EpsinR and gadkin are two components of intracellular clathrin-coated vesicles whose precise functions are unclear. Rapid depletion of each protein from the available pool using the knocksideways method strongly inhibited the production of intracellular clathrin-coated vesicles, providing new insights into the functions of both proteins.
Abstract
The precise functions of most of the proteins that participate in clathrin-mediated intracellular trafficking are unknown. We investigated two such proteins, epsinR and gadkin, using the knocksideways method, which rapidly depletes proteins from the available pool by trapping them onto mitochondria. Although epsinR is known to be an N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE)-specific adaptor, the epsinR knocksideways blocked the production of the entire population of intracellular clathrin-coated vesicles (CCVs), suggesting a more global function. Using the epsinR knocksideways data, we were able to estimate the copy number of all major intracellular CCV proteins. Both sides of the vesicle are densely covered, indicating that CCVs sort their cargo by molecular crowding. Trapping of gadkin onto mitochondria also blocked the production of intracellular CCVs but by a different mechanism: vesicles became cross-linked to mitochondria and pulled out toward the cell periphery. Both phenotypes provide new insights into the regulation of intracellular CCV formation, which could not have been found using more conventional approaches.
INTRODUCTION
Clathrin-coated vesicles bud from both the plasma membrane and intracellular membranes. The pathway from the plasma membrane, known as clathrin-mediated endocytosis, has been more intensively investigated than the intracellular pathway and is now much better understood. Clathrin-mediated endocytosis makes use of both the adaptor protein 2 (AP-2) complex and a number of accessory proteins, many of which bind to the appendage domains of the two large subunits of AP-2. Some of these accessory proteins (e.g., Dab2, ARH, epsin, β-arrestin, and CALM) act as cargo-selective adaptors; others facilitate vesicle scission (e.g., dynamin), uncoating (e.g., auxilin, synaptojanin), and interactions with the actin cytoskeleton (e.g., Hip1R; Traub and Bonifacino, 2013; Kirchhausen et al., 2014). In contrast, although we know that clathrin-mediated intracellular trafficking uses the AP-1 adaptor complex together with accessory proteins that bind to its appendages, for the most part we do not know what these accessory proteins actually do.
One way of investigating clathrin-mediated intracellular trafficking is by proteomic analyses of clathrin-coated vesicle (CCV)–enriched fractions. CCV fractions from HeLa cells are dominated by trans-Golgi network (TGN)– and endosome-derived CCVs (Borner et al., 2012), and we have used different fractionation protocols, knockdowns, and knocksideways to learn more about the functions of different coat components (Borner et al., 2012, 2014; Hirst et al., 2012). The knocksideways method is particularly powerful because it uses a small molecule, rapamycin, to rapidly deplete the available pool of a protein of interest by linking it to a construct called Mitotrap, which localizes to mitochondria. Trapping of the protein onto mitochondria occurs within minutes, so the cells do not have time to up-regulate other pathways in order to compensate (Robinson et al., 2010). We have previously used this approach to investigate the functions of AP-1 and the alternative intracellular adaptor GGA2 (Hirst et al., 2012). The AP-1 knocksideways caused a global change in CCV composition, with nearly 100 proteins depleted twofold or more. These proteins included alternative adaptors and accessory proteins (e.g., GGAs, epsinR, gadkin), as well as >40 different transmembrane and lumenal cargo proteins, indicating that AP-1 normally acts as a linchpin for intracellular CCV formation. The GGA2 knocksideways produced a very different phenotype. Many fewer proteins were depleted, and nearly all of these were either lysosomal hydrolases or their receptors, indicating that GGAs function as cargo-selective adaptors for receptor-mediated trafficking of hydrolases.
Using the same approach, we have begun to investigate the roles of some of the other accessory proteins for intracellular CCVs, starting with epsinR and gadkin. Both of these proteins have been shown to bind competitively to the same site on the AP-1 γ appendage (Neubrand et al., 2005), but there is limited information about their functions.
EpsinR (also known as CLINT1, epsin4, and enthoprotin) has an epsin N-terminal homology (ENTH) domain, which binds to a cargo protein, vti1b (Hirst et al., 2004; Miller et al., 2007), followed by a long, unstructured region containing binding sites for AP-1 and clathrin (Figure 1A). Knocking down epsinR in HeLa cells causes vti1b to relocate, consistent with a function as a cargo-selective adaptor (Hirst et al., 2004; Miller et al., 2007). Effects have also been reported on the trafficking of the cation-independent mannose 6-phosphate receptor and TGN46, suggesting a more general role (Saint Pol et al., 2004), although the epsinR-knockdown phenotype is relatively mild compared with, for instance, the clathrin-knockdown phenotype (Motley et al., 2003). Animals and fungi (i.e., opisthokonts) also have “conventional” epsins, which contribute to clathrin-mediated endocytosis; however, most other eukaryotes have only a single type of epsin, which may fulfill the functions of both epsinR and epsin, acting on both the AP-1 pathway and the AP-2 pathway (Gabernet-Castello et al., 2009; Brady et al., 2010).
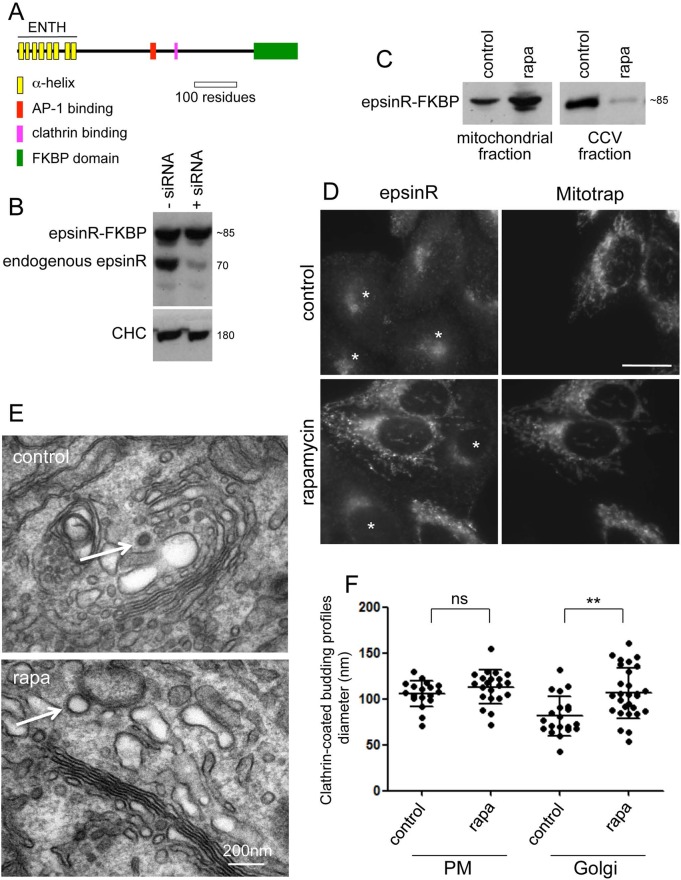
FIGURE 1:
Phenotype of epsinR knocksideways. (A) Schematic diagram of FKBP-tagged epsinR. (B) Western blots of epsinR-FKBP–expressing cells. The cells were treated with siRNA to deplete endogenous epsinR, and the blots were probed with anti-epsinR, using anti-clathrin as a loading control. The epsinR-FKBP construct is expressed at a similar level to endogenous epsinR and is siRNA resistant. (C) Addition of rapamycin (rapa) to the epsinR-FKBP–expressing cells causes the construct to accumulate in a mitochondrial fraction (isolated using magnetic beads) and become depleted from our CCV fraction. (D) Relocation to mitochondria demonstrated by immunofluorescence. A mixed population of epsinR-FKBP–expressing cells, from which endogenous epsinR had been depleted using siRNA, and wild-type cells (asterisks) were treated with rapamycin and labeled for total epsinR. Scale bar, 20 μm. (E) Electron micrographs of epsinR-FKBP–expressing cells, depleted of endogenous epsinR, either with or without rapamycin treatment. In the rapamycin-treated cells, enlarged clathrin-coated structures can be seen in the Golgi region (arrows). (F) The diameters of clathrin-coated budding profiles, associated with either the plasma membrane or the Golgi region, were measured in electron micrographs of control and rapamycin-treated cells. Data were collected from two separate experiments, analyzing at least five separate cells for each experiment. At least 20 profiles were measured for each condition. Rapamycin treatment causes an increase in the diameter of clathrin-coated structures in the Golgi region from 85.3 ± 27.6 to 108.6 ± 29.1 nm (SD; p = 0.0059 for the Golgi region; p = 0.2246 for the plasma membrane; ns, not significant).
Gadkin (also called AP1AR, γ-BAR, and C4orf16) has a much more recent evolutionary history than epsinR/epsin and is found only in certain animals. The protein is S-palmitoylated, which facilitates its association with membranes (Maritzen et al., 2010), and thus its recruitment is very different from that of other intracellular CCV coat components such as epsinR, AP-1, and GGAs, which cycle on and off the membrane in an ARF1-dependent manner. In addition, there is no evidence that gadkin acts as a cargo adaptor; instead, it provides a molecular link between AP-1 and the cytoskeleton through its ability to interact with both the microtubule motor kinesin-1 (Schmidt et al., 2009) and the actin-nucleating ARP2/3 complex (Maritzen et al., 2012). However, there are still questions about gadkin's precise function.
Here we used knocksideways together with proteomics to learn more about the roles of epsinR and gadkin. We speculated that the epsinR knocksideways would show a cargo-selective phenotype, mainly affecting vti1b, and that the phenotype of the gadkin knocksideways would also be relatively mild. However, in both cases, the knocksideways had surprisingly far-reaching consequences, providing new insights into the functions of both proteins.
RESULTS
EpsinR knocksideways cells
To set up a knocksideways system for epsinR, we synthesized a short interfering RNA (siRNA)–resistant construct with an FKBP domain at its 3′ end (Figure 1A) and stably transfected the construct into Mitotrap-expressing HeLa cells. Western blots of a clonal cell line showed that the construct was expressed at a similar level to endogenous epsinR and that only endogenous epsinR was depleted when the cells were treated with siRNA (Figure 1B). Addition of rapamycin to the cells, followed by subcellular fractionation to generate mitochondria-enriched and CCV-enriched preparations, showed that the drug caused the construct to accumulate in the mitochondrial fraction, with a concomitant loss from the CCV fraction (Figure 1C). To observe rerouting to mitochondria by immunofluorescence, we used siRNA to knock down endogenous epsinR in the epsinR-FKBP/Mitotrap–expressing cells and then mixed these cells with control HeLa cells and treated the mixed population with rapamycin. Figure 1D shows that in the absence of rapamycin, epsinR-FKBP is concentrated in the juxtanuclear region of the cell, similar to endogenous epsinR in control cells (asterisks), but that addition of rapamycin causes the construct to colocalize with Mitotrap. Other proteins associated with intracellular CCVs, including AP-1, GGA2, and clathrin, did not relocate to mitochondria when epsinR was knocked sideways (Supplemental Figure S1).
In a conventional epsinR knockdown, the morphology of intracellular clathrin-coated structures changes and the diameters become wider (Hirst et al., 2004). To investigate whether the same phenotype occurs in epsinR knocksideways cells, we collected electron microscopic images of the cells both with and without rapamycin treatment and measured the diameters of clathrin-coated structures at the plasma membrane and in the Golgi region (Figure 1, E and F). Although we observed no changes at the plasma membrane, the mean diameter of clathrin-coated structures near the Golgi apparatus increased by ∼30%, indicating that this morphological change occurs rapidly upon loss of epsinR.
Subcellular fractionation of epsinR knocksideways cells
To look for changes in the overall protein composition of CCVs in epsinR knocksideways cells, we carried out stable isotope labeling by amino acids in cell culture (SILAC)–based mass spectrometry on CCV fractions from control and rapamycin-treated cells after first depleting endogenous epsinR with siRNA. Figure 2 shows a list of proteins depleted twofold or more from the knocksideways CCVs, based on three independent experiments, with known CCV components indicated in color. We had anticipated that with the exception of epsinR itself and vti1b, most coat and cargo proteins would be relatively unaffected. Surprisingly, however, the epsinR knocksideways had a global effect on intracellular CCVs, similar to the effect of an AP-1 knocksideways. The top hits included not only epsinR, but also GGA2, AP-1, and clathrin, indicating that, like AP-1, epsinR normally acts as a linchpin for the formation of an entire population of CCVs. Moreover, although vti1b was depleted from the preparation, several cargo proteins that depend on AP-1 and/or GGAs (e.g., hydrolase receptors and carboxypeptidase D) were depleted even more strongly. It is not clear why the abundant intracellular clathrin-coated structures we observed by electron microscopy were not recovered in our CCV fraction, but the altered morphology is consistent with some sort of abnormality in their ability to dissociate from membranes.
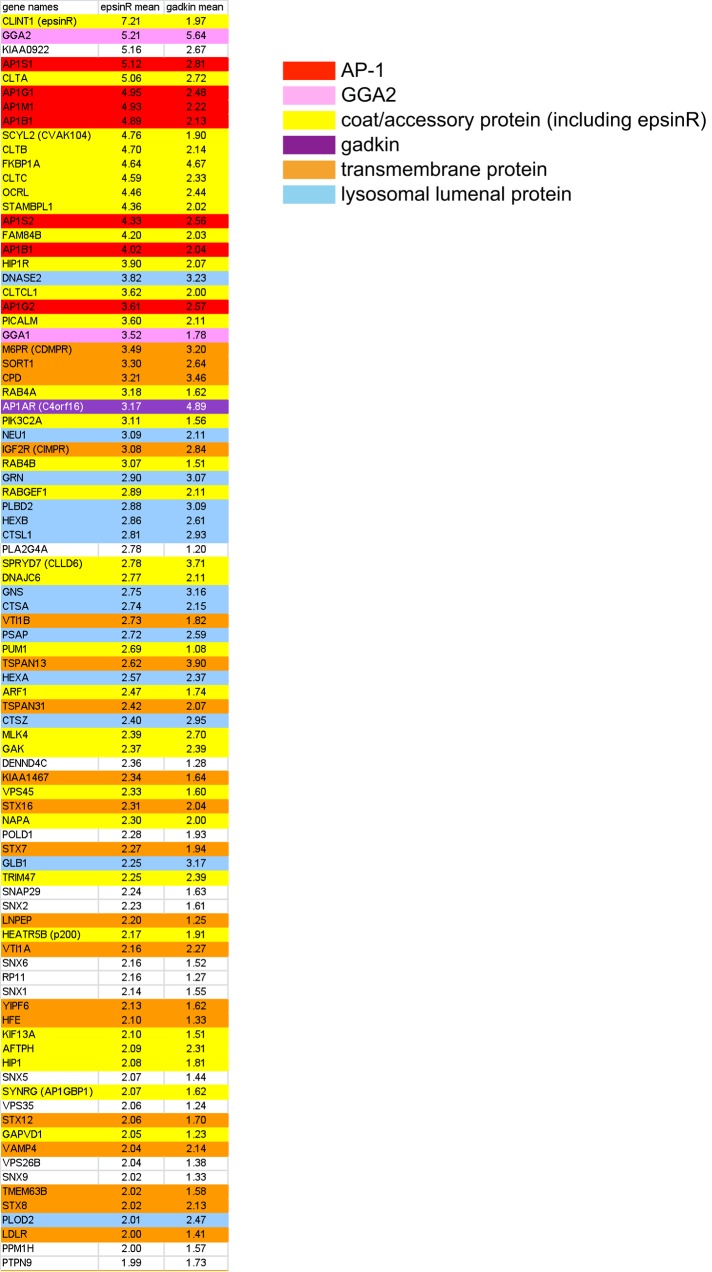
FIGURE 2:
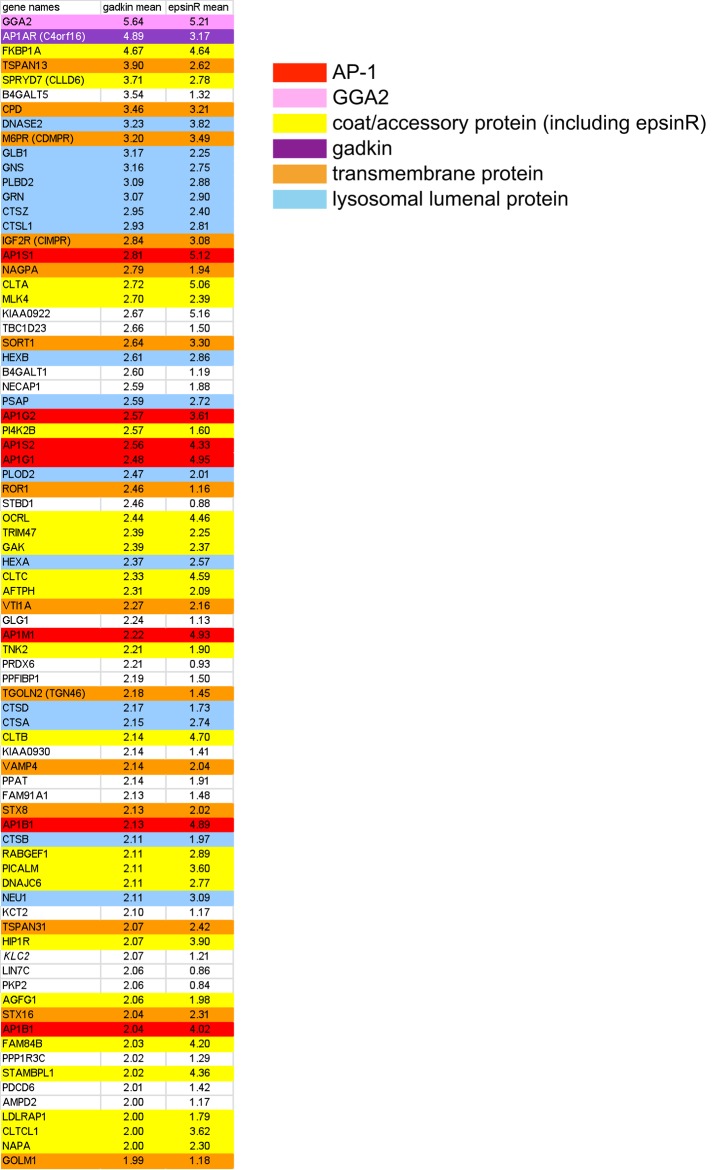
Proteomics of CCVs from SILAC-labeled epsinR knocksideways cells. The fold change (control:knocksideways) was calculated for each protein, and proteins were ranked from the highest to lowest ratio. Proteins with ratios of ≥2.0 are shown in order of rank for the epsinR knocksideways. For ease of visualization, known CCV components are color coded.
Using the data sets from our epsinR knocksideways and our earlier AP-1 knocksideways (Hirst et al., 2012) together with additional data sets from our recent fractionation profiling study (Borner et al., 2014), we were able estimate the copy number of different types of machinery and cargo proteins in intracellular CCVs. The fractionation profiling data set gave us the relative abundance of each protein in our CCV-enriched fraction; however, many of these proteins are not actually associated with CCVs, and even those that are may also be associated with other membranes or mainly or exclusively with plasma membrane–derived CCVs. Thus we used the knocksideways data sets to estimate the fraction of a particular protein that was associated with the population of CCVs that depends on AP-1 and epsinR. For instance, if a protein was depleted fourfold by the AP-1 or epsinR knocksideways, this indicates that 80% of the protein is associated with intracellular CCVs and 20% is not, whereas for a protein depleted only twofold, 50% would be associated with intracellular CCVs.
Table 1 shows the approximate copy number of all major coat and cargo proteins for a CCV for which the clathrin coat has the same geometry as a soccer ball, consisting of 20 hexagons and 12 pentagons. Such a CCV would be coated with 60 clathrin triskelions (i.e., 180 monomers of clathrin heavy chain) and have a diameter of ∼100 nm, which is somewhat larger than the majority of CCVs in our fraction (average diameter, 84 nm; Hirst et al., 2008) but still within the normal range. We then used these data to construct three-dimensional (3D) physical models of an intracellular CCV (Figure 3). Consistent with our previous study (Borner et al., 2012), in which we used a different method to estimate copy number (Exponentially Modified Protein Abundance Index [emPAI] vs. Intensity-Based Absolute Quantification [iBAQ]), we calculate that there is close to one AP complex for every clathrin triskelion. This ratio also agrees with a cryo–electron tomography study of CCVs isolated from brain, in which an ultrastructural method was used to determine the number of AP complexes relative to clathrin (Heymann et al., 2013). Also consistent with our previous study, we find that epsinR is even more abundant than AP-1, although it has a much smaller “footprint” on the vesicle (Figure 3, A and B). Other peripheral membrane proteins are present in fewer copies than epsinR or AP-1. There are 20–25 copies of ARF1, but most of it is likely to be associated with adaptors such as AP-1 and GGAs. A protein of unknown function, CVAK104, which consists of a folded N-terminal domain followed by a long, unstructured region containing clathrin and AP-1–binding sites, is relatively abundant, with ∼13 copies per CCV. In contrast, there are predicted to be only one or two copies of GGA2 associated with CCVs, and GGAs 1 and 3 are usually undetectable in our CCV-enriched fractions. However, the association of GGAs with membranes is extremely labile (Hirst et al., 2001), and so there might be many more GGAs associated with intracellular CCVs in vivo, which are lost during the homogenization and fractionation steps.
TABLE 1:
Estimated copy number for all major CCV proteins.
| Protein/gene name | Copy number (from AP-1 ks) | Copy number (from epsinR ks) |
|---|---|---|
| Clathrin | ||
| Clathrin heavy chain/CHC17/CLTC | 180 | 180 |
| CHC22/CLTCL1 | 4.4 | 4.0 |
| Clathrin light chain A/CLTA | 97.2 | 97.3 |
| Clathrin light chain B/CLTB | 43.1 | 42.3 |
| AP-1 | ||
| AP-1 γ/AP1G1 | 60.7 | 47.8 |
| AP-1 β/AP1B1 | 62 | 50.7 |
| AP-1 μ1A/AP1M1 | 48.9 | 38.7 |
| AP-1 σ1A/AP1S1 | 29.3 | 23.1 |
| AP-1 σ1B/AP1S2 | 3.5 | 2.7 |
| Other peripheral membrane proteins | ||
| EpsinR/CLINT1 | 74.7 | 73.4 |
| ARF1 | 25.7 | 20.8 |
| CVAK104/SCYL2 | 13.1 | 13.0 |
| Rab4A | 11.9 | 9.2 |
| OCRL | 11.6 | 11.4 |
| Rab6A | 8.1 | 4.0 |
| PICALM | 7.6 | 8.2 |
| VPS45 | 6.7 | 4.8 |
| STAMBPL1 | 6.4 | 6.1 |
| PIK3C2A | 5.7 | 6.5 |
| Auxilin 2/GAK | 5.5 | 4.2 |
| Hip1R | 5.4 | 5.3 |
| DNAJC6 | 4.4 | 3.9 |
| Gadkin/C4orf16 | 3.4 | 2.4 |
| CLLD6/SPRYD7 | 3.3 | 2.5 |
| Dynamin 2/DNM2 | 2.6 | 2.9 |
| FAM84B | 2.3 | 1.9 |
| TRIM47 | 2.0 | 1.8 |
| Aftiphilin/AFTPH | 1.7 | 0.8 |
| Rab4B | 1.6 | 1.1 |
| GGA2 | 1.5 | 1.2 |
| γ-Synergin/AP1GBP1 | 1.4 | 0.7 |
| PI4K2B | 1.4 | 0.6 |
| p200/HEATR5B | 1.3 | 0.7 |
| LDLRAP1/ARH | 1.2 | 0.8 |
| AGFG1/Hrb | 1.0 | 0.8 |
| RABGEF1 | 0.7 | 0.6 |
| Kif13A | 0.6 | 0.4 |
| Integral membrane proteins | ||
| Cation-dependent M6P receptor/M6PR | 44.7 | 42.1 |
| Transferrin receptor/TFRC | 31.8 | 23.8 |
| VAMP2 (R-SNARE) | 23.4 | 15.6 |
| Cation-independent M6P receptor/IGF2R | 16.2 | 13.3 |
| VAMP8 (R-SNARE) | 11.2 | 10.0 |
| Syntaxin 10/STX10 (Qc-SNARE) | 11.0 | 6.7 |
| VAMP3 (R-SNARE) | 10.3 | 7.9 |
| Vti1b (Qb-SNARE) | 8.2 | 6.5 |
| Syntaxin 6/STX6 (Qc-SNARE) | 6.3 | 3.8 |
| Syntaxin 8/STX8 (Qc-SNARE) | 5.4 | 3.6 |
| SNAP29 (Qbc-SNARE) | 4.9 | 5.1 |
| Vti1a (Qb-SNARE) | 4.7 | 3.1 |
| YIPF6 | 4.0 | 2.7 |
| Syntaxin 16/STX16 (Qa-SNARE) | 3.7 | 2.6 |
| IRAP/LNPEP | 3.5 | 2.5 |
| Syntaxin 12/STX12 (Qa-SNARE) | 3.2 | 2.6 |
| TSPAN13 | 3.2 | 2.2 |
| Carboxypeptidase D/CPD | 2.9 | 1.9 |
| Sortilin/SORT1 | 2.2 | 1.8 |
| VAMP4 (R-SNARE) | 2.1 | 1.3 |
| Lumenal proteins/hydrolases | ||
| Cathepsin Z/CTSZ | 10.9 | 9.2 |
| N-acetylglucosamine-6-sulfatase/GNS | 5.6 | 5.3 |
| DNASE2 | 2.6 | 2.4 |
| Prosaposin/PSAP | 2.5 | 2.8 |
| Hexosaminidase B/HEXB | 1.7 | 2.5 |
| Palmitoyl-protein thioesterase 1/PPT1 | 0.8 | 0.6 |
| Hexosaminidase A/HEXA | 0.6 | 1.3 |
| Granulin/GRN | 0.6 | 0.9 |
| Phospholipase B domain 2/PLBD2 | 0.5 | 0.5 |
The numbers were deduced from our fractionation profiling data (Borner et al., 2014), our earlier AP-1 knocksideways (ks) data (Hirst et al., 2012), and the epsinR ks data from the present study.
FIGURE 3:
Models of intracellular CCVs. (A) Model of the outside of a CCV, using copy number data from Table 1 and structural data from the literature. (B) Model of three known adaptors (AP-1, epsinR, and GGA2) and one candidate adaptor (CVAK104) on the vesicle. (C) Model of the inside of a CCV. (D) Comparison of the models with cryo–electron tomography images of CCVs isolated from HeLa cells. See also Supplemental Movies M1–M3. Scale bar, 100 nm.
The knocksideways data were particularly useful for determining the copy number of cargo proteins because at steady state, such proteins generally reside mainly in stable compartments rather than in transport intermediates, and so it was essential to factor in the relative amount of the protein actually associated with intracellular CCVs. The most abundant integral membrane cargo protein is the cation-dependent mannose 6-phosphate receptor (CDMPR/M6PR), present in >40 copies. The cation-independent mannose 6-phosphate receptor (CIMPR/IGF2R) is also abundant, with ∼15 copies per CCV. The CDMPR has a relatively small lumenal domain, consisting mainly of a ∼150-residue carbohydrate-binding module, but the lumenal domain of the cation-independent mannose 6-phsophate receptor consists of 15 copies of this module in tandem and thus takes up much of the space inside the vesicle (Figure 3C). Each CCV also contains on average ∼20–30 copies of various types of lysosomal hydrolases. However, if some of the CCVs are involved in anterograde traffic and some in retrograde traffic, the retrograde CCVs may be essentially devoid of hydrolases, whereas the anterograde CCVs may have saturated or near-saturated receptors.
Another abundant cargo protein is the transferrin receptor (TfR), estimated to be present in 20–30 copies per CCV. Although we did not previously highlight the TfR as an intracellular CCV cargo protein because in all of our knocksideways studies it was less than twofold depleted, nevertheless it was depleted 1.87-fold in the AP-1 knocksideways and 1.73-fold in the epsinR knocksideways, indicating that ∼40% of the TfR in our preparation is in intracellular CCVs. These data are consistent with previous reports that the TfR is a cargo protein not only for endocytic CCVs, but also for intracellular CCVs (Futter et al., 1998; Gravotta et al., 2012).
The other major transmembrane cargo proteins in intracellular CCVs are soluble N-ethylmaleimide–sensitive factor attachment protein receptors (SNAREs). The most abundant R-SNAREs in the preparation, VAMPs 2, 3, and 8, are only moderately depleted upon AP-1 or epsinR knocksideways (1.5- to 1.9-fold), but even after introducing the correction factor, each one is found to be present in 10–20 copies per intracellular CCV. VAMP4 is almost exclusively associated with AP-1/epsinR–dependent CCVs, but it is much less abundant than the other three VAMPs. Each CCV also contains ∼40 copies of Q-SNAREs. Although SNAREs are relatively small proteins, with essentially no lumenal domains, their sheer number (∼64–90 monomers) must add to the crowding on the cytoplasmic side of the vesicle.
In cryo–electron tomographic slices of HeLa cell CCVs sectioned through the center, the cargo appears as electron densities within the lumen of the vesicle (Figure 3D and Supplemental Movies M1–M3). Some CCVs appear to be fuller than others, suggesting that although we can model an “average” CCV based on our proteomics data, the actual population is likely to be much more heterogeneous.
Gadkin knocksideways cells
Gadkin differs from epsinR in many ways. It is much less abundant in CCVs (only two to four copies), it is found in animals only, and there is no evidence that it acts as a cargo adaptor; instead, it provides a connection between AP-1 and the cytoskeleton. There is no structural information available for gadkin; however, various binding sites and features have been mapped and are indicated in Figure 4A. Three cysteine residues near the N-terminus have been shown to be palmitoylated (Maritzen et al., 2010), and there are several predicted α-helical domains. Binding sites for kinesin-1 and Arp2/3 map to two overlapping motifs centered on a crucial tryptophan residue (Schmidt et al., 2009; Maritzen et al., 2012), and the major binding site for AP-1, which also contains a tryptophan, is some 50 residues downstream (Maritzen et al., 2010). Again, we synthesized a construct with an FKBP domain at the 3′ end, which was stably transfected into Mitotrap-expressing cells. Unlike epsinR-FKBP, gadkin-FKBP was found to be expressed at considerably higher levels than the endogenous protein (Figure 4B). In addition, unlike most other coat components, which cycle on and off the membrane, both endogenous gadkin and the FKBP-tagged construct were constitutively membrane associated, with no detectable cytosolic pool (Figure 4B), presumably because the palmitic acid acts as an efficient membrane anchor. Both endogenous and tagged gadkin were also found to be enriched in CCVs (Figure 4, B and C).
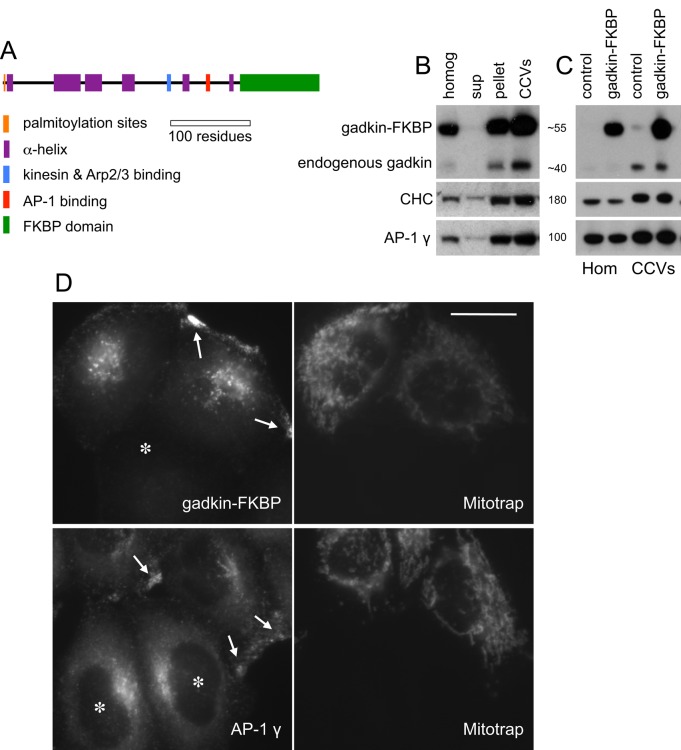
FIGURE 4:
Expression of a gadkin-FKBP construct. (A) Schematic diagram of FKBP-tagged gadkin. (B) Expression and fractionation of gadkin-FKBP. The construct is highly overexpressed and enriched in CCVs. Unlike clathrin (CHC) and AP-1, it cannot be detected in a high-speed supernatant, indicating that it has no cytosolic pool but is entirely membrane associated. (C) Homogenates and CCV fractions from control and gadkin-FKBP–expressing cells in the absence of rapamycin. Gadkin, clathrin, and AP-1 are all enriched similarly in the CCV fraction, whether or not the cells are expressing gadkin-FKBP. (D) Cells expressing gadkin-FKBP and Mitotrap were mixed with nonexpressing HeLa cells and labeled for either gadkin or AP-1. Gadkin-FKBP localizes in a punctate pattern in the perinuclear region of the cell and is also concentrated at the cell periphery. In wild-type cells (marked with asterisks), AP-1 is mainly localized in the perinuclear region of the cell, but in gadkin-FKBP–expressing cells (which coexpress Mitotrap), there is a second pool at the cell periphery (arrows). Scale bar, 20 μm.
Consistent with previous reports (Neubrand et al., 2005; Schmidt et al., 2009), we found that overexpression of the gadkin construct led to partial accumulation of both the construct itself and endogenous AP-1 at the cell periphery (Figure 4D). However, this did not impair the ability of endogenous gadkin or AP-1 to be incorporated into CCVs (Figure 4, B and C). Thus, in spite of the altered appearance of the AP-1 compartment, CCV formation is not compromised.
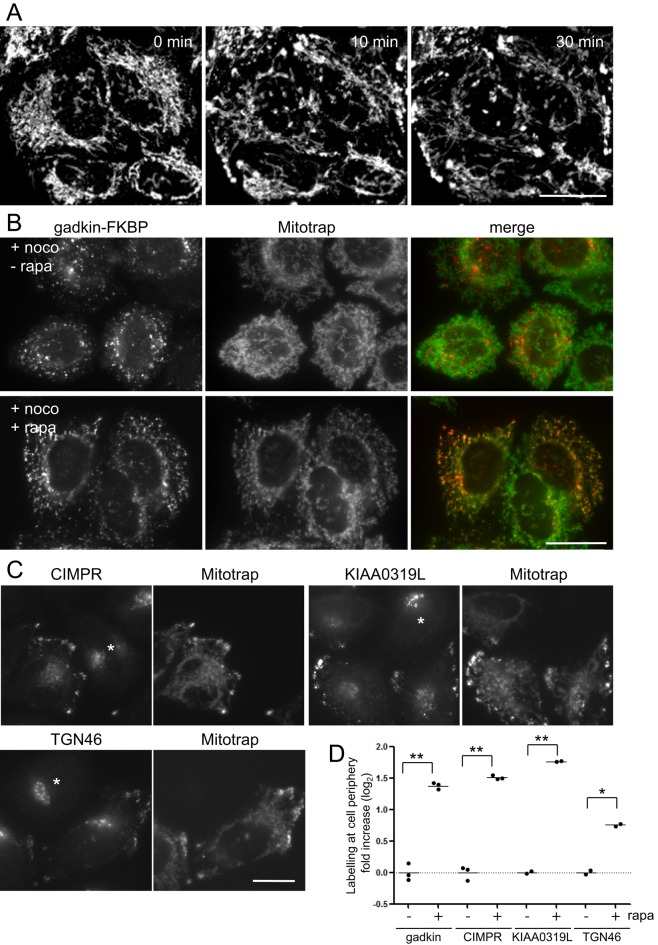
Although there was a certain amount of gadkin-FKBP and AP-1 at the cell periphery even in the absence of rapamycin, addition of the drug caused a twofold to threefold increase in the peripheral labeling of both proteins (Figure 5, A-C). Rapamycin treatment also resulted in strong colocalization between Mitotrap and both gadkin-FKBP and AP-1 (Figure 5, A and B). What was most striking, however, was the dramatic redistribution of mitochondria to the cell periphery. None of our other FKBP-tagged constructs has ever caused the mitochondria to relocate. However, the ability of gadkin to interact with kinesin-1, which is a plus end–directed motor, suggests that when attached to mitochondria, the gadkin may be able to pull the mitochondria along microtubules out toward the perimeter of the cell, where the microtubule plus ends are located.
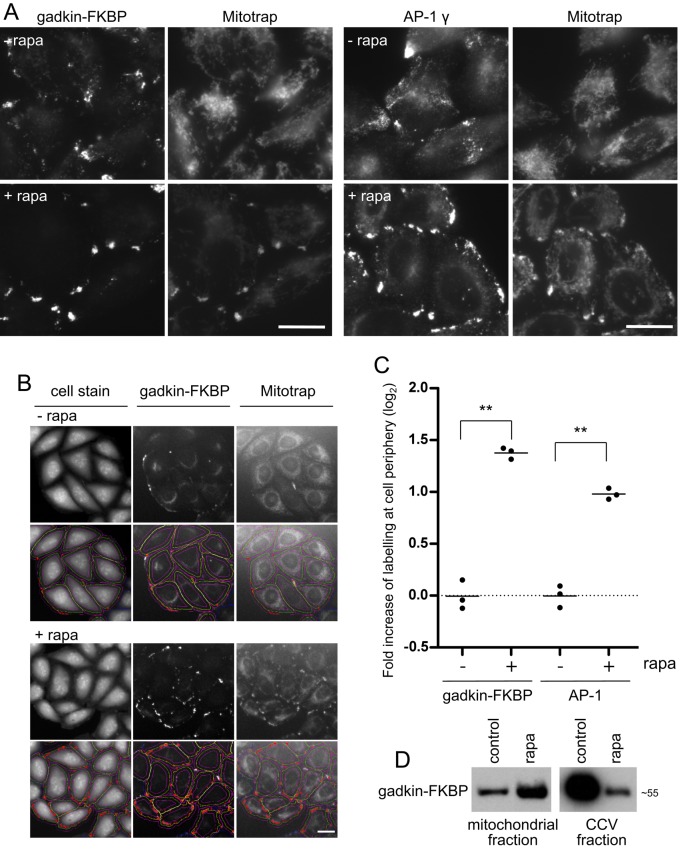
FIGURE 5:
Phenotype of gadkin knocksideways. (A) Rapamycin increases the accumulation of both gadkin-FKBP and AP-1 at the cell periphery and also causes accumulation of mitochondria at the cell periphery, where they colocalize with both gadkin-FKBP and AP-1. Scale bar, 20 μm. (B) Automated microscopy images. The cell stain was used to define the boundaries of the cells, and the peripheral labeling was quantified in both control and rapamycin-treated cells. Scale bar, 20 μm. (C) Quantification of the automated microscopy, with 500 cells quantified for each experiment and repeated three times. The y-axis is a log2 scale. p = 0.0067 for gadkin and 0.0085 for AP-1. (D) After treatment with rapamycin, gadkin-FKBP is enriched in the mitochondrial fraction and depleted from the CCV fraction.
Subcellular fractionation of gadkin knocksideways cells
Subcellular fractionation into mitochondria-enriched and CCV-enriched fractions showed that the addition of rapamycin caused the gadkin-FKBP to accumulate in the mitochondrial fraction and become depleted from the CCV fraction (Figure 5D). To look for other changes in the CCV fraction, we turned again to mass spectrometry. Figure 6 lists all of the proteins depleted twofold or more in a gadkin knocksideways. Once again, there was a strong effect on both accessory proteins and cargoes. Gadkin itself and GGA2 were at the top of the list, both depleted approximately fivefold, and 76 other proteins were also substantially lost (Figure 6).
FIGURE 6:
Proteomics of CCVs from SILAC-labeled gadkin knocksideways cells. The fold change (control:knocksideways) was calculated for each protein, and proteins were ranked from the highest to lowest ratio. Proteins with ratios of ≥2.0 are shown in order of rank for gadkin knocksideways, with known CCV components in color.
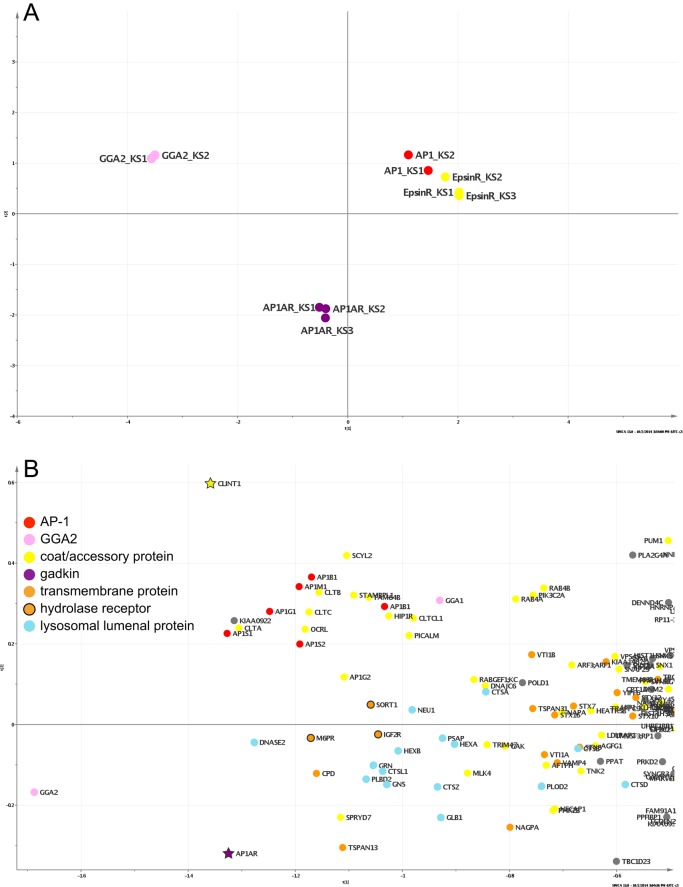
We were surprised by the global effects of both the epsinR knocksideways and the gadkin knocksideways. However, because in both cases AP-1 itself was strongly depleted, one possibility was that the phenotypes were an indirect consequence of the loss of AP-1. To try to tease apart potential differences between the different knocksideways profiles, we analyzed the data from both the present study and our earlier knocksideways study (Hirst et al., 2012) using principal component analysis (PCA). PCA allows one to compare multiple data sets, and therefore it can reveal patterns of similarities and differences that are otherwise difficult to discern. We compared ∼900 proteins identified in common among the knocksideways experiments on AP-1 (two data sets), GGA2 (two data sets), epsinR (three data sets), and gadkin (AP1AR; three data sets), of which ∼100 were likely to be genuine CCV proteins. The coclustering of the AP-1 and epsinR data sets in the PCA plot (Figure 7A) indicates that knocking either one of them sideways had very similar effects on the CCV proteome, consistent with epsinR being a component of the core machinery like AP-1. In contrast, the gadkin knocksideways phenotype was clearly distinguishable from both the AP-1/epsinR knocksideways phenotype and the GGA2 knocksideways phenotype.
FIGURE 7:
PCA of the SILAC data. (A) PCA was used to compare the global effects of epsinR, gadkin, AP-1, and GGA2 knocksideways on CCV composition. Ten data sets were included: the three epsinR and three gadkin data sets from this study and the two AP-1 and GGA2 data sets from our previous study (Hirst et al., 2012). The behavior of 875 proteins common to all 10 sets was used to group individual experiments according to overall similarity. Repeat experiments of the same knocksideways cluster closely, and the AP-1 and epsinR knocksideways experiments cluster, showing that they affect CCV composition in a similar way. The GGA2 and gadkin knocksideways experiments have different effects and cluster separately. (B) PCA on the epsinR and gadkin knocksideways data sets alone, comparing how individual proteins are affected across the six data sets. Some proteins (e.g., hydrolases) are shifted more toward gadkin (purple star); others (e.g., AP-1 subunits) are shifted more toward epsinR (yellow star).
A PCA comparison of individual proteins from just the gadkin and epsinR knocksideways data sets (Figure 7B) highlighted differences in the behavior of several CCV components. Proteins that were more strongly affected by the gadkin knocksideways are shifted toward the bottom of the plot, and those that were more strongly affected by the epsinR knocksideways are shifted toward the top. Lysosomal hydrolases tend to be in the lower portion of the plot, whereas their three receptors (the two mannose 6-phosphate receptors and sortilin) cluster in the middle. Similarly, the raw data sets (Figures 2 and 6 and Supplemental Figure S2) show that the gadkin knocksideways had equivalent effects on hydrolases and their receptors, whereas the epsinR knocksideways had a stronger effect on the receptors. We found a similar trend in our earlier study on GGA2 and AP-1 (Hirst et al., 2012) and interpreted it to mean that GGA2 facilitates the trafficking of hydrolase–receptor complexes, whereas AP-1 facilitates the trafficking not only of hydrolase–receptor complexes (in conjunction with GGA2) but also of empty receptors. These two pathways go in opposite directions: hydrolase–receptor complexes travel in the anterograde direction, from TGN to endosomes, and empty receptors travel in the retrograde direction, from endosomes to TGN. If, as is suggested by Figure 7A, AP-1 and epsinR consistently act together, then the different phenotype of the gadkin knocksideways suggests a preferential association of gadkin with the anterograde population of CCVs, which fits in well with gadkin's role in microtubule-mediated transport of vesicles away from the Golgi region toward the cell periphery.
Relocation of mitochondria and vesicles by gadkin knocksideways
To investigate the kinetics of the gadkin knocksideways, we used live-cell imaging to follow the behavior of Mitotrap, which has a yellow fluorescent protein (YFP) tag. The addition of rapamycin to the gadkin-FKBP cells caused the Mitotrap-labeled mitochondria to fan out toward the cell periphery, a process that was well underway by 10 min and essentially complete by 30 min (Figure 8A and Supplemental Movie M4). Gadkin's ability to interact with kinesin-1 (Schmidt et al., 2009) suggested that this relocation was dependent on microtubules, and, indeed, the microtubule-depolymerizing agent nocodazole prevented the rapamycin-induced relocation of both gadkin-FKBP and mitochondria to the cell periphery, although rerouting of gadkin-FKBP to mitochondria still occurred (Figure 8B).
FIGURE 8:
Movement of mitochondria and membranes to the cell periphery. (A) Gadkin knocksideways cells were treated with rapamycin and imaged on a confocal microscope over 30 min; stills from Supplemental Movie M4. Mitochondria (labeled with Mitotrap, which has a YFP reporter) relocate to the cell periphery. Scale bar, 20 μm. (B) Gadkin knocksideways cells were treated with nocodazole to depolymerize microtubules. In the presence of rapamycin, gadkin-FKBP still reroutes onto mitochondria, but neither the gadkin-FKBP nor the mitochondria reroute to the cell periphery. Scale bar, 20 μm. (C) Wild-type cells (asterisks) and gadkin-FKBP/Mitotrap–expressing cells were mixed, treated with rapamycin, and labeled with antibodies against the indicated cargo proteins, all of which relocate to the cell periphery in the knocksideways cells. Scale bar, 20 μm. (D) Quantification of the relocation by automated microscopy, with the amount of label in the cell periphery quantified in 500 cells in three independent experiments and normalized to 1 to allow increases to be compared (see legend to Figure 5). The y-axis is a log2 scale. p = 0.0067 for gadkin, 0.0029 for CIMPR, 0.0085 for KIAA03197, and 0.0465 for TGN46.
Because gadkin is constitutively membrane associated, we speculated that the rapamycin treatment might be causing an entire compartment or transport intermediate to be rerouted to mitochondria and then to move with mitochondria to the cell periphery. We tested this first by localizing a number of endogenous AP-1 cargo proteins, including CIMPR, KIAA0319L, TGN46, ATP7A, ATP7B, and TfR, as well as two mCherry-tagged proteins, CIMPR and carboxypeptidase D (CPD). All of these proteins were found to relocate to the cell periphery in response to rapamycin, and we also saw relocation of AP-1-associated machinery such as GGA2 (Figure 8, C and D, Supplemental Figures S3 and S4, and Supplemental Movies M5 and M6). In contrast, several other endosome- or Golgi-associated transmembrane proteins, which are not cargo for AP-1, showed no relocation, and we also saw no effects on AP-2, AP-3, or clathrin (Supplemental Figure S5).
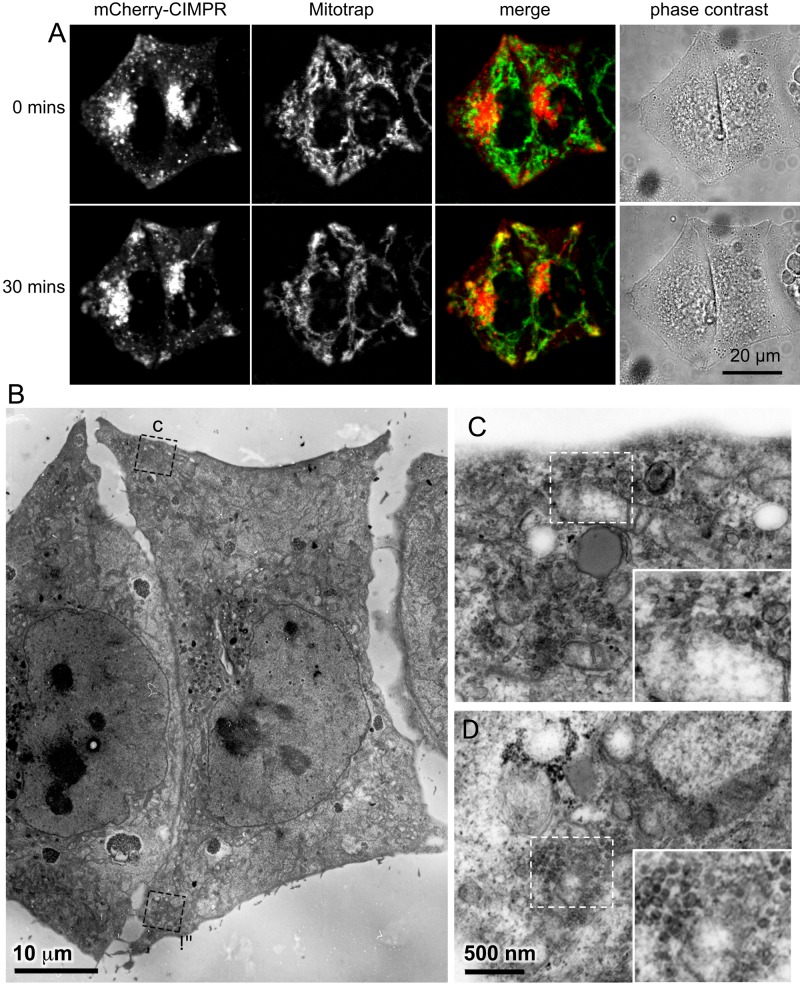
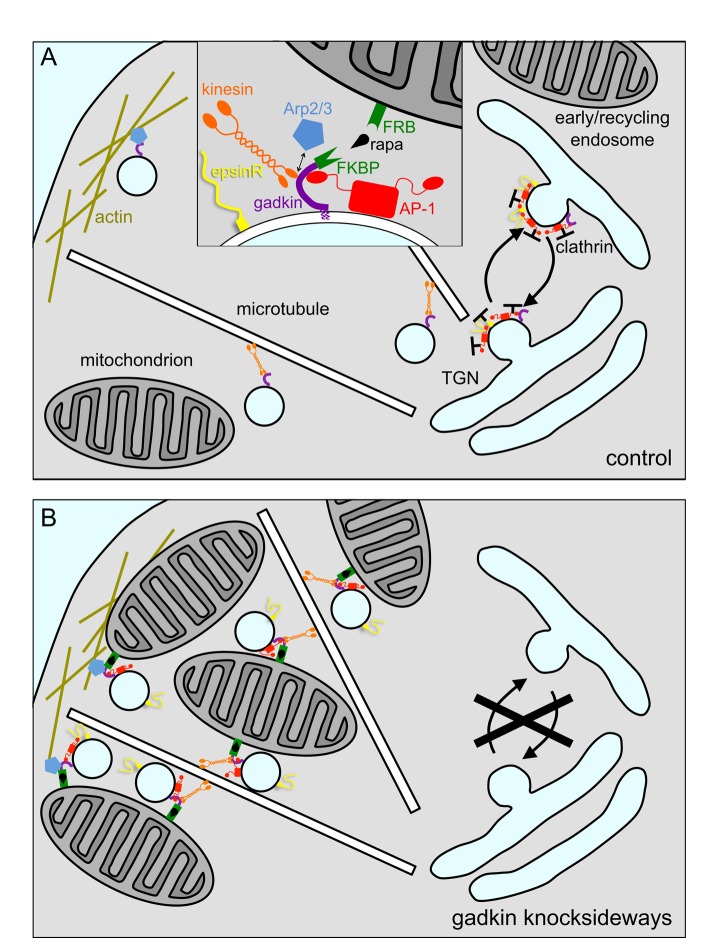
To observe the relocating compartment at the ultrastructural level, we used correlative light and electron microscopy (CLEM) on cells expressing mCherry-tagged CIMPR. First, cells were imaged at frequent intervals after the addition of rapamycin, and both the Mitotrap and the tagged CIMPR were seen to move out toward the cell periphery (Figure 9A). After 30 min, the cells were fixed and processed for electron microscopy, and cells that had been imaged while alive were identified in thin sections (Figure 9B). Peripheral regions of the cells were found to contain not only abundant mitochondria, but also clusters of vesicles surrounding the mitochondria (Figure 9, C and D). These vesicles had diameters ranging from 30 to 50 nm, similar in size to the vesicles of CCVs, except that the mitochondria-associated vesicles did not appear to have clathrin coats. Thus, given the ability of gadkin to interact with both kinesin-1 and Arp2/3 (Maritzen et al., 2012), we propose that in the knocksideways situation, gadkin-FKBP transports both vesicles and mitochondria along microtubules out to the cell periphery and then attaches the vesicles and mitochondria to cortical actin filaments (Figure 10).
FIGURE 9:
Correlative light and electron microscopy of the gadkin knocksideways. (A) Cells coexpressing gadkin-FKBP and Mitotrap were transiently transfected with mCherry-CIMPR, rapamycin was added at 0 min, and the cells were imaged for up to 30 min. (B) The cells shown in A were prepared for electron microscopy. Higher-magnification images were taken for the regions in the periphery outlined by dotted lines (C, D). These regions contain mitochondria surrounded by vesicles.
FIGURE 10:
Schematic diagram of how the gadkin knocksideways might affect trafficking. (A) In control cells, gadkin associates with membranes via its palmitic acids and interacts with the appendage of AP-1; however, there is a large excess of AP-1 over gadkin. After vesicle budding, the gadkin uses its kinesin-1–binding site to pull the vesicles along microtubules and its Arp2/3 binding site to anchor the vesicles in the actin cortex. The inset is modified from Maritzen and Haucke (2010). (B) In the gadkin knocksideways situation, gadkin-FKBP binds essentially all of the membrane-associated AP-1 and also binds to mitochondria, cross-linking the mitochondria to vesicles, with AP-1 and other machinery (but not clathrin) still attached. This may then deplete the machinery from the TGN and endosomes, causing a block in CCV formation.
DISCUSSION
The advantage of the knocksideways method over conventional knockdowns is that it is extremely rapid and thus can provide unique insights into the function of a protein of interest. Before carrying out the present study, we predicted that the epsinR knocksideways would have a cargo-selective phenotype similar to our previous GGA2 knocksideways but affecting vti1b rather than hydrolases and their receptors, and that the gadkin knocksideways would have no effect on the production of CCVs but might affect the ability of the vesicles to move along microtubule tracks. However, in both cases, we saw much more global phenotypes than we had anticipated.
EpsinR
Although epsinR has been shown to be a cargo adaptor, the present study indicates that it is an order of magnitude more abundant in CCVs than its known cargo protein, vti1b, suggesting that it may have other functions as well. Knocking down epsinR in cultured cells has produced variable results, but in general there does not appear to be a very strong trafficking phenotype (Hirst et al., 2003, 2004; Saint-Pol et al., 2004), although whether this is because the cells are able to compensate using other pathways or because the cells can get by with only very small amounts of epsinR is unclear.
There is only one report of an epsinR mutant in a vertebrate: an insertional mutagenesis study in zebrafish, in which epsinR was identified as a causative gene for hyperproliferation of the epidermis (Dodd et al., 2009), although the precise mechanism is unknown because the insertion was within an intron. Epidermal hyperproliferation has also been observed in patients with homozygous mutations in AP1S1, one of three related genes encoding AP-1 small subunits (Montpetit et al., 2008), and in patients who are heterozygous for a mutation in AAGAB, which encodes a binding partner for cytosolic AP-1 and AP-2 (Giehl et al., 2012; Pohler et al., 2012). Together these studies point to a role for the AP-1/epsinR pathway in epidermal development, possibly by damping down signals that would otherwise cause excessive cell proliferation.
Why are the epsinR knocksideways cells unable to produce isolatable intracellular CCVs when they have abundant clathrin-coated budding profiles in the Golgi region? We do not yet know the answer, but there are some interesting parallels between epsinR and CALM, an accessory protein for clathrin-mediated endocytosis. Both epsinR and CALM are even more abundant in HeLa cell CCVs than AP-1 or AP-2, respectively (Borner et al., 2012); both have ENTH-like domains (Evans and Owen, 2002); both can bind SNAREs (Miller et al., 2007, 2011); and, in both cases, knockdowns cause widening of clathrin-coated structures (Meyerholz et al., 2005). A recent study showed that the first 18 residues of CALM form an amphipathic helix, which senses and promotes membrane curvature (Miller et al., 2015), and epsinR also has a predicted N-terminal membrane-binding amphipathic helix (Miller et al., 2007). In the case of CALM, the membrane-inserting helix, together with CALM's small footprint on the vesicle relative to AP-2 and the types of cargo proteins sorted by CALM (i.e., SNAREs, which have no real lumenal domain) combine to reduce the size of clathrin-coated pits and drive CCV closure. Thus, in cells in which CALM is depleted and AP-2 is the major adaptor, the pits are larger and a higher percentage of them are open, whereas in cells in which AP-2 is depleted and CALM is the major adaptor, the pits are not only greatly reduced in number, but are also smaller. It is tempting to speculate that a similar relationship may exist between epsinR and AP-1.
In the case of the CALM knockdown, CCVs are still able to form from the plasma membrane, although less efficiently than in control cells, whereas the present study shows a near block in intracellular CCV formation when epsinR is knocked sideways. However, it is important to remember that knocking down epsinR or AP-1 produces a much weaker phenotype than knocking them sideways, and the same may turn out to be true for CALM. Moreover, another treatment that causes widening of clathrin-coated pits is cholesterol depletion, and in this case, clathrin-mediated endocytosis is completely abolished (Rodal et al., 1999; Subtil et al., 1999; Kozik et al., 2013). Further ultrastructural studies, including 3D reconstruction, should help to pinpoint precisely what happens to clathrin-coated structures when epsinR is knocked sideways and why CCV formation is inhibited.
Like other adaptors for CCVs, epsinR binds clathrin. The best characterized of the adaptors, AP-2, was recently shown to use an autoinhibitory mechanism to render its clathrin- binding site inaccessible when in its “locked” (i.e., cytosolic) conformation (Kelly et al., 2014). Of interest, none of the adaptors we investigated using the knocksideways system, including AP-2, AP-1, GGA2, and epsinR, appears to be capable of recruiting clathrin onto mitochondria (Robinson et al., 2010; Hirst et al., 2012; Supplemental Figure S1). This suggests that they all have regulatory switches to mask their clathrin-binding sites unless they are “productively” associated with a membrane.
One outcome of our epsinR knocksideways study, together with our previous knocksideways study on AP-1, is that we can estimate the copy number of every protein associated with intracellular CCVs. These numbers show that the cytoplasmic surface of the vesicle is extremely crowded with machinery, which is consistent with previous studies (Borner et al., 2012; Heymann et al., 2013), but the finding that the lumenal side of the vesicle is equally crowded was more surprising. A cryo–electron tomography study on brain CCVs reported that the vesicles have “small payloads” (Heymann et al., 2013), that is, the authors concluded that the CCVs contained relatively little cargo because of a lack of density in the vesicle lumen. In contrast, AP-1/epsinR–positive CCVs from HeLa cells are full of cargo: a 100-nm CCV contains on average ∼200 transmembrane proteins, many of which have most of their mass on the noncytoplasmic side of the membrane. However, if one assumes that most of the brain CCVs are synaptic vesicle precursors containing a similar complement of membrane proteins to the synaptic vesicles themselves, then a vesicle with a diameter of 41.6 nm would contain ∼150 integral membrane proteins (Takamorim et al., 2006), which is actually slightly higher than the density of integral membrane proteins we calculated for an intracellular CCV with an ∼50-nm vesicle inside. The major difference is that most of the transmembrane proteins of synaptic vesicles are either SNAREs or polytopic proteins, with little mass on the noncytoplasmic side, whereas intracellular CCVs contain a number of cargo proteins with large lumenal domains, several of which also have bulky ligands bound to them.
These ligands include ∼25 copies of lysosomal hydrolases, and in our previous knocksideways study, we proposed that GGAs were the main adaptors for anterograde transport of hydrolase–receptor complexes (Hirst et al., 2012). If this hypothesis is correct, then the prediction is that CCVs ought to contain similar copy numbers of GGAs and hydrolases. However, we calculate only one to two GGAs per CCV. Because we previously showed that GGAs are rapidly lost from the membrane once cells are broken open (Hirst et al., 2001), we suspect that our GGA copy number is a gross underestimate; otherwise, it is difficult to imagine how an adaptor of such low abundance could carry such a high load of cargo.
Transferrin receptors are also abundant and are likely to be sorted by AP-1 via their YXXΦ motif. However, SNAREs are even more abundant than transferrin receptors, and most do not have recognizable sorting motifs. We previously showed that R-SNAREs can be sorted at the plasma membrane by CALM (Miller et al., 2011), and our data suggest that CALM is also associated with intracellular CCVs. However, CALM behaves mainly as a plasma membrane adaptor, and although our data suggest that CALM is also associated with intracellular CCVs, there are not enough copies to bind all of the R-SNAREs (∼8 vs. ∼40 copies; Table 1). Another possibility is that epsinR may be able to bind to other SNAREs in addition to vti1b (Chidambaram et al., 2008) or that CVAK104, which is fairly abundant and has an adaptor-like structure, might be a SNARE adaptor. Of interest, we previously showed by Western blotting that conventional knockdowns of CVAK104 result in reduced levels of syntaxin 8 and vti1b associated with CCVs (Borner et al., 2007).
The high density of proteins that we observe, not only on the cytoplasmic side of the membrane, but also on the lumenal side and within the phospholipid bilayer, provides strong support for a model that was first proposed nearly 35 years ago (Pearse and Bretscher, 1981): that selected cargo proteins are so tightly packed in newly forming CCVs that unwanted proteins are simply “squeezed out” because there is not enough room for them.
Gadkin
Unlike the epsinR knocksideways phenotype, we suspect that the gadkin knocksideways phenotype is a dominant-negative effect rather a than loss of function. First, the construct was expressed at considerably higher levels than wild-type gadkin (our proteomics data suggest ∼70-fold overexpression), and second, a gadkin-knockout mouse has been generated and found to have almost no phenotype (Maritzen et al., 2012), whereas AP-1 knockouts are embryonic lethal (Zizioli et al., 1999; Meyer et al., 2000), which argues against an essential role for gadkin in AP-1 function. Nevertheless, the unusual nature of the gadkin knocksideways phenotype is informative.
Gadkin is palmitoylated, and the lack of any detectable cytosolic pool suggests that this modification converts the protein into a constitutively membrane-associated form, almost like an integral membrane protein. Thus, although we had assumed that the knocksideways system would only work on proteins that cycle rapidly back and forth between a cytosolic pool and an “anchored” pool, our results with gadkin indicate that membrane proteins can also be rerouted to mitochondria and that this rerouting can produce dramatic effects on the cell. Wong and Munro (2014) also recently demonstrated that vesicles can be captured by mitochondria, using a somewhat different system in which different types of golgins were attached to mitochondria, although in their study, the mitochondria did not relocate. Other, more recent studies used inducible heterodimerization of cytoskeletal motors and membrane-associated proteins to reposition different membrane compartments (Bentley et al., 2015; van Bergeijk et al., 2015). Thus this type of approach holds great promise as a means of investigating and manipulating the dynamics of vesicles and organelles.
Overexpression of gadkin on its own also has a phenotype: it causes AP-1 to redistribute to the cell periphery and stabilizes the membrane association of AP-1. This phenotype presumably occurs because normally there are only two to three copies of gadkin per CCV, which is not enough to “pin down” greater than ∼5% of the AP-1, but if the gadkin is expressed at a 70-fold higher level, all of the AP-1 will be affected. However, in the absence of rapamycin, this redistribution and increased stability of AP-1 do not affect CCV production, and so the AP-1 must still be cycling on and off the membrane to some extent. But once rapamycin is added, CCV production is inhibited and at the same time, a population of vesicles moves out to the cell periphery.
What is the nature of these vesicles? They are similar in size to CCVs, but both immunofluorescence and electron microscopy indicate that they are clathrin-negative. However, they have AP-1 and other machinery associated with them and are laden with AP-1–dependent cargo proteins without containing resident proteins of the organelles from which CCVs bud, such as sialyl transferase. Therefore, we suspect that they are pinched-off CCVs that have lost their clathrin coats but retained much of their other machinery. How this correlates with the cessation of CCV production is less clear because AP-1 is partially distributed to the cell periphery in the gadkin-FKBP–expressing cells whether or not rapamycin is added, although the redistribution is more pronounced in the presence of rapamycin. However, we suspect that the rapamycin-induced cross-linking prevents one or more key components of the machinery from being recycled, and these components could include integral as well as peripheral membrane proteins.
Although the gadkin knocksideways, like the epsinR and AP-1 knocksideways, blocks intracellular CCV formation, its phenotype is distinct, with some proteins affected more than others. For instance, lysosomal hydrolases were strongly affected by the gadkin knocksideways, whereas the transferrin receptor was affected very little. This may be because gadkin preferentially associates with anterograde rather than retrograde vesicles. Two mechanisms have been shown to be important for gadkin localization: palmitoylation and AP-1 binding (Maritzen et al., 2010). However, palmitoylated proteins can associate with many different membranes, and AP-1 facilitates the formation of retrograde as well as anterograde CCVs, so it remains to be determined how gadkin can distinguish between these two populations or between the two membranes from which they bud.
Thus, knocking either epsinR or gadkin sideways effectively blocks the production of intracellular CCVs but by very different mechanisms, providing insights into the physiological functions of the two proteins. However, because in both cases the knocksideways produced global effects on CCV formation, the knocksideways phenotypes raise additional questions about where, when, and how the two proteins act in clathrin-mediated intracellular trafficking. Promising approaches for future studies include live-cell imaging with 3D tracking to try to define the order of events in intracellular CCV formation (Kural et al., 2012) and genome editing to remove proteins completely instead of only partially (Kim and Kim, 2014). For instance, if the epsinR gene is deleted, can cells still make intracellular CCVs? If so, which cargo proteins are affected? Presumably vti1b will be unable to get into CCVs, but will other cargo proteins be affected as well? By combining the knocksideways approach with gene knockouts, it should be possible to uncover the functions of each of the many components of intracellular CCVs.
MATERIALS AND METHODS
Antibodies and constructs
Antibodies used in this study include in-house antibodies against clathrin (Simpson et al., 1996) and epsinR (Hirst et al., 2003) and commercial antibodies against EEA1 (E41120; BD Transduction Labs, Oxford, UK), LAMP1 (sc18821; Santa Cruz Biotechnology, Heidelberg, Germany), GM130 (51-9001978; BD Transduction Labs), CIMPR (ab2733; Abcam, Cambridge, UK), AP-1 γ (mAb100.3), AP-1 μ1 (AP50; 611351; BD Transduction Labs; although raised against μ2, this antibody cross-reacts with μ1), and gadkin (AP1AR; ab122296; Abcam). The rabbit anti-TGN46 and mouse anti-GGA2 were kind gifts from Matthew Seaman (Cambridge Institute for Medical Research, Cambridge, United Kingdom) and Doug Brooks (Women's and Children's Hospital, North Adelaide, Australia), respectively. Horseradish peroxidase–labeled secondary antibodies were purchased from Sigma-Aldrich (Gillingham, UK), and fluorescently labeled secondary antibodies were from Invitrogen (Grand Island, NY). mCherry-CPD and mCherry-CIMPR were made by transferring the tails of CPD and CIMPR from CD8-CPD and CD8-CIMPR constructs described in Harasaki et al. (2005) into an mCherry vector (Clontech, Saint-Germain-en-Laye, France). The mCherry-tagged clathrin light-chain plasmid was obtained from Addgene. pLXIN(epsinR-FKBP) and pLXIN(gadkin-FKBP) were constructed synthetically, as this substantially reduced the manipulations. Various internal restriction enzyme sites were removed, cloning sites (5′ Xho1 and 3′ Not1) were added, the stop codons were mutated, and silent mutations for siRNA resistance were introduced. The synthetic constructs were transferred into the pLXIN vector containing an FKBP domain–coding sequence between the Not1 and BamH1 sites for the C-terminal tag (both proteins). For gadkin-FKBP and epsinR-FKBP, a hemagglutinin (HA) tag was inserted as a linker between the target protein and FKBP, which allowed expression levels relative to Mitotrap to be assessed (Mitotrap also contains an HA linker). Both pLXIN[Xho1-Not1]HA-FKBP and pLXIN[Xho1-Not1]myc-FKBP exist as cassettes for rapid cloning of other knocksideways constructs. Triple myc-tagged rat α-2,6-sialyltransferase was expressed from SMH4, a derivative of STM (Munro, 1991).
RNA interference
Knockdowns were performed using the following On-Target Plus SMARTpool siRNA reagents from Dharmacon (UK), with a nontargeting SMARTpool siRNA (D-001810-10) used as a control. The siRNAs were as follows: for CLINT1 (epsinR), LU-021406-00 (duplex 5, GCUCCUAGCUUACCUCAUA; duplex 6, CAGCAGCCAUCACUGAAUA; duplex 7, AUUCAGAGAUCGAGUCUAA; duplex 8, UGGUAAGGAUCAAGGUAUA); and for gadkin, LU-015504-02 (duplex 17, GCACUUAAGUAUAGCAACA; duplex 18, GAGGUGAGCACUUAACAAU; duplex 19; GGAAGACAUUCUACGGGCA; duplex 20, CUAUAUUUCCAAUGAACGA). All siRNAs were used at a concentration of 25 nM, and for SILAC experiments, knockdowns were performed with a single-hit 96-h protocol (to reduce the effect of the unlabeled amino acids in Optimem) using Oligofectamine (Invitrogen) and Optimem, following the manufacturer's instructions. Knockdown efficiencies were determined by Western blotting and showed >85% depletion of the target proteins.
Tissue culture
HeLaM cells (Tiwari et al., 1987) were grown in DMEM (Sigma-Aldrich) supplemented with 10% (vol/vol) fetal calf serum (Sigma-Aldrich), 2 mM l-glutamine, 50 U/ml penicillin, and 50 μg/ml streptomycin. The construction of a stable cell line expressing just the Mitotrap construct (consisting of a mitochondrial targeting signal, YFP, and an FRB domain) was previously described (Robinson et al., 2010; Hirst et al., 2012). These cells were used to control for spurious rerouting of proteins that contain FKBP domains and for nonspecific rapamycin effects and were also the starting point for transfecting in pLXIN(epsinR-FKBP) or pLXIN(gadkin-FKBP). Clonal cell lines were derived by G418 selection and selected on the basis on expression of both Mitotrap and target protein–FKBP and by the ability of the construct to reroute to mitochondria after addition of rapamycin.
For proteomics, cells were grown in SILAC medium supplemented with 10% (vol/vol) dialyzed fetal calf serum (10,000 molecular weight cut-off; Invitrogen), penicillin/streptomycin (Sigma-Aldrich), and either “heavy” amino acids (l-arginine-13C615N4:HCl [50 mg/l] and l-lysine-13C615N2:2HCl [100 mg/l]; Cambridge Isotope Laboratories) or the equivalent “light” amino acids. Cells were grown for at least 7 d to achieve metabolic labeling, and the average incorporation efficiency was ∼95%, as determined by mass spectrometry. In all experiments, the cells to be treated with rapamycin were grown in heavy SILAC medium.
Fluorescence microscopy
For immunofluorescence microscopy, cells were plated into glass-bottom dishes (MatTek) and treated where indicated with 200 ng/ml rapamycin for 30 min or 16.6 mM nocodazole for 2 h or fed with 10 μg/ml Alexa Fluor 594–labeled transferrin (Tf-594) for 5–15 min. The cells were then fixed with 3% formaldehyde, permeabilized with 0.1% Triton X-100, and labeled as indicated. The cells were imaged with a Zeiss Axiovert 200 inverted microscope using a Zeiss Plan Achromat 63× oil immersion objective (numerical aperture, 1.4), a Hamamatsu OCRA-ER2 camera, and ImproVision Openlab software.
For live-cell microscopy, cells were plated into glass-bottom dishes, incubated in CO2-independent medium, and imaged on a Zeiss LSM710 confocal microscope with Zeiss ZEN software. Movie images were captured every 10 s for a period of up to 30 min and run at 10 frames/s.
To quantify knockdown phenotypes, we used an automated ArrayScan VTI microscope (Cellomics/Thermo-Fisher) and the SpotDetector V4 assay algorithm. Cells were plated onto 96-well PerkinElmer microplates and treated with and without rapamycin for 30 min, fixed, and then stained with various antibodies, followed by Alexa Fluor 647–donkey anti-mouse immunoglobulin G and blue whole-cell stain (Invitrogen). The cells were imaged with a modified Zeiss Axiovert 200M inverted microscope, a Zeiss 40×/0.5 Achroplan objective, and a Hamamatsu OCRA-ER camera, and 500 cells were quantified for each condition using ARRAYSCAN software and repeated three times. The blue whole stain allowed a mask to be drawn around the cell and the intensity of labeling at the cell periphery to be quantified.
Electron microscopy
For conventional electron microscopy (EM), gadkin knocksideways cells were treated with or without rapamycin for 30 min and then fixed by adding an equal volume of freshly prepared 4% paraformaldehyde/5% gluteraldehyde in 0.1 M cacodylate buffer, pH 7.2. For epsinR knocksideways, cells were first depleted of the endogenous epsinR using a 72-h siRNA knockdown protocol and then treated with or without rapamycin for 30 min before fixation as described. After 2 min, the solution was removed and replaced by 2% paraformaldehyde/2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.2, for 1 h at room temperature. Cells were then secondarily fixed with 1% osmium tetroxide, followed by incubation with 1% tannic acid to enhance contrast. Cells were dehydrated using increasing percentages of ethanol before being embedded onto Epoxy resin (Agar Scientific, United Kingdom) stubs. Coverslips were cured overnight at 65°C. Ultrathin sections were cut using a diamond knife mounted to a Reichert ultracut S ultramicrotome, and floating sections were collected onto copper grids. Grids were poststained with drops of lead citrate. Sections were viewed on a FEI Tecnai transmission electron microscope (Eindhoven, Netherlands) at a working voltage of 80 kV. For epsinR knocksideways, clathrin-coated vesicles in the vicinity of the Golgi or within 500 nm of the plasma membrane were quantified using ImageJ software (National Institutes of Health, Bethesda, MD). For CLEM, gadkin knocksideways cells were transfected with mCherry-CIMPR and then plated onto gridded glass-bottom dishes (MatTek). The cells were then imaged (mCherry-CIMPR and YFP-Mitotrap) at different times during a 30-min treatment with rapamycin and then fixed for EM as described. A resin stub was embedded over the area of interest and cured overnight at 65°C, and then the dish was removed from the resin stub using liquid nitrogen.
For cryo–electron tomography, CCV pellets were homogenized in buffer A (0.1 M MES, pH 6.5, 0.2 mM EGTA, and 0.5 mM MgCl2) and mixed with 10-nm colloid gold particles before application of to holey carbon grids. The grids were briefly blotted with filter paper and plunged into liquid ethane slush. Tomographic tilt series were collected using a Tecnai F30 microscope (FEI) operating at 200 kV and University of California, San Francisco, tomography software. A typical data set was from −60 to +60 deg with 2-deg increments. The nominal magnification was set at 20,000×, resulting in a final pixel size of 7.15 Å on a 2k × 2k Tietz charge-coupled device (F224HD). The defocus was set to ∼7 μm, and the final accumulative dose on the specimen was ∼50e−/Å2. The program package IMOD was used to align the tomographic tilt series, and the aligned image stack was normalized using Bsoft software. The final reconstructed volume was generated using the Spider program. Volumes containing clathrin-coated structures were displayed using IMOD, and individual assemblies were boxed out from the 3D reconstructed tomograms and presented as a series of parallel slices. The slice thickness is 14.3 Å. Images were contrasted in Photoshop (Adobe).
Isolation of mitochondria
To isolate mitochondria, we used a standard isolation kit (Mitenyl) with minor modifications. In brief, 5 × 106 cells were scraped off a dish with 1.5 ml of lysis buffer and homogenized by 18 strokes of syringe and needle (21 gauge). The lysate was centrifuged at 100 × g for 1 min to remove some nuclei, and then 1 ml of homogenate was mixed with 9 ml of separation buffer and incubated with 50 μl of TOM-22 coated micromagnetic beads for 1 h. The magnetic beads were collected by passing the sample through a column placed in a magnetic field, washed with 4 ml of separation buffer (repeated three times), and eluted in 1.5 ml of separation buffer. The mitochondria were then concentrated into 100 μl of storage buffer by spinning at 13,000 × g for 2 min.
Knocksideways and proteomics
Where indicated, cells grown in both SILAC heavy and SILAC light medium were first treated with siRNAs to deplete the endogenous versions of the target proteins. SILAC heavy cells were then treated with 200 ng/ml rapamycin for 30 min at 37°C (for both gadkin and epsinR knocksideways, this was determined to be the optimal time based on microscopy imaging of the rerouting onto mitochondria) and then CCVs were isolated in parallel from the SILAC heavy and SILAC light cells, maintaining rapamycin in the buffers where appropriate, as described in detail in Hirst et al. (2012). The SILAC heavy and SILAC light samples were mixed at equal protein concentrations with a maximum combined total of 50 μg, loaded in a single lane onto a preparative 1.5-mm 10% acrylamide gel, and run so that the sample separated into a 5-cm strip. The gel was then washed, stained with Coomassie blue, and cut into 10 slices. Proteins were reduced, alkylated with iodoacetamide (A3221; Sigma-Aldrich), and in-gel digested with trypsin, and the sample was analyzed by liquid chromatography–tandem mass spectrometry in an Orbitrap mass spectrometer (Q-Exactive; Borner et al., 2012).
Data analysis
For both the epsinR knocksideways and the gadkin knocksideways, data sets were produced of three independent biological repeats. The raw data files were processed using MaxQuant (with requantify and match between runs features enabled). The primary output for each SILAC comparison of CCVs was a list of identified proteins, a ratio of relative abundance (heavy/light ratio), and the number of quantification events (count).
The raw data, shown in Supplemental Table S1, were formatted as follows:
Reverse hits, proteins identified only by site, and common contaminants were removed.
Proteins with no gene names were removed.
Ratios were linearly normalized based on total intensities, assuming equal protein quantities in both heavy and light samples (essentially as in Borner et al., 2012).
Only the 1390 proteins identified in all six data sets were kept (Master Data, Supplemental Table S1).
Means and medians of changes for epsinR or gadkin knocksideways were determined. For each protein, the ratio of median/mean was calculated for both epsinR and gadkin knocksideways. Seven proteins showed median/mean ratios >2 or <0.5, indicative of outlier quantification in at least one experiment. They were removed from the data set, leaving 1383 proteins.
A data set was produced from a control cell line expressing only Mitotrap, and proteins that were affected >1.5- or <0.67-fold were removed from the gadkin and epsinR knocksideways data sets, resulting in a final list of 1370 proteins. This was to eliminate proteins that were affected purely by the addition of rapamycin.
In all cases, the rapamycin-treated cells were labeled with SILAC heavy amino acids and the controls with SILAC light amino acids. The Master Data in Supplemental Table S1 show the heavy/light ratio and thus levels of protein left after knocksideways. For the tables in Figures 2 and 6, these data were averaged across the three biological repeats and inverted to show mean fold-depletion caused by knocksideways (see Inverted Averages, Supplemental Table S1, for a complete list).
For PCA, SILAC ratio data were log transformed and centered as in Borner et al. (2012). PCA was performed in SIMCA-P 13.0.3 (Umetrics).
Statistical analysis was performed in Excel (Microsoft) and Prism (GraphPad Software).
Protein copy number and 3D physical models
Three data sets were used to estimate the copy number of each protein in intracellular CCVs: our recent fractionation profiling data set (Borner et al., 2014), our AP-1 knocksideways data set (Hirst et al., 2012), and the epsinR data set obtained in the present study. These should all be comparable because CCV-enriched fractions were prepared using an identical protocol in all three studies.
The fractionation profiling data set was used to determine the copy number of every protein in the CCV-enriched fraction relative to clathrin heavy chain (CLTC), which was assigned an iBAQ abundance score of 1,000,000. Thus, for instance, the abundance score for the cation-dependent mannose 6-phosphate receptor (M6PR) was 256,176. The AP-1 and epsinR knocksideways data sets gave a fold-depletion value for every protein; for example, CLTC was depleted 2.83-fold in the AP-1 knocksideways and 4.59-fold in the epsinR knocksideways, giving an abundance score for intracellular CCVs of 646,643 and 782,135 respectively (1,000,000 divided by 2.83 or 4.94 gives the amount of CLTC not affected by the knocksideways; this number is then subtracted from 1,000,000). M6PR was depleted 2.68- and 3.49-fold in the AP-1 and epsinR knocksideways, respectively, giving abundance scores of 160,588 and 182,773. To determine how many copies of each protein were associated with an intracellular CCV, the CCV was assumed to contain 180 copies of CLTC and the other numbers were adjusted accordingly. Thus, from the AP-1 knockdown, we calculated that there were 44.7 copies of M6PR, and from the epsinR knockdown, we calculated 42.1 copies.
These numbers were used to construct 3D physical models of intracellular CCVs, using either solid or hollow polystyrene balls for the vesicles, a Buckminsterfullerene construction kit (ezmolecule.com) for the clathrin, and painted balsa wood or polystyrene for the other proteins. The flexible linkers of the adaptors were made out of 0.5-mm copper wire. Where available, x-ray crystallography structures were used to determine the sizes and shapes of the proteins; otherwise, the proteins were assumed to be globular and the sizes calculated from the number of amino acids, using www.calctool.org/CALC/prof/bio/protein_size.
Supplementary Material
Abbreviations used:
- AP-2
adaptor protein 2
- CCV
clathrin-coated vesicle
- CLEM
correlative light and electron microscopy
- ENTH
epsin N-terminal homology
- SILAC
stable isotope labeling by amino acids in cell culture
- SNARE
N-ethylmaleimide–sensitive factor attachment protein receptor
- TGN
trans-Golgi network.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-04-0245) on July 15, 2015.
*These are to be considered joint second authors.
Present addresses: †Department of Proteomics and Signal Transduction, Max-Planck Institute of Biochemistry, 82152 Martinsried, Germany
‡Departments of Biochemistry/Biophysics and Pharmaceutical Chemistry, University of California, San Francisco, San Francisco, CA 94133
§Bioelectron Microscopy Core Facility, École Polytechnique Fédérale de Lausanne, 1015 Lausanne, Switzerland.
REFERENCES
- Bentley M, Decker H, Luisi J, Banker G. A novel assay reveals preferential binding between Rabs, kinesins, and specific endosomal subpopulations. J Cell Biol. 2015;208:273–281. doi: 10.1083/jcb.201408056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GHH, Antrobus R, Hirst J, Bhumbra GS, Kozik P, Jackson LP, Sahlender DA, Robinson MS. Multivariate proteomic profiling identifies novel accessory proteins of coated vesicles. J Cell Biol. 2012;197:141–160. doi: 10.1083/jcb.201111049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GHH, Hirst J, Hein MY, Edgar J, Mann M, Robinson MS. Fractionation profiling: a fast and versatile approach for mapping vesicle proteomes and protein-protein interactions. Mol Biol Cell. 2014;25:3178–3194. doi: 10.1091/mbc.E14-07-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GHH, Rana AA, Forster R, Harbour M, Smith JC, Robinson MS. CVAK104 is a novel regulator of clathrin-mediated SNARE sorting. Traffic. 2007;8:893–903. doi: 10.1111/j.1600-0854.2007.00576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady RJ, Damer CK, Heuser JE, O'Halloran TJ. Regulation of Hip1r by epsin controls the temporal and spatial coupling of actin filaments to clathrin-coated pits. J Cell Sci. 2010;123:3652–1361. doi: 10.1242/jcs.066852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidambaram S, Zimmermann J, von Mollard GF. ENTH domain proteins are cargo adaptors for multiple SNARE proteins at the TGN endosome. J Cell Sci. 2008;121:329–338. doi: 10.1242/jcs.012708. [DOI] [PubMed] [Google Scholar]
- Dodd ME, Hatzold J, Mathias JR, Walters KB, Bennin DA, Rhodes J, Kanki JP, Look AT, Hammerschmidt M, Huttenlocher A. The ENTH domain protein Clint1 is required for epidermal homeostasis in zebrafish. Development. 2009;136:2591–2600. doi: 10.1242/dev.038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PR, Owen DJ. Endocytosis and vesicle trafficking. Curr Opin Struct Biol. 2002;12:814–821. doi: 10.1016/s0959-440x(02)00395-0. [DOI] [PubMed] [Google Scholar]
- Futter CE, Gibson A, Allchin EH, Maxwell S, Ruddock LJ, Odorizzi G, Domingo D, Trowbridge IS, Hopkins CR. In polarized MDBK cells basolateral vesicles arise from clathrin-gamma-adaptin-coated domains on endosomal tubules. J Cell Biol. 1998;141:611–623. doi: 10.1083/jcb.141.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabernet-Castello C, Dacks JB, Field MC. The single ENTH-domain protein of trypanosomes; endocytic functions and evolutionary relationship with epsin. Traffic. 2009;10:894–911. doi: 10.1111/j.1600-0854.2009.00910.x. [DOI] [PubMed] [Google Scholar]
- Giehl KA, Eckstein GN, Pasternack SM, Praetzel-Wunder S, Ruzicka T, Lichtner P, Seidl K, Rogers M, Graf E, Langbein L, et al. Nonsense mutations in AAGAB cause punctate palmoplantar keratoderma type Buschke-Fischer-Brauer. Am J Hum Genet. 2012;91:754–759. doi: 10.1016/j.ajhg.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravotta D, Carvajal-Gonzalez JM, Mattera R, Deborde S, Banfelder JR, Bonifacino JS, Rodriguez-Boulan E. The clathrin adaptor AP-1A mediates basolateral polarity. Dev Cell. 2012;22:811–823. doi: 10.1016/j.devcel.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harasaki K, Lubben NB, Harbour M, Taylor MJ, Robinson MS. Sorting of major cargo glycoproteins into clathrin-coated vesicles. Traffic. 2005;6:1014–1026. doi: 10.1111/j.1600-0854.2005.00341.x. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Winkler DC, Yim YI, Eisenberg E, Greene LE, Steven AC. Clathrin-coated vesicles from brain have small payloads: a cryo-electron tomographic study. J Struct Biol. 2013;184:43–51. doi: 10.1016/j.jsb.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Borner GHH, Antrobus R, Peden AA, Hodson NA, Sahlender DA, Robinson MS. Distinct and overlapping roles for AP-1 and GGAs revealed by the “knocksideways” system. Curr Biol. 2012;22:1711–1716. doi: 10.1016/j.cub.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Lindsay M, Robinson MS. GGAs: Roles of the different domains and comparison with AP-1 and clathrin. Mol Biol Cell. 2001;12:3573–3588. doi: 10.1091/mbc.12.11.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Miller SE, Taylor MJ, von Mollard GF, Robinson MS. EpsinR is an adaptor for the SNARE protein vti1b. Mol Biol Cell. 2004;15:5593–5602. doi: 10.1091/mbc.E04-06-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Motley A, Harasaki K, Peak Chew SY, Robinson MS. EpsinR: an ENTH domain-containing protein that interacts with AP-1. Mol Biol Cell. 2003;14:625–641. doi: 10.1091/mbc.E02-09-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Sahlender DA, Li S, Lubben NB, Borner GHH, Robinson MS. Auxilin depletion causes self-assembly of clathrin into membraneless cages in vivo. Traffic. 2008;9:1354–1371. doi: 10.1111/j.1600-0854.2008.00764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly BT, Graham SC, Liska N, Dannhauser PN, Höning S, Ungewickell EJ, Owen DJ. AP2 controls clathrin polymerization with a membrane-activated switch. Science. 2014;345:459–463. doi: 10.1126/science.1254836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T, Owen DJ, Harrison SC. Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harb Perspect Biol. 2014;6:a016725. doi: 10.1101/cshperspect.a016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozik P, Hodson NA, Sahlender DA, Simecek N, Soromani C, Wu J, Collinson LM, Robinson MS. A human genome-wide screen for regulators of clathrin-coated vesicle formation reveals an unexpected role for the V-ATPase. Nat Cell Biol. 2013;15:50–60. doi: 10.1038/ncb2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kural C, Tacheva-Grigorova SK, Boulant S, Cocucci E, Baust T, Duarte D, Kirchhausen T. Dynamics of intracellular clathrin/AP1- and clathrin/AP3-containing carriers. Cell Rep. 2012;2:1111–1119. doi: 10.1016/j.celrep.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maritzen T, Haucke V. Gadkin: a novel link between endosomal vesicles and microtubule tracks. Commun Integr Biol. 2010;3:299–302. doi: 10.4161/cib.3.4.11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maritzen T, Schmidt MR, Kukhtina V, Higman VA, Strauss H, Volkmer R, Oschkinat H, Dotti CG, Haucke V. A novel subtype of AP-1-binding motif within the palmitoylated trans-Golgi network/endosomal accessory protein Gadkin/gamma-BAR. J Biol Chem. 2010;285:4074–4086. doi: 10.1074/jbc.M109.049197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maritzen T, Zech T, Schmidt MR, Krause E, Machesky LM, Haucke V. Gadkin negatively regulates cell spreading and motility via sequestration of the actin-nucleating ARP2/3 complex. Proc Natl Acad Sci USA. 2012;109:10382–10387. doi: 10.1073/pnas.1206468109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C, Zizioli D, Lausmann S, Eskelinen EL, Hamann J, Saftig P, von Figura K, Schu P. mu1A-adaptin-deficient mice: lethality, loss of AP-1 binding and rerouting of mannose 6-phsophate receptors. EMBO J. 2000;19:2193–2203. doi: 10.1093/emboj/19.10.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerholz A, Hinrichsen L, Groos S, Esk PC, Brandes G, Ungewickell EJ. Effect of clathrin assembly lymphoid myeloid leukemia protein depletion on clathrin coat formation. Traffic. 2005;6:1225–1234. doi: 10.1111/j.1600-0854.2005.00355.x. [DOI] [PubMed] [Google Scholar]
- Miller SE, Collins BM, McCoy AJ, Robinson MS, Owen DJ. A SNARE-adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles. Nature. 2007;450:570–574. doi: 10.1038/nature06353. [DOI] [PubMed] [Google Scholar]
- Miller SE, Mathiasen S, Bright NA, Pierre F, Kelly BT, Kladt N, Schauss A, Merrifield CJ, Stamou D, Höning S, Owen DJ. CALM regulates clathrin-coated vesicle size and maturation by directly sensing and driving membrane curvature. Dev Cell. 2015;33:163–175. doi: 10.1016/j.devcel.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SE, Sahlender DA, Graham SC, Höning S, Robinson MS, Peden AA, Owen DJ. The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM. Cell. 2011;147:1118–1131. doi: 10.1016/j.cell.2011.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montpetit A, Côté S, Brustein E, Drouin CA, Lapointe L, Boudreau M, Meloche C, Drouin R, Hudson TJ, Drapeau P, Cossette P. Disruption of AP1S1, causing a novel neurocutaneous syndrome, perturbs development of the skin and spinal cord. PLoS Genet. 2008;4:e1000296. doi: 10.1371/journal.pgen.1000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley AM, Bright NA, Seaman MNJ, Robinson MS. Clathrin-mediated endocytosis in AP-2-depleted cells. J. Cell Biol. 2003;162:909–918. doi: 10.1083/jcb.200305145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. Sequences within and adjacent to the transmembrane segment of alpha-2,6-sialyltransferase specify Golgi retention. EMBO J. 1991;10:3577–3588. doi: 10.1002/j.1460-2075.1991.tb04924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubrand VE, Will RD, Möbius W, Poustka A, Wiemann S, Schu P, Dotti CG, Pepperkok R, Simpson JC. Gamma-BAR, a novel AP-1-interacting protein involved in post-Golgi trafficking. EMBO J. 2005;24:1122–1133. doi: 10.1038/sj.emboj.7600600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse BMF, Bretscher MS. Membrane recycling by coated vesicles. Annu Rev Biochem. 1981;50:85–101. doi: 10.1146/annurev.bi.50.070181.000505. [DOI] [PubMed] [Google Scholar]
- Pohler E, Mamai O, Hirst J, Zamiri M, Horn H, Nomura T, Irvine AD, Moran B, Wilson NJ, Smith FJ, et al. Haploinsufficiency for AAGAB causes clinically heterogeneous forms of punctate palmoplantar keratoderma. Nat Genet. 2012;44:1272–1276. doi: 10.1038/ng.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS, Sahlender DA, Foster SD. Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria. Dev Cell. 2010;18:324–331. doi: 10.1016/j.devcel.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal SK, Skretting G, Garred O, Vilhardt F, van Deurs B, Sandvig K. Extraction of cholesterol with methyl-beta-cyclodextrin perturbs formation of clathrin-coated endocytic vesicles. Mol Biol Cell. 1999;10:961–974. doi: 10.1091/mbc.10.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Pol A, Yélamos B, Amessou M, Mills IG, Dugast M, Tenza D, Schu P, Antony C, McMahon HT, Lamaze C, Johannes L. Clathrin adaptor epsinR is required for retrograde sorting on early endosomal membranes. Dev Cell. 2004;6:525–538. doi: 10.1016/s1534-5807(04)00100-5. [DOI] [PubMed] [Google Scholar]
- Schmidt MR, Maritzen T, Kukhtina V, Higman VA, Doglio L, Barak NN, Strauss H, Oschkinat H, Dotti CG, Haucke V. Regulation of endosomal membrane traffic by a Gadkin/AP-1/kinesin KIF5 complex. Proc Natl Acad Sci USA. 2009;106:15344–15349. doi: 10.1073/pnas.0904268106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson F, Bright NA, West MA, Newman LS, Darnell RB, Robinson MS. A novel adaptor-related protein complex. J Cell Biol. 1996;133:749–760. doi: 10.1083/jcb.133.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtil A, Gaidarov I, Kobylarz K, Lampson MA, Keen JH, McGraw TE. Acute cholesterol depletion inhibits clathrin-coated pit budding. Proc Natl Acad Sci USA. 1999;96:6775–6780. doi: 10.1073/pnas.96.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamorim S, Holt M, Stenius K, Lemke EA, Grønborg M, Riedel D, Urlaub H, Schenck S, Brügger B, Ringler P, et al. Mol anatomy trafficking organelle Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Tiwari RK, Kusari J, Sen GC. Functional equivalents of interferon-mediated signals needed for induction of an mRNA can be generated by double-stranded RNA and growth factors. EMBO J. 1987;6:3373–3378. doi: 10.1002/j.1460-2075.1987.tb02659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub LM, Bonifacino JS. Cargo recognition in clathrin-mediated endocytosis. Cold Spring Harb Perspect Biol. 2013;5:a016790. doi: 10.1101/cshperspect.a016790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bergeijk, Adrian M, Hoogenraad CC, Kapitein LC. Optogenetic control of organelle transport and positioning. Nature. 2015;518:111–114. doi: 10.1038/nature14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M, Munro S. The specificity of vesicle traffic to the Golgi is encoded in the golgin coiled-coil proteins. Science. 2014;346:1256898. doi: 10.1126/science.1256898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zizioli D, Meyer C, Guhde G, Saftig P, von Figura K, Schu P. Early embryonic death of mice deficient in g-adaptin. J Biol Chem. 1999;274:5385–5390. doi: 10.1074/jbc.274.9.5385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.