Abstract
The replisome is important for DNA replication checkpoint activation, but how specific components of the replisome coordinate with ATR to activate Chk1 in human cells remains largely unknown. Here, we demonstrate that And-1, a replisome component, acts together with ATR to activate Chk1. And-1 is phosphorylated at T826 by ATR following replication stress, and this phosphorylation is required for And-1 to accumulate at the damage sites, where And-1 promotes the interaction between Claspin and Chk1, thereby stimulating efficient Chk1 activation by ATR. Significantly, And-1 binds directly to ssDNA and facilitates the association of Claspin with ssDNA. Furthermore, And-1 associates with replication forks and is required for the recovery of stalled forks. These studies establish a novel ATR–And-1 axis as an important regulator for efficient Chk1 activation and reveal a novel mechanism of how the replisome regulates the replication checkpoint and genomic stability.
Keywords: And-1, ATR, Chk1, Claspin, replication stress
Introduction
To maintain genomic stability, cells have evolved multiple DNA damage checkpoint pathways to coordinate cellular responses to DNA damage (Ciccia & Elledge, 2010). Among these signaling pathways, the DNA replication checkpoint is activated in response to a wide spectrum of DNA damage and DNA replication stress (Cimprich & Cortez, 2008). Central to the replication checkpoint are two protein kinases termed ATR and Chk1, which regulate multiple physiological processes, including inhibition of DNA synthesis, DNA repair, cell cycle delay, apoptosis, and transcription. Each of these processes is essential for the maintenance of genomic stability.
DNA lesions induced by replication stress lead to replication fork stalling, which in turn activates ATR and Chk1. Activation of this pathway requires both single-stranded DNA (ssDNA) and primer–template junctions that are likely found at the stalled replication forks (MacDougall et al, 2007). Single-stranded DNA is generated when DNA helicase and DNA polymerase activities become uncoupled from one another due to either physical obstructions or nucleotide deficiencies that block DNA polymerase progression (Byun et al, 2005). The replication and checkpoint protein TopBP1 cooperates with the BACH1/FANCJ helicase to promote loading of replication protein A (RPA) onto the exposed ssDNA at the stalled replication forks (Gong et al, 2010). ATR kinase is then recruited to the stalled replication forks through a specific interaction between RPA and the ATR-interacting protein ATRIP (Zou & Elledge, 2003). Primer–template junctions at stalled replication forks serve as a substrate for the Rad17/RFC-dependent loading of the Rad9/Hus1/Rad1 protein complex. TopBP1 promotes the kinase activity of ATR at the stalled replication forks by interacting with Rad9 (Kumagai et al, 2006; Delacroix et al, 2007). Once activated, ATR phosphorylates Chk1 in a process involving multiple replisome proteins, including Claspin, Timeless, and Tipin. These replisome proteins act coordinately to bring Chk1 to stalled forks so that Chk1 can be activated by ATR (Kumagai & Dunphy, 2000; Chini & Chen, 2003; Chou & Elledge, 2006; Unsal-Kacmaz et al, 2007; Yoshizawa-Sugata & Masai, 2007; Burrows & Elledge, 2008). Although Timeless, Tipin, and Claspin are critical for Chk1 activation, the mechanism by which these replisome proteins coordinate with ATR to activate Chk1 in mammalian cells remains largely unknown.
Timeless and Tipin form a stable heterodimeric complex that binds to replication forks and is required for efficient DNA replication in unperturbed cells (Gotter et al, 2007; Yoshizawa-Sugata & Masai, 2007; Errico et al, 2009; Numata et al, 2010). Like Timeless–Tipin, Claspin is also important for efficient DNA replication in unperturbed cells (Lee et al, 2003; Sar et al, 2004; Petermann et al, 2008). Recent studies indicate that Tipin interacts with RPA and that this interaction stabilizes the association of Timeless–Tipin and Claspin with RPA-coated ssDNA to promote Claspin-mediated phosphorylation of Chk1 by ATR (Kemp et al, 2010). Interestingly, Claspin also physically interacts with Chk1 (Kumagai & Dunphy, 2000; Uno & Masai, 2011). Since Claspin interacts with both ATR and Chk1 (Kumagai & Dunphy, 2000; Chini & Chen, 2003), it is most likely that Claspin functions as an adaptor to bring Chk1 to ATR for Chk1 phosphorylation (Kumagai et al, 2004). Lastly, as important components of the replisome, both Timeless–Tipin and Claspin are essential to maintain fork stability in both unperturbed cells and cells undergoing replication stress (Errico et al, 2007; Scorah & McGowan, 2009; Leman et al, 2010).
Previous studies by others and us showed that the acidic nucleoplasmic DNA-binding protein 1 (And-1) is a replisome component and is required for efficient DNA replication in unperturbed cells (Zhu et al, 2007; Errico et al, 2009; Gambus et al, 2009; Im et al, 2009; Yoshizawa-Sugata & Masai, 2009; Bermudez et al, 2010; Li et al, 2012b). Ctf4/Mcl1, the ortholog of And-1 in yeast, is required for chromosome transmission fidelity, sister chromatin cohesion, DNA damage repair, maintenance of genome integrity, and the regulation of telomere replication (Kouprina et al, 1992; Hanna et al, 2001; Williams & McIntosh, 2002; Mayer et al, 2004; Tsutsui et al, 2005). Recent studies indicate that And-1 interacts with Tipin in Xenopus egg extracts and with Claspin in human cells (Errico et al, 2009; Li et al, 2012b), suggesting that And-1 may regulate DNA replication checkpoint activation in response to replication stress. This hypothesis is consistent with the effects of siRNA suppression of And-1 in human cells (Yoshizawa-Sugata & Masai, 2009), even though attempts to elucidate the mechanism by which And-1 affects the DNA damage response yielded inconclusive results (Yoshizawa-Sugata & Masai, 2009).
Here, we have identified, for the first time, a novel ATR–And-1 functional link that is essential for Chk1 activation in response to DNA replication stress in human cells. We found that And-1 is recruited to DNA damage sites in response to DNA replication stress. In particular, downregulation of And-1 impairs the interaction of Claspin with Chk1, and considerably reduces Chk1 phosphorylation after DNA damage. We also found that And-1 is phosphorylated at T826 by ATR and that this phosphorylation is required for its interaction with Claspin and for efficient Chk1 activation. Significantly, in vitro analyses indicated that And-1 directly binds to ssDNA and that And-1 phosphorylation at T826 enhances its affinity to ssDNA and the association of Claspin with ssDNA. Lastly, we found that And-1 is critical for the recovery of stalled replication forks. These results collectively demonstrate an important role of And-1 in the maintenance of genomic stability after replication stress.
Results
And-1 associates with proteins involved in the DNA replication checkpoint
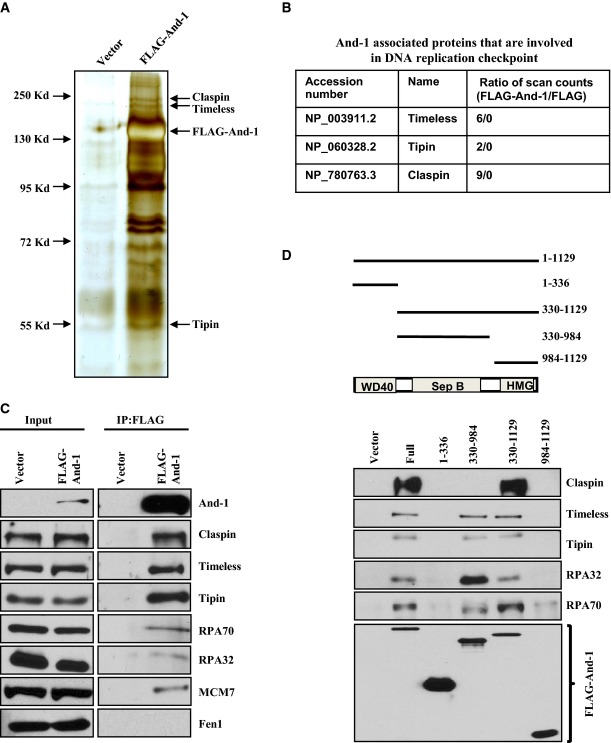
We previously demonstrated that And-1 is an important component of the replisome for DNA replication in S-phase (Zhu et al, 2007). Given that replisome components often play critical roles in DNA replication checkpoint regulation following replication stress (Ciccia & Elledge, 2010), we hypothesized that And-1 plays a role in the regulation of the replication checkpoint by interacting with other replication checkpoint proteins. To test this possibility, we conducted mass spectrometry analyses to identify specific And-1-associated proteins that are known to regulate DNA replication checkpoint. To rule out the possibility that the protein–protein interactions are mediated by non-specific associations with chromatin, we included ethidium bromide and DNase I in the cell lysis buffer to disrupt protein–DNA interactions. We performed affinity purification using anti-FLAG beads incubated with cell extracts from HCT116 cells stably expressing FLAG-And-1 or FLAG. The purified FLAG-associated proteins were then resolved by gel electrophoresis followed by silver staining, which showed multiple proteins unique to the FLAG-And-1 immunoprecipitations (IPs) (Fig1A). Mass spectrometry analyses of these unique proteins and subsequent database searching enabled us to identify the replisome proteins, Claspin, Timeless, and Tipin, in the FLAG-And-1 but not in FLAG-IPs (Fig1B). We next conducted co-immunoprecipitation (co-IP) assays to further confirm these interactions in HCT116 cell lines. We identified all these three proteins as well as the ssDNA-binding proteins, RPA70 and RPA32, which are also known to be critical for activation of the replication checkpoint (Fig1C and Supplementary Fig S1) (Zou & Elledge, 2003). Thus, And-1 forms complexes with multiple replication checkpoint proteins in a manner independent of chromatin structure.
Figure 1.
And-1 forms complexes with DNA replication checkpoint proteins
- Purification of And-1-associated proteins. Cell extracts from HCT116 cells stably expressing FLAG-And-1 protein or FLAG alone were subjected to immunoprecipitation using anti-FLAG beads. FLAG immunoprecipitates (IPs) were resolved by SDS–PAGE followed by silver staining. Replication checkpoint proteins that are specifically presented more in FLAG-And-1 complexes than in control FLAG complexes were indicated.
- Identification of And-1-associated proteins that are involved in DNA replication checkpoint by mass spectrometry.
- And-1 interacts with multiple replication checkpoint proteins. Co-immunoprecipitation (co-IP) assays were performed using HCT116 cell lines expressing FLAG-And-1 or FLAG alone. FLAG-IPs were resolved by SDS–PAGE and immunoblotted for the indicated proteins.
- The associations of And-1 or its mutants with replication checkpoint proteins. Upper panel, schematic of the And-1 protein domains and deletion mutants used for protein–protein interactions; lower panel, FLAG-And-1 and its mutants were expressed in 293T cells. FLAG-IPs were resolved by SDS–PAGE and immunoblotted for the indicated proteins.
Source data are available online for this figure.
And-1 contains WD40 repeats at its N-terminus, a SepB domain in the middle of the proteins, and a HMG domain at its C-terminus (Fig1D). We next tested whether a particular domain of And-1 mediates the interactions of And-1 with these replication checkpoint proteins. To this end, we performed co-IPs using cell extracts from 293T cells expressing full-length And-1 or its truncated mutants from transfected plasmids (Fig1D). The results showed that the interactions of And-1 with Timeless, Tipin, RPA70, and RPA32 were detected in IPs from full-length And-1, And-1(330–984), and And-1(330–1129), suggesting that the SepB domain is important for these associations. Surprisingly, Claspin was detected in IPs from And-1(330–1129) but not from And-1(330–984) or And-1(984–1129), indicating that both the SepB and HMG domains of And-1 are required for its interaction with Claspin.
And-1 is recruited to DNA damage sites in response to replication stress
If And-1 is involved in the DNA damage response in cells with replication stress, it is likely that And-1 is recruited to damage sites. To test this possibility, we exposed HCT116 cells to UV irradiation through polycarbonate isopore membrane filters to induce focal DNA damage in the nucleus. After pre-extraction of the cells with Triton X-100, And-1 was found to co-localize with γ-H2AX, RPA70, RPA32, and ATR (Fig2A and B). Thus, And-1 is recruited to DNA damage sites following replication stress caused by UV.
Figure 2.
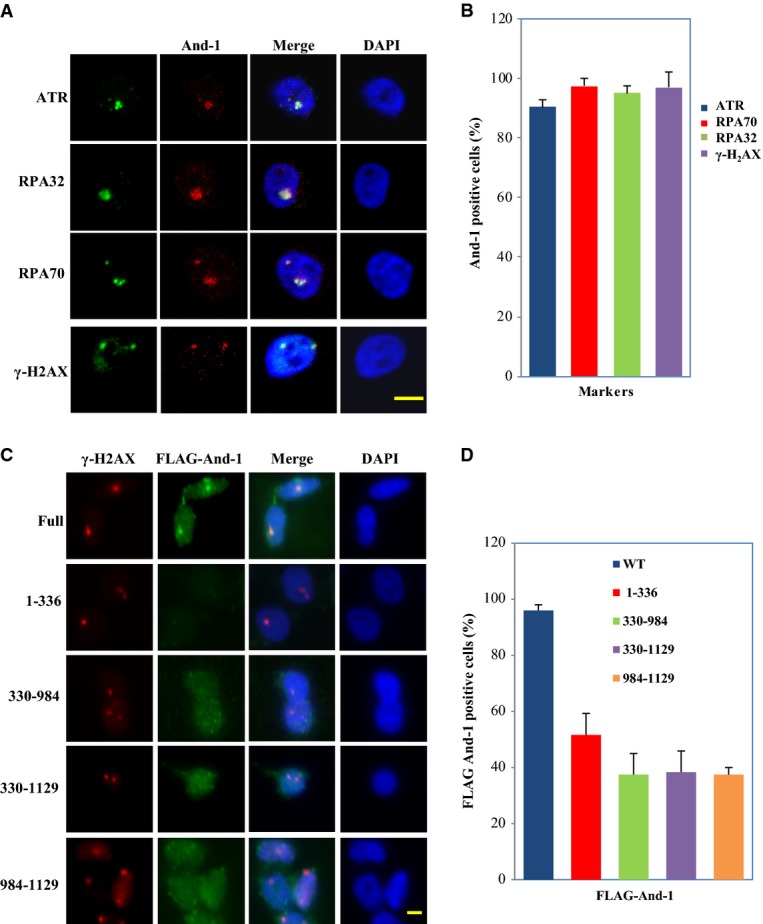
And-1 is localized to DNA damage sites
- And-1 co-localizes with γ-H2AX, RPA70, RPA32, and ATR at localized damage areas induced by UV light. HCT116 cells were covered with a 5-μm polycarbonate isopore membrane filter and subjected to 100 J/m2 UV irradiation. Two hours after irradiation, harvested cells were immunostained for the indicated proteins. Scale bar, 5 μm.
- Cells described in (A) were quantified in at least 50 cells in three separate experiments. The data were presented as percentage of And-1-positive cells co-localized with γ-H2AX, RPA70, RPA32, and ATR (mean ± SD).
- Localization of wild-type And-1 and its mutants at damage sites. HCT116 cells stably expressing FLAG-And-1 or indicated FLAG-tagged mutants were treated as in (A). Scale bar, 5 μm.
- Cells described in (C) were quantified in at least 50 cells in three separate experiments. The data were presented as percentage of And-1-positive cells co-localized with γ-H2AX (mean ± SD).
To determine which domains of And-1 are specifically responsible for And-1 association with DNA damage sites, we generated HCT116 cell lines stably expressing FLAG-tagged full-length And-1 or a series of truncated And-1 mutants. The full-length And-1 co-localized with γ-H2AX at the damaged sites, whereas the localization of And-1 mutants And-1(1–336), And-1(330–984), And-1(984–1129), and And-1(330–1129) at DNA damage sites was significantly reduced (Fig2C and D). Thus, the integrity of And-1 protein structure is critical for stable localization of And-1 at DNA damage sites.
Treatment of cells with hydroxyurea (HU) is also known to induce γ-H2AX and RPA70 foci formation (Ward & Chen, 2001; Zou et al, 2003). We also observed the formation of HU-induced And-1 foci HCT116 cells that co-localized with γ-H2AX and RPA70 foci (Supplementary Fig S2A).
And-1 facilitates phosphorylation of Chk1 proteins
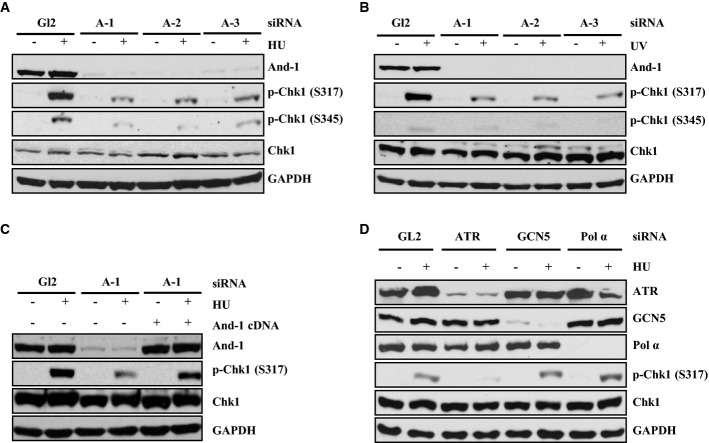
The fact that And-1 forms complexes with multiple replication checkpoint proteins and is recruited to DNA damage sites leads us to postulate that And-1 affects Chk1 phosphorylation. To test this hypothesis, HCT116 cells were transfected with three independent siRNAs to deplete And-1 and then treated with HU. Depletion of And-1 by each And-1 siRNA significantly reduced Chk1 phosphorylation at S317 and S345 in cells following HU treatment without affecting the overall protein levels of Chk1 (Fig3A). Depletion of And-1 also reduced Chk1 phosphorylation in cells exposed to UV (Fig3B). In order to rule out the possibility that the observed effect of And-1 siRNAs on Chk1 is an off-target effect of the siRNAs, we expressed an A-1 siRNA-resistant And-1 cDNA in U2OS cells and found that ectopic expression of And-1 restored Chk1 phosphorylation in the cells after HU treatment even in the presence of the A-1 siRNA (Fig3C). Taken together, our data reveal that And-1 is required for efficient phosphorylation of Chk1 in response to either HU- or UV-induced DNA damage.
Figure 3.
And-1 is required for efficient replication checkpoint activation
- And-1 depletion reduces Chk1 phosphorylation in cells treated with HU. HCT116 cells transfected with three independent siRNAs for 2 days were harvested after 10-mM HU treatment for 2 h. Harvested cells were lysed and immunoblotted for the indicated proteins.
- And-1 depletion reduces Chk1 phosphorylation in cells treated with UV (100 J). HCT116 cells transfected with siRNA as in (A) were harvested after UV treatment. Harvested cells were treated as in (A).
- Expression of siRNA-resistant And-1 restores Chk1 phosphorylation. U2OS cells expressing empty vector or siRNA-resistant And-1 were transfected with indicated siRNAs, followed by HU treatment as in (A).
- Depletion of ATR but not GCN5 or polymerase-α reduces Chk1 phosphorylation. HCT116 cells transfected with the indicated siRNAs were treated as in (A).
Source data are available online for this figure.
To further investigate the dynamics of Chk1 activation in And-1-depleted cells, we conducted detailed time-course experiments using cells treated with either HU or UV. And-1 depletion greatly reduced Chk1 phosphorylation as early as 15 min after either HU or UV treatment compared to control siRNA-treated cells (Supplementary Fig S3E). Although Chk1 phosphorylation levels increased over time following either HU or UV treatments, the strength of the signal was severely compromised in And-1-depleted cells compared to cells treated with control siRNA (Supplementary Fig S3E).
In previous studies, we observed that the fraction of BrdU-positive S-phase cells was reduced 72 h after And-1 siRNA transfection (Zhu et al, 2007). These data suggested that the observed reduction of phosphorylated Chk1 by And-1 depletion might be due to a decreased S-phase cell population. We therefore analyzed the cell cycle progression using the EdU-click reaction method in cells depleted of And-1 for different lengths of time. At 48 h after And-1 siRNA treatment, we observed neither cell cycle progression defects nor DNA damage (Supplementary Fig S3A–D). Thus, reduced Chk1 phosphorylation in the And-1-depleted cells in response to DNA replication stress was not due to a reduction of the S-phase cell population.
Because we recently showed that And-1 is required to maintain the stability of DNA polymerase-α and GCN5 (Zhu et al, 2007; Li et al, 2012a), the reduced phosphorylation of Chk1 by And-1 depletion might have resulted from the degradation of either DNA polymerase-α or GCN5. To investigate this possibility, we depleted DNA polymerase-α, GCN5, or ATR with siRNAs and then challenged the cells with HU. In agreement with previous studies (Cortez et al, 2001), depletion of ATR impaired Chk1 phosphorylation (Fig3D). However, neither depletion of GCN5 nor depletion of DNA polymerase-α had any effect on Chk1 phosphorylation (Fig3D). Therefore, the observed effect of And-1 depletion on Chk1 phosphorylation involves neither DNA polymerase-α nor GCN5.
And-1 facilitates the interaction between Claspin and Chk1 in response to DNA replication stress
Given that And-1 is recruited to DNA damage sites, we speculated that the interactions of And-1 with other components of the replication checkpoint machinery might be enhanced in response to DNA damage. To test this possibility, 293T cells transfected with FLAG-tagged And-1 plasmid were treated with or without HU. Cell extracts were then used for co-IPs and Western blotting analyses. The results showed that the interactions of And-1 with Claspin, Timeless, Tipin, RPA70, and RPA32 were significantly enhanced after HU treatments (Fig4A).
Figure 4.
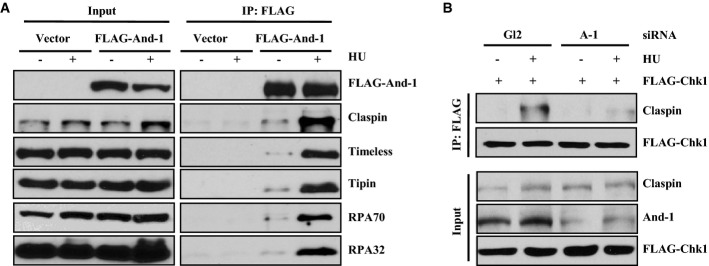
And-1 is required for Claspin to interact with Chk1 in response to replication stress
- The interactions of And-1 with Claspin, Timeless, Tipin, RPA70, and RPA32 are increased after HU treatment. 293T cells transfected with FLAG vector or FLAG-And-1 plasmids were treated with 1 mM HU overnight. FLAG-And-1 and FLAG-IPs were resolved by SDS–PAGE and immunoblotted for the indicated proteins.
- And-1 is required for the interaction between Claspin and Chk1 in cells with replication stress. U2OS cells transiently expressing FLAG-Chk1 were transfected with siAnd-1 for 2 days and harvested after HU (10 mM) treatment for 2 h. Harvested cells were lysed and immunoprecipitated with anti-FLAG agarose beads. FLAG-IPs were resolved by SDS–PAGE and immunoblotted for the indicated proteins.
Source data are available online for this figure.
Claspin interacts with Chk1, and this interaction is critical for Chk1 activation in response to replication stress (Chini & Chen, 2003, 2006). Since And-1 interacts with Claspin and is required for Chk1 activation, we next investigated whether And-1 could regulate the interaction between Claspin and Chk1. We used siRNA to deplete And-1 in cells expressing FLAG-tagged Chk1 and then treated the cells with HU. Subsequent Western blotting analyses of FLAG immunoprecipitates from cell extracts revealed that, consistent with a previous report (Chini & Chen, 2003), FLAG-Chk1 interacted with Claspin and this interaction was enhanced after HU treatment (Fig4B). However, the interaction between Claspin and Chk1 was dramatically reduced in And-1-depleted cells after HU treatment (Fig4B). Therefore, And-1 appears to facilitate a stronger interaction between Claspin and Chk1 in cells undergoing replication stress.
And-1 is phosphorylated by ATR in response to DNA replication stress
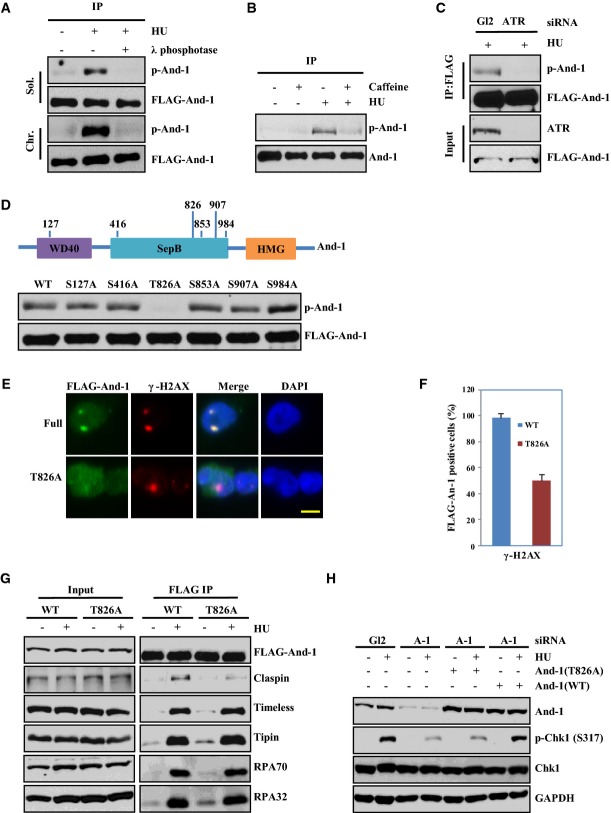
Through analysis of the amino acid sequence of And-1 using the protein phosphorylation prediction program GPS2.1, we identified six putative ATR/ATM phosphorylation sites (Fig5D). To determine whether And-1 is phosphorylated after replication stress, we treated U2OS cells stably expressing FLAG-And-1 with HU. Cell extracts were immunoprecipitated with anti-FLAG beads and then immuno-blotted with an ATR/ATM phosphorylation-specific substrate antibody [p(S/T) ATR/ATM substrate antibody] (Matsuoka et al, 2007). The antibody did not detect any signal of And-1 phosphorylation in the FLAG-And-1 IP samples from cells in the absence of HU treatment; however, it detected a strong λ phosphatase-sensitive FLAG-And-1 protein band in the FLAG-And-1 IP samples from cells treated with HU, indicating that And-1 is phosphorylated by ATR or ATM (Fig5A). To determine whether ATR is the kinase responsible for And-1 phosphorylation following HU treatment, the phosphorylation status of And-1 was examined in cells treated with ATR siRNA or caffeine, an inhibitor of ATR (Sarkaria et al, 1999). Downregulation of ATR by siRNA or inhibition of ATR with caffeine significantly reduced the phosphorylation of And-1 in cells treated with HU (Fig5B and C), indicating that ATR is the primary kinase that phosphorylates And-1 following HU treatment. To identify the exact phosphorylation site(s) in And-1 by ATR, we mutated the serine or threonine residues to alanine in each of the six putative phosphorylation sites by site-directed mutagenesis. We then expressed wild-type And-1 or the And-1 mutants in 293T cells and treated the cells with HU. The results revealed that only the T826A mutant was no longer phosphorylated after HU treatment, indicating that ATR phosphorylates And-1 specifically at T826 in response to replication stress (Fig5D).
Figure 5.
And-1 is phosphorylated at T826 by ATR in response to replication stress
- And-1 is phosphorylated after HU treatment. U2OS cells stably expressing FLAG-And-1 were harvested after treatment with 10 mM HU for 2 h. Harvested cells were processed for chromatin fractionation assays. Soluble and chromatin fractions were immunoprecipitated with anti-FLAG beads. After washing, immunobeads were treated with or without λ phosphatase. FLAG-IPs were resolved by SDS–PAGE and immunoblotted for the indicated proteins. A phosphor-(Ser/Thr) ATM/ATR substrate antibody was used to detect phosphorylation of And-1.
- And-1 phosphorylation is reduced after caffeine treatment. HCT116 cells stably expressing FLAG-And-1 were pre-treated with buffer or 5 mM caffeine for 2 h before exposed to 10 mM HU for another 2 h. FLAG-IPs from whole-cell extracts were resolved by SDS–PAGE and immunoblotted for the indicated proteins.
- And-1 phosphorylation is reduced in cells with downregulated ATR. HCT116 cells stably expressing FLAG-And-1 were transfected with siGL2 or siATR and treated with 10 mM HU for 2 h before harvest. FLAG-IPs were resolved by SDS–PAGE and immunoblotted for the indicated proteins.
- And-1 is phosphorylated at T826 in response to HU treatment. Upper panel, schematic of the And-1 protein domains with putative ATM/ATR phosphorylation sites predicted by GSP2.1; lower panel, 293T cells expressing FLAG-And-1 with indicated single point mutation were treated with HU, harvested, and treated as in (C).
- Phosphorylation of And-1 at T826 is required for its localization onto damage sites. HCT116 cells stably expressing FLAG-And-1 or mutant FLAG-And-1(T826A) were irradiated with UV as in Fig2B. Scale bar, 5 μm.
- Cells described in (E) were quantified in at least 50 cells in three separate experiments. The data were presented as percentage of FLAG-positive cells co-localized with γ-H2AX (mean ± SD).
- T826 of And-1 is required for the elevated interaction between And-1 and Claspin in response to HU treatment. 293T cells expressing FLAG-And-1 or FLAG-And-1(T826A) from plasmids were treated with HU as in Fig4A before harvest. FLAG-IPs were resolved by SDS–PAGE and immunoblotted for the indicated proteins.
- T826 of And-1 is required for efficient Chk1 phosphorylation in response to HU treatment. 293T cells stably expressing siRNA-resistant wild-type And-1 or mutant And-1(T826A) were transfected with siAnd-1. Cells were harvested after HU treatment (10 mM) for 2 h and immunoblotted for the indicated proteins.
Source data are available online for this figure.
Efficient activation of the DNA replication checkpoint requires phosphorylation of And-1 at T826
To determine the biological significance of And-1 phosphorylation at T826 during the DNA damage response, we first determined whether T826 was required for And-1 to associate with DNA damage sites. We constructed HCT116 cell lines stably expressing FLAG-And-1 or FLAG-And-1(T826A). These cells were exposed to UV irradiation and then subjected to Triton X-100 extraction followed by immunostaining to detect FLAG-And-1 or FLAG-And-1(T826A). The results showed that the localization of And-1(T826A) but not wild-type And-1 to UV-induced damage sites was impaired, indicating that phosphorylation at T826 is important for And-1 localization to DNA damage sites (Fig5E and F).
To explore the functional role of And-1 phosphorylation at T826 in replication checkpoint signaling, we investigated whether phosphorylation at T826 regulates the associations of And-1 with other replication checkpoint proteins. Consistent with the results in Fig4A, HU treatment significantly increased the interactions of wild-type And-1 with Claspin, Timeless, Tipin, and RPA. Strikingly, although And-1(T826A) retained the full capability to interact with Timeless, Tipin, and RPA in cells after HU treatment, its ability to interact with Claspin was greatly weakened (Fig5G). Thus, phosphorylation of And-1 at T826 is required for And-1 to form a stable interaction with Claspin in response to replication stress.
The above observations prompted us to test the effect of And-1 phosphorylation at T826 on the activation of Chk1. To this end, we generated 293T cell lines expressing either siRNA-resistant wild-type And-1 or siRNA-resistant mutant And-1(T826A) proteins. These cells were treated with And-1 siRNA to suppress the expression of the endogenous And-1 before the cells were challenged with HU. Cells expressing the siRNA-resistant wild-type And-1 had higher levels of phosphorylated Chk1 compared to the And-1-depleted control cells, whereas cells expressing the siRNA-resistant And-1(T826A) mutant did not show an increase in Chk1 phosphorylation after HU treatment (Fig5H). We next tested whether And-1 phosphorylation is required for the Claspin–Chk1 interaction. We found that expression of And-1 WT but not mutant And-1(T826A) in U2OS cells transfected with And-1 siRNA restored Claspin–Chk1 interaction (Supplementary Fig S4A). Taken together, these results collectively indicate that phosphorylation of And-1 at T826 is important not only for Claspin–And-1 interaction but also for efficient Chk1 activation.
And-1 binds to ssDNA and promotes the association of Claspin with ssDNA
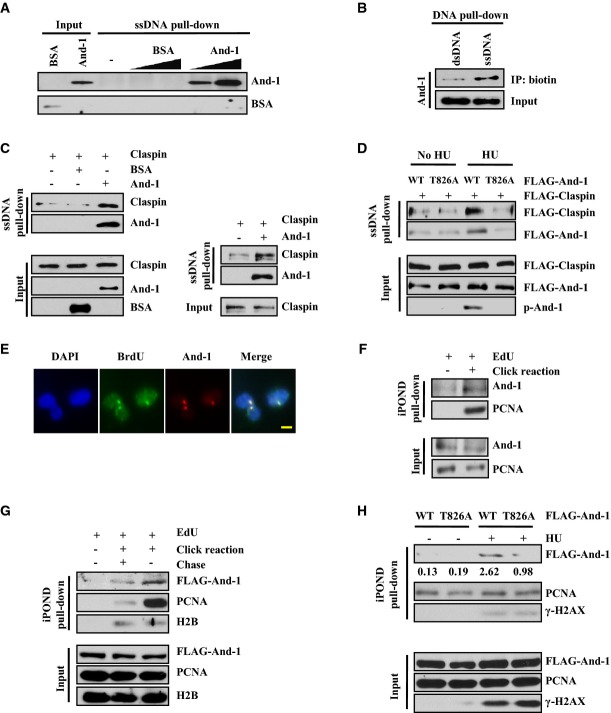
Since And-1 contains an HMG DNA-binding domain and ssDNA is generated at stalled replication forks (Lopes et al, 2001; Sogo et al, 2002), we used an in vitro ssDNA binding assay to determine whether And-1 could bind to ssDNA. We purified recombinant human And-1 proteins from Sf9 insect cells as previously described (Zhu et al, 2007). The purified And-1 protein was incubated with biotinylated ssDNA immobilized on streptavidin-conjugated magnetic beads. After extensive washing (Unsal-Kacmaz & Sancar, 2004), the ssDNA-bound proteins were analyzed by immunoblot. To rule out non-specific protein binding, BSA was used as a control in incubations with the ssDNA beads. As expected, BSA did not bind to ssDNA (Fig6A). Strikingly, And-1 bound to ssDNA in a concentration-dependent manner (Fig6A). We next determined whether And-1 preferably binds to ssDNA or binds equally well to both ssDNA and dsDNA. To this end, we compared the association of And-1 with ssDNA and biotinylated dsDNA obtained by annealing biotinylated ssDNA with its complementary oligomers. The results showed that more recombinant And-1 protein was pulled down with the ssDNA than with the same molar amount of dsDNA (Fig6B). Thus, we conclude that And-1 binds preferentially to ssDNA.
Figure 6.
And-1 promotes the association of Claspin with ssDNA
- And-1 binds to ssDNA in a dose-dependent manner. Immobilized 80-mer ssDNA was incubated in reactions with buffer, BSA (200 or 400 ng), or recombinant And-1 (200 or 400 ng) for 30 min at room temperature. The beads were washed, and bound proteins were resolved by SDS–PAGE and immunoblotted for the indicated proteins.
- And-1 binds preferably to ssDNA rather than dsDNA. ssDNA was annealed with buffer or its complementary oligonucleotide before immobilized onto magnetic beads. Immobilized ssDNA or dsDNA was incubated with recombinant And-1. The beads were washed and treated as in (A).
- And-1 promotes the association of Claspin with ssDNA. Immobilized 80-mer ssDNA was incubated in reactions with BSA or And-1 for 30 min at room temperature followed by incubation with recombinant Claspin at 4°C for 2 h. Beads were washed and treated as in (A). Recombinant Claspin proteins purified from 293T cells (left) or insect cells (right).
- Phosphorylation of And-1 at T826 enhances the association of Claspin with ssDNA. And-1 or And-1(T826) recombinant proteins were purified from 293T cells treated with or without HU, and were first incubated with ssDNA and then with recombinant FLAG-Claspin proteins as described in (C).
- And-1 co-localizes with ssDNA. HCT116 cells were labeled with BrdU 10 μM for 48 h, covered with a 5-μm polycarbonate isopore membrane filter, and subjected to 100 J/m2 UV irradiation. Harvested cells were immunostained for And-1 and BrdU without DNA denaturation. The co-localization of And-1 with BrdU was observed in 97% of BrdU foci-positive cells. Data are from three independent experiments. Scale bar, 5 μm.
- Endogenous And-1 is associated with replication forks. Asynchronous 293T cells were harvested for iPOND assay (see Materials and Methods for detail).
- And-1 is a bona fide replication fork protein. U2OS cell line constitutionally expressing FLAG-And-1 was labeled with EdU, or chased with regular growth medium for 2 h after EdU labeling to be used as chromatin pull-down control.
- And-1 is accumulated on stalled replication forks. 293T cells transfected with wild-type or T826A mutant FLAG-And-1 plasmids were treated with 10 mM HU for 2 h before harvested for iPOND assay. The intensity of Western blots was quantified with ImageJ software and indicated below the blots. The average relative intensity of bands is from three independent experiments.
Source data are available online for this figure.
Claspin alone displays little affinity for ssDNA and a Tipin–RPA interaction was shown to stabilize the association of Claspin with ssDNA, thereby promoting Claspin-mediated Chk1 activation (Sar et al, 2004; Kemp et al, 2010; Uno & Masai, 2011). Given that And-1 directly binds to ssDNA (Fig6A) and that And-1 interacts with Timeless–Tipin and Claspin (Fig1 and Supplementary Fig S1), we determined whether And-1 facilitates the association of Claspin with ssDNA. To address this issue, purified recombinant Claspin proteins were incubated with either ssDNA or And-1-bound ssDNA. The ssDNA-bound proteins were then analyzed by SDS–PAGE. In agreement with previous reports (Sar et al, 2004; Kemp et al, 2010; Uno & Masai, 2011), Claspin itself had little affinity for ssDNA (Fig6C). However, the presence of And-1 significantly increased the association of Claspin with ssDNA (Fig6C).
Given that the increased interaction between And-1 and Claspin following HU treatment is dependent on And-1 phosphorylation at T826 (Fig5F), we hypothesized that And-1 phosphorylation at T826 could play a positive role in the association of Claspin with ssDNA. To test this hypothesis, we purified wild-type And-1 or And-1(T826A) mutant recombinant proteins from 293T cells treated with or without HU. And-1 proteins were first incubated with ssDNA, and then, the And-1-bound ssDNA was incubated with purified FLAG-Claspin protein. ssDNA-associated proteins were examined by gel electrophoresis as described above. Wild-type And-1 and And-1(T826A) proteins purified from cells without HU treatment were unphosphorylated and displayed similar affinity to ssDNA (Fig6D); however, wild-type And-1 proteins purified from 293T cells treated with HU were phosphorylated and exhibited a stronger association with ssDNA than the And-1(T826A) mutant, indicating that And-1 phosphorylation at T826 enhances its affinity to ssDNA. Strikingly, the presence of phosphorylated wild-type And-1, but not And-1(T826A) mutant, significantly increased the association of Claspin with ssDNA (Fig6D). Taken together, we conclude that And-1 is an ssDNA-binding protein that can promote the association of Claspin with ssDNA in a manner dependent on And-1 phosphorylation at T826. Accordingly, we also found that And-1 co-localized with ssDNA accumulated BrdU foci in vivo (Fig6E).
We next applied the iPOND assay to examine the association of And-1 with DNA replication forks in vivo (Kliszczak et al, 2011; Sirbu et al, 2011, 2012). We found that endogenous And-1 is associated with replication forks (Fig6F). To test whether And-1 is a bona fide replication fork protein, we chased the cells with regular growth medium for 2 h after EdU label to create an EdU-labeled chromatin control. We detected more And-1 proteins on active replication forks than on the newly synthesized chromatin (Fig6G), suggesting that And-1 is indeed a replication fork-associated protein. To examine whether And-1 phosphorylation at T826 regulates its association with replication forks, we performed iPOND assays in 293T cells expressing either the wild-type And-1 or mutant And-1(T826A). EdU-labeled cells were treated with 10 mM HU for 2 h before analysis by iPOND. We found that HU treatment increased the amount of wild-type And-1 but not mutant And-1(T826A) associated with replication forks (Fig6H). These data strongly suggest that And-1 is indeed a replisome protein that accumulates at stalled replication forks after replication stress in a manner dependent on its phosphorylation at T826.
And-1 stabilizes DNA replication forks during DNA replication stress
Claspin has been shown to regulate replication fork stability (Scorah & McGowan, 2009). Given that And-1 regulates Claspin, we next asked whether And-1 could also be involved in the stabilization of stalled replication forks in response to replication stress. To test this possibility, we analyzed the ability of replication forks to recover after HU treatment using the DNA combing technique as we described previously (Fig7A) (Li et al, 2012b). Specifically, we measured the efficiency of fork recovery from a short-term (2 h) HU treatment between IdU and CldU pulse labeling of DNA in siGl2- or siAnd-1-treated HCT116 cells (Fig7B). DNA fibers were then stretched and immunostained to detect IdU (red) and CldU (green). This assay allowed us to monitor replication fork recovery throughout the genome. If a stalled fork fails to recover after HU treatment, it should contain only red IdU-labeled track; if a stalled fork recovers after the removal of HU, then it should contain both IdU- and CldU-labeled tracks. To quantify replication fork recovery, the amount of stalled forks (IdU signal only) was normalized against the total number of replication forks (IdU plus CldU signal). In the absence of HU treatment, we did not detect any difference in the number of stalled or collapsed replication forks between control siRNA- and siAnd-1-treated cells. Consistent with a previous report (Petermann et al, 2010), HU treatment slightly increased the percentage of forks that failed to incorporate CldU after HU block in the control siRNA-treated cells. However, depletion of And-1 increased the fraction of forks that failed to recover from HU treatment by 2- to 3-fold (Fig7C), indicating that And-1 helps to maintain the stability of the stalled replication forks and prevent them from collapsing during replication stress.
Figure 7.
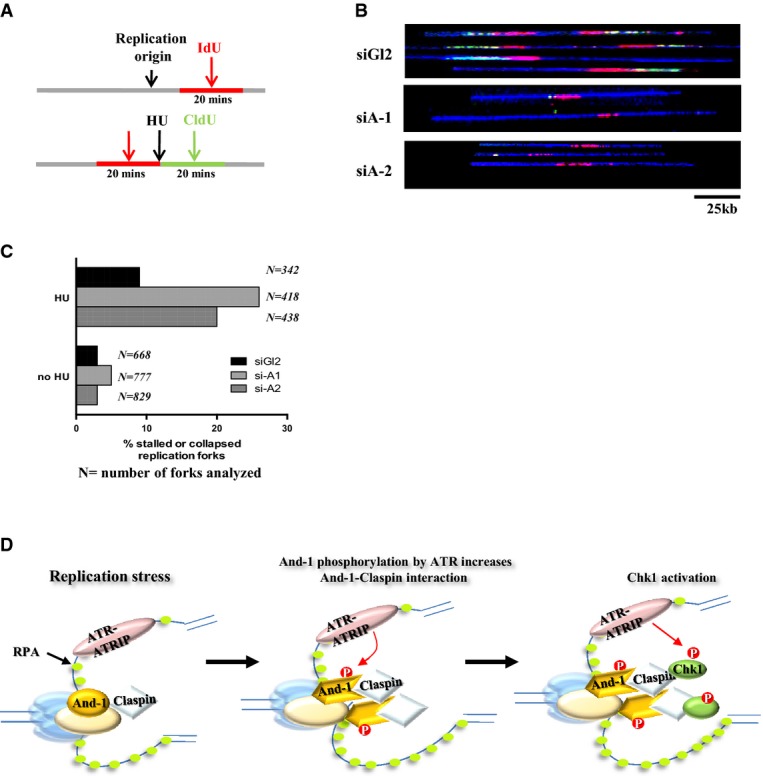
And-1 is required for recovery of stalled replication forks after replication stress
- Schematic of cell preparation for single-molecule analysis. Cells transfected with siGl2 or two independent And-1 siRNAs for 36 h were labeled with IdU (red) for 20 min followed by a 2-h block with 2 mM HU. After removal of HU, cells were labeled with CldU (green) for 20 min and then harvested for DNA combing analyses.
- And-1 depletion reduces the incorporation of CldU (green) at stalled replication forks after HU treatment. A representative example of the DNA fibers obtained by combing from HU-treated cells is represented. Note, stalled or collapsed forks after HU treatment failed to incorporate CldU. The full DNA fiber is shown in blue.
- The percentage of stalled or collapsed forks (IdU tracks only) is increased in And-1-depleted cells.
- A working model for And-1 to regulate replication checkpoint pathway. In response to replication stress, And-1 phosphorylation by ATR facilitates the accumulation of And-1 at stalled forks and enhances the association of And-1 with Claspin, resulting in the recruitment of Claspin–Chk1 to stalled replication forks for its activation.
Discussion
Although the replisome components, Claspin and Timeless–Tipin, have been shown to facilitate Chk1 phosphorylation by the ATR signaling pathway, the mechanisms by which these proteins coordinate with ATR to activate Chk1 remain speculative. Here, we show that an additional replisome component, And-1, is critical for Chk1 activation by linking Claspin–Chk1 to ATR at DNA damage sites. Our data demonstrated that And-1 promotes the association of Claspin with Chk1 at stalled replication forks. Specifically, And-1 phosphorylation at T826 by ATR is essential for establishing a functional link between Claspin and And-1, which is important for efficient Chk1 activation. Our in vitro analyses indicate that And-1 directly binds to ssDNA and promotes the association of Claspin with ssDNA. Based on these observations, we propose that ATR-mediated And-1 phosphorylation at stalled replication forks leads to its accumulation at damage sites, where And-1 promotes the interaction of Claspin with Chk1 and the recruitment of Claspin–Chk1 to the stalled forks for Chk1 activation by ATR (Fig7D).
A previous study by Yoshizawa-Sugata & Masai (2009) proposed that And-1 regulates DNA replication checkpoint by maintaining the stability of Chk1. In their study, they observed DNA damage and Chk1 degradation 72 h after And-1 siRNA transfection in cells (Yoshizawa-Sugata & Masai, 2009). In our study, we treated cells with And-1 siRNA for 48 h and did not detect degradation of Chk1 protein within this period of time (Fig3). Although we did observe DNA damage 3 days after And-1 depletion, we did not observe DNA damage in cells harvested 48 h after siRNA transfection (Supplementary Fig S3D). Given that DNA damage itself can induce Chk1 protein degradation (Zhang et al, 2005, 2009; Leung-Pineda et al, 2009), it is possible that the degradation of Chk1 that was observed by Yoshizawa-Sugata et al resulted from DNA damage due to an extended period of And-1 depletion.
Claspin by itself displays little affinity for ssDNA (Sar et al, 2004; Kemp et al, 2010; Uno & Masai, 2011), and the stable association of Claspin with ssDNA is facilitated by an interaction between Tipin and RPA (Kemp et al, 2010). Our studies revealed that And-1 is another factor that regulates the association of Claspin with ssDNA. This conclusion is strongly supported by the fact that And-1 forms complexes with both Claspin and Timeless–Tipin. Since And-1 phosphorylation at T826 is not required for its ability to interact with Timeless–Tipin but is essential for its interaction with Claspin in response to replication stress (Fig5F), it is more likely that ATR-mediated And-1 phosphorylation at T826 specifically regulates the interplay between And-1 and Claspin, which is critical for Chk1 activation. However, we cannot rule out the possibility that And-1 phosphorylation may affect other proteins that contribute to the association of Claspin with ssDNA in response to replication stress.
Claspin is a Chk1-interacting protein that brings Chk1 to stalled replication forks for its phosphorylation by ATR via protein–protein interactions (Chini & Chen, 2003; Liu et al, 2006; Lindsey-Boltz et al, 2009). In support of this view, we and others have found that the interaction between Claspin and Chk1 is increased in response to replication fork stalling (Fig4B) (Chini & Chen, 2003). However, in the absence of And-1, the interaction between Chk1 and Claspin is significantly reduced in cells under replication stress, indicating that And-1 is a critical factor that mediates the interaction between Chk1 and Claspin at stalled replication forks (Fig4B). Because the interaction between And-1 and Claspin is increased in a manner dependent on its phosphorylation at T826 by ATR in response to replication stress (Fig4A), phosphorylated And-1 likely functions as a linker to facilitate the recruitment of Chk1 to the stalled replication forks via Claspin. This provides a direct route for Chk1 activation by ATR at DNA damage sites.
And-1 contains a HMG DNA-binding domain, and And-1 alone binds ssDNA preferentially over dsDNA (Fig6). These observations further support a role of And-1 in the regulation of DNA replication checkpoint because ssDNA is generated during replication checkpoint activation in response to stalled forks (Lopes et al, 2001; Sogo et al, 2002). It should be noted that in contrast to our results, a recent study by Bermudez et al (2010) reported that And-1 associates weakly with ssDNA. This difference may be due to difference in the lengths or sequences of the oligonucleotides that were used. It is possible that the longer nucleotide we used in this study is a more appropriate binding substrate of And-1. Previous studies have demonstrated that Timeless–Tipin can promote the association of Claspin with ssDNA in vitro (Kemp et al, 2010). Because And-1 also interacts with Timeless–Tipin, it will be interesting to investigate whether there is a functional link between And-1 and Timeless–Tipin that regulates the association of Claspin with ssDNA.
Our data indicate that And-1 phosphorylation at T826 is critical for its localization at damage sites and interaction with Claspin as well as efficient Chk1 activation. These data suggest that And-1 phosphorylation by ATR is an important event for Chk1 activation in cells undergoing replication stress. So far, we do not know how And-1 T826 phosphorylation regulates its ability to interact with Claspin. A possible explanation is that ATR-mediated And-1 phosphorylation may change the conformation of And-1 and spatially orient the SepB and HMG domains to allow them to interact with Claspin and promote the formation of Claspin–Chk1 complexes at stalled replication forks. Future structural studies will yield molecular insights into the mechanism by which And-1 phosphorylation by ATR regulates And-1–Claspin interaction.
Materials and Methods
Cells culture and cell line construction
HCT116, U2OS, and 293T cells were grown in DMEM medium supplemented with 10% FBS at 37°C in 5% CO2. To make stable cell lines, And-1 full-length and truncated mutants were digested from pEFF-N vector at BamHI and NotI sites and subcloned into pMSCVpuro FLAG retroviral vector. And-1 full-length and And-1(T826A) cDNA sequence were amplified by PCR with primers containing the BamHI and SalI sites and then digested with BamHI and SalI. The digested And-1 WT and And-1 T826A cDNA sequences were ligated into pBABEpuro retroviral vector at BamHI and SalI sites. Retroviruses were produced by transfecting Phoenix A cells with the retroviral plasmids. HCT116, U2OS, and 293T cells were used as parent cell lines. HCT116 FLAG-And-1, HCT116 FLAG-And-1 (T826A) siAnd-1-resistant, HCT116 FLAG-And-1-truncated (1–330, 336–984, 984–1129, and 336–1129), U2OS FLAG-And-1, 293T And-1 siAnd-1-resistant, and 293T And-1 (T826A) siAnd-1-resistant cell lines were generated by retroviral infection, and single-cell clones were isolated by antibiotic selection.
Co-immunoprecipitation
Co-IP was performed as we described previously (Zhu et al, 2007; Li et al, 2012a).
Plasmids and antibodies
The following antibodies were used: Chk1 (sc-7898, sc-8408), RPA70 (sc-28304), ATR (sc-1887), MCM7 (sc-9966), GCN5 (sc-20698), H2B (sc-10808), and pol-α (sc-5921) were from Santa Cruz Biotechnology. P-Chk1-317 (2344S), p-Chk1-345 (2341S), rabbit γ-H2AX (9718), Claspin (2800), and p(S/T) ATR/ATM substrate antibody (2909S) were from Cell Signaling. Mouse γ-H2AX (05–636) was from Millipore. BrdU antibody (555627) was from BD Pharmigen. Anti-FLAG (F7425) and GAPDH (G9545) were from Sigma. RPA32 (ab2175) was from Abcam. Timeless and Tipin antibodies were gifts from Dr. Eishi Noguchi. And-1 antibody was raised as previously described (Li et al, 2011). pcDNA3.1 FLAG-Claspin plasmid was a gift from Dr. Michele Pagano (Addgene plasmid #12659). pEFF-N FLAG-Claspin was constructed by inserting the Claspin cDNA into pEFF-N FLAG vector using restriction sites BamHI and XhoI. FLAG-And-1 (full), FLAG-And-1 mutants (1–336), (330–1129) and (984–1129) plasmids were constructed as previously described (Zhu et al, 2007). A fragment of And-1 encoding amino acids 336–984 was cloned into pEFF-N FLAG vector for the expression of FLAG-And-1 (336–984). Single amino acid mutant And-1 plasmids were generated using QuikChange II XL Site-Directed Mutagenesis kit (Agilent Technologies, Cat no. 200521). And-1 and And-1 T826A sequences were digested from pEFF-FLAG-And-1 plasmids and subcloned into the pcDNA3 vector at the BamHI and NotI site. GST-And-1 plasmids were generated as described previously (Zhu et al, 2007).
Mass spectrometry and silver staining
293T cells were transfected with pEFF-FLAG-And-1 plasmid, and the cells were harvested and immunoprecipitated as described in the Co-IP procedure. Silver staining was carried out with the SilverQuest™ silver staining kit (Invitrogen). Mass spectrometry was performed as described earlier (Li et al, 2012b).
Plasmid transfection and siRNA interference
Both control vector and protein expression plasmids were transfected into cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s manual. siRNA oligonucleotides were made to the following target sequences (sense): And-1-1 (AAGCAGGCAUCUGCAGCAUCCdTdT), And-1-2 (AGGAAAACAUGCCUGCCACdTdT), And-1-3 (GGUGUAGGUAACAGGACAUdTdT), ATR (CCUCCGUCAUGUUGCUUGAdTdT), and Claspin (GGAAAGAAAGGCAGCCAGAdTdT). Control GL2 was as described earlier (Zhu et al 2004). siRNA transfection was performed twice with siRNA oligonucleotide duplexes using Lipofectamine™ RNAiMAX (Invitrogen) according to the manufacturer’s instructions. Cells were harvested for analyses 48 h after the first transfection.
Protein purification
Recombinant And-1 proteins were expressed and purified in the Sf9 cell line as previously reported (Zhu et al, 2007). Recombinant Claspin proteins were purified as previously reported (Kemp et al, 2010). To purify recombinant proteins from human cells, FLAG-Claspin, FLAG-And-1 WT, and FLAG-And-1 T826A proteins were expressed in 293T cells for 48 h. Cells were treated with or without 1 mM HU for 15 h before harvest. Cell extracts were treated as in IP. After extensive wash, bound proteins were eluted from the beads by detergent-free IP lysis buffer containing 200 μg/ml FLAG peptide (Sigma F3290).
Immobilized DNA pull-down assays
DNA pull-down assays were performed as described previously (Sar et al, 2004; Unsal-Kacmaz & Sancar, 2004; Kemp et al, 2010; Sercin & Kemp, 2011). The sequence of the 80-mer ssDNA oligomer was previously reported (Unsal-Kacmaz & Sancar, 2004). Both the biotinylated ssDNA oligomer and its non-labeled complement oligomer were synthesized by Invitrogen. The streptavidin-coupled magnetic beads were from New England Biolabs (S1420S). For each sample, 20 μl beads and 20 pmol of biotin-labeled oligomer were used. For ssDNA-binding assays, ssDNA was immobilized on streptavidin-coupled magnetic beads in binding buffer [0.5 M NaCl, 20 mM Tris–HCl (pH 7.5), and 1 mM EDTA]. For dsDNA-binding assays, 20 pmol of biotinylated ssDNA was annealed with either buffer or 80 pmol complement oligomer before being immobilized on beads. After extensive washing, recombinant And-1 or BSA was incubated with DNA-conjugated beads in PBS for 30 min at room temperature. After washing off the residual primary binding proteins with PBS containing 0.01% NP-40, recombinant Claspin proteins were incubated with And-1- or BSA-treated beads for 2 h at 4°C. The beads were then collected after extensive washing, and bound proteins were eluted by SDS loading buffer. The protein samples were separated by SDS–PAGE and analyzed by immunoblotting.
Immunofluorescence
Cells attached to coverslips were pre-extracted with 0.2% Triton X-100 in PBS buffer for 5 min on ice. Cells were fixed with PBS containing 4% paraformaldehyde for 10 min at room temperature and rinsed three times with PBS containing 0.02% Tween-20. Cells were blocked and incubated with primary antibody for 1 h at room temperature. Afterward, the cells were washed three times with PBS containing 0.02% Tween-20 and incubated in secondary antibody [anti-rabbit Alexa Fluor-594 and anti-mouse Alexa Fluor-488 1:1,000 (R37114, Life Technologies)] for 40 min. After washing, cells were mounted in Fluoromount G (SouthernBiotech, 0100-20) containing DAPI. Slides were imaged using Nikon Eclipse 80i microscope or Carl Zeiss LSM 510 confocal system. When UV was applied, the cells on coverslips were covered with 5.0 μm isopore membrane (Millipore, TMTP02500) before being subjected to 100 J/m2 UV irradiation (UVP, CL-1000).
iPOND
Replication forks were pulled down following the iPOND protocol (Kliszczak et al, 2011; Sirbu et al, 2011, 2012). Briefly, 293T cells were labeled with 10 μM EdU (Life Technologies, A10044) for 30 min followed by 10 mM HU for 2 h to induce stalled replication forks as indicated. Cross-linking was performed with 0.2% formaldehyde (Sigma, F8775) in PBS at 4°C for 10 min and quenched with glycine. Cells were collected and permeabelized with 0.2% Triton X-100 in PBS on ice for 10 min. Click reaction was performed at RT for 2 h in PBS solution containing 10 μM biotin azide (Life technologies, B10184), 10 mM sodium ascorbate, and 2 mM CuSO4 (Fisher, C489). DMSO was used to replace biotin azide in the control. Cells were washed and lysed in lysis buffer [150 mM NaCl, 1.0% IGEPAL® CA-630, 0.5% sodium deoxycholate, 0.5% (wt/vol) SDS in 50 mM Tris (pH 8.0)] containing protease and phosphatase inhibitors. Cells were sonicated until the solution was clear and centrifuged to remove any residues. Equal amounts of protein were used in replication fork pull downs with Streptavidin agarose (Novagen, 69203-3) at 4°C overnight. The beads were then collected after extensive washing, and bound proteins were eluted by SDS loading buffer. The protein samples were separated by SDS–PAGE and analyzed by immunoblotting.
DNA combing
DNA combing was performed following the protocol published previously (Li et al, 2012b). Briefly, 36 h after siRNA transfection, cells were pulsed with 100 μM iodo-deoxyuridine (IdU) for 20 min. The cells were washed twice with warm PBS and supplied with fresh medium containing 2 mM HU for 2 h. HU was released by washing cells twice with warm PBS, and the cells were pulsed with 200 μM chloro-deoxyuridine (CldU) for 20 min in fresh medium. CldU was washed off with PBS, and cells were trypsinized and collected. The cells were embedded into agarose plugs and treated with proteinase K to extract DNA. The plugs were then melted and treated with β-agarase, and the resulted DNA solution was used for DNA combing. DNA fibers were stretched on silanized coverslips (Microsurfaces Inc), and immunostaining was performed to detect IdU and CldU signals.
Acknowledgments
We thank Drs. A. Dutta, J. Chen, E. Noguchi, and A. Sancar for reagents. This work was partially supported by funding from the National Institutes of Health (CA177898 and CA184717). Wenge Zhu was supported by a Research Scholar Grant, RSG-13-214-01-DMC, from the American Cancer Society. This work was partially supported by core grants NICHD/NINDS 2R24HD050846-06 (National Center for Medical Rehabilitation Research), NICHD 5P30HD040677-10 (Intellectual and Developmental Disabilities Research Center), and NIH NCATS UL1RR031988 (CTSI-CN). The authors would like to thank Dr. Kristy J. Brown for her kind assistance in collection and processing of the mass spectrometry samples.
Author contributions
Experiments were performed by JH, YL, CR, HX and MGK; ZH and MLD participated in parts of manuscript writing. Experimental design, interpretation of data, and manuscript writing were performed by JH and WZ.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Materials and Methods, Supplementary Figure Legends
Source Data for Supplementary Figure S1
Source Data for Supplementary Figure S3
Source Data for Supplementary Figure S4
Review Process File
Source Data for Figure 1
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
References
- Bermudez VP, Farina A, Tappin I, Hurwitz J. Influence of the human cohesion establishment factor Ctf4/AND-1 on DNA replication. J Biol Chem. 2010;285:9493–9505. doi: 10.1074/jbc.M109.093609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows AE, Elledge SJ. How ATR turns on: topBP1 goes on ATRIP with ATR. Genes Dev. 2008;22:1416–1421. doi: 10.1101/gad.1685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini CC, Chen J. Human claspin is required for replication checkpoint control. J Biol Chem. 2003;278:30057–30062. doi: 10.1074/jbc.M301136200. [DOI] [PubMed] [Google Scholar]
- Chini CC, Chen J. Repeated phosphopeptide motifs in human Claspin are phosphorylated by Chk1 and mediate Claspin function. J Biol Chem. 2006;281:33276–33282. doi: 10.1074/jbc.M604373200. [DOI] [PubMed] [Google Scholar]
- Chou DM, Elledge SJ. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc Natl Acad Sci USA. 2006;103:18143–18147. doi: 10.1073/pnas.0609251103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico A, Costanzo V, Hunt T. Tipin is required for stalled replication forks to resume DNA replication after removal of aphidicolin in Xenopus egg extracts. Proc Natl Acad Sci USA. 2007;104:14929–14934. doi: 10.1073/pnas.0706347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico A, Cosentino C, Rivera T, Losada A, Schwob E, Hunt T, Costanzo V. Tipin/Tim1/And1 protein complex promotes Pol alpha chromatin binding and sister chromatid cohesion. EMBO J. 2009;28:3681–3692. doi: 10.1038/emboj.2009.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, van Deursen F, Polychronopoulos D, Foltman M, Jones RC, Edmondson RD, Calzada A, Labib K. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009;28:2992–3004. doi: 10.1038/emboj.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Kim JE, Leung CC, Glover JN, Chen J. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol Cell. 2010;37:438–446. doi: 10.1016/j.molcel.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter AL, Suppa C, Emanuel BS. Mammalian TIMELESS and Tipin are evolutionarily conserved replication fork-associated factors. J Mol Biol. 2007;366:36–52. doi: 10.1016/j.jmb.2006.10.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna JS, Kroll ES, Lundblad V, Spencer FA. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol. 2001;21:3144–3158. doi: 10.1128/MCB.21.9.3144-3158.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im JS, Ki SH, Farina A, Jung DS, Hurwitz J, Lee JK. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc Natl Acad Sci USA. 2009;106:15628–15632. doi: 10.1073/pnas.0908039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp MG, Akan Z, Yilmaz S, Grillo M, Smith-Roe SL, Kang TH, Cordeiro-Stone M, Kaufmann WK, Abraham RT, Sancar A, Unsal-Kacmaz K. Tipin-replication protein A interaction mediates Chk1 phosphorylation by ATR in response to genotoxic stress. J Biol Chem. 2010;285:16562–16571. doi: 10.1074/jbc.M110.110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliszczak AE, Rainey MD, Harhen B, Boisvert FM, Santocanale C. DNA mediated chromatin pull-down for the study of chromatin replication. Sci Rep. 2011;1:95. doi: 10.1038/srep00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina EK, Bannikov V, Bliskovsky V, Gizatullin R, Kirillov A, Shestopalov B, Zakharyev V, Hieter P, Spencer F. CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5736–5747. doi: 10.1128/mcb.12.12.5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Kim SM, Dunphy WG. Claspin and the activated form of ATR-ATRIP collaborate in the activation of Chk1. J Biol Chem. 2004;279:49599–49608. doi: 10.1074/jbc.M408353200. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Lee J, Kumagai A, Dunphy WG. Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol Cell. 2003;11:329–340. doi: 10.1016/s1097-2765(03)00045-5. [DOI] [PubMed] [Google Scholar]
- Leman AR, Noguchi C, Lee CY, Noguchi E. Human Timeless and Tipin stabilize replication forks and facilitate sister-chromatid cohesion. J Cell Sci. 2010;123:660–670. doi: 10.1242/jcs.057984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung-Pineda V, Huh J, Piwnica-Worms H. DDB1 targets Chk1 to the Cul4 E3 ligase complex in normal cycling cells and in cells experiencing replication stress. Cancer Res. 2009;69:2630–2637. doi: 10.1158/0008-5472.CAN-08-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YM, Jaramillo-Lambert A, Hao J, Yang Y, Zhu WG. The stability of histone acetyltransferase general control non-derepressible (Gcn) 5 is regulated by cullin4-RING E3 ubiquitin ligase. J Biol Chem. 2011;286:41344–41352. doi: 10.1074/jbc.M111.290767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jaramillo-Lambert AN, Yang Y, Williams R, Lee NH, Zhu W. And-1 is required for the stability of histone acetyltransferase Gcn5. Oncogene. 2012a;31:643–652. doi: 10.1038/onc.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xiao H, de Renty C, Jaramillo-Lambert A, Han Z, Depamphilis ML, Brown CJ, Zhu W. The involvement of acidic nucleoplasmic DNA-binding protein (And-1) in the regulation of pre-replicative complex (pre-RC) assembly in human Cells. J Biol Chem. 2012b;287:42469–42479. doi: 10.1074/jbc.M112.404277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey-Boltz LA, Sercin O, Choi JH, Sancar A. Reconstitution of human claspin-mediated phosphorylation of Chk1 by the ATR (ataxia telangiectasia-mutated and rad3-related) checkpoint kinase. J Biol Chem. 2009;284:33107–33114. doi: 10.1074/jbc.M109.064485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, Lukas J. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol. 2006;26:6056–6064. doi: 10.1128/MCB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Pot I, Chang M, Xu H, Aneliunas V, Kwok T, Newitt R, Aebersold R, Boone C, Brown GW, Hieter P. Identification of protein complexes required for efficient sister chromatid cohesion. Mol Biol Cell. 2004;15:1736–1745. doi: 10.1091/mbc.E03-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata Y, Ishihara S, Hasegawa N, Nozaki N, Ishimi Y. Interaction of human MCM2-7 proteins with TIM, TIPIN and Rb. J Biochem. 2010;147:917–927. doi: 10.1093/jb/mvq028. [DOI] [PubMed] [Google Scholar]
- Petermann E, Helleday T, Caldecott KW. Claspin promotes normal replication fork rates in human cells. Mol Biol Cell. 2008;19:2373–2378. doi: 10.1091/mbc.E07-10-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sar F, Lindsey-Boltz LA, Subramanian D, Croteau DL, Hutsell SQ, Griffith JD, Sancar A. Human claspin is a ring-shaped DNA-binding protein with high affinity to branched DNA structures. J Biol Chem. 2004;279:39289–39295. doi: 10.1074/jbc.M405793200. [DOI] [PubMed] [Google Scholar]
- Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM, Abraham RT. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- Scorah J, McGowan CH. Claspin and Chk1 regulate replication fork stability by different mechanisms. Cell Cycle. 2009;8:1036–1043. doi: 10.4161/cc.8.7.8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sercin O, Kemp MG. Characterization of functional domains in human Claspin. Cell Cycle. 2011;10:1599–1606. doi: 10.4161/cc.10.10.15562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011;25:1320–1327. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu BM, Couch FB, Cortez D. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat Protoc. 2012;7:594–605. doi: 10.1038/nprot.2012.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- Tsutsui Y, Morishita T, Natsume T, Yamashita K, Iwasaki H, Yamao F, Shinagawa H. Genetic and physical interactions between Schizosaccharomyces pombe Mcl1 and Rad2, Dna2 and DNA polymerase alpha: evidence for a multifunctional role of Mcl1 in DNA replication and repair. Curr Genet. 2005;48:34–43. doi: 10.1007/s00294-005-0584-2. [DOI] [PubMed] [Google Scholar]
- Uno S, Masai H. Efficient expression and purification of human replication fork-stabilizing factor, Claspin, from mammalian cells: DNA-binding activity and novel protein interactions. Genes Cells. 2011;16:842–856. doi: 10.1111/j.1365-2443.2011.01535.x. [DOI] [PubMed] [Google Scholar]
- Unsal-Kacmaz K, Sancar A. Quaternary structure of ATR and effects of ATRIP and replication protein A on its DNA binding and kinase activities. Mol Cell Biol. 2004;24:1292–1300. doi: 10.1128/MCB.24.3.1292-1300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsal-Kacmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol Cell Biol. 2007;27:3131–3142. doi: 10.1128/MCB.02190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- Williams DR, McIntosh JR. mcl1+, the Schizosaccharomyces pombe homologue of CTF4, is important for chromosome replication, cohesion, and segregation. Eukaryot Cell. 2002;1:758–773. doi: 10.1128/EC.1.5.758-773.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa-Sugata N, Masai H. Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint. J Biol Chem. 2007;282:2729–2740. doi: 10.1074/jbc.M605596200. [DOI] [PubMed] [Google Scholar]
- Yoshizawa-Sugata N, Masai H. Roles of human AND-1 in chromosome transactions in S phase. J Biol Chem. 2009;284:20718–20728. doi: 10.1074/jbc.M806711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Otterness DM, Chiang GG, Xie W, Liu YC, Mercurio F, Abraham RT. Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol Cell. 2005;19:607–618. doi: 10.1016/j.molcel.2005.07.019. [DOI] [PubMed] [Google Scholar]
- Zhang YW, Brognard J, Coughlin C, You Z, Dolled-Filhart M, Aslanian A, Manning G, Abraham RT, Hunter T. The F box protein Fbx6 regulates Chk1 stability and cellular sensitivity to replication stress. Mol Cell. 2009;35:442–453. doi: 10.1016/j.molcel.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Chen Y, Dutta A. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol Cell Biol. 2004;24:7140–7150. doi: 10.1128/MCB.24.16.7140-7150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Ukomadu C, Jha S, Senga T, Dhar SK, Wohlschlegel JA, Nutt LK, Kornbluth S, Dutta A. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev. 2007;21:2288–2299. doi: 10.1101/gad.1585607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci USA. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Materials and Methods, Supplementary Figure Legends
Source Data for Supplementary Figure S1
Source Data for Supplementary Figure S3
Source Data for Supplementary Figure S4
Review Process File
Source Data for Figure 1
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6