Abstract
There is a common interest for studying xeno-nucleic acid systems in the fields of synthetic biology and the origin of life, in particular, those with an engineered backbone and possessing novel properties. Along this line, we have investigated xylonucleic acid (XyloNA) containing a potentially prebiotic xylose sugar (a 3′-epimer of ribose) in its backbone. Herein, we report for the first time the synthesis of four XyloNA nucleotide building blocks and the assembly of XyloNA oligonucleotides containing all the natural nucleobases. A detailed investigation of pairing and structural properties of XyloNAs in comparison to DNA/RNA has been performed by thermal UV-melting, CD, and solution state NMR spectroscopic studies. XyloNA has been shown to be an orthogonal self-pairing system which adopts a slightly right-handed extended helical geometry. Our study on one hand, provides understanding for superior structure-function (-pairing) properties of DNA/RNA over XyloNA for selection as an informational polymer in the prebiotic context, while on the other hand, finds potential of XyloNA as an orthogonal genetic system for application in synthetic biology.
INTRODUCTION
DNA and RNA are the two monopolistic biopolymers that are capable of storage and dissemination of genetic information in the extant biology. While the nucleobases of DNA and RNA are at the very core of the helical structure providing crucial stabilization by Watson–Crick type hydrogen bonding and stacking interactions, the unique sugar-phosphate backbone is significantly providing an optimal structural and dynamic framework. These two natural nucleic acid versions differ in the sugar moiety in their backbone i.e. deoxy-d-ribose for DNA and d-ribose for RNA (Figure 1), thereby remarkably influencing their structural properties and biological functions (1). Recently, a number of sugar-modified nucleic acid variants has been revealed as new genetic polymers, (2) some of them are endowed with catalytic activity (for e.g. FANA and HNA) (3). The structure of these artificial nucleic acids, however, mimics natural nucleic acid helicity (4). In the context of studies on chemical etiology of nucleic acid structure (5–7) and the origin of life, alternative nucleic acids with more deviating backbone structures have been investigated. Some of them are having different double helical geometry than that of DNA/RNA, (5–8) others with a similar geometry, and thus shown as potential direct RNA precursors (9–11) (like TNA and PNA) that might have been evolved into the existing nucleic acid. A relatively much less explored area in alternative nucleic acid engineering is the search and study of a complementary pair of synthetic nucleic acids with novel backbone and helical geometries which can support life i.e. perform catalysis like RNA and store genetic information like DNA. This might assist in the design principles of an orthogonal nucleic acid pair for application in xenobiology (orthogonal life) (12–14). Likewise, it may also expand our knowledge in the context of prebiotic origin and evolution of nucleic acids.
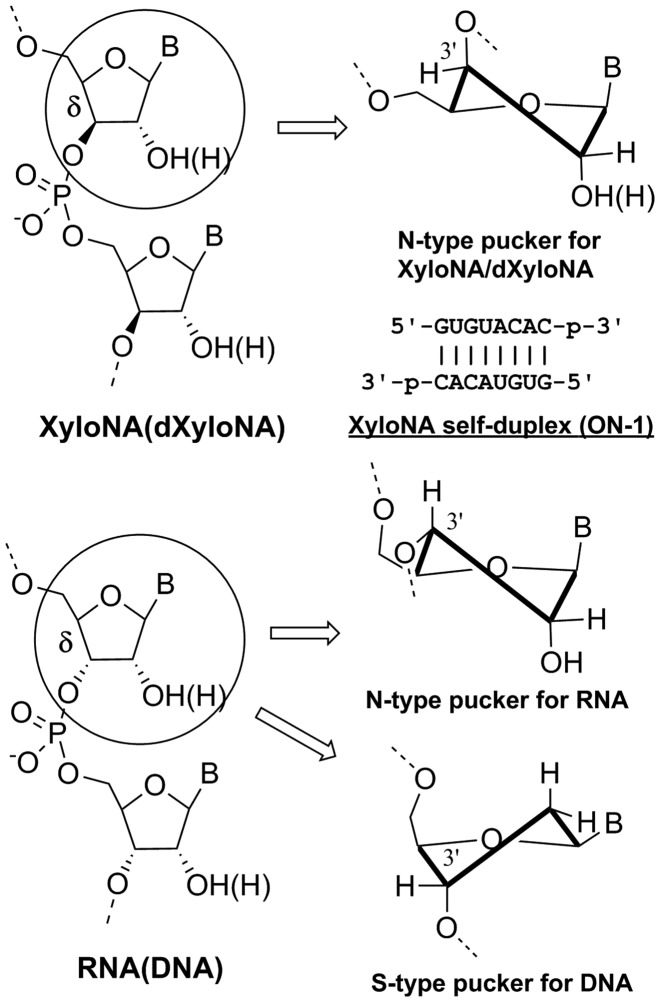
Figure 1.
Chemical structure of the sugar-phosphate backbone in XyloNA (dXyloNA) compared with RNA (DNA). Sequence and base-pairing are shown for a fully-modified self-complementary XyloNA duplex (ON-1) investigated (by CD and NMR) in this study. Types of sugar puckering are indicated as exist and characterized by NMR spectroscopy in different natural and xylose-based nucleic acid systems.
In this context, we have studied the physicochemical properties of a nucleic acid containing d-xylose (wood sugar), a prebiotic pentofuranosyl sugar. When compared with ribose, the xylose sugar has an inverted 3′-carbon center; hence xylose nucleic acid (XyloNA) and its cousin deoxyxylonucleic acid (dXyloNA) possess a diastereoisomeric backbone (Figure 1) in comparison with the natural RNA and DNA respectively. Previously, dXyloNA (15–18) and locked XyloNA (19–22) were studied in the perspective of potential therapeutic applications, but without success. Recently, we have shown the potential heterogeneous helical structure and conformational malleability of dXyloNA (23,24). In the present study, we describe the systematic exploration on self-pairing versus cross-pairing ability of XyloNA and its helical structural characterization by circular dichroism (CD) and solution NMR spectroscopy that illuminate on the inherent structural properties of xylonucleic acid backbone (Figure 1). In particular, the effect of 2′-hydroxyl groups is shown in tuning the structural dynamics in comparison with the previously studied dXyloNA (24) system.
MATERIALS AND METHODS
General
For all reactions, analytical grade solvents were used. All moisture sensitive reactions were carried out under argon or nitrogen atmosphere in oven dried glasswares (120°C). 1,2-O-Isopropylidene-α-d-xylofuranose was purchased from Acros Organics. Dry acetonitrile and 1,2-dichloroethane were obtained by distillation over CaH2. All other dry solvents and reagents were purchased from commercial sources. Pre-coated aluminum sheets (Fluka) were used for TLC. Flash silica column chromatography was performed on silica gel 60 A, 40–60 μM (Acros Organics). NMR spectra were recorded on Bruker Avance II spectrometers; 300 MHz with a 5mm broad band probe, 500 MHz with a TXI-HCP Z gradient probe, or on a 600 MHz NMR equipped with a 5 mm TCI-HCN Z gradient cryo-probe. The spectra were processed with Bruker Topspin 2.1 software. Chemical shifts (δ) were reported in parts per million (ppm). The 1H and 13C chemical shifts were referenced relative to TMS peak (δ = 0.00 ppm). 31P NMR chemical shifts were referenced to an external 85% H3PO4 standard (δ = 0.00 ppm). 1H and 13C resonance assignments were made by using 2D correlation experiments (COSY, gHSQC and gHMBC). High resolution mass spectra were acquired on a quadrupole orthogonal acceleration time-of-flight mass spectrometer (Synapt G2 HDMS, Waters, Milford, MA, USA).
Synthetic procedures and the characterization data for the intermediates of XyloNA building blocks synthesis are provided in the supplementary information.
General procedure for the phosphitylation reaction
Tritilated nucleoside (1 mmol) was coevaporated with dry THF (3×) and further dried under high vacuum for 1 h. Under an argon atmosphere, the compound was dissolved in anhydrous CH2Cl2 (10 ml) and N,N-diisopropyl ethylamine (5 mmol) was added. To this solution, 2-cyanoethyl-N,N-diisopropylchlorophosphoramidite (2.5 mmol) was added dropwise and the reaction mixture was stirred at room temperature for 4 h. Methanol (0.5 ml) was added dropwise to the reaction mixture at 0°C and the solution was stirred for another 15 min at room temperature. Then volatile organic solvents were removed under reduced pressure at room temperature and the crude product was purified by flash column chromatography.
1-{2′-O-Acetyl-3′-O-[2-cyanoethoxy(diisopropylamino)phosphanyl]-5′-O-(4,4′-dimethoxytrityl)-β-d-xylofuranosyl}uracil (6)
Tritilated nucleoside 5 (1.3 g, 2.2 mmol) was converted into the corresponding amidite by following the aforementioned general procedure for phosphitylation reaction. The crude product was purified by flash column chromatography using solvent system 25–75% EtOAc in hexane containing 0.5% triethylamine (Rf = 0.49, 0.64 for 50% EtOAc in CH2Cl2). Compound 6 was isolated as white foam containing a mixture of two diastereomers in 87% yield (1.54 g). 1H NMR (500 MHz, acetone-d6): δ 10.06 (br s, H3), 7.63, 7.58 (d, J = 8.2 Hz, 1H, H6), 7.54–6.87 (m, 13H, Ar-H), 6.00, 5.99 (s, 1H, H1′), 5.54, 5.50 (d, J = 8.2 Hz, 1H, H5), 5.39, 5.23 (s, 1H, H2′), 4.53–4.49 (m, 1H, H4′), 4.43–4.40 (m, 1H, H3′), 3.94–3.70 (m, 2H, OCH2CH2CN), 3.80, 3.79 (s, 6H, 2 OCH3), 3.69–3.31 (m, 2H, H5′,5″), 3.54–3.43 (m, 2H, 2×CH(CH3)2), 2.73–2.56 (m, 2H, OCH2CH2CN), 2.12 (s, 3H, COCH3), 1.15, 1.13, 1.02, 0.97 (d, 12H, J = 5.0, 4.7, 6.7, 6.7 Hz, 2×CH(CH3)2); 13C NMR (125 MHz, acetone-d6): δ 169.9, 169.8 (COCH3), 163.4, 163.3 (C4), 159.7 (Ar-C), 151.1 (C2), 146.0 (Ar-C), 141.0, 140.8 (C6), 136.9–127.6 (Ar-C), 118.8, 118.7 (CN), 114.0 (Ar-C), 102.3, 102.2 (C5), 90.1, 90.0 (C1′), 87.3, 87.2 (tert C of DMTr), 83.6, 83.3 (d, JCP = 3.5, 9.0 Hz, C4′), 81.4, 81.3 (d, JCP = 3.0, 0.0 Hz, C2′), 77.3, 75.9 (d, JCP = 18.3, 12.7 Hz, C3′), 63.7, 63.5 (C5′), 59.8, 59.5 (d, JCP = 22.3, 21.2 Hz, OCH2CH2CN), 55.6, 55.5 (OCH3), 43.9, 43.8 (d, JCP = 4.0, 3.8 Hz, CH(CH3)2), 25.1, 24.9, 24.7 (d, JCP = 8.6, 7.4, 7.0 Hz, CH(CH3)2), 20.8, 20.7 (COCH3), 20.5, 20.4 (d, JCP = 7.6, 7.7 Hz, OCH2CH2CN); 31P NMR (202 MHz, acetone-d6): δ 152.71, 148.94; HRMS (ESI+), calcd for C41H50N4O10P [M+H]+ 789.3259, found 789.3268.
1-{2′-O-Acetyl-3′-O-[2-cyanoethoxy(diisopropylamino)phosphanyl]-5′-O-(4,4′-dimethoxytrityl)-β-d-xylofuranosyl}-4-N-benzoylcytosine (10)
Tritilated nucleoside 9 (1.35 g, 2.0 mmol) was converted into the corresponding amidite by following the general procedure for phosphitylation. The crude product was purified by flash column chromatography using solvent system 20–40% acetone in hexane containing 0.5% triethylamine (Rf = 0.58, 0.66 for 60% acetone in hexane). Compound 10 was isolated as white foam containing a mixture of two diastereoisomers in 81% yield (1.4 g). 1H NMR (500 MHz, acetone-d6): δ 9.78 (br s, NH), 8.16, 8.15 (d, J = 6.9, 6.5 Hz, 2H, o H-Bz), 7.97, 7.96 (d, J = 7.4 Hz, 1H, H6), 7.68–7.64 (m, 1H, p H-Bz), 7.58–7.56 (m, 2H, m H-Bz), 7.44–7.33 (d, J = 7.4 Hz, 1H, H5), 7.58–6.90 (m, 13H, Ar-H of DMTr), 5.94, 5.91 (s, 1H, H1′), 5.53, 5.34 (s, 1H, H2′), 4.72–4.65 (m, 1H, H4′), 4.39–4.37 (m, 1H, H3′), 3.82, 3.81 (s, 6H, 2 OCH3), 3.82–3.54 (m, 2H, OCH2CH2CN), 3.82–3.33 (m, 2H, H5′,5″), 3.40–3.33 (m, 2H, 2×CH(CH3)2), 2.71–2.56 (m, 2H, OCH2CH2CN), 2.15 (s, 3H, COCH3), 1.09, 1.04, 0.92 (d, 12H, J = 6.8 Hz, 2×CH(CH3)2); 13C NMR (125 MHz, acetone-d6): δ 169.8, 169.7 (COCH3), 164.0, 163.9 (C4), 159.8 (Ar-C), 155.2 (C2), 146.1 (Ar-C), 145.2 (C6), 136.8–127.7 (Ar-C), 118.8, 118.7 (CN), 114.0, 113.9 (Ar-C), 96.3 (C5), 92.1, 91.9 (C1′), 87.3, 87.2 (tert C of DMTr), 84.9, 84.8 (d, JCP = 4.5 Hz, C4′), 80.8, 80.2 (d, JCP = 3.3, 0.0 Hz, C2′), 77.0, 75.8 (d, JCP = 19.3, 11.6 Hz, C3′), 63.9, 63.7 (C5′), 59.9, 59.5 (d, JCP = 23.1, 22.9 Hz, OCH2CH2CN), 55.6 (OCH3), 43.9, 43.8 (d, JCP = 9.4, 8.8 Hz, CH(CH3)2), 25.1, 24.9, 24.6 (d, JCP = 8.2, 7.3, 4.9 Hz, CH(CH3)2), 20.8, 20.7 (COCH3), 20.6, 20.2 (d, JCP = 7.5, 8.2 Hz, OCH2CH2CN); 31P NMR (202 MHz, acetone-d6): δ 152.80, 148.52; HRMS (ESI+), calcd for C48H55N5O10P [M+H]+ 892.3681, found 892.3669.
9-{2′-O-Acetyl-3′-O-[2-cyanoethoxy(diisopropylamino)phosphanyl]-5′-O-(4,4′-dimethoxytrityl)-β-d-xylofuranosyl}-6-N-benzoyladenine (14)
Tritilated nucleoside 13 (2.28 g, 3.2 mmol) was converted into the corresponding amidite by following the general procedure for phosphitylation. The crude product was purified by flash column chromatography using solvent system 45–85% EtOAc in hexane containing 0.5% triethylamine (Rf = 0.42, 0.54 for 90% EtOAc in hexane). Compound 14 was isolated as white foam containing a mixture of two diastereoisomers in 84% yield (2.46 g). 1H NMR (500 MHz, acetone-d6): δ 10.00 (br s, NH), 8.67, 8.65 (s, 1H, H2), 8.30, 8.29 (s, 1H, H8), 8.14, 8.12 (d, J = 6.2, 6.9 Hz, 2H, o H-Bz), 7.66–7.63 (m, 1H, p H-Bz), 7.57–7.53 (m, 2H, m H-Bz), 7.57–6.85 (m, 13H, Ar-H of DMTr), 6.40, 6.39 (s, 1H, H1′), 5.82, 5.68 (s, 1H, H2′), 4.70–4.69 (m, 1H, H4′), 4.59–4.53 (m, 1H, H3′), 3.79, 3.78, 3.77 (s, 6H, 2 OCH3), 3.74–3.54 (m, 2H, OCH2CH2CN), 3.74–3.35 (m, 2H, H5′,5″), 3.42–3.35 (m, 2H, 2×CH(CH3)2), 2.65–2.52 (m, 2H, OCH2CH2CN), 2.18, 2.17 (s, 3H, COCH3), 1.11, 1.08, 0.95, 0.92 (d, 12H, J = 6.9, 7.2, 6.9, 6.7 Hz, 2×CH(CH3)2); 13C NMR (125 MHz, acetone-d6): δ 169.9 (COCH3), 159.7 (Ar-C), 152.8 (C4, C2), 151.2 (C6), 146.1 (Ar-C), 142.5, 142.4 (C8), 136.9–127.6 (Ar-C), 125.9, 125.7 (C5), 118.8, 118.7 (CN), 113.9 (Ar-C), 89.1, 88.7 (C1′), 87.3, 87.2 (tert C of DMTr), 84.0, 83.9 (d, JCP = 3.4, 0.0 Hz, C4′), 81.3, 81.2 (d, JCP = 2.5, 0.0 Hz, C2′), 77.3, 76.0 (d, JCP = 18.1, 12.5 Hz, C3′), 64.2, 64.0 (C5′), 59.8, 59.5 (d, JCP = 21.9, 20.8 Hz, OCH2CH2CN), 55.6 (OCH3), 43.9, 43.8 (d, JCP = 12.5, 12.1 Hz, CH(CH3)2), 25.1, 24.9, 24.7, 24.6 (d, JCP = 8.0, 7.5, 8.2, 8.0 Hz, CH(CH3)2), 20.8, 20.7 (COCH3), 20.5, 20.4 (d, JCP = 7.5 Hz, OCH2CH2CN); 31P NMR (202 MHz, acetone-d6): δ 152.14, 148.98; HRMS (ESI+), calcd for C49H54N7O9PNa [M+Na]+ 938.3613, found 938.3616.
9-{2′-O-Acetyl-3′-O-[2-cyanoethoxy(diisopropylamino)phosphanyl]-5′-O-(4,4′-dimethoxytrityl)-β-d-xylofuranosyl}-2-N-acetylguanine (20)
Tritilated nucleoside 19 (0.87 g, 1.3 mmol) was converted into the corresponding amidite by following the general procedure for phosphitylation. The crude product was purified by flash column chromatography using solvent system 25–60% acetone in hexane containing 0.5% triethylamine (Rf = 0.60, 0.60 for 60% acetone in hexane). Compound 20 was isolated as white foam containing a mixture of two diastereoisomers in 88% yield (1.0 g). 1H NMR (500 MHz, acetone-d6): δ 12.04 (br s, 1H, H1), 10.54 (br s, 1H, NHAc), 7.92, 7.89 (s, 1H, H8), 7.52–6.84 (m, 13H, Ar-H), 6.06, 6.04 (s, 1H, H1′), 5.62, 5.47 (s, 1H, H2′), 4.58 (m, 1H, H4′), 4.53–4.50 (m, 1H, H3′), 3.79, 3.78 (s, 6H, 2 OCH3), 3.85–3.63 (m, 2H, OCH2CH2CN), 3.69–3.29 (m, 2H, H5′,5″), 3.47–3.40 (m, 2H, 2×CH(CH3)2), 2.71–2.51 (m, 2H, OCH2CH2CN), 2.31 (s, 3H, NHCOCH3), 2.15 (s, 3H, OCOCH3), 1.13, 1.10, 0.97, 0.95 (d, 12H, J = 6.8, 6.8, 5.2, 5.4 Hz, 2×CH(CH3)2); 13C NMR (125 MHz, acetone-d6): δ 174.0, 173.9 (NHCOCH3), 169.9, 169.8 (OCOCH3), 159.7 (Ar-C), 155.8, 155.7 (C6), 149.2, 149.1 (C4, C2), 146.1, 146.0 (Ar-C), 137.9, 137.8 (C8), 136.9–127.6 (Ar-C), 121.9, 121.7 (C5), 118.8, 118.7 (CN), 113.9 (Ar-C), 88.6, 88.3 (C1′), 87.2, 87.2 (tert C of DMTr), 83.9, 83.7 (d, JCP = 3.0, 9.0 Hz, C4′), 81.8, 81.7 (d, JCP = 3.1, 0.0 Hz, C2′), 77.6, 76.2 (d, JCP = 18.8, 11.3 Hz, C3′), 64.1, 64.0 (C5′), 59.7, 59.4 (d, JCP = 22.5, 21.0 Hz, OCH2CH2CN), 55.5 (OCH3), 43.9, 43.8 (d, JCP = 12.4, 12.1 Hz, CH(CH3)2), 25.1, 24.9, 24.7, 24.6 (d, JCP = 8.3, 7.3, 6.9, 9.0 Hz, CH(CH3)2), 24.2 (NHCOCH3), 20.9, 20.8 (OCOCH3), 20.5, 20.4 (d, JCP = 7.7 Hz, OCH2CH2CN); 31P NMR (202 MHz, acetone-d6): δ 152.68, 148.48; HRMS (ESI+), calcd for C44H53N7O10P [M+H]+ 870.3586, found 870.3594.
Synthetic procedure and characterization data for XyloNA oligonucleotides, experimental procedures for thermal (UV-) melting, circular dichroism (CD) and NMR spectroscopic studies are provided in the supplementary information.
NMR-derived structural restraints
Distance restraints were derived from NOESY spectra recorded with 50, 100, 150 and 300 ms mixing times. By using the computer-aided resonance assignment (CARA) program, (25) the cross-peaks in NOESY spectra were assigned and the cross-peak volumes were obtained by integration. Based on the NOE build-up curves, inter-proton distances were calculated. The calibration of NOE cross-relaxation rates was performed against the average of all the isolated H5-H6 cross-peaks as a reference with distance 2.45 Å. Upper and lower bounds were set to 20% of the calculated distances in the structure calculation. Hydrogen-bonding restraints were used as distance restraints in all structure calculation.
All xylose sugar rings were restrained to North (N)-type sugar puckering in structure calculations [dihedral restraints applied: H1′-C1′-C2′-H2′ (99.2±20°), H2′-C2′-C3′-O3′ (35±25°), O3′-C3′-C4′-H4′ (−157 ± 25°)].
Backbone torsion angles β, γ and ϵ were restrained as follows. The β torsion angles (C4′-C5′-O5′-P5′) were restrained to trans regions in the structure calculations as determined by experimental vicinal hetero-nuclear phosphorus proton (3JP5′-H5′, 3JP5′-H5″) couplings (Supplementary Table S1, in the supporting information) derived from the 31P coupled and decoupled DQF-COSY spectrum and based on a standard Karplus plot (26). For purine and pyrimidine nucleotides of XyloNA1 duplex, the β torsion angle regions 180 ± 75° and 200 ± 40° were restrained respectively. The γ torsion angles (C3′-C4′-C5′-O5′) were restrained from gauche – to anticlinal – region in the structure calculations as determined by vicinal proton-proton (3JH4′-H5′, 3JH4′-H5″) couplings (Supplementary Table S1) derived from the 31P decoupled DQF-COSY spectrum (26). For purine and pyrimidine nucleotides of XyloNA1 duplex, the γ torsion angles were restrained to regions 210±35° and 270±45° respectively. The ϵ torsion angles (P3′-O3′-C3′-C4′) were restrained to the trans region (ϵ = 180±60°) in all the residues of XyloNA1 based on the observed cross-peaks for long range phosphorus proton couplings (4JH4′-P3′) in 1H-31P HETCOR spectra (Supplementary Figure S1, in the supporting information), indicative for a W-shaped conformation of the P3′-O3′-C3′-C4′- H4′ fragment in the backbone.
No torsion angle restraints were applied on the backbone dihedral angles α, and ζ.
Structure calculations
All structure calculations were performed using the X-PLOR NIH program (27). The standard topology and parameter file (topallhdg.dna, parallhdg.dna) for DNA were adapted for Xylose residue by introducing inversion in the 3′-chiral improper angle. The torsion-angle molecular dynamics protocol used was largely identical to that proposed for the DNA duplex. A set of 100 structures was generated by torsion-angle molecular dynamics, starting from two extended strands with random structures by using all structural restraints. After the torsion angle molecular dynamics round, the majority of structures were converged to very similar structures with similar total energies and with no violations of the NOE and dihedral restraints. The 20 lowest energy structures were used for further refinement during a ‘gentle molecular dynamics’ round by using the X-PLOR NIH script refine_gentle.inp. The final refinement was started with 20 ps constant-temperature molecular dynamics simulation at 300 K (20 000 steps of 0.001 ps) which gradually introduces the van der Waals radii, and was followed by a 200-step conjugate-gradient energy minimization on an average structure of the last 10 of 20 ps molecular dynamics simulation. Torsion angles and helical parameters of the obtained solution structures were determined using CURVES 5.3 (28). Finally, visual representations of the molecules were obtained by using the UCSF Chimera 1.10rc program.
RESULTS
Synthesis of XyloNA building blocks
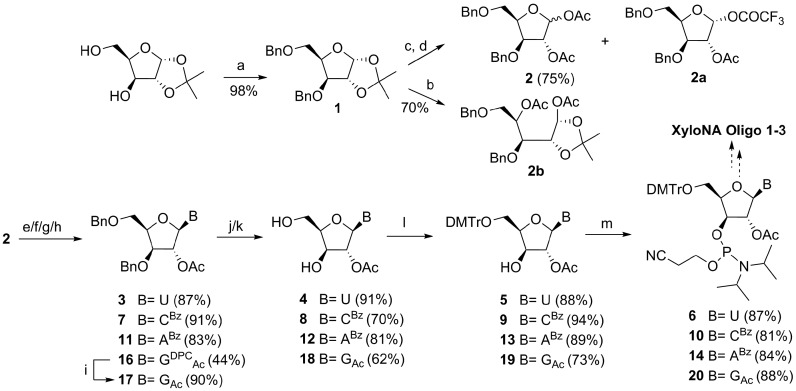
In the chemistry part, we have set out to implement an easy synthetic route (Scheme 1) for the XyloNA building blocks. The commercially available 1,2-acetonide of xylofuranose was benzylated to have compound 1. The subsequent steps to obtain protected glycone (2) are acid-catalyzed acetonide deprotection and acetylation. Since the one-pot transformation solely produced a ring-opened byproduct 2b, we have followed a two-step procedure to synthesize diacetate 2 in a reasonably good yield. Although the formation of the 1-fluoroacetate side product 2a could not be completely avoided in the latter procedure, its formation was reduced to some extent by decreasing the percentage of trifluoroacetic acid (TFA). Fluoroacetate sugar 2a could not afford the desired nucleoside 3 in a classical glycosylation reaction since it formed several decomposition products as observed by TLC. While standard Vorbrüggen-type glycosylation reaction was performed for pyrimidine nucleosides (3 and 7), for purines non-standard conditions were necessary. Thus, the reaction of N-benzoyladenine with the protected sugar 2 was performed in the presence of SnCl4 without silylation at room temperature, where the desired N9-β-isomer (11) was majorly formed as a kinetically controlled product. Nonetheless, depending on the reaction time (<2 h) a minor amount of N9-α-isomer (11a in SI) was formed and separated from the N9-β-stereoisomer 11 by column chromatography. For guanine, N2-acetyl-O6-diphenylcarbamoyl (DPC) protection (15 in SI) was employed for the regioselective glycosylation at the N9 position, (29) since glycosylation without DPC protection (15a in SI) resulted in the formation of an inseparable (1:1) mixture of N7 (17a in SI) and N9 (17) regioisomers. Then the DPC protecting group was selectively removed by treating with 90% aqueous TFA as it was no longer necessary and thought to procure complication in the subsequent steps. The removal of the benzyl (-Bn) groups was found to be difficult due to the sluggish reaction rate under different hydrogenation conditions, with potential problems like reduction of the pyrimidine ring and solvolysis of the acetyl and benzoyl groups were additionally encountered. These problems were efficiently addressed by using the Lewis-acid (BCl3) mediated debenzylation reaction. The debenzylated xylonucleosides were tritylated and phosphitylated to the corresponding amidites (6, 10, 14 and 20) using the standard reaction conditions (Scheme 1). Three fully modified oligonucleotide sequences (Table 1, XyloNA1–3) containing β-d-xylofuranosyl nucleotides were assembled on the solid support using synthesized phosphoramidites.
Scheme 1.
Schematic representation for the synthesis of four XyloNA nucleotide building blocks. Reagents and conditions: (a) BnBr, NaH, dry THF, 0°C–RT, 16 h; (b) AcOH, Ac2O, cat. H2SO4/CF3SO3H, 0°C–RT, 2 h; (c) 60% TFA in H2O, 0°C–RT, 4 h; (d) Ac2O, dry pyridine, 0°C–RT, 4 h; (e) for 3: uracil, BSA, CH3CN, 80°C, 1 h, then sugar 2, SnCl4, 50°C, 2 h; (f) for 7: N4-benzoylcytosine, BSA, CH3CN, 80°C, 45 min, then 2, SnCl4, 80°C, 1.5 h; (g) for 11: N6-benzoyladenine, 2, SnCl4, CH3CN, RT, 2 h; (h) for 16: N2-acetyl-O6-DPC guanine (15), BSA, DCE, 80°C, 30 min, then 2 in dry toluene, TMSOTf, 80°C, 1 h; (i) 90% TFA in H2O, RT, 2 h; (j) for 4: method A-Pd/C 10%, MeOH, H2, RT, 20 h or method B- BCl3, dry CH2Cl2, –78°C to –10°C, 3 h, then EtOH, –78/–40°C to RT, 30 min, 55%; for 8 and 12: method B, quenching with a mixture of EtOH and Et3N; (k) for 18: Pd/C 10%, Pd(OH)2/C 20%, H2O, MeOH, H2, RT, 72 h; (l) DMTr-Cl, dry pyridine, RT, 16 h; (m) NC(CH2)2OP(Cl)N(iPr)2, DIPEA, dry CH2Cl2, RT, 4 h.
Table 1. Thermal melting data for the evaluation of self-pairing and cross-pairing abilities of XyloNA/dXyloNA vs. natural DNA/RNA systems.
| Oligo. no. | Type of pairing | Sequence | Tm (°C) |
|---|---|---|---|
| ON-1 | XyloNA1:XyloNA1 | 5′-x(GUGUACAC)-3′ | 67 |
| 3′-(CACAUGUG)x-5′ | |||
| ON-2a | dXyloNA:dXyloNA | 5′-dx(GTGTACAC)-3′ | 57 |
| 3′-(CACATGTG)dx-5′ | |||
| ON-3 | RNA:RNA | 5′-r(GUGUACAC)-3′ | 41 |
| 3′-(CACAUGUG)r-5′ | |||
| ON-4a | DNA:DNA | 5′-d(GTGTACAC)-3′ | 26 |
| 3′-(CACATGTG)d-5′ | |||
| ON-5 | XyloNA2:RNA | 5′-x(UGCUACGC)-3′ | n.o. |
| 3′-(ACGAUGCG)r-5′ | |||
| ON-6 | XyloNA2:DNA | 5′-x(UGCUACGC)-3′ | n.o. |
| 3′-(ACGATGCG)d-5′ | |||
| ON-7 | RNA:RNA | 5′-r(UGCUACGC)-3′ | 50 |
| 3′-(ACGAUGCG)r-5′ | |||
| ON-8 | RNA:DNA | 5′-r(UGCUACGC)-3′ | n.c. |
| 3′-(ACGATGCG)d-5′ | |||
| ON-9 | XyloNA3:RNA | xU12:rA12 | 37–50 |
| ON-10 | XyloNA3:DNA | xU12:dA12 | 35–38 |
Melting points were determined in a buffer solution (pH 7.5) containing 0.1 M NaCl, 20 mM KH2PO4, and 0.1 mM Na2EDTA. For the self-complementary sequences (ON-1 to ON-4) 8 μM and for other sequences (ON-5 to ON-10) 4 μM concentration of each strand have been used.
[a] Ref. (24); n.o., no cross-pairing observed; n.c., Tm could not be calculated due to an unfavorable melting profile. Melting profiles for ON-3, ON-7, and ON-8 are provided in the SI (Supplementary Figure S2–S4).
Thermal (UV-) melting study
The fully-modified self-complementary XyloNA oligonucleotide (ON-1, Figure 1) was initially investigated for its capability in forming a duplex structure by thermal (UV-) melting study. This data indicates that XyloNA is able to form a self-paired duplex with a melting temperature (Tm) of 67°C and is associated with a typical sigmoidal cooperative melting profile (Figure 2A). In order to compare the thermal stability of duplexes possessing different backbones with the same sequence context (Table 1), we have synthesized and analyzed ON-1 to ON-4. The relative thermal stability of different duplexes decreased in the order (Table 1, ON1–4) of XyloNA:XyloNA>dXyloNA:dXyloNA>RNA:RNA>DNA:DNA. Notably, the xylose-based backbone modification (XyloNA/dXyloNA) significantly increased the duplex stability by 1.6–2.2-fold in comparison with the corresponding ribose-based DNA/RNA pair. In particular, the effect of the 2′-hydroxyl moiety is instrumental in increasing the duplex stability of XyloNA versus dXyloNA, as also seen in the case of natural RNA vs. DNA system. A non self-complementary fully-modified XyloNA oligonucleotide with mixed sequence context (Table 1, XyloNA2) was additionally synthesized and investigated for its potential to cross-pair with the natural DNA/RNA complements. These studies highlight that the XyloNA system could neither pair with DNA nor with RNA (Table 1 and Figure 2B, C), and thus demonstrating autonomous self-pairing property of XyloNA.
Figure 2.
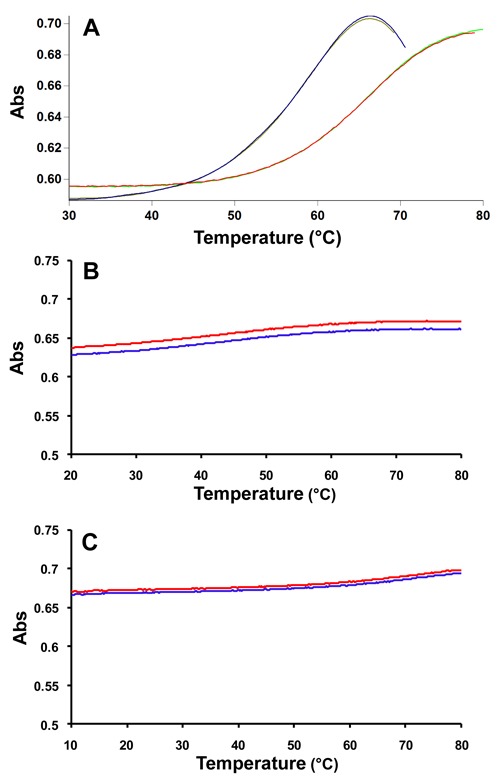
Thermal (UV-) melting profile of (A) XyloNA1 self-duplex (ON-1), sigmoidal up- and down-curves are shown along with first derivative plots for Tm determination; (B) XyloNA2:RNA hybrid (ON-5) and (C) XyloNA2:DNA hybrid (ON-6), up- and down-curves are shown; no sigmoidal behavior was observed and thus Tm could not be determined.
In order to investigate if XyloNA is universally orthogonal or not in pairing with other DNA/RNA sequence, we have also constructed a homo-pyrimidine sequence (XyloNA3, xU12) and evaluated its cross pairing ability with natural DNA (dA12) and RNA (rA12) complements. As shown in Figure 3, xU12 is indeed capable of pairing with both dA12 and rA12 strands (Table 1). In both the cases triplex formation could be confirmed based on the following observations (Figure 3): (i) a strong hysteresis effect (ΔTm = 13°C between heating and cooling curves) was observed (Figure 3) in normal melting buffer, in particular with rA12, which can be considered unusual for a duplex; (ii) these structures are highly stabilized in a Mg2+ buffer without any significant hysteresis effect; since the extent of cationic stabilization is usually more pronounced in a triplex than a duplex; (iii) moreover, a temperature-dependent hypochromism was observed in 284 and 295 nm (Figure 3) which represents the melting of Hoogsteen paired third-strand from a triplex structure (30–32). Melting temperatures of both the strands of these triplexes are identical and represent a single transition at 260 nm while melting of the third strand can only be visible at 284/295 nm. Melting curves for 1:0.5, 1:1, and 1:2 (dA12/rA12:xU12) mixing stoichiometries are appeared to be very similar (data not shown). In all these cases, a triplex is formed and when heating, it is directly converted to single strands. Therefore, a stable duplex could not be identified and this observation is in contrary with the corresponding natural triplexes (30). However, these results are in agreement with the previously reported data for analogous study on dXyloNA system. Initially, it was assumed that homo-pyrimidine dXyloNA (dxT12) forms a duplex with the complementary natural DNA/RNA; (15,33) however, later on Wengel et al. (20) confirmed it as a triplex. Additionally, Gottikh et al. reported that pyrimidine dXyloNA sequences are in general prone to form triplexes with natural purine DNA/RNA sequences having more or less similar stability to that of the natural counterparts (34–36). Notably, no duplex formation was observed in all of these reported studies.
Figure 3.
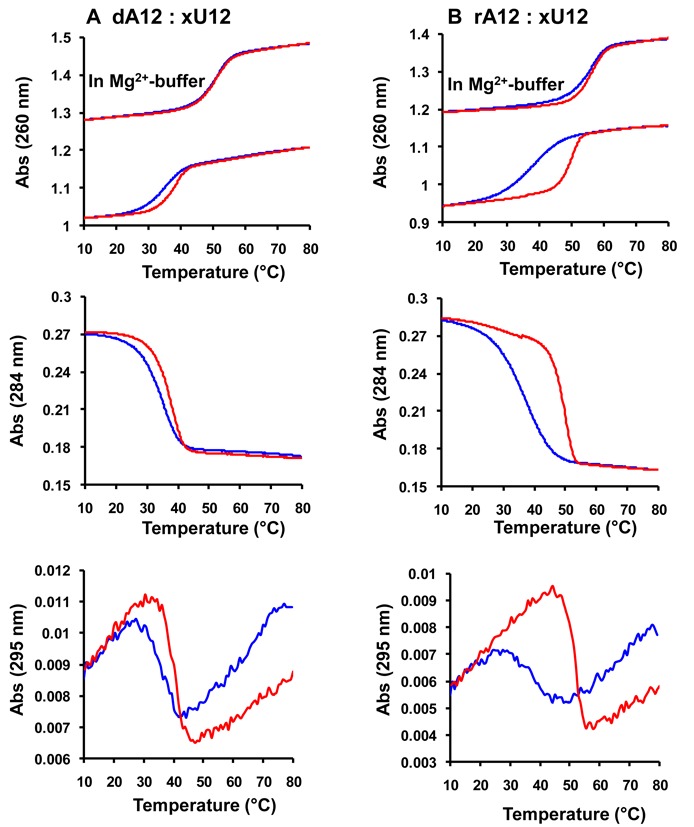
Thermal (UV-) melting profile for (A) pairing of XyloNA3 (xU12) with dA12; (B) pairing of XyloNA3 (xU12) with rA12. Sigmoidal melting curves are shown in different wavelengths (260, 284 and 295 nm). Heating- and cooling-curves are highlighted in red and blue respectively. Melting experiments were generally performed in 20 mM KH2PO4 buffer (pH 7.5) containing 0.1 M NaCl, and 0.1 mM Na2EDTA or in a Mg2+-buffer, as indicated above, containing 10 mM Tris·HCl (pH 7.5) and 50 mM MgCl2. Melting curves at 295 nm are from a faster heating-cooling experiments. Data presented here are only for a 1:1 mixing stoichiometry with 4 μM concentration of individual strand.
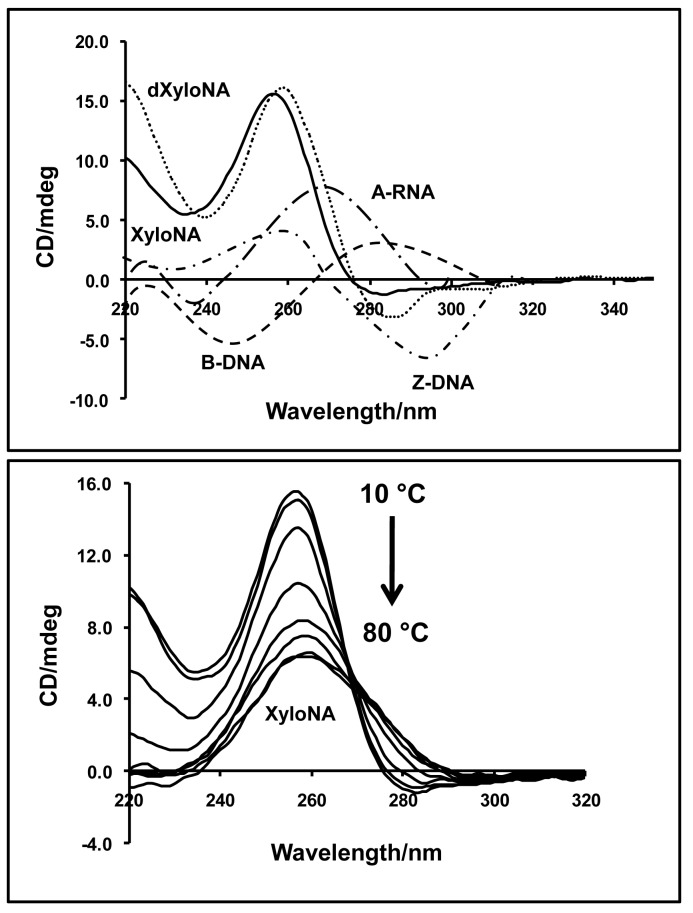
Circular dichroism (CD) study
To gain insights into the helical conformation and dynamics of double-stranded (ds) XyloNA (ON-1), we have performed circular dichroism (CD) studies. The CD spectra of XyloNA appear to be different from the known CD spectra of various helical nucleic acid structures i.e. A-, B- and Z-type helices, (1) as shown in Figure 4. However, the CD spectrum of XyloNA was found to be very similar to that of our previously studied analogous dXyloNA system in water (24). A characteristic positive Cotton (effect) band at 256 nm was observed in the CD spectrum, which showed a gradual melting effect with increasing temperature (Figure 4, bottom). These observations suggest that the XyloNA duplex has formed a slightly right-handed extended ladder-like structure, as characterized by NMR (described in the following section), and is in line with our previous observation for dXyloNA (24). The repeated appearance of this type of CD spectrum can be considered as a finger-print for the right-handed extended duplex structure (RE-type helix), and might facilitate in the future cataloging of non-natural nucleic acid conformation by CD spectroscopy. Unlike dXyloNA duplex (24), the CD spectra of XyloNA in saline buffer (Supplementary Figure S5) appeared to be identical with the spectra in pure water. This suggests that the presence of the 2′-hydroxyl group in the XyloNA backbone has resulted in a reduced conformational flexibility compared to the dXyloNA system. In our previous study, the latter has been identified to be conformationally malleable since it exists in two different helical conformations in saline aqueous/buffer solution (24).
Figure 4.
Top: Circular dichroism (CD) spectra of XyloNA and dXyloNA duplexes in water (12 μM, pH 7.0) at 10°C. For comparison, the CD spectra of the right-handed B-DNA, A-RNA and the left-handed Z-DNA duplexes are shown and illustrated. Bottom: Temperature dependent CD spectra of the XyloNA duplex showing a typical cooperative melting profile.
Structure elucidation by solution state NMR spectroscopy
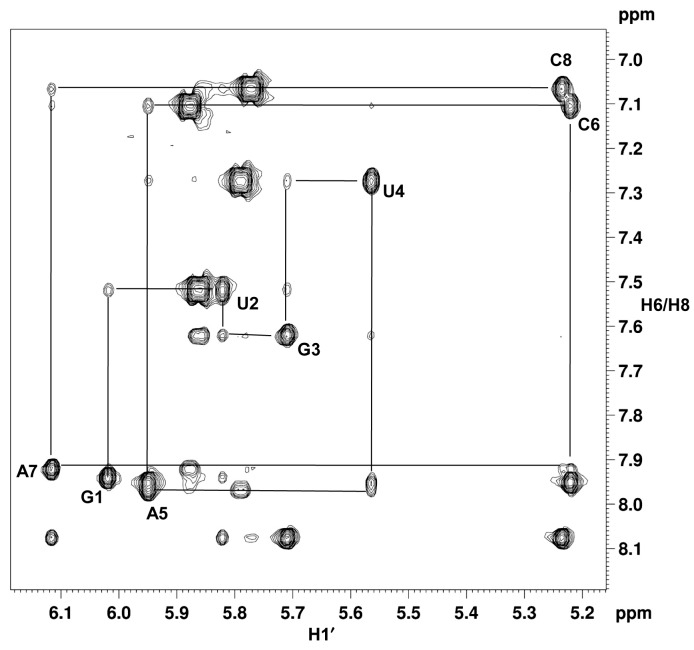
To examine the nature of the helical structure adopted by the dsXyloNA (ON-1), we have undertaken solution state NMR structural analysis. Appearance of a single set of resonances in the NMR spectra suggests the formation of a unique type of duplex conformation (Figure 5A and B). The duplex forms Watson–Crick type base-pairing with anti-parallel strand orientation as evidenced by the hydrogen-bonded imino-proton signals in the NMR spectrum recorded in H2O (Figure 5A and Supplementary Figure S6). All the xylose sugar rings are in N-type puckering as suggested by the appearance of singlet anomeric-proton peaks in the 1H NMR spectrum (Figure 5B). Unlike native DNA/RNA double helices, unique diagonal cross-strand (n+1 to n-1) NOE interactions between anomeric (H1′) protons were observed in XyloNA self-duplex, as depicted in Figure 5C. Although less pronounced, a classical NOE-walk (Figure 6) between aromatic and anomeric protons was observed, that is generally more prominent in the natural DNA/RNA helices. During simulations, the structures were converged (structure statistics in SI, Supplementary Table S2) to a family of structures (Figure 7B-D and E) with similar geometries and energies. The aforementioned diagonal NOE distance restraints were also reflected in the final structure as depicted in Figure 7A. Diagnostic helical parameters and backbone dihedral angles were calculated and compared (Table 2) with the natural DNA/RNA helices. Atomic coordinates for 20 refined structures, structural restraints, and assigned chemical shifts have been deposited in the RCSB Protein Data Bank with PDB ID 2N4J. Unlike the natural B-DNA and A-RNA helices, XyloNA duplex like dXyloNA (24) has adopted a slightly right-handed extended ladder-like structure (Figure 7B–D) with significantly impaired helicity. This has been reflected by the relatively low helical twist (10.7° versus 32.7°) and high base-pair rise (5.1 Ǻ versus 2.8 Ǻ). Both the major and minor grooves are widened to an extent that they are almost indistinguishable (Figure 7B and D). Nucleobases are highly inclined with respect to the backbone, as shown by a high inclination value (Table 2). Thus, it is predominantly forming a zipper like interstrand stacking interaction to provide duplex stabilization, in contrary to a dominant intrastrand stacking as found in native DNA/RNA helices. This interstrand interaction could be a cause for higher thermodynamic stability in XyloNA. High and dissimilar inclination together with largely reduced helicity of XyloNA compared with DNA/RNA might be the reasons for its inability in cross-pairing with the DNA/RNA complements (37). A comparison of the torsion angles (Table 2) between XyloNA and DNA/RNA suggests a cooperative adjustment in different dihedral angles across the sugar-phosphate backbone. The chiral inversion in the 3′-carbon center of the sugar ring in XyloNA with respect to DNA/RNA primarily changed the endo-cyclic torsion angle δ to synclinal− from gauche+ or anticlinal+. This leads to a chain of inversions in α, γ and ζ torsion angles from an almost mirror-image domain (opposite in sign and magnitude, shown in Table 2) while retaining both the torsion angles β and ϵ in the anti domain. These distinctive features allow a rationalization of the structural orthogonality of XyloNA in comparison with the natural DNA/RNA.
Figure 5.
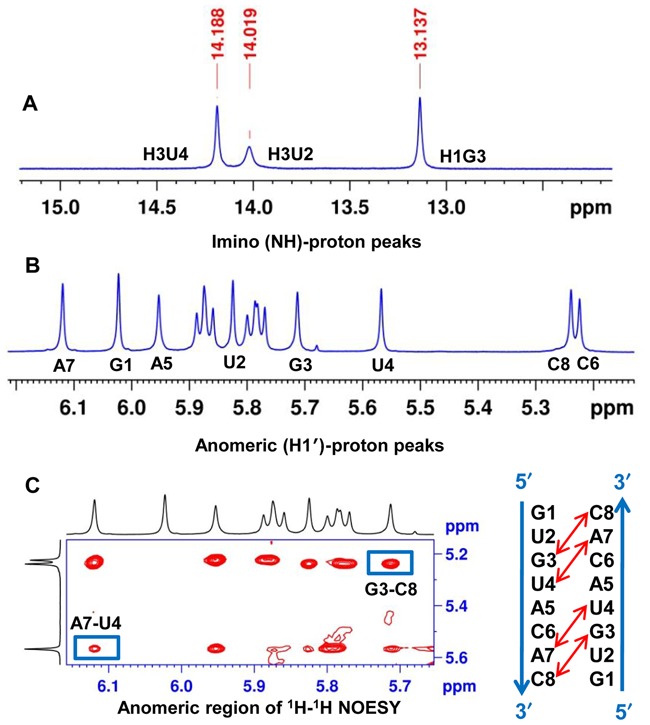
(A) Hydrogen-bonded imino-proton peaks of the XyloNA duplex (ON-1) representing Watson–Crick type base-pairing; (B) Appearance of eight singlet anomeric-proton peaks (G1 to C8) in the 1H NMR spectrum suggesting N-type puckering for the xylose sugar in XyloNA; (C) a unique diagonal cross-strand NOE interaction(s) between H1′ protons observed (boxed and indicated by red arrow) in the 2D NOESY spectrum of XyloNA duplex. All the above spectra were recorded in a 600 MHz spectrometer with a 1.5 mM XyloNA (ON-1) sample (pH 7.2) at 10°C.
Figure 6.
Expansion of the NOESY spectrum (recorded with 300 ms mixing time, in a 600 MHz spectrometer) of XyloNA (ON-1: 5′-G1UGUACAC8–3′) duplex (1.5 mM, pH 7.2) in D2O at 10°C, showing NOE walk between H6/H8-H1′ intra-residue and H1′(n)-H6/H8(n+1) inter-residue protons.
Figure 7.
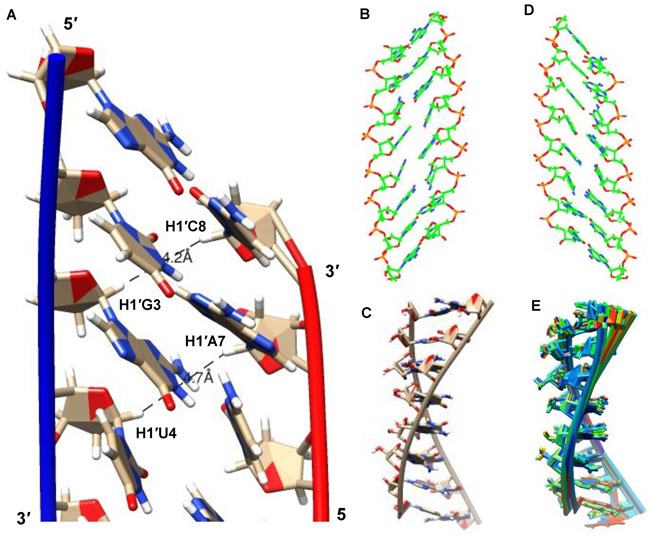
Structural representations of the XyloNA duplex as determined by NMR structural analysis, (A) a close view of the minimum energy closest-to-average structure showing base stacking interactions and the proximity of the diagonal anomeric protons as observed in the NOESY spectra, (B) view from the minor groove, (C) side view with highlighted backbone, (D) view from the major groove, and (E) an overlay of 20 refined structures representing convergence in the structure calculation.
Table 2. Torsion angles and key helical parameters of natural B-DNA and A-RNA duplexes are in comparison with the synthetic XyloNA and dXyloNA duplex structures derived by NMR structural analysisa.
| B-DNA | A-RNA | dXyloNA | XyloNA | |
|---|---|---|---|---|
| Torsion angles [°] | ||||
| α | −47 | −62 | 53 ± 18 | 47 ± 20 |
| β | −146 | 180 | −156 ± 14 | −130 ± 14 |
| γ | 36 | 47 | −122 ± 8 (A/G) | −100 ± 25 (A/G) |
| −64 ± 4 (C/T) | −91 ± 9 (C/U) | |||
| δ | 156 | 83 | −25 ± 3 | −21 ± 6 |
| ϵ | 155 | −152 | 146 ± 13 | 132 ± 16 |
| ζ | −95 | −73 | 75 ± 12 | 86 ± 21 |
| χ | −98 | −166 | −161 ± 6 | −165 ± 17 |
| Helical parameters | ||||
| Twist [°] | 36.0 | 32.7 | 2.7 ± 0.5 | 10.7 ± 1.7 |
| Inclination [°] | −5.9 | 15.8 | −52.2 ± 0.4 | −45. 2± 0.9 |
| Inclination-ηb [°] | 0 | −30 | 51 | 41 |
| Rise [Å] | 3.4 | 2.8 | 5.8 ± 0.06 | 5.1 ± 0.05 |
aStandard values for DNA, RNA and parameters for dXyloNA are from (24). Average and standard deviations (±) were calculated for the XyloNA duplex from the output data generated by using the CURVES program for an ensemble of 20 refined NMR structures. Backbone-base inclination angles (ηb) were calculated by using the ‘Inclination’ program, (37).
DISCUSSION
As outlined above, our endeavor in synthesizing and characterizing a xeno-nucleic acid analogue by appending the potentially prebiotic xylose sugar in the backbone has resulted in the creation of a novel xylonucleic acid (XyloNA) system with interesting structural and biophysical properties. Like RNA, XyloNA could have been formed and existed in the prebiotic era since xylose has shown to be a product in the formose reaction (38), the most plausible prebiotic route of sugar synthesis. This suggests a possibility that XyloNA/dXyloNA could have been selected by natural evolution as a genetic information system. There are, however, several structure-function properties of the xylose-based information system (as revealed by our studies) that make it less suitable for this purpose. Although XyloNA/dXyloNA could form base-paired duplexes, it lacks the very helical nature and thus the compactness possessed by the natural helices. Moreover, XyloNA/dXyloNA systems, by virtue of their predominant interstrand stacking property, are thermodynamically more stable than the natural ones. The higher duplex stability might be a plausible disadvantage since it would be rather difficult to unwind the helices for accessing the genetic information in the prebiotic and/or biotic polymerization. Previously it was emphasized that natural DNA/RNA structures are selected based on the principle of optimization rather than maximization of duplex stability and conformational flexibility (5). The highly inclined bases along with reduced helicality in XyloNA/dXyloNA strand make them ineligible to cross-pair with the natural nucleic acids in the general sequence context, as evidenced by the melting studies (Table 1). Since unable to cross-pair, it lacks communication with the RNA world, and therefore XyloNA could not be considered as a direct prebiotic RNA progenitor (39). These unfavorable properties together might have contributed and favored the selection of RNA rather than XyloNA as an informational polymer in the prebiotic nucleic acid evolution. Besides structure-pairing properties, there is a prebiotic generational issue for any proposed primordial RNA analogues. Conformational rigidity and altered helical geometry of XyloNA in comparison with RNA may potentially influence the prebiotic polymerization fidelity, linkage regio-chemistry (3′-5′ versus 2′-5′ isomers) and differential chemical stability (40) of the competing regioisomers that can form, as shown previously in the case of polymerization of different natural and non-natural systems (41–44). In the prebiotic synthesis of nucleotide building blocks, there can also be a different outcome with a xylose nucleoside containing 2′, 3′-trans diol functionality than a ribonucleoside with 2′,3′-cis diol functionality (22,45). These aspects remain to be studied to realize the full prebiotic potential of XyloNA.
Previously, six-membered sugar-modified homo-DNA/pyranosyl-RNA (p-RNA) (8) and acyclic glycol nucleic acid (GNA) (46) have also adopted a similar extended ladder-like structure. Interestingly, we could here demonstrate that an extended helical structure could also be derived by introducing a pentofuranosyl (Xylo) sugar. Xylose nucleic acid (XyloNA) is found to be conformationally less flexible (as seen in RNA compared to DNA), due to the 2′-hydroxyl effect, than the previously studied analogous dXyloNA counterpart (24). In contrary, the structural compactness and flexibility of XyloNA and dXyloNA are not as favorable as in the case of natural (DNA/RNA) systems. DNA (usually B-form) and RNA (A-form) can shape into different types of helical conformations by adopting different sugar pucker (1) (as shown in Figure 1), as well as by essentially changing the important endo-cyclic δ-torsion angle in the backbone. A similar structural polymorphism could not be observed in the xylose-based nucleic acids due to an identical δ value and sugar pucker for both XyloNA and dXyloNA, and thereby limiting their conformational diversity. Torsion angle δ is found here to be crucial in determining the helical twist and backbone flexibility versus rigidity as previously suggested for DNA/RNA (47). Chiral inversion in the C3′ of furanose sugar in XyloNA/dXyloNA is constraining the sugar conformation and δ torsion angle in comparison with DNA/RNA, as a result forming a ladder-like structure with increased duplex stability and to some extent rigidifying the backbone conformation (16). Although dXyloNA was previously shown to be structurally somewhat malleable in its duplex conformation than natural DNA duplex (23,24). In DNA/RNA, duplex stability and backbone flexibility seem to have optimally selected and supported by their very unique double helical structures. Considering the favorable properties that are required for an efficient storage and transfer of genetic information, natural DNA/RNA structure is unique and superior to that of xylose-based (XyloNA/dXyloNA) nucleic acids.
Although xylonucleic acid could not supersede the structure and pairing properties of natural nucleic acids, its backbone could support essential self-pairing ability to form double helix with slightly right-handed extended conformation. It is anticipated that XyloNA should be chemically and enzymatically more stable than RNA due to an inverted C-3′ orientation of the phosphate linkage, and thus making the phosphodiester moiety inaccessible by the neighboring 2′-hydroxyl nucleophile. By virtue of its unique structure, it could demonstrate novel orthogonal (inability in pairing with DNA/RNA) self-pairing properties. Triplex formation with purine DNA/RNA (as shown in melting study) is a special case with pyrimidine XyloNA/dXyloNA sequences. From an applicability point of view, if XyloNA/dXyloNA is not a universal orthogonal system, most importantly XyloNA/dXyloNA (as shown for XyloNA2) should be an orthogonal information system in a mixed sequence context which is mostly the case with natural unbiased genomic sequences. In general, xylose or deoxyxylose nucleic acid by virtue of their inability to cross talk with the natural nucleic acids could in principle be used for construction of an orthogonal episome for exploration in synthetic biology (12–14). Along this line, the main bottleneck for implementing such a XyloNA-based technology is the discovery of a XyloNA polymerase which can efficiently polymerize XyloNA nucleotide precursors. In our previous study, dXylo nucleoside triphosphates were shown to be partially recognized by some mutant DNA polymerases, like for example Taq mutant M1, for up to four nucleotides incorporation (24). Recently, polymerases have been evolved that can copy or transfer the genetic information from the non-natural nucleic acid backbones, those mimicking the natural helical structure (2,3). Polymerase evolution for XyloNA like deviant nucleic acid system poses a major challenge in the hands of evolutionary molecular biologist.
CONCLUSION
Studies on artificial nucleic acid analogues from the structural neighborhood of DNA/RNA are inspired by motivations like gaining knowledge on the structural uniqueness of DNA/RNA, and understanding the primordial ancestors that are lost in the evolutionary past. Among them, those with novel properties could be useful in synthetic biology. Herein, we have investigated such an isomeric RNA analogue, XyloNA derived from prebiotic xylose sugar. A convergent synthetic route was implemented to access XyloNA building blocks. Structural and pairing properties of the synthesized XyloNAs are elucidated by employing biophysical techniques. The helical form of DNA/RNA is found to be structurally unique in a sense that they possess optimal duplex stability and conformational flexibility in comparison with that of XyloNA. Unlike DNA/RNA, XyloNA adopts an altered extended helical geometry, and thus unable to cross-pair with the unbiased sequences of DNA/RNA. Based on the preliminary structure-function understanding available so far, it seems that RNA may have gained an evolutionary advantage over XyloNA. On the other hand, the autonomous pairing potential of XyloNA could further be exploited as an orthogonal information system in synthetic biology.
Supplementary Material
Acknowledgments
Dr Mohitosh Maiti is a postdoctoral fellow (1200113N) of FWO Vlaanderen, Belgium. Technical support by Mr Guy Schepers is gratefully acknowledged. We thank Dr Puneet Srivastava for scientific discussion and Chantal Biernaux for secretarial assistance. This article is dedicated to Professor A. Eschenmoser on the occasion of his 90th birthday.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council under the European Union's Seventh Framework Programme [FP7/2007–2013] /ERC Grant agreement [ERC-2012-ADG_20120216/320683]; FWO Vlaanderen, Belgium (G078014N to P.H., O6260 to MoM); Mass spectrometry was made possible by the support of the Hercules Foundation of the Flemish Government [20100225-7]. Funding for open access charge: FWO Vlaanderen, Belgium [O6260 of MoM].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bloomfield V.A., Crothers D.M., Tinoco I. Nucleic Acids: Structures, Properties, and Functions. University Science Books; 2000. [Google Scholar]
- 2.Pinheiro V.B., Taylor A.I., Cozens C., Abramov M., Renders M., Zhang S., Chaput J.C., Wengel J., Peak-Chew S.Y., McLaughlin S.H., et al. Synthetic genetic polymers capable of heredity and evolution. Science. 2012;336:341–344. doi: 10.1126/science.1217622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor A.I., Pinheiro V.B., Smola M.J., Morgunov A.S., Peak-Chew S., Cozens C., Weeks K.M., Herdewijn P., Holliger P. Catalysts from synthetic genetic polymers. Nature. 2015;518:427–430. doi: 10.1038/nature13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egli M., Herdewijn P., editors. Chemistry and Biology of Artificial Nucleic Acids. Weinheim: VHCA, Zurich & WILEY-VCH Verlag; 2012. [Google Scholar]
- 5.Eschenmoser A. Chemical etiology of nucleic acid structure. Science. 1999;284:2118–2124. doi: 10.1126/science.284.5423.2118. [DOI] [PubMed] [Google Scholar]
- 6.Beier M., Reck F., Wagner T., Krishnamurthy R., Eschenmoser A. Chemical etiology of nucleic acid structure: comparing pentopyranosyl-(2′–>4′) oligonucleotides with RNA. Science. 1999;283:699–703. doi: 10.1126/science.283.5402.699. [DOI] [PubMed] [Google Scholar]
- 7.Schoning K., Scholz P., Guntha S., Wu X., Krishnamurthy R., Eschenmoser A. Chemical etiology of nucleic acid structure: the α-threofuranosyl-(3′–>2′) oligonucleotide system. Science. 2000;290:1347–1351. doi: 10.1126/science.290.5495.1347. [DOI] [PubMed] [Google Scholar]
- 8.Ebert M.O., Jaun B. Oligonucleotides with sugars other than ribo- and 2′-deoxyribofuranose in the backbone: the solution structures determined by NMR in the context of the ‘Etiology of nucleic acids’ project of Albert Eschenmoser. Chem. Biodivers. 2010;7:2103–2128. doi: 10.1002/cbdv.201000096. [DOI] [PubMed] [Google Scholar]
- 9.Yu H., Zhang S., Chaput J.C. Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat. Chem. 2012;4:183–187. doi: 10.1038/nchem.1241. [DOI] [PubMed] [Google Scholar]
- 10.Egholm M., Buchardt O., Christensen L., Behrens C., Freier S.M., Driver D.A., Berg R.H., Kim S.K., Norden B., Nielsen P.E. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 11.Nelson K.E., Levy M., Miller S.L. Peptide nucleic acids rather than RNA may have been the first genetic molecule. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3868–3871. doi: 10.1073/pnas.97.8.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herdewijn P., Marliere P. Toward safe genetically modified organisms through the chemical diversification of nucleic acids. Chem. Biodivers. 2009;6:791–808. doi: 10.1002/cbdv.200900083. [DOI] [PubMed] [Google Scholar]
- 13.Marliere P. The farther, the safer: a manifesto for securely navigating synthetic species away from the old living world. Syst. Synth. Biol. 2009;3:77–84. doi: 10.1007/s11693-009-9040-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt M. Xenobiology: a new form of life as the ultimate biosafety tool. BioEssays. 2010;32:322–331. doi: 10.1002/bies.200900147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosemeyer H., Seela F. 1-(2′-Deoxy-β-D-xylofuranosyl)thymine Building Blocks for Solid-Phase Synthesis and Properties of Oligo(2′-Deoxyxylonucleotides) Helv. Chim. Acta. 1991;74:748–760. [Google Scholar]
- 16.Rosemeyer H., Seela F. Xylose-DNA: Sequence-Dependent Duplex Stability as Function of Backbone Configuration. Nucleosides Nucleotides. 1995;14:1041–1045. [Google Scholar]
- 17.Seela F., Heckel M., Rosemeyer H. Xylose-DNA Containing the Four Natural Bases. Helv. Chim. Acta. 1996;79:1451–1461. [Google Scholar]
- 18.Schoppe A., Hinz H.J., Rosemeyer H., Seela F. Xylose-DNA: comparison of the thermodynamic stability of oligo(2′-deoxyxylonucleotide) and oligo(2′-deoxyribonucleotide) duplexes. Eur. J. Biochem. 1996;239:33–41. doi: 10.1111/j.1432-1033.1996.0033u.x. [DOI] [PubMed] [Google Scholar]
- 19.Rajwanshi V.K., Hakansson A.E., Dahl B.M., Wengel J. LNA stereoisomers: xylo-LNA (β-D-xylo configured locked nucleic acid) and α-L-LNA (α-L-ribo configured locked nucleic acid) Chem. Commun. 1999:1395–1396. [Google Scholar]
- 20.Poopeiko N.E., Juhl M., Vester B., Sorensen M.D., Wengel J. Xylo-Configured oligonucleotides (XNA, xylo nucleic acid): synthesis of conformationally restricted derivatives and hybridization towards DNA and RNA complements. Bioorg. Med. Chem. Lett. 2003;13:2285–2290. doi: 10.1016/s0960-894x(03)00441-4. [DOI] [PubMed] [Google Scholar]
- 21.Ravindra Babu B., Raunak, Poopeiko N.E., Juhl M., Bond A.D., Parmar V.S., Wengel J. XNA (xylo Nucleic Acid): A Summary and New Derivatives. Eur. J. Org. Chem. 2005;2005:2297–2321. [Google Scholar]
- 22.Seela F., Wörner K., Rosemeyer H. 1-(2′-Deoxy-β-D-xylofuranosyl)cytosine: Base pairing of oligonucleotides with a configurationally altered sugar-phosphate backbone. Helv. Chim. Acta. 1994;77:883–896. [Google Scholar]
- 23.Ramaswamy A., Froeyen M., Herdewijn P., Ceulemans A. Helical structure of xylose-DNA. J. Am. Chem. Soc. 2010;132:587–595. doi: 10.1021/ja9065877. [DOI] [PubMed] [Google Scholar]
- 24.Maiti M., Siegmund V., Abramov M., Lescrinier E., Rosemeyer H., Froeyen M., Ramaswamy A., Ceulemans A., Marx A., Herdewijn P. Solution structure and conformational dynamics of deoxyxylonucleic acids (dXNA): an orthogonal nucleic acid candidate. Chem. Eur. J. 2012;18:869–879. doi: 10.1002/chem.201102509. [DOI] [PubMed] [Google Scholar]
- 25.Keller R. The Computer Aided Resonance Assignment Tutorial. 1st ed. Cantina, Goldau: 2004. [Google Scholar]
- 26.Wijmenga S.S., van Buuren B.N.M. The use of NMR methods for conformational studies of nucleic acids. Prog. Nucl. Magn. Reson. Spectrosc. 1998;32:287–387. [Google Scholar]
- 27.Schwieters C.D., Kuszewski J.J., Tjandra N., Clore G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 28.Ravishanker G., Swaminathan S., Beveridge D.L., Lavery R., Sklenar H. Conformational and helicoidal analysis of 30 PS of molecular dynamics on the d(CGCGAATTCGCG) double helix: ‘curves’, dials and windows. J. Biomol. Struct. Dyn. 1989;6:669–699. doi: 10.1080/07391102.1989.10507729. [DOI] [PubMed] [Google Scholar]
- 29.Robins M.J., Zou R., Guo Z., Wnuk S.F. Nucleic acid related compounds. 93. A solution for the historic problem of regioselective sugar-base coupling to produce 9-glycosylguanines or 7-glycosylguanines. J. Org. Chem. 1996;61:9207–9212. [Google Scholar]
- 30.Pilch D.S., Levenson C., Shafer R.H. Structural analysis of the (dA)10.2(dT)10 triple helix. Proc. Natl. Acad. Sci. U.S.A. 1990;87:1942–1946. doi: 10.1073/pnas.87.5.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mergny J.L., Li J., Lacroix L., Amrane S., Chaires J.B. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyoshi D., Nakamura K., Tateishi-Karimata H., Ohmichi T., Sugimoto N. Hydration of Watson-Crick base pairs and dehydration of Hoogsteen base pairs inducing structural polymorphism under molecular crowding conditions. J. Am. Chem. Soc. 2009;131:3522–3531. doi: 10.1021/ja805972a. [DOI] [PubMed] [Google Scholar]
- 33.Alekseev Y.I., Gottikh M.B., Romanova E.A., Shabarova Z.A. Interaction of Escherichia coli RNase H with modified oligonucleotide duplexes .II. Duplexes formed by oligonucleotides consisting of (1-β-D-2′-deoxy-threo-pentofuranosyl)thymine residues. Mol. Biol. 1996;30:206–208. [PubMed] [Google Scholar]
- 34.Gottikh M.B., Alekseev Y.I., Perminov A.V., Pinskaya M.D., Shabarova Z.A. Oligodeoxyxylonucleotides Form Stable Triplexes with Single-Stranded DNA. Nucleosides Nucleotides. 1999;18:1625–1627. [Google Scholar]
- 35.Ivanov S.A., Alekseev Y.I., Gottikh M.B. Pyrimidine oligodeoxyxylonucleotides form triplexes with purine DNA in neutral medium. Mol. Biol. 2002;36:160–170. [PubMed] [Google Scholar]
- 36.Ivanov S., Alekseev Y., Bertrand J.R., Malvy C., Gottikh M.B. Formation of stable triplexes between purine RNA and pyrimidine oligodeoxyxylonucleotides. Nucleic Acids Res. 2003;31:4256–4263. doi: 10.1093/nar/gkg443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pallan P.S., Lubini P., Bolli M., Egli M. Backbone-base inclination as a fundamental determinant of nucleic acid self- and cross-pairing. Nucleic Acids Res. 2007;35:6611–6624. doi: 10.1093/nar/gkm612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Müller D., Pitsch S., Kittaka A., Wagner E., Wintner C.E., Eschenmoser A., Ohlofjgewidmet G. Chemie von α-Aminonitrilen. Aldomerisierung von Glycolaldehyd-phosphat zu racemischen Hexose-2,4,6-triphosphaten und (in Gegenwart von Formaldehyd) racemischen Pentose-2,4-diphosphaten: rac-Allose-2,4,6-triphosphat und rac-Ribose-2,4-diphosphat sind die Reaktionshauptprodukte. Helv. Chim. Acta. 1990;73:1410–1468. [Google Scholar]
- 39.Orgel L.E. Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. Biol. 2004;39:99–123. doi: 10.1080/10409230490460765. [DOI] [PubMed] [Google Scholar]
- 40.Usher D.A. RNA double helix and the evolution of the 3′,5′ linkage. Nat. New Biol. 1972;235:207–208. doi: 10.1038/newbio235207a0. [DOI] [PubMed] [Google Scholar]
- 41.Rohatgi R., Bartel D.P., Szostak J.W. Nonenzymatic, template-directed ligation of oligoribonucleotides is highly regioselective for the formation of 3′-5′ phosphodiester bonds. J. Am. Chem. Soc. 1996;118:3340–3344. doi: 10.1021/ja9537134. [DOI] [PubMed] [Google Scholar]
- 42.Kozlov I.A., Politis P.K., Van Aerschot A., Busson R., Herdewijn P., Orgel L.E. Nonenzymatic synthesis of RNA and DNA oligomers on hexitol nucleic acid templates: the importance of the A structure. J. Am. Chem. Soc. 1999;121:2653–2656. doi: 10.1021/ja983958r. [DOI] [PubMed] [Google Scholar]
- 43.Schrum J.P., Ricardo A., Krishnamurthy M., Blain J.C., Szostak J.W. Efficient and rapid template-directed nucleic acid copying using 2′-amino-2′,3′-dideoxyribonucleoside-5′-phosphorimidazolide monomers. J. Am. Chem. Soc. 2009;131:14560–14570. doi: 10.1021/ja906557v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schrum J.P., Zhu T.F., Szostak J.W. The origins of cellular life. Cold Spring Harb. Perspect. Biol. 2010;2:a002212. doi: 10.1101/cshperspect.a002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banfalvi G. Why ribose was selected as the sugar component of nucleic acids. DNA Cell Biol. 2006;25:189–196. doi: 10.1089/dna.2006.25.189. [DOI] [PubMed] [Google Scholar]
- 46.Meggers E., Zhang L. Synthesis and properties of the simplified nucleic acid glycol nucleic acid. Acc. Chem. Res. 2010;43:1092–1102. doi: 10.1021/ar900292q. [DOI] [PubMed] [Google Scholar]
- 47.Eschenmoser A., Dobler M. Warum Pentose- und nicht Hexose-Nucleinsäuren?? Teil I. Einleitung und Problemstellung, Konformationsanalyse für Oligonucleotid-Ketten aus 2′,3′-Dideoxyglucopyranosyl-Bausteinen (‘Homo-DNS’) sowie Betrachtungen zur Konformation von A- und B-DNS. Helv. Chim. Acta. 1992;75:218–259. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.