive inhibitor of glutamine PRPP amidotransferase (PurF). Most competitive inhibitors bind to the active site, preventing substrate binding; thus, it is likely that feruloyl amide binds to the glutamine-binding site in PurF. The amide-containing side chain in feruloyl amide is structurally similar to the glutamine side chain. This structural similarity may be an important determinant of the inhibitory effects of feruloyl amide against glutamine amidotransferases and help explain why ferulic acid is not such a strong inhibitor of nucleotide biosynthesis; additional studies are required to test these hypotheses. Nonetheless, a strategy that could be used to overcome the inhibitory effects of phenolic amides would be to directly engineer glutamine amidotransferase enzymes (e.g., PurF, CarAB, PyrG, or GuaA) to be less susceptible to inhibition by these compounds.
Phenolic carboxylates and their derivatives are ubiquitous in plants. They are present in fruits, in leaves, and as constituents of plant cell walls (42, 43). Plant phenolic compounds have long been known to have antimicrobial properties, and their production constitutes a defense mechanism to combat growth of cell wall-degrading microbes and intracellular pathogens (44–46). Conversely, some plant pathogens and plant-associated microbes have evolved the ability to break down phenolic compounds such as feruloyl and coumaroyl derivatives (47–50). The mechanisms by which plant phenolics inhibit microbial growth remain largely unexplored, but it seems reasonable to suggest that at least some of them might have inhibitory mechanisms similar to the ones presented here for feruloyl amide, coumaroyl amide, or ferulic acid.
In conclusion, we have shown that feruloyl amide and coumaroyl amide are potent inhibitors of de novo purine and pyrimidine biosynthesis in E. coli. These biosynthetic pathways and their enzymes are very highly conserved across microorganisms. It is therefore likely that these phenolic amides will have similar inhibitory mechanisms in other biofuel producers. We expect that the results presented here will be useful in guiding future strategies to overcome the toxicity of lignin-derived inhibitors and that the metabolomics-based approach that we have outlined can be applied to investigate the toxicity mechanisms of other microbial inhibitors.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yaoping Zhang for providing the phenolic compounds used in this study, Kuanqing Liu for helpful discussion, and Timothy Donohue and Robert Landick for reading the manuscript.
This work was supported by the Department of Energy, Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-FC02-07ER64494).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01324-15.
REFERENCES
- 1.Schubert C. 2006. Can biofuels finally take center stage? Nat Biotechnol 24:777–784. doi: 10.1038/nbt0706-777. [DOI] [PubMed] [Google Scholar]
- 2.Kumar P, Barrett DM, Delwiche MJ, Stroeve P. 2009. Methods for pretreatment of lignocellulosic biomass for efficient hydrolysis and biofuel production. Ind Eng Chem Res 48:3713–3729. doi: 10.1021/ie801542g. [DOI] [Google Scholar]
- 3.Hendriks ATWM, Zeeman G. 2009. Pretreatments to enhance the digestibility of lignocellulosic biomass. Bioresour Technol 100:10–18. doi: 10.1016/j.biortech.2008.05.027. [DOI] [PubMed] [Google Scholar]
- 4.da Costa Sousa L, Chundawat SPS, Balan V, Dale BE. 2009. ‘Cradle-to-grave’ assessment of existing lignocellulose pretreatment technologies. Curr Opin Biotechnol 20:339–347. doi: 10.1016/j.copbio.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Panagiotou G, Olsson L. 2007. Effect of compounds released during pretreatment of wheat straw on microbial growth and enzymatic hydrolysis rates. Biotechnol Bioeng 96:250–258. doi: 10.1002/bit.21100. [DOI] [PubMed] [Google Scholar]
- 6.Klinke HB, Thomsen AB, Ahring BK. 2004. Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl Microbiol Biotechnol 66:10–26. doi: 10.1007/s00253-004-1642-2. [DOI] [PubMed] [Google Scholar]
- 7.Palmqvist E, Grage H, Meinander NQ, Hahn-Hägerdal B. 1999. Main and interaction effects of acetic acid, furfural, and p-hydroxybenzoic acid on growth and ethanol productivity of yeasts. Biotechnol Bioeng 63:46–55. [DOI] [PubMed] [Google Scholar]
- 8.Palmqvist E, Almeida JS, Hahn-Hägerdal B. 1999. Influence of furfural on anaerobic glycolytic kinetics of Saccharomyces cerevisiae in batch culture. Biotechnol Bioeng 62:447–454. [DOI] [PubMed] [Google Scholar]
- 9.Franden M, Pilath H, Mohagheghi A, Pienkos P, Zhang M. 2013. Inhibition of growth of Zymomonas mobilis by model compounds found in lignocellulosic hydrolysates. Biotechnol Biofuels 6:99. doi: 10.1186/1754-6834-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwalbach MS, Keating DH, Tremaine M, Marner WD, Zhang Y, Bothfeld W, Higbee A, Grass JA, Cotten C, Reed JL, da Costa Sousa L, Jin M, Balan V, Ellinger J, Dale B, Kiley PJ, Landick R. 2012. Complex physiology and compound stress responses during fermentation of alkali-pretreated corn stover hydrolysate by an Escherichia coli ethanologen. Appl Environ Microbiol 78:3442–3457. doi: 10.1128/AEM.07329-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller EN, Jarboe LR, Turner PC, Pharkya P, Yomano LP, York SW, Nunn D, Shanmugam KT, Ingram LO. 2009. Furfural inhibits growth by limiting sulfur assimilation in ethanologenic Escherichia coli strain LY180. Appl Environ Microbiol 75:6132–6141. doi: 10.1128/AEM.01187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Yomano LP, Lee JY, York SW, Zheng H, Mullinnix MT, Shanmugam KT, Ingram LO. 2013. Engineering furfural tolerance in Escherichia coli improves the fermentation of lignocellulosic sugars into renewable chemicals. Proc Natl Acad Sci U S A 110:4021–4026. doi: 10.1073/pnas.1217958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson CM, Yang S, Rodriguez M, Ma Q, Johnson CM, Dice L, Xu Y, Brown SD. 2013. Clostridium thermocellum transcriptomic profiles after exposure to furfural or heat stress. Biotechnol Biofuels 6:131. doi: 10.1186/1754-6834-6-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Miller EN, Yomano LP, Zhang X, Shanmugam KT, Ingram LO. 2011. Increased furfural tolerance due to overexpression of NADH-dependent oxidoreductase FucO in Escherichia coli strains engineered for the production of ethanol and lactate. Appl Environ Microbiol 77:5132–5140. doi: 10.1128/AEM.05008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller EN, Turner PC, Jarboe LR, Ingram LO. 2010. Genetic changes that increase 5-hydroxymethyl furfural resistance in ethanol-producing Escherichia coli LY180. Biotechnol Lett 32:661–667. doi: 10.1007/s10529-010-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller EN, Jarboe LR, Yomano LP, York SW, Shanmugam KT, Ingram LO. 2009. Silencing of NADPH-dependent oxidoreductase genes (yqhD and dkgA) in furfural-resistant ethanologenic Escherichia coli. Appl Environ Microbiol 75:4315–4323. doi: 10.1128/AEM.00567-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Almeida JRM, Röder A, Modig T, Laadan B, Lidén G, Gorwa-Grauslund M. 2008. NADH- vs NADPH-coupled reduction of 5-hydroxymethyl furfural (HMF) and its implications on product distribution in Saccharomyces cerevisiae. Appl Biochem Biotechnol 78:939–945. [DOI] [PubMed] [Google Scholar]
- 18.Lewis Liu Z, Moon J, Andersh BJ, Slininger PJ, Weber S. 2008. Multiple gene-mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and 5-hydroxymethylfurfural by Saccharomyces cerevisiae. Appl Biochem Biotechnol 81:743–753. [DOI] [PubMed] [Google Scholar]
- 19.Wahlbom C, Hahn-Hagerdal B. 2002. Furfural, 5-hydroxymethyl furfural, and acetoin act as external electron acceptors during anaerobic fermentation of xylose in recombinant Saccharomyces cerevisiae. Biotechnol Bioeng 78:172–178. doi: 10.1002/bit.10188. [DOI] [PubMed] [Google Scholar]
- 20.Turner PC, Miller EN, Jarboe LR, Baggett CL, Shanmugam KT, Ingram LO. 2011. YqhC regulates transcription of the adjacent Escherichia coli genes yqhD and dkgA that are involved in furfural tolerance. J Ind Microbiol Biotechnol 38:431–439. doi: 10.1007/s10295-010-0787-5. [DOI] [PubMed] [Google Scholar]
- 21.Liu ZL, Moon J. 2009. A novel NADPH-dependent aldehyde reductase gene from Saccharomyces cerevisiae NRRL Y-12632 involved in the detoxification of aldehyde inhibitors derived from lignocellulosic biomass conversion. Gene 446:1–10. doi: 10.1016/j.gene.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 22.Zaldivar J, Martinez A, Ingram LO. 1999. Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol Bioeng 65:24–33. doi:. [DOI] [PubMed] [Google Scholar]
- 23.Zaldivar J, Martinez A, Ingram LO. 2000. Effect of alcohol compounds found in hemicellulose hydrolysate on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol Bioeng 68:524–530. [DOI] [PubMed] [Google Scholar]
- 24.Laadan B, Almeida JRM, Rådström P, Hahn-Hägerdal B, Gorwa-Grauslund M. 2008. Identification of an NADH-dependent 5-hydroxymethylfurfural-reducing alcohol dehydrogenase in Saccharomyces cerevisiae. Yeast 25:191–198. doi: 10.1002/yea.1578. [DOI] [PubMed] [Google Scholar]
- 25.Almeida J, Bertilsson M, Hahn-Hagerdal B, Liden G, Gorwa-Grauslund M. 2009. Carbon fluxes of xylose-consuming Saccharomyces cerevisiae strains are affected differently by NADH and NADPH usage in HMF reduction. Appl Microbiol Biotechnol 84:751–61. doi: 10.1007/s00253-009-2053-1. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Han B, Ezeji TC. 2012. Biotransformation of furfural and 5-hydroxymethyl furfural (HMF) by Clostridium acetobutylicum ATCC 824 during butanol fermentation. Nat Biotechnol 29:345–351. [DOI] [PubMed] [Google Scholar]
- 27.Ujor V, Agu C, Gopalan V, Ezeji T. 2014. Glycerol supplementation of the growth medium enhances in situ detoxification of furfural by Clostridium beijerinckii during butanol fermentation. Appl Microbiol Biotechnol 98:6511–6521. doi: 10.1007/s00253-014-5802-8. [DOI] [PubMed] [Google Scholar]
- 28.Kim Y, Hendrickson R, Mosier NS, Ladisch MR, Bals B, Balan V, Dale BE. 2008. Enzyme hydrolysis and ethanol fermentation of liquid hot water and AFEX pretreated distillers' grains at high-solids loadings. Bioresour Technol 99:5206–5215. doi: 10.1016/j.biortech.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 29.Almeida JRM, Modig T, Petersson A, Hahn-Hägerdal B, Lidén G, Gorwa-Grauslund MF. 2007. Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J Chem Technol Biotechnol 82:340–349. doi: 10.1002/jctb.1676. [DOI] [Google Scholar]
- 30.Keating DH, Zhang Y, Ong IM, McIlwain S, Morales EH, Grass JA, Tremaine M, Bothfeld W, Higbee A, Ulbrich A, Balloon AJ, Westphall MS, Aldrich J, Lipton MS, Kim J, Moskvin OV, Bukhman YV, Coon JJ, Kiley PJ, Bates DM, Landick R. 2014. Aromatic inhibitors derived from ammonia-pretreated lignocellulose hinder bacterial ethanologenesis by activating regulatory circuits controlling inhibitor efflux and detoxification. Front Microbiol 5:402. doi: 10.3389/fmicb.2014.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clasquin MF, Melamud E, Rabinowitz JD. 2012. LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr Protoc Bioinformatics 37:14.11.11–14.11.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melamud E, Vastag L, Rabinowitz JD. 2010. Metabolomic analysis and visualization engine for LC-MS data. Anal Chem 82:9818–9826. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res 12:291–299. [DOI] [PubMed] [Google Scholar]
- 34.Liu K, Myers AR, Pisithkul T, Claas KR, Satyshur KA, Amador-Noguez D, Keck JL, Wang JD. 2015. Molecular mechanism and evolution of guanylate kinase regulation by (p)ppGpp. Mol Cell 57:735–749. doi: 10.1016/j.molcel.2014.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parawira W, Tekere M. 2011. Biotechnological strategies to overcome inhibitors in lignocellulose hydrolysates for ethanol production: review. Crit Rev Biotechnol 31:20–31. doi: 10.3109/07388551003757816. [DOI] [PubMed] [Google Scholar]
- 36.Rizzi M, Schindelin H. 2002. Structural biology of enzymes involved in NAD and molybdenum cofactor biosynthesis. Curr Opin Struct Biol 12:709–720. doi: 10.1016/S0959-440X(02)00385-8. [DOI] [PubMed] [Google Scholar]
- 37.Hible G, Renault L, Schaeffer F, Christova P, Zoe Radulescu A, Evrin C, Gilles A-M, Cherfils J. 2005. Calorimetric and crystallographic analysis of the oligomeric structure of Escherichia coli GMP kinase. J Mol Biol 352:1044–1059. doi: 10.1016/j.jmb.2005.07.042. [DOI] [PubMed] [Google Scholar]
- 38.Palmqvist E, Hahn-Hägerdal B. 2000. Fermentation of lignocellulosic hydrolysates. II. Inhibitors and mechanisms of inhibition. Bioresour Technol 74:25–33. [Google Scholar]
- 39.Yang S, Land ML, Klingeman DM, Pelletier DA, Lu T-YS, Martin SL, Guo H-B, Smith JC, Brown SD. 2010. Paradigm for industrial strain improvement identifies sodium acetate tolerance loci in Zymomonas mobilis and Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 107:10395–10400. doi: 10.1073/pnas.0914506107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casey E, Sedlak M, Ho NWY, Mosier NS. 2010. Effect of acetic acid and pH on the cofermentation of glucose and xylose to ethanol by a genetically engineered strain of Saccharomyces cerevisiae. FEMS Yeast Res 10:385–393. doi: 10.1111/j.1567-1364.2010.00623.x. [DOI] [PubMed] [Google Scholar]
- 41.Narendranath NV, Thomas KC, Ingledew WM. 2001. Effects of acetic acid and lactic acid on the growth of Saccharomyces cerevisiae in a minimal medium. J Ind Microbiol Biotechnol 26:171–177. doi: 10.1038/sj.jim.7000090. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, Wei N, Huang J, Tan Y, Jin D. 2012. A new feruloyl amide derivative from the fruits of Tribulus terrestris. Nat Prod Res 26:1922–1925. doi: 10.1080/14786419.2011.643886. [DOI] [PubMed] [Google Scholar]
- 43.Mueller-Harvey I, Hartley RD, Harris PJ, Curzon EH. 1986. Linkage of p-coumaroyl and feruloyl groups to cell-wall polysaccharides of barley straw. Carbohydr Res 148:71–85. doi: 10.1016/0008-6215(86)80038-6. [DOI] [Google Scholar]
- 44.Tawata S, Taira S, Kobamoto N, Zhu J, Ishihara M, Toyama S. 1996. Synthesis and antifungal activity of cinnamic acid esters. Biosci Biotechnol Biochem 60:909–910. doi: 10.1271/bbb.60.909. [DOI] [PubMed] [Google Scholar]
- 45.Kwon Y-S, Kobayashi A, Kajiyama S-I, Kawazu K, Kanzaki H, Kim C-M. 1997. Antimicrobial constituents of Angelica dahurica roots. Phytochemistry 44:887–889. doi: 10.1016/S0031-9422(96)00634-6. [DOI] [PubMed] [Google Scholar]
- 46.Medina A, Jakobsen I, Egsgaard H. 2011. Sugar beet waste and its component ferulic acid inhibits external mycelium of arbuscular mycorrhizal fungus. Soil Biol Biochem 43:1456–1463. doi: 10.1016/j.soilbio.2011.03.016. [DOI] [Google Scholar]
- 47.McCrae SI, Leith KM, Gordon AH, Wood TM. 1994. Xylan-degrading enzyme system produced by the fungus Aspergillus awamori: isolation and characterization of a feruloyl esterase and a p-coumaroyl esterase. Enzyme Microb Technol 16:826–834. doi: 10.1016/0141-0229(94)90055-8. [DOI] [Google Scholar]
- 48.Huang Z, Dostal L, Rosazza JP. 1994. Purification and characterization of a ferulic acid decarboxylase from Pseudomonas fluorescens. J Bacteriol 176:5912–5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lesage-Meessen L, Delattre M, Haon M, Thibault J-F, Ceccaldi BC, Brunerie P, Asther M. 1996. A two-step bioconversion process for vanillin production from ferulic acid combining Aspergillus niger and Pycnoporus cinnabarinus. J Biotechnol 50:107–113. doi: 10.1016/0168-1656(96)01552-0. [DOI] [PubMed] [Google Scholar]
- 50.Brunati M, Marinelli F, Bertolini C, Gandolfi R, Daffonchio D, Molinari F. 2004. Biotransformations of cinnamic and ferulic acid with actinomycetes. Enzyme Microb Technol 34:3–9. doi: 10.1016/j.enzmictec.2003.04.001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.