Abstract
Herein we employ Myh11-CreERT2 ROSA floxed STOP eYFP Apoe−/− smooth muscle cell (SMC) lineage tracing mice to show that traditional methods for detecting SMCs based on immuno-staining fail to detect > 80% of SMC-derived cells within advanced atherosclerotic lesions. These unidentified SMC-derived cells exhibit phenotypes of other cell lineages including macrophages (Mϕs), and mesenchymal stem cells (MSCs). SMC-specific conditional knockout (KO) of Krüppel-like factor 4 (KLF4) resulted in reduced numbers of SMC-derived MSC-, and Mϕ-like cells, marked reductions in lesion size, and increases in multiple indices of plaque stability, including an increase in fibrous cap thickness. Results of in vivo KLF4 ChIP-Seq analyses, and studies in cultured SMC treated with cholesterol identified > 800 KLF4 target genes including many that regulate pro-inflammatory responses of SMC. Results indicate that the contribution of SMCs within atherosclerotic plaques has been greatly underestimated, and that KLF4-dependent transitions in SMC phenotype are critical in lesion pathogenesis.
Keywords: smooth muscle cell, krüppel-like factor 4, Mϕ, phenotypic transitions, atherosclerosis, plaque pathogenesis
Introduction
Atherosclerosis is a disease of chronic inflammation and is the leading cause of morbidity and mortality worldwide. There is general consensus that the majority of coronary syndromes are the result of rupture of unstable plaques and associated thrombotic events1-3. Plaque instability has been associated with disruption of the fibrous cap, an atheroprotective layer of smooth muscle α-actin (ACTA2) positive cells that cover the atherosclerotic plaque3-7; large numbers of cells positive for Mϕ markers such as LGALS3; and the presence of a large foam cell laden necrotic core within the plaque2-4, 8. Indeed, these data are interpreted as evidence that plaques which contain a high ratio of Mϕs relative to SMCs are less stable, particularly those that have a thin ACTA2+ fibrous cap presumed to be composed primarily of phenotypically modulated SMCs9, 10.
Although ACTA2+ and LGALS3+ cells are assumed to be of SMC and myeloid lineage respectively, there is extensive ambiguity about cell lineage origin within atherosclerotic plaques11. These ambiguities were originally based on in vitro studies showing that SMCs down-regulate SMC markers and activate Mϕ markers following cholesterol loading12; and that Mϕs activate SMC genes following treatment with factors known to be present within lesions including thrombin13. However, the most compelling evidence that SMCs and Mϕs are being misidentified within human advanced coronary lesions comes from studies of cross gender bone marrow transplant subjects showing that > 10% of ACTA2+ cells within lesions are of hematopoietic stem cell (HSC) and not SMC origin14. Consistent with these human data, studies by Iwata et al.15 demonstrated that a substantial fraction of cells within lesions of Apoe−/− mice that express SMC markers, including ACTA2 but not MYH11, are of HSC not SMC origin.
Conversely, there is also extensive evidence suggesting that many SMC-derived cells within advanced lesions of Apoe−/− mice lack detectible expression of conventional SMC markers like ACTA2 (ref. 16), and/or activate expression of Mϕ markers17. This includes in vivo studies from our lab showing large numbers or ACTA2−, MYH11−, and TAGLN− cells within advanced lesions of Apoe−/− Western-diet fed mice that retain expression of a mutant Tagln lacZ transgene resistant to down-regulation compared to the wild type transgene16. Unfortunately, these studies are not definitive since we could not rule out the possibility that non-SMCs present within lesions may activate the mutant G/C repressor mutant Tagln lacZ transgene.
As such, despite decades of atherosclerosis research, we still do not know which cells within lesions are SMC-derived and to what extent they contribute to lesion pathogenesis. Recent studies by Feil et al.17 using a Tagln ERT2Cre LacZ SMC lineage tracing Apoe−/− mouse model provide evidence that SMC-derived cells within advanced lesions activate some Mϕ markers including LGALS3 and CD68. Unfortunately, as highlighted in an editorial on this paper by Swirski et al.18 the labeling efficiency of SMC in these studies was only 11%, precluding them from determining what fraction of Mϕ-like cells within lesions are derived from SMC and most importantly, how these cells might contribute to lesion pathogenesis. A recent study19 highlights the magnitude of the “SMC/Mϕ mis-identification problem” with respect to understanding human disease in that they showed that 50% of foam cells within advanced human coronary artery lesions express the SMC marker ACTA2. However, the majority of these ACTA2+ foam cells also expressed the Mϕ marker CD68 and represented 40% of all CD68+ lesion cells. Given clear evidence that Mϕs can activate SMC markers and vice versa it is unclear if these cells are derived from SMCs, Mϕs, or other cell type.
The preceding observations clearly establish that there is major ambiguity in identifying which cells within atherosclerotic lesions are SMC- versus Mϕ-derived. However, in the final analysis, the most critical questions are: What regulates phenotypic transitions of SMCs, Mϕs, and other cell types within lesions? What is the function of these cells? How do these phenotypic transitions impact overall disease pathogenesis? To begin to address these questions, we developed Apoe−/− mice where we could simultaneously lineage trace SMCs and study the effects of SMC-specific conditional KO of the stem cell pluripotency gene, KLF4, which we and others have previously shown plays a key role in regulating phenotypic transitions of SMC in vivo during development20 and following carotid ligation injury21, as well as in vitro in cultured SMCs treated with PDGFBB22, 23, PDGFDD24, and oxidized phospholipids25.
Results
Most atherosclerotic plaque SMCs are not identified by ACTA2
SMCs are distinguished from other cell types by expression of a unique repertoire of genes including Acta2, Tagln, and Myh11, which are coordinately down-regulated, at least in vitro, during SMC phenotypic switching such that they may be undetectable using traditional immunohistochemical staining methods11, 26. Therefore, in order to rigorously analyze the overall contributions of SMCs to lesion pathogenesis, we utilized a Myh11-CreERT2 ROSA floxed STOP eYFP Apoe−/− (SMC YFP+/+ Apoe−/−) mouse model previously described27 which labels > 95% of medial SMCs within large arteries (Supplementary Fig. 1a). To ensure the fidelity and SMC specificity of this lineage tracing model we completed a number of further validation studies beyond those shown in our previous studies27 including showing: 1) SMC-specific YFP labeling within all tissue specimens examined with no detectable expression of the eYFP indicator gene in the absence of tamoxifen (Supplementary Figs. 1a, 1b, and 2a); 2) no detectable YFP+ cells within blood or bone marrow preparations based on flow cytometry (Supplementary Fig. 2a); 3) no evidence of YFP+ cells within lesions of Western diet fed SMC YFP+/+ Apoe−/− mice not given tamoxifen (Supplementary Fig. 2b); 4) no detectable YFP+ cells in the blood of SMC YFP+/+ Apoe−/− mice given a high fat diet for 18 weeks (Supplementary Fig. 2c); and 5) YFP+ labeling of approximately 60% of freshly enzymatically dissociated cells from the ascending, descending thoracic aorta plus the abdominal aorta to the iliac bifurcation based on flow cytometry (Supplementary Fig. 2d). Since Cre excision is permanent, this SMC lineage tracing model system provides permanently lineage tagging of virtually all mature (MYH11+) arterial SMCs that exist at the time of tamoxifen injection and subsequent determination of what they or their progeny become irrespective of continued expression of ACTA2, MYH11, or other SMC-marker genes.
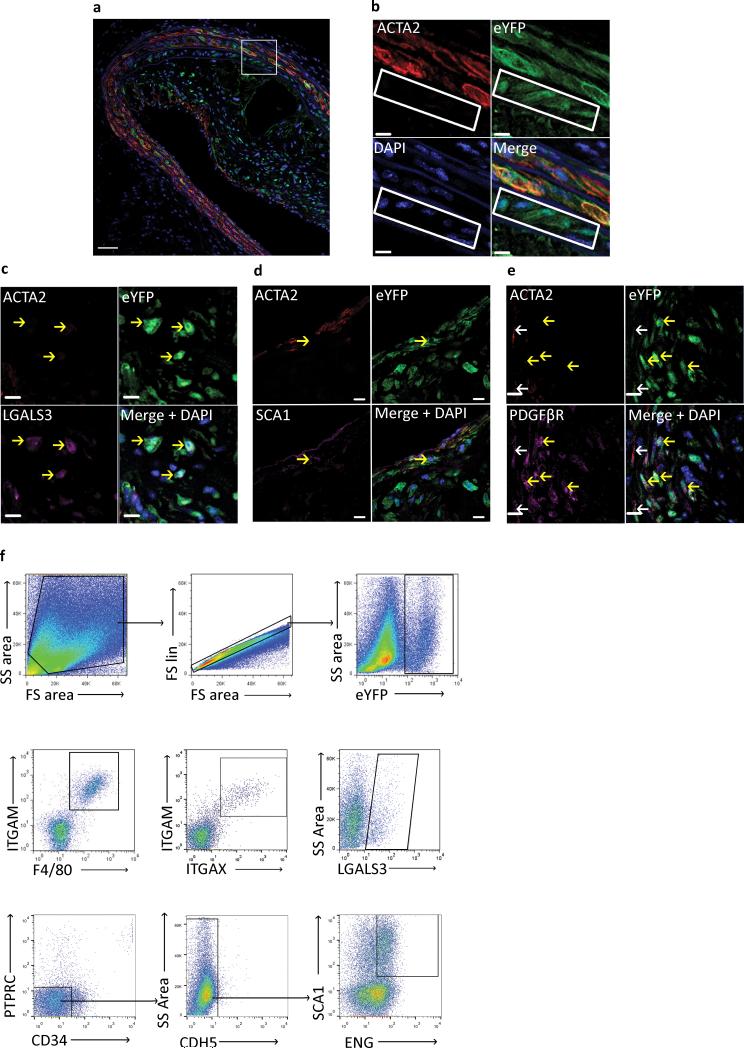
We harvested brachiocephalic arteries (BCAs) from SMC YFP+/+ Apoe−/− mice and immunostained them for YFP, ACTA2, and LGALS3 following 18 weeks of Western diet. Confocal microscopy z-stacks of the collected tissues were acquired and analyzed to accurately profile individual cells (Fig. 1a-e). Remarkably, ~82% of SMCs within atherosclerotic lesions (YFP+DAPI+ cells) were ACTA2 negative (Fig. 1a, Table 1), indicating that the majority of SMCs within the lesion cannot be identified using traditional SMC markers. Results also showed that phenotypically modulated SMCs (YFP+ACTA2−/DAPI+) comprised approximately 30% of the total cells within lesions (Fig. 1a, Table 1).
Figure 1. SMC lineage tracing provides evidence for large populations of phenotypically modulated SMCs within BCAs of 18 Week WD fed SMC eYFP+/+ Apoe−/− mice.
Immunofluorescence staining of representative BCAs from 18 week Western diet fed SMC eYFP+/+ Apoe−/− mice. Results show that differentiated SMCs present at the time of tamoxifen injection (YFP+) subsequently give rise to multiple phenotypes including: (a, b) phenotypically modulated SMCs (YFP+ACTA2− cells highlighted in white rectangle), (c) Mϕ-like SMCs (YFP+ACTA2−LGALS3+), (d) mesenchymal stem cell-like SMCs (YFP+ACTA2−SCA1+), and (e) MF-like cells (YFP+ACTA2+PDGFβR+). Scale bar represents 50μm (a) and 10μm (b-e). (c-e) Yellow arrows indicate de-differentiated (YFP+/ACTA2−) SMCs, white arrows indicate differentiated (YFP+/ACTA2+) SMCs. Samples were either fixed and embedded in paraffin (a-c, e) or Neg40 (d). (f) Flow cytometry of single cell suspension from 18 Week WD fed SMC eYFP+/+ Apoe−/− mouse aortas to further assess phenotypically modulated SMC populations. The gating strategy included: forward versus side scatter gate; a singlets gate; and YFP+ cell gate. Sub-populations of YFP+ cells were found to be double positive for ITGAM (CD11b) and F4/80, ITGAM (CD11b) and ITGAX (CD11c), LGALS3 (mac2), and double positive for SCA1 and ENG after going through lin− gating, n = 6 animals.
SMCs within atherosclerotic plaques express markers of Mϕs, MSCs, and MFs
When characterizing these phenotypically modulated SMCs we discovered YFP+ populations within lesions that expressed markers of Mϕs (LGALS3) (Fig. 1c), MSCs (SCA1) (Fig. 1d), and MFs (ACTA2 and PDGFβR) (Fig. 1e). From this data we estimate the following distribution of SMC-derived cells within lesions: 30% Mϕ-like (YFP+ACTA2−LGALS3+); 7% MSC-like (YFP+ACTA2−SCA1+); 12% MF-like (YFP+ACTA2+PDGFβR+), 32–51% an unknown phenotype (YFP+ACTA2−LGALS3−SCA1−) (Supplementary Table 1). In addition, these analyses showed that 36% of LGALS3+ cells within advanced atherosclerotic lesions were YFP+, indicating that ~1/3 of cells that would normally be classified as Mϕs in most previous studies in the field originated from SMCs, not myeloid cells as previously assumed.
These initial studies were performed in paraffin embedded samples to maintain the ultrastructure of the plaque and to determine the location of various SMC-derived cells within lesions. However, this technique limits the number of simultaneous markers examined. To further characterize the various SMC phenotypes, we performed flow cytometric analyses of freshly dissociated cells from the aortic root through the iliac bifurcation. Results demonstrated that substantial numbers of SMC-derived cells express multiple additional Mϕ/hematopoietic markers (Fig. 1f) including YFP+ cells that co-express ITGAM (CD11b) a monocyte/Mϕ marker, and F4/80 a marker associated with mature Mϕs, as well as the dendritic cell marker ITGAX (CD11c). In addition, flow cytometric analyses showed that 13% of SMCs were dual positive for the MSC markers SCA1 and ENG (CD105) (Fig. 1f). When analyzing YFP+ MSCs through traditional negative gating strategies (CD45−CD34−CDH5−) we find that up to 45% of MSC-like cells within the aortas of 18 week Western diet fed SMC YFP+/+ Apoe−/− mice are YFP+ (Supplementary Fig. 3a) although it is unclear if all of these are located within lesions or if they also contribute to adventitial SCA1+ cells previously described by several groups28-32. Gating strategies for all flow cytometry experiments were determined based on fluorescence minus one (FMO) controls (Supplementary Fig. 3b, c).
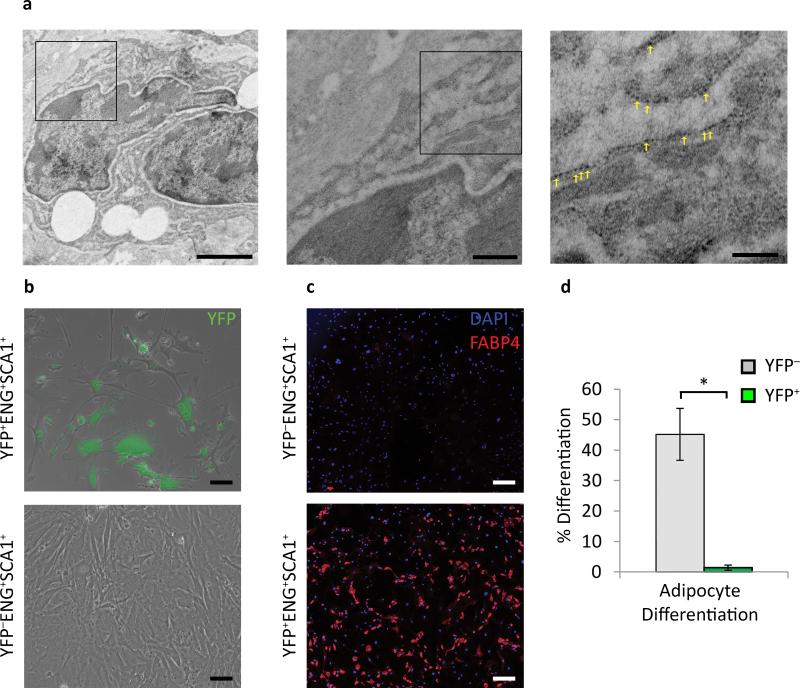
To further assess the morphological and possible functional properties of SMC derived Mϕ-like cells in vivo we analyzed BCA lesions from SMC YFP+/+ Apoe−/− mice by transmission electron microscopy combined with detection of YFP expression by immunogold labeling. As seen in Fig. 2a and Supplementary Fig. 4, we identified YFP+ cells containing multiple large lipid vacuoles that appear to be phagocytic. In addition, we flow sorted SMC-derived MSCs (YFP+ENG+SCA1+), non-SMC derived MSCs (YFP−ENG+SCA1+), and SMC non-MSCs (YFP+ENG−SCA1−) from 18 week Western diet fed lineage tracing mice to test their ability to differentiate into multiple lineages including adipocytes and osteoblasts. After two passages in MSC maintenance media, the SMC non-MSCs became unhealthy and died (data not shown). The SMC-derived MSCs managed to survive in MSC maintenance media but appeared senescent and grew very slowly (Fig. 2b). They also failed to differentiate into either adipocytes (Fig. 2c, d) or osteoblasts (data not shown) when exposed to these respective differentiation medias. In contrast, the YFP− (non-SMC-derived) MSCs grew well and showed high efficiency differentiation into adipocytes (Fig. 2c, d) and osteoblasts (data not shown). These data indicate that although a subset of SMC-derived cells within atherosclerotic lesions express multiple markers of MSCs, we have no evidence they are pluripotent and thus do not appear to function as MSCs.
Figure 2. Phenotypically modulated Mϕ-like SMCs take on some functional characteristics of Mϕs, but MSC-like SMCs show impaired growth and are not multi-potential.
(a) Immuno-TEM of BCAs from 18 week Western diet SMC eYFP+/+ Apoe−/− mice using a 10 nm gold bead conjugated secondary antibody revealed lipid laden YFP+ cells engulfing neighboring cells. Yellow arrows indicate immuno-gold beads. Scale bars (from left to right): 2 μm, 0.5 μm, 0.2 μm. (b) Isolated YFP+ and YFP− MSCs (SCA1+ENG+) in mesenchymal stem cell media 10 days post plating, green is native YFP signal. (c) YFP+ and YFP− MSCs (SCA1+ENG+) after adipogeneisis differentiation based on adipocyte marker FABP4 staining. Scale bar = 200 μm. (d) quantification of adipogenesis from five fields of view per group. Error bars = S.D. * P < 0.05 by Student's t-test.
SMCs within human atheromas express the Mϕ marker CD68
To independently detect phenotypically modulated SMCs that express LGALS3, and to validate a method for detecting these cells in human lesions, we utilized a novel in situ hybridization proximity ligation assay (ISH-PLA) recently developed by our lab27. This technique permits identification of phenotypically modulated SMCs within fixed tissues based on detection of H3K4dime of the Myh11 promoter (PLA+), a SMC-specific epigenetic signature that persists in cells that have no detectable expression of SMC markers27, 33. We first validated the method by showing that YFP+LGALS3+ SMCs within our lineage tracing mice also retained this SMC-specific epigenetic signature (Supplementary Fig. 5a). We also showed that neither cultured RAW 264-7 mouse Mϕ cells (Supplementary Fig. 5b) or human monocytes (Supplementary Fig. 5c) exhibited H3K4dime of Myh11 when exposed to POVPC, an oxidative product of LDL that activates monocytes/Mϕs34.
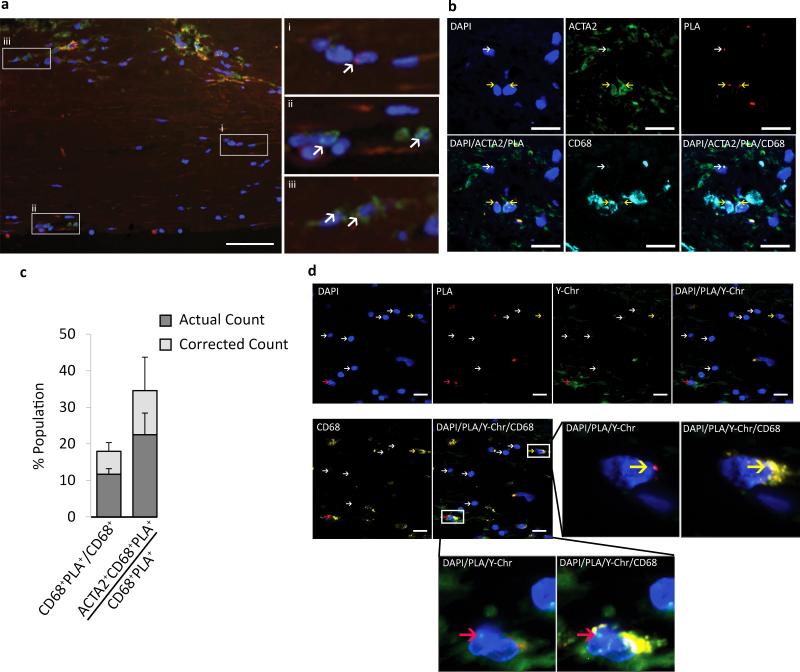
To determine if SMC transition to a Mϕ-like state in human lesions, we stained human coronary artery atherosclerotic lesions for CD68 and ACTA2 as well as ISH-PLA detection of the SMC-specific epigenetic marker MYH11 H3K4dime. Multiple human coronary artery lesion sections from 12 human subjects were analyzed (Supplementary Fig. 5d). We found 18% of CD68+ cells with advanced coronary artery lesions in humans were positive for the SMC-specific MYH11 H3K4dime epigenetic signature based on ISH-PLA assays (Fig. 3a-c), indicating that they were of SMC origin. To further validate these findings, we performed ISH-PLA analysis of MYH11 H3K4dime in coronary artery samples from men that had received a cross gender heart transplant (Supplementary Fig. 6) and found MYH11 H3K4dime PLA+ CD68+ cells that were Y-chromosome negative (Fig. 3d), consistent with these Mϕ-like cells being of SMC and not hematopoietic origin. Importantly, we never saw cells that were MYH11 H3K4dime PLA+ and Y-chromosome+ (Fig. 3d and unpublished data) thus clearly demonstrating that myeloid cells do not acquire the MYH11 H3K4diMe SMC epigenetic signature even in the context of human atherosclerotic lesions.
Figure 3. SMCs within human coronary artery lesions express the Mϕ marker CD68.
(a) SMCs within advanced atherosclerotic lesion specimens were identified based on PLA detection of the SMC specific stable epigenetic signature H3K4dime on the MYH1127. MYH11 H3K4dime PLA+ cells exhibit a punctate red dot within the nucleus while the non-nuclear amorphous red staining is autofluorescence or non-specific background. (a) Samples were also immuno-stained for CD68 (green), and DAPI (blue). Results showed three distinct cell populations highlighted in enlarged panels to the right and indicated with white arrows: (i) MYH11 H3K4dime PLA+ SMCs that are CD68−, (ii) MYH11 H3K4dime PLA−CD68+ (HSC-derived Mϕs), and (iii) H3K4dime MYH11 H3K4dime PLA+CD68+ SMC-derived Mϕ-like cells. Scale bar = 100 μm. (b) Shoulder regions within plaques [stained with DAPI (blue), ACTA2 (green), PLA (red), and CD68 (cyan)] exhibited a high incidence of SMC-derived Mϕ-like cells (MYH11 H3K4dime PLA+CD68+) (yellow arrows) and several phenotypically modulated SMCs negative for CD68 (MYH11 H3K4dime PLA+ACTA2− CD68−) (white arrows). Scale bar = 50 μm. (c) Quantitative analysis of SMC-derived Mϕ-like cells within human coronary lesions based on MYH11 H3K4dime ISH-PLA +/− adjustment for the efficiency of PLA27. Error bars = S.E.M. for 12 independent samples of human atherosclerosis in the right coronary artery. (d) Combined epigenetic SMC and genetic HSC lineage tracing analyses of cross gender human heart transplant samples. Coronary artery specimens from a male patient who received a female heart were processed for MYH11 H3K4dime PLA (red), Y-chromosome FISH (green), and CD68 staining (yellow). Results show cells that were MYH11 H3K4dime PLA+ Y-chromosome− and CD68+ (yellow arrows) reflecting a SMC-derived Mϕ-like cell not of hematopoietic origin (top). In contrast, Mϕs of hematopoietic origin are MYH11 H3K4dime PLA−Y-chromosome+CD68+ (red arrows) (bottom). Scale bar = 50 μm.
KLF4 plays a critical role in regulating SMC phenotype and overall plaque pathogenesis
We have previously shown that KLF4, an ESC and iPS cell pluripotency factor35, is required for SMC phenotypic switching in several in vitro25, 36 models. However, as yet there is no evidence that SMC phenotypic transitions within atherosclerotic lesions are KLF4 dependent, and if so, what role these transitions play in lesion pathogenesis.
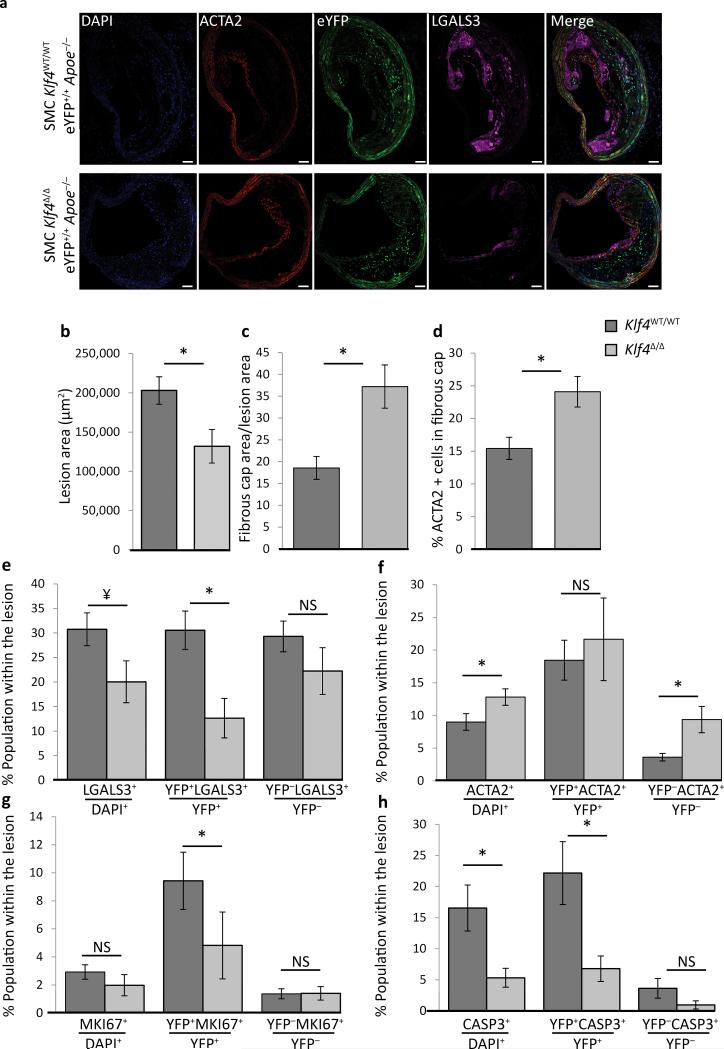
Consistent with our hypothesis that KLF4 regulates phenotypic transitions of SMCs within atherosclerotic lesions, we observed large numbers of YFP+ cells within BCA lesions from 18 week Western diet fed SMC YFP+/+ Apoe−/− mice that expressed KLF4 (Supplementary Fig. 7a). To determine if KLF4 regulates SMC phenotypic transitions and overall lesion pathogenesis, we crossed the SMC YFP+/+ Apoe−/− mouse with Klf4FL/FL mice. We observed highly efficacious recombination of the floxed Klf4 alleles (Klf4Δ/Δ) after tamoxifen treatment (Supplementary Fig. 7b, c) including what we estimate to be nearly 100% recombination within SMCs in the aorta when corrected for > 40% non-SMC DNA present in these samples based on flow cytometric evaluations (Supplementary Fig. 2b). After 18 weeks of Western diet feeding SMC YFP+/+ Klf4WT/WT Apoe−/− and SMC YFP+/+ Klf4Δ/Δ Apoe−/− mice (Fig. 4a) we determined that loss of Klf4 exclusively in SMCs resulted in a nearly 50% reduction in lesion size (Fig. 4b) and multiple changes consistent with increased plaque stability including a > 2-fold increase in fibrous cap area (Fig. 4c), an increase in ACTA2+ cells within the fibrous cap (Fig. 4d), and a reduced number of LGALS3+ cells (Fig. 4e).
Figure 4. SMC specific Klf4 conditional KO in Apoe−/− mice fed a high fat diet for 18 weeks resulted in decreased lesion size and increased indices of plaque stability.
SMC eYFP+/+ Apoe−/− mice crossed to Klf4FL/FL mice were tamoxifen treated and given Western diet for 18 weeks prior to analysis. (a) Representative image demonstrating the changes in lesion morphology and cell composition. Results showed the following: (b) decreased total lesion area, (c) increased fibrous cap area (defined as the region of the lesion within 30 μm of the luminal surface) relative to the size of the lesion, (d) increased ACTA2+ cells within the fibrous cap, (e) an overall decrease in the fraction of LGALS3+ cells, and a large decrease in the fraction of SMC-derived LGALS3+ cells (YFP+LGALS3+), (f) an increase in the ACTA2+ population within the lesion, (g) decreased proliferation of SMC-derived cells (YFP+MKI67+), and (h) decreased overall cell death, mostly due to a decrease in SMC-derived apoptosis (YFP+CASP3+). *P < 0.05, ¥ P = 0.07, analysis completed by 2-way ANOVA comparing genotyping and distance from start of the BCA with a Tukey post-test, error bars are based on S.E.M. Quantification is based on analysis of five 71415 μm2 regions in each of three sections (d-g), or two sections (h, i), per mouse spanning a 600μm length of the BCA. Scale bar = 50 μm. SMC YFP+/+ Klf4WT/WTApoe−/− n = 11, SMC YFP+/+ Klf4Δ/ΔApoe−/− n = 8.
SMC lineage tracing analyses showed that loss of Klf4 within SMCs did not result in a change in the overall number of SMCs (YFP+ cells) within the lesions (Supplementary Fig. 8a), but resulted in major changes in SMC phenotypic transitions. This included a 53% decrease in the number of Mϕ-like SMCs (YFP+LGALS3+/YFP+) within the lesion (Fig. 4e), a 70% decrease in the MSC-like SMCs within the media underlying lesions (Supplementary Fig. 8b), but no change in the MSC-like SMCs in the lesion itself (Supplementary Fig. 8c). Consistent with these results, flow cytometric analyses showed a decrease in the total number of SMC-derived MSC-like cells (YFP+SCA1+ENG+CDH5−PTPRC−CD34−) (Supplementary Fig. 8d) but no change in either the total MSC population (Supplementary Fig. 8e) or the total number of YFP+ cells (Supplementary Fig. 8f). SMC specific Klf4 KO mice also showed an increase in the total number of ACTA2+ cells within the fibrous cap (Fig. 4d), and within lesions (Fig. 4f), but reduced proliferation of SMC-derived cells (Fig. 4g) and marked reduction in the YFP+ SMC apoptosis (Fig. 4h). These changes were not associated with changes in medial area, lumen area (Supplementary Fig. 8g), percent YFP+PDGFβR+ SMC (Supplementary Fig. 8h), or YFP+ACTA2+ SMC (Fig. 4f). In addition, we did not observe changes in cholesterol, or triglyceride levels (Supplementary Fig. 8i).
KLF4 modulates phenotypic transitions and functional properties of SMCs
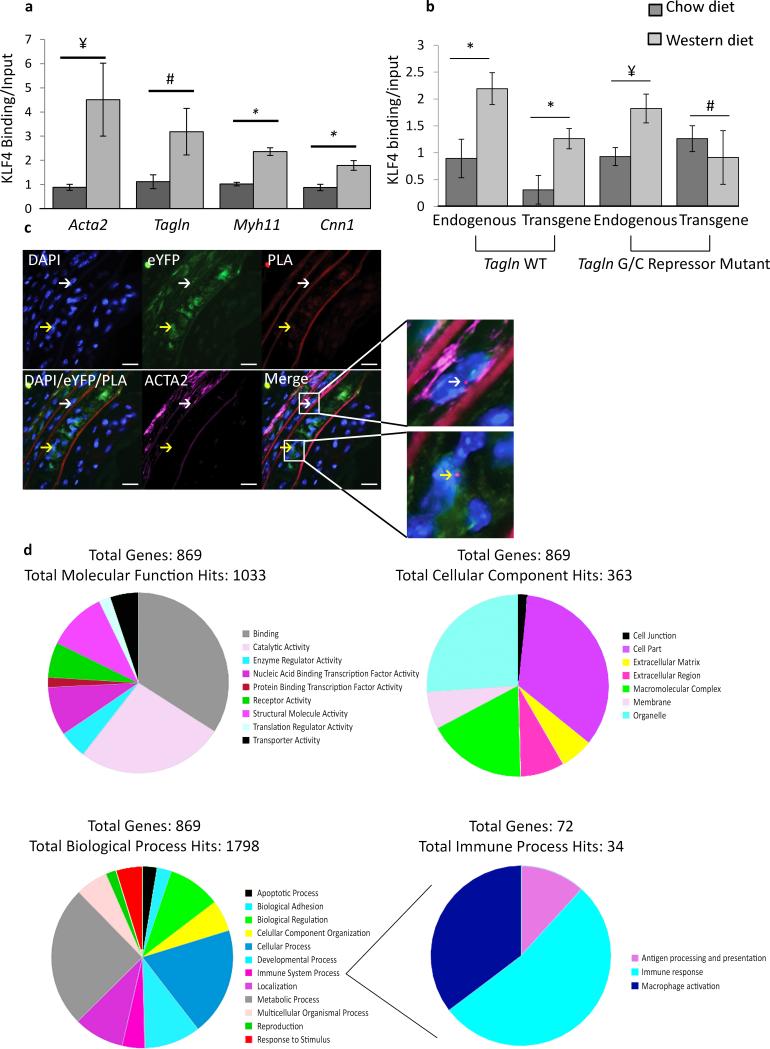
We have previously presented evidence that Klf4 is induced in cultured SMCs by treatment with oxidized phospholipids36 and suppresses expression of SMC marker genes through several mechanisms including binding to the G/C repressor element found in most SMC marker gene promoters including Acta2, Tagln, and Myh11, and inhibiting binding of SRF to CARG elements16, 37, 38. To determine if similar mechanisms contribute to suppression of SMC marker genes within atherosclerotic lesions in vivo, we performed Chromatin Immunoprecipitation (ChIP) assays on chromatin extracted from BCA regions of Apoe−/− transgenic mice containing either a wild type Tagln promoter driving lacZ or a Tagln promoter with a mutation of the GC repressor element (Tagln GC repressor mutant lacZ mouse). Results showed marked enrichment of KLF4 binding to Acta2, Tagln, Myh11, and Cnn1 endogenous promoters in Western diet fed animals (Fig. 5a). Moreover, we showed that enhanced KLF4 binding to the Tagln promoter was dependent on the G/C repressor element (Fig. 5b) indicating this repression is mediated through the GC repressor. Finally, we showed that KLF4 is bound to the Tagln promoter within individual phenotypically modulated SMC (YFP+ACTA2− cells) within Apoe−/− lesions using ISH-PLA (Fig. 5c). Taken together, results provide compelling evidence that coordinate suppression of expression of SMC marker genes is mediated by direct binding of KLF4 to the promoters of SMC marker genes.
Figure 5. KLF4 binds to > 800 genes within SMC in advanced atherosclerotic lesions.
(a) in vivo ChIP assays on BCAs obtained from 18 week Western diet fed Apoe−/− mice show increased binding of KLF4 to SM marker gene promoters compared to Apoe−/− mice fed a chow diet. P-values based on one-way ANOVA with Tukey post-test. ¥ P = 0.07, # P = 0.11, * P < 0.05. Error bars are based on S.E.M. (b) KLF4 binding to the Tagln promoter in response to Western diet treatment is dependent on the GC repressor element.Apoe−/− mice were crossed to either a Tagln wild type (WT) transgene or Tagln G/C repressor mutant transgene and KLF4 ChIP analyses performed on chromatin extracted from the BCA region. P-values based on two-way ANOVA with Tukey post-test. ¥ P = 0.06, # P = 0.43, * P < 0.05. Error bars are based on S.E.M. n = 3 independent pooled groups of 5 mice per treatment group. (c) KLF4 binding to the G/C repressor element of the Tagln promoter in vivo was determined by Klf4-Tagln ISH-PLA. The white arrow indicates a cell where KLF4 is bound to the Tagln promoter of a differentiated SMC (top), while the yellow arrow identifies a phenotypically modulated SMC (bottom). Scale bar = 10 μm. (d) Aortic segments from the aortic root and aortic arch up to the carotid bifurcations from 18 week WD fed SMC Klf4WT/WT eYFP+/+ Apoe−/− (n = 14), SMC Klf4Δ/Δ eYFP+/+ Apoe−/− (n = 14), and 8 week old chow fed SMC Klf4WT/WT eYFP+/+ Apoe−/− (n = 15) mice were utilized for ChIP-Seq analysis of KLF4 binding targets. 869 targets were enriched in Western diet treated SMC Klf4WT/WT eYFP+/+ Apoe−/− mice as compared to SMC Klf4Δ/Δ eYFP+/+ Apoe−/− mice, and thus represent putative SMC KLF4 target genes.
We21, 23 and others39-42 have shown that KLF4 can act as either a transcriptional repressor or activator depending on the cell type and gene locus. To more fully define the repertoire of KLF4 target genes that mediate SMC phenotypic switching, we performed KLF4 ChIP-seq analyses on chromatin samples derived from the BCA from our SMC YFP+/+ Klf4 WT/WT Apoe−/− versus SMC YFP+/+ Klf4Δ/Δ Apoe−/− mice. This required pooling of BCA samples from 14 mice per group to obtain sufficient DNA. Remarkably, results showed 869 KLF4 target genes (Fig. 5d, Supplementary Table 2) that were selectively enriched in Klf4WT/WT versus Klf4Δ/Δ Apoe−/− Western diet fed mice indicating they represent putative KLF4 target genes selective for SMCs. This included the SMC marker genes Acta2 and Tagln, thus validating the fidelity of our ChIP-Seq analyses. In addition, results provided evidence of enhanced KLF4 enrichment within gene regulatory pathways likely to be important in the pathogenesis of lesions including enrichment of gene families associated with phagocytosis, apoptosis, cell migration, and inflammation (Fig. 5d, Supplementary Table 2), which likely contributed to the beneficial overall effects of loss of Klf4 within SMCs on lesion size and pathogenesis. KLF4 also bound regions near Itgal (CD11a), Itgax (CD11b), Itgam (CD11c) (Fig. 5d, Supplementary Table 2), and Arg1 (data not shown) in Klf4WT/WT mice, but not Klf4Δ/Δ mice. We also noted 459 KLF4 targets in Klf4Δ/Δ Apoe−/− samples that were not present in Klf4WT/WT Apoe−/− samples (Supplementary Fig. 9, Supplementary Table 3).
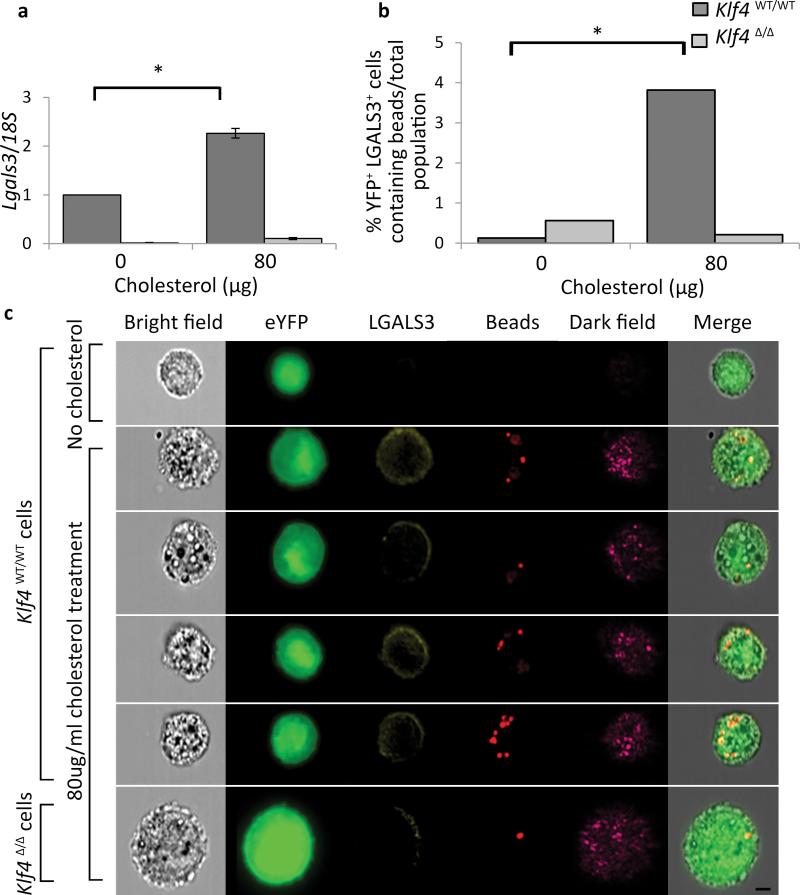
Given observations that SMC specific loss of Klf4 was associated with a marked reduction in SMC-derived Mϕ-like cells (Fig. 4f), we determined if KLF4 is required for transition of cultured SMC to Mϕ-like state in vitro following cholesterol loading12. Due to recent controversies about the origins and purity of SMCs cultured from mouse vessels43, 44, we utilized our SMC-lineage tracing mice to harvest SMC populations from 8-week old tamoxifen injected SMC YFP+/+ Apoe−/− mice. Flow cytometry results showed > 98% YFP+ cells (Supplementary Fig. 10a), indicating they are derived from mature differentiated SMCs in vivo and not a stem cell source as has been speculated43, 44. Cholesterol loading of cultured SMCs resulted in increased expression of Lgals3 (Fig. 6a), and increased Klf4 expression (Supplementary Fig. 10b), which were abolished in aortic SMCs derived from SMC YFP+/+ Klf4Δ/Δ mice (Fig. 6a). Cholesterol loading was also associated with increased phagocytosis that was KLF4 dependent (Fig. 6b, c). Finally, we showed that cholesterol loading resulted in activation of expression of the pro-inflammatory cytokines MCP1, CXCR1, sTNFα, (Supplementary Fig. 10c-e), Lgals3 (Fig. 6a), and the MSC markers Sca1 and Eng (Supplementary Fig. 10f, g), all through KLF4-dependent mechanisms. In contrast, although cholesterol loading increased Abca1 expression this was not KLF4-dependent (Supplementary Fig. 10h).
Figure 6. KLF4-dependent activation of Lgals3, MSC markers, and phagocytotic activity in cholesterol loaded cultured SMC.
(a-c) Aortic SMCs were isolated from SMC YFP+/+ Klf4 WT/WT and SMC YFP+/+ Klf4Δ/Δ mice and sorted using a FACSVantage SE DIVA to ensure a pure SMC (YFP+) cell population. (a) Induction of Lgals3 mRNA expression following cholesterol loading (80 μg/mL cholesterol for 72 hours) was decreased in cells derived from SMC YFP+/+ Klf4Δ/Δ as compared to cells derived from SMC YFP+/+ Klf4 WT/WT mice. P-values based on two-way ANOVA with Tukey post-test. *P < 0.05. Data normalized to Klf4 WT/WT 0 ug/mL cholesterol. Error bars based on S.E.M. n = 3 independent experiments. (b-c) Klf4WT/WT and Klf4Δ/Δ cells were incubated with 0.8 μm polysterene beads for 1.5 hours after 72 hours of cholesterol loading to induce a Mϕ-like state. Cells were then analyzed on an Amnis ImagestreamX Mark II to assess expression of YFP, LGALS3, and to identify bead uptake. (b) Quantification of bead uptake was performed using Amnis IDEAS software. After normalization to the total population, values were subjected to Fisher's exact test, which showed that the Klf4WT/WT YFP+LGALS3+ cells contained significantly (P < 0.05) more beads than the Klf4Δ/Δ YFP+LGALS3+ cells (representative experiment from n = 2). (c) Representative images from the Amnis IDEAS software, YFP (green), LGALS3 (yellow), beads (red), Dark Field (magenta). Scale bar = 5 μm.
Global heterozygous KO of Klf4 alters plaque pathogenesis
Previous studies using VE-cadherin-Cre Klf4FL/FL Apoe−/− and LysMcre/cre Klf4FL/FL Apoe−/− mouse models provided evidence that KLF4 plays an atheroprotective role in endothelial cells (ECs) and myeloid cells respectively with KO resulting in increased lesion size and changes consistent with enhanced inflammation41, 42. Indeed, we have confirmed the latter findings by showing that LysM-cre-dependent KO of Klf4 was associated with increased lesion size (Supplementary Fig. 11a) and Sudan IV lipid staining (Supplementary Fig. 11b) in LysMcre/cre ROSA stop floxed eYFP Klf4 Apoe−/− mice. However, unlike previous studies we saw no changes in triglycerides or total cholesterol (Supplementary Fig. 11c). We also showed a reduced number of LysM-cre derived LGALS3+ cells (Supplementary Fig. 11d) based on YFP identification. However, these data are equivocal regarding what cell populations are affected by the loss of Klf4 in this model since cells other than myeloid cells, including SMC, may activate LysM-cre.
Since loss of Klf4 in SMCs had the opposite effects, a key unresolved question is whether global conditional loss of Klf4 in all cell types would be beneficial or detrimental. To test this, we generated tamoxifen-inducible homozygous (ERT-Cre+ Klf4FL/FLApoe−/−) and heterozygous Klf4 (ERT-Cre+ Klf4FL/WT Apoe−/−) KO mice, with the latter representing a model of partial inhibition of KLF4 across all cell types thus mimicking potential therapeutic approaches of partial suppression of KLF4. Unfortunately, mice with conditional global homozygous Klf4 KO had to be euthanized at 8–10 weeks of Western diet feeding due to excessive weight loss, and development of skin lesions, presumably because KLF4 regulates proliferation and differentiation of epithelial cells45-48. Tamoxifen treated ERT-Cre+ Klf4FL/WT Apoe−/− mice (ERT-Cre+ Klf4Δ/WT Apoe−/−) demonstrated 50% recombination, the maximum possible recombination for heterozygous KO, in the aorta, liver, and colon (Supplementary Fig. 12a, b), but exhibited no changes in body weight, heart weight, or cholesterol and triglyceride levels when compared to ERT-Cre− Klf4FL/WT Apoe−/− littermate control mice (Supplementary Fig. 12c).
Of major interest, ERT-Cre+ Klf4Δ/WT Apoe−/− mice had similar changes in atherosclerotic lesions as observed in our SMC specific conditional Klf4 KO mice including a 30% decrease in lesion size (Supplementary Fig. 13a), and exhibited several indices of increased plaque stability, including increased ACTA2 cap coverage (Supplementary Fig. 13b), and decreased LGALS3+ lesion area (Supplementary Fig. 13c). In addition, they also showed decreased intra-plaque hemorrhage (Supplementary Fig. 13d), and decreased apoptosis and proliferation (Supplementary Fig. 13e, f). Taken together, these results indicate that loss of one Klf4 allele globally in all cell types has beneficial overall effects on plaque development, including the ideal therapeutic goal of smaller more stable lesions.
Discussion
Despite numerous papers showing that cultured SMCs down-regulate expression of SMC differentiation marker genes following exposure to environmental cues present in atherosclerotic lesions including cholesterol12, POVPC25, PDGFBB49, and IL-1β50, the field has relied almost entirely on detection of these markers to ascertain if a given cell within a lesion is a SMC. Indeed, this practice has contributed to the well-established dogma in the field that the role of SMCs within plaques is rather limited, albeit presumed to be beneficial by virtue of phenotypically modulated SMCs secreting extracellular matrix and contributing to fibrous cap formation. Herein we report definitive evidence that > 80% of phenotypically modulated SMCs within BCAs have previously gone undetected and comprise ~30% of the total cellular composition of the lesion. Moreover, we show that phenotypically modulated SMCs transition to multiple phenotypes within lesions including cells that express markers of Mϕs, MSCs, and/or MFs. However, of greatest significance, we show that these transitions are functionally important in that selective loss of Klf4 within SMCs resulted in reduced lesion size, increased fibrous cap thickness, and major reductions in the fraction of SMC-derived Mϕ- and MSC-like cells but an increase in ACTA2+ cells within the fibrous cap. In addition, we show that cholesterol loading of cultured SMCs induced KLF4-dependent activation of Mϕ and MSC markers; expression of proinflammatory cytokines; and increased phagocytosis. Finally, in vivo KLF4 ChIP-seq analyses identified > 800 putative KLF4 regulated genes within SMC including many associated with pro-inflammatory processes. Taken together results provide compelling evidence that transitions in SMC phenotype play a critical role in lesion development, plaque composition, and stability; and for the first time establish that therapeutic targeting aimed at promoting beneficial changes in SMC phenotype may be a viable means of treating advanced atherosclerosis.
A key question is how did loss of Klf4 within SMCs result in marked reductions in overall lesion size as well as multiple changes consistent with increased plaque stability? Of major significance, we show that these effects were not due to a change in the number of SMC-derived cells within lesions. Recent studies by the Fisher lab51 found that while some SMCs will express markers of Mϕs after cholesterol loading in vitro, principal component analyses of microarray data from these cells revealed that they are distinctly different from classical monocytes, Mϕs, and dendritic cells; and have reduced phagocytic capacity51. Interestingly, we also showed that SMC derived MSC-like lesion cells appeared to be dysfunctional. As such, we postulate that loss of Klf4 within SMCs resulted in phenotypic transitions that are favorable for plaque pathogenesis including loss of SMC-derived cells with “proinflammatory” Mϕ-like properties, and gain of SMC-derived cells that contribute to plaque stabilization through mechanisms and functions yet to be defined. Although it has long been postulated that SMCs within lesions play a beneficial role (see reviews9-11, 26, 52, 53), our studies show this is an over-simplification and can vary dramatically depending on the nature of the SMC phenotypic transitions. A critical challenge for future studies will be to identify the environmental cues within advanced atherosclerotic lesions that regulate phenotypic transitions of SMCs, as well as each of the major cell types within lesions, and to determine how these might be manipulated therapeutically to reduce plaque burden and increase plaque stability.
Online Methods
Mice
Animal protocols were approved by the University of Virginia Animal Care and Use Committee.
Male Myh11-CreERT2, LysMcre/cre, Apoe−/−, ROSA26 STOP-flox eYFP+/+, Klf4FL/FL, ERT-Cre, and Tagln G/C Repressor mutant mice were used in this study. ROSA26 STOP-flox eYFP+/+ and Apoe−/− mice were obtained from Jackson Laboratories. Myh11-CreERT2, ROSA26 STOP-flox eYFP, Klf4FL/FL, and ERT-Cre mice were genotyped by PCR as previously described56-59. Cre recombinase was activated in male mice with a series of ten 1 mg tamoxifen intraperitoneal injections (Sigma T-5648) from 6 to 8 weeks of age for a total of 10 mg of tamoxifen per 25 g mouse for both the Myh11-CreERT2 and ERT-Cre mouse models. Tagln G/C Repressor mutant ApoE−/− mouse was previously generated and described by our lab60. Male littermate controls were utilized for all studies. Next, mice were fed a high-fat diet containing 21% milk fat and 0.15% cholesterol (Harlan Teklad) for 18 weeks starting at the end of tamoxifen treatment. Mice were euthanized by CO2 asphyxiation and then perfused via the left ventricle as follows: 5 mL PBS, 10 mL 4% paraformaldehyde, 5mL PBS. Brachiocephalic arteries were carefully dissected and fixed for and additional hour in 4% paraformaldehyde before they were embedded in paraffin. Assays for determining total plasma cholesterol and triglyceride levels (Abbott Laboratories) were performed by the University of Virginia Clinical Pathology Laboratory. Mice were allocated to experimental groups based on genotyping and then randomized for the various experimental measurements. Any animal with triglyceride or cholesterol levels outside of three standard deviations were excluded from all future analyses.
Analysis of Atherosclerotic Plaques
Paraffin-embedded brachiocephalic arteries (BCAs) were serially sectioned at 10 μm thickness from the aortic arch to the right subclavian artery. For immunofluorescent analysis of LGALS3+ SMCs within the, BCA three sections were taken 300 μm apart, spanning the length of the BCA. Slides were stained with antibodies to GFP (Abcam ab6673), ACTA2 (Sigma F3777), LGALS3 (Cedarlane CL8942AP), MKI67 (Abcam ab15580), CASP3 (Cell Signaling 9661S), KLF4 (R&D Systems AF3158), MYH11 (Kamiya Biomedical Company MC-352), PDGFβR (Abcam ab32570), and SCA1 (Ly6A/E) (Abcam ab51317). Using a Zeiss LSM700 confocal microscope a series of 8 z-stack images of 1 μm thickness were acquired for further analysis. Five 14283 μm2 locations within each z-stack of every BCA were analyzed using Zen 2009 Light Edition Software for the presence of immunofluorescent staining coinciding with a single DAPI+ nucleus to determine the average cell populations within each lesion. Every plane of the z-stack was used to assess the co-localization of cellular markers within a single cell. The region of the lesion within 30 μm of the luminal boundary, as determined using Zen 2009 Light Edition Software, was analyzed to determine the cellular composition of the lesion cap, the area within this region was compared to the entire area of the atherosclerotic lesion to determine cap area/lesion area. Morphometric analyses of lesion size were completed using ImagePro Plus as described in Alexander et al61. Researchers were blinded to the genotype of the animals until the end of the analysis.
Immuno-Transmission Electron Microscopy
18 week WD fed SMC YFP+/+ Apoe−/− mice were euthanized by CO2 asphyxiation after 18 weeks of Western diet treatment and perfusion fixed with 0.5% gluteraldehyde (Electron Microscopy Sciences 16300), 4% Paraformaldehyde (Electron Microscopy Sciences 15700) in 1XPBS. Brachiocephalic arteries were isolated, frozen in liquid nitrogen, and sent to the University of Virginia Advanced Microscopy Core for processing. Grids were stained with an antibody to GFP (Abcam ab6673), and followed with a rabbit anti-goat secondary conjugated to 10 nm gold beads (Electron Microscopy Sciences 25229). Images were captured using a JEOL 1230 transmission electron microscope with ultra-high resolution capture camera.
Flow Cytometry
SMC YFP+/+ Apoe−/− mice were euthanized by CO2 asphyxiation after 18 weeks of Western diet treatment. Mice were then perfused with 10mL of PBS and the aorta from the iliac bifurcation to the aortic root was gently cleaned of fat and fascia before removal from the animal. Once cleaned, the tissue was placed into an enzyme cocktail containing 4U/mL Liberase TM (Roche 05401119001), 0.1mg/mL DNaseI, and 60U/mL Hyaluronidase in RPMI-1640. Once immersed in the digestion cocktail, the tissue was cut longitudinally, minced, and placed in a 37°C incubator for 1.5 hours. Cells were run through a 70 μm strainer and spun down at 500g for 5 min. Cells were resuspended in red blood cell lysis buffer (BD PharmLyse 555899) for two minutes and then inactivated using serum containing media, spun down again, and resuspended in 200 μL 1XPBS. Mϕ marker expressing SMCs were identified using antibodies to F4/80 (eBioscience 17-4801), PTPRC (eBioscience 47-0451), ITGAM (eBioscience 45-0112), DAPI (Invitrogen), LGALS3 (BioLegend 125405), and ITGAX (eBioscience 25-0114). MSCs were identified using negative gating for PTPRC (eBioscience 12-0451-82), CDH5 (eBioscience 17-1441), and CD34 (BioLegend 128611); and positive gating for ENG (eBioscience 48-1051-82) and SCA1 (eBioscience 45-5981-82). All samples were run on a Beckman Coulter CyAn ADP LX flow cytometer equipped with 405nm, 488nm, and 633nm lasers. Mesenchymal stem cells were isolated using a Becton Dickinson Influx Cell Sorter.
Human Specimens
De-identified coronary arteries specimens from patients (n = 12) collected during autopsy. These specimens were processed, fixed in paraformaldehyde and paraffin-embedded blocks were cut into 5μm sections. Coronary artery specimen was from a male patient whom received a heart from a female donor (n = 1). The institutional review board at University of Virginia approved the use of all autopsy specimens.
ISH-PLA
ISH-PLA was performed as previously described57. Briefly, human MYH11, mouse Myh11, and mouse Tagln probes were generated by Nick Translation (Roche) using biotin-14-dATP (Invitrogen). Biotin labeled probes (40 ng/slide) underwent denaturation in Hybridization Buffer (2× SSC, 50% high grade formamide, 10% dextran sulfate, 1μg of human or mouse Cot-1 DNA) for 5 min at 80°C. After immunostaining for ACTA2 (Sigma F3777), CD68 (Santa Cruz sc20060 KP1 clone), slides were dehydrated in ethanol series and incubated in 1mM EDTA (pH 8.0) for 20 min. Then, samples were incubated with pepsin (0.5%) in buffer (0.05M Tris, 2mM CaCl2, 0.01M EDTA, 0.01M NaCl) at 37°C for 20 min, as previously described62. Hybridization mixture containing biotin labeled probes or 5-TAMRA-dUTP labeled Y chromosome probe (Clone RP11-88F4, Empire Genomics) was applied on sections. Sections were incubated at 80°C for 5 min, followed by 16–24h incubation at 37°C. Hybridization was followed by multiple washes in 2× SSC, 0.1% NP-40 buffer. PLA was performed directly after ISH following manufacturer's instructions (Olink) and as previously described (Nature Methods ref) Sections were incubated with mouse H3K4dime (5 μg/mL, clone CMA303 Millipore) and rabbit Biotin (5 μg/mL, # ab53494, Abcam) antibodies overnight at 4°C. The PLA amplification was performed using the Duolink detection kit Orange, 555nm. Finally, mounting medium with DAPI was used to coverslip the slides. Images were acquired with an Olympus BX41 fitted with a Q imaging Retiga 2000R camera. Image acquisition was performed using Q Capture Pro software (Media Cybernetics & QImaging Inc). Settings were fixed at the beginning of both acquisition and analysis steps and were unchanged. Brightness and contrast were equally adjusted after merging. Image analysis was performed with Image J. To estimate the percentage of SMC-derived Mϕ-like cells, the number of CD68+/PLA+ cells in CD68 positive staining areas of human coronary lesions (n = 12). We previously rigorously estimated the efficiency of the ISH-PLA method in mouse and human tissues that reaches 65%57. Thus, the percentage of the CD68+/PLA+ cells counted in human lesions is corrected considering the incomplete efficiency of ISH-PLA.
ChIP Assays
Cell culture ChIP was performed as previously described63. Cells were fixed with 1% paraformaldehyde for 10 min at room temperature. The cross-linked chromatin was sonicated to shear chromatin into fragments of 200–600 base pairs. The sheared chromatin was immunoprecipitated with 2 μg of dimeH3K4 (clone CMA303, Millipore), or KLF4 (Santa Cruz sc20691) while negative control was incubated with mouse or rabbit IgG. Immune complexes were captured with magnetic bead-coupled protein G (Millipore). After elution and purification of the genomic DNA (gDNA), real time PCR was performed on IP and non-immunoprecipitated INPUT gDNA. Primer sets used for Myh11, Arg1 and Cox2 promoters were previously described57, 64. Results are expressed as the percentage of IP/INPUT. In vivo KLF4 ChIP assays were performed using flash frozen BCAs from mice that had been fed either 18 weeks of high-fat or standard chow (Harlan 7012) beginning at 8 weeks of age as previously described65, 66.
KLF4 Chromatin Immunoprecipitation Sequencing (ChIP-seq)
Segments of the aorta from the arch to the aortic root and up to the carotid bifurcations were isolated from 18 week WD fed SMC Klf4WT/WT eYFP+/+ Apoe−/− (n = 14), SMC Klf4Δ/Δ eYFP+/+ Apoe−/− (n = 14), and 8 week old chow fed SMC Klf4WT/WT eYFP+/+ Apoe−/− (n = 15) were snap frozen in liquid nitrogen and then processed as mentioned in ChIP methods section for KLF4 binding. DNA library preparation and deep sequencing was performed by HudsonAlpha Institute for Biotechnology via Illumina TruSeq Chip Library Kit according to the manufacturer's protocol. Quality control and quantification of DNA and library were performed using Agilent 2100 Bioanalyser and Kapa Library Quant Kit according to manufacturer's protocol.
ChIP-seq data processing
The sequencing reads from Illumina HiSeq were aligned to the mouse genome (mm10) using the BOWTIE alignment tool67. These aligned reads were then processed and converted into bam/bai (http://genome.ucsc.edu/goldenPath/help/bam.html) format, and then loaded in the Integrative Genomics Viewer (http://www.broadinstitute.org/igv/) for visualization. The processing steps involved removing duplicate reads and format conversions using SAMtools68 suite. The reads were also converted to BED format (http://genome.ucsc.edu/FAQ/FAQformat#format1) for further data analysis processes such as peak calling. Peak finding was performed using MACS14 (ref. 69) with ChIP-seq BED files as input, default parameters (P = 1 × 10−5). Once the peaks were obtained for all the tracks, the common peaks and genes between these tracks were removed by BEDtools70 intersect command. The outcome of the above filtering steps resulted in signals present, theoretically, in KLF4 bound to genes within SMC. Functional annotation was performed using PANTHER71 and Gene Ontology Consortium (http://geneontology.org/) and statistical over representation test was performed using PANTHER71. GEO accession number GSE65812.
Cell Culture Studies
Primary mouse aortic SMCs were isolated from 8 week old SMC-lineage tracing mice after a series of 10 tamoxifen injections using a previously described protocol72. Cells isolated from SMC YFP+/+ Klf4WT/WT and SMC YFP+/+ Klf4Δ/Δ mice were passaged three times before undergoing a cell selective sort for YFP+. Cholesterol assays were performed using water-soluble cholesterol from Sigma (C4951-30MG) as previously published73 with minor modification: Cholesterol was reconstituted in DMEM-F12 media from Gibco containing; 0.2% FBS (Gibco); 100 U/mL penicillin/streptomycin (Gibco); 1.6 mmol/l l-glutamine, (Gibco). Cells were allowed to grow to ~70% confluency before being switched to 0, 20, 40, or 80 μg/mL cholesterol containing media. After 72hrs cells were harvested for mRNA, protein, or ChIP analysis. Human coronary SMC were purchased from Lonza and cultured in SMC maintaining media (Lonza). RAW264.7 mouse Mϕs and human monocytes were cultured as previously described.
Bead Uptake Assays
Primary mouse aortic SMCs were treated with cholesterol as aforementioned. After 72hrs cells were switched back to DF10 media containing 1.5% v/v 0.84μm polysterene beads (Spherotech FP-0870-2) for 1.5 hours. Cells were then harvested, stained with LGALS3 (BioLegend 125405) and run on an Amnis ImageStreamX Mark II using a 60X objective. Data was analyzed using Amnis IDEAS software.
Mesenchymal Stem Cell Differentiation
Cells were isolated from 12, 18 week Western diet fed SMC YFP+/+ Apoe−/− mice as described in the flow cytometry section. Next, cells were stained with markers for lin− (CD34− BioLegend 128611, PTPRC− eBioscience 56-0451-80, CDH5 – eBioscience 17-1441-80); and positive gating for ENG (eBioscience 48-1051-82) and SCA1 (eBioscience 45-5981-82). YFP was detected by native fluorescence. Cells were then sorted into four populations (YFP+SCA1+ENG+, YFP−SCA1+ENG+, YFP+SCA1−ENG−, and YFP−SCA1−ENG−) using a Becton Dickinson Influx Cell Sorter. Cells were then placed at in StemXVivo MSC media (R&D Systems CCM004) media at a density of 30,000 cells/mm2 and were passed at 70% confluency (changing media every 2–3 days). YFP+, SCA1+, ENG+ cells were p2 for differentiation experiment, and YFP−, SCA1+, ENG+ cells were p4 at the start of the differentiation experiment. Differentiation experiment was conducted using a kit from R&D systems (mouse mesenchymal stem cell functional differentiation kit, SC010). Cells were differentiated in the appropriate media for 14 days (changing media every 2–3 days), then fixed and immunocytochemistry utilizing antibodies from the kit and performed according to the protocol provided.
Statistics
Fisher's exact test was used for categorical data. Two-way ANOVA with Tukey post-hoc tests were used for multiple group comparisons determined to have normal distribution by the Kolmogorov-Smirnov test. Two analyses showed statistical difference across multiple locations, ACTA2/DAPI, and YFP/DAPI, using 2-way ANOVA. All other analyses across multiple locations were therefore not reported in the manuscript. Non-parametric data were analyzed using the Wilcoxon rank sum test. There were no significant interactions between genotype and location for any of the end points analyzed by the 2-way ANOVA. P < 0.05 was considered significant. SAS v9.3 with Enterprise Guide v5.1 software (SAS Institute Inc.) was used for all statistical analyses.
Supplementary Material
Acknowledgements
We would like to acknowledge the valuable contributions of technicians M. McCanna and M.Bevard for their assistance in histological cutting and staining, R. Tripathi for assistance with cell culture work; J. Lannigan and M. Solga from the Flow Cytometry Core at UVA for their help designing flow cytometry panels and help running the cytometers; S. Guilot at the Advanced Microscopy Facility at UVA for her help running the TEM, J. Roithmayr, N. Hendley, M. Quetsch and M. Goodwin for their assistance in immunofluorescent image analysis; Y. Babiy for designing the cartoon in Supplementary Fig. 6, and W. Evans for assistance in processing statistical analyses. We would also like to thank N. McGinn from the Department of Pathology, University of Virginia Hospital for the de-identified coronary arteries specimens, C. Murray from the University of Washington for the coronary artery heart transplant specimen, S. Offermanns from the Max Planck Institute54 for the Myh11-CreERT2 mice, Kaestner for the Klf4FL/FL (ref. 47) mice, G. Randolph for the LysMcre/cre mice, and A. Berns from the Netherlands Cancer Institute, Amsterdam, Netherlands for the transgenic tamoxifen-inducible Cre (ERT-Cre) recombinase mice55. This work was supported by US National Institutes of Health R01 grants HL057353, HL098538, and HL087867 to G.K.O., HL112904 to ACS, as well as a pilot grant from AstraZeneca Pharmaceutical as part of a University of Virginia – AstraZeneca Research Alliance; and Mid-Atlantic American Heart Association fellowship grants 11PRE7170008, and 13POST17080043 to L.S.S., and D.G. respectively.
Footnotes
Contributions
L.S.S. conducted experiments, performed data analysis; generated most of the experimental mice; performed immunostaining, image analysis and flow cytometry; and was primary writer of the manuscript. D.G. performed in vitro ChIP analysis and conceived and performed all the ISH-PLA experiments. M.S. generated Tagln wild type lacZ Apoe−/−, performed in vivo ChIP assays, performed immunohistochemistry, and data analysis. O.A.C. was involved in designing experiments and data analysis; assisted in image analysis, cell culture and animal experiments throughout the project. A.C.S. and B.I. participated in designing immuno-TEM protocol and analysis of images. R.M.H. optimized MSC staining IF protocols, conducted MSC differentiation experiments, and helped design cartoons. G.F.A. conducted data analysis of ChIP-seq experiments. P.S. assisted in cell culture experiments and analysis of data. A.A.C.N. peformed PDGFβR staining and analysis, E.S.G. assisted in animal experiments, conducted statistical analyses and performed immunohistochemistry and data analysis. L.S.S., D.G., M.S., O.A.C., E.S.G., B.I., G.J.R. and G.K.O. participated in making final manuscript revisions.
G.K.O. supervised the entire project and played a major role in experimental design, data interpretation, and writing the manuscript.
GEO accession number for KLF4 ChIP-seq data, GSE65812.
Competing financial interests
The authors declare no competing financial interests.
Reference List
- 1.Saffitz JE, Schwartz CJ. Coronary atherosclerosis and thrombosis underlying acute myocardial infarction. Cardiol. Clin. 1987;5:21–30. [PubMed] [Google Scholar]
- 2.Libby P, Aikawa M. Stabilization of atherosclerotic plaques: new mechanisms and clinical targets. Nat. Med. 2002;8:1257–1262. doi: 10.1038/nm1102-1257. [DOI] [PubMed] [Google Scholar]
- 3.Falk E, Nakano M, Benton JF, Finn AV, Virmani R. Update on acute coronary syndromes: the pathologists' view. Eur. Heart J. 2012 doi: 10.1093/eurheartj/ehs411. [DOI] [PubMed] [Google Scholar]
- 4.Falk E, Shah PK, Fuster V. Coronary plaque disruption. Circulation. 1995;92:657–671. doi: 10.1161/01.cir.92.3.657. [DOI] [PubMed] [Google Scholar]
- 5.Virmani R, Kolodgie FD, Burke AP, Farb A, Schwartz SM. Lessons from sudden coronary death: a comprehensive morphological classification scheme for atherosclerotic lesions. Arterioscler. Thromb. Vasc. Biol. 2000;20:1262–1275. doi: 10.1161/01.atv.20.5.1262. [DOI] [PubMed] [Google Scholar]
- 6.Lee RT, Libby P. The unstable atheroma. Arterioscler. Thromb. Vasc. Biol. 1997;17:1859–1867. doi: 10.1161/01.atv.17.10.1859. [DOI] [PubMed] [Google Scholar]
- 7.Ross R. Atherosclerosis--an inflammatory disease. N. Engl. J. Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 8.Libby P. Inflammation in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2012;32:2045–2051. doi: 10.1161/ATVBAHA.108.179705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glass CK, Witztum JL. Atherosclerosis. the road ahead. Cell. 2001;104:503–516. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 10.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–325. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 11.Gomez D, Owens GK. Smooth muscle cell phenotypic switching in atherosclerosis. Cardiovasc. Res. 2012;95:156–164. doi: 10.1093/cvr/cvs115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rong JX, Shapiro M, Trogan E, Fisher EA. Transdifferentiation of mouse aortic smooth muscle cells to a macrophage-like state after cholesterol loading. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13531–13536. doi: 10.1073/pnas.1735526100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin K, et al. Thrombin stimulates smooth muscle cell differentiation from peripheral blood mononuclear cells via protease-activated receptor-1, RhoA, and myocardin. Circ. Res. 2009;105:214–218. doi: 10.1161/CIRCRESAHA.109.199984. [DOI] [PubMed] [Google Scholar]
- 14.Caplice NM, et al. Smooth muscle cells in human coronary atherosclerosis can originate from cells administered at marrow transplantation. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4754–4759. doi: 10.1073/pnas.0730743100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwata H, et al. Bone marrow-derived cells contribute to vascular inflammation but do not differentiate into smooth muscle cell lineages. Circulation. 2010;122:2048–2057. doi: 10.1161/CIRCULATIONAHA.110.965202. [DOI] [PubMed] [Google Scholar]
- 16.Wamhoff BR, et al. A G/C element mediates repression of the SM22alpha promoter within phenotypically modulated smooth muscle cells in experimental atherosclerosis. Circ. Res. 2004;95:981–988. doi: 10.1161/01.RES.0000147961.09840.fb. [DOI] [PubMed] [Google Scholar]
- 17.Feil S, et al. Transdifferentiation of vascular smooth muscle cells to macrophage-like cells during atherogenesis. Circ. Res. 2014;115:662–667. doi: 10.1161/CIRCRESAHA.115.304634. [DOI] [PubMed] [Google Scholar]
- 18.Swirski FK, Nahrendorf M. Do vascular smooth muscle cells differentiate to macrophages in atherosclerotic lesions? Circ. Res. 2014;115:605–606. doi: 10.1161/CIRCRESAHA.114.304925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allahverdian S, Chehroudi AC, McManus BM, Abraham T, Francis GA. Contribution of Intimal Smooth Muscle Cells to Cholesterol Accumulation and Macrophage-Like Cells in Human Atherosclerosis. 2014 doi: 10.1161/CIRCULATIONAHA.113.005015. [DOI] [PubMed] [Google Scholar]
- 20.Cordes KR, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida T, Kaestner KH, Owens GK. Conditional deletion of Kruppel-like factor 4 delays downregulation of smooth muscle cell differentiation markers but accelerates neointimal formation following vascular injury. Circ. Res. 2008;102:1548–1557. doi: 10.1161/CIRCRESAHA.108.176974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deaton RA, Gan Q, Owens GK. Sp1-dependent activation of KLF4 is required for PDGF-BB-induced phenotypic modulation of smooth muscle. Am. J. Physiol Heart Circ. Physiol. 2009;296:H1027–H1037. doi: 10.1152/ajpheart.01230.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshida T, Gan Q, Owens GK. Kruppel-like factor 4, Elk-1, and histone deacetylases cooperatively suppress smooth muscle cell differentiation markers in response to oxidized phospholipids. Am. J. Physiol Cell Physiol. 2008;295:C1175–C1182. doi: 10.1152/ajpcell.00288.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas JA, et al. PDGF-DD, a novel mediator of smooth muscle cell phenotypic modulation, is upregulated in endothelial cells exposed to atherosclerosis-prone flow patterns. Am. J. Physiol Heart Circ. Physiol. 2009;296:H442–H452. doi: 10.1152/ajpheart.00165.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pidkovka NA, et al. Oxidized phospholipids induce phenotypic switching of vascular smooth muscle cells in vivo and in vitro. Circ. Res. 2007;101:792–801. doi: 10.1161/CIRCRESAHA.107.152736. [DOI] [PubMed] [Google Scholar]
- 26.Alexander MR, Owens GK. Epigenetic Control of Smooth Muscle Cell Differentiation and Phenotypic Switchingin Vascular Development and Disease. Annu. Rev. Physiol. 2011 doi: 10.1146/annurev-physiol-012110-142315. [DOI] [PubMed] [Google Scholar]
- 27.Gomez D, Shankman LS, Nguyen AT, Owens GK. Detection of histone modifications at specific gene loci in single cells in histological sections. Nat. Methods. 2013;10:171–177. doi: 10.1038/nmeth.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein D, Benchellal M, Kleff V, Jakob HG, Ergun S. Hox genes are involved in vascular wall-resident multipotent stem cell differentiation into smooth muscle cells. Sci. Rep. 2013;3:2178. doi: 10.1038/srep02178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein D, et al. Vascular wall-resident CD44+ multipotent stem cells give rise to pericytes and smooth muscle cells and contribute to new vessel maturation. PLoS. One. 2011;6:e20540. doi: 10.1371/journal.pone.0020540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao Q, et al. Sca-1+ progenitors derived from embryonic stem cells differentiate into endothelial cells capable of vascular repair after arterial injury. Arterioscler. Thromb. Vasc. Biol. 2006;26:2244–2251. doi: 10.1161/01.ATV.0000240251.50215.50. [DOI] [PubMed] [Google Scholar]
- 31.Xiao Q, Zeng L, Zhang Z, Hu Y, Xu Q. Stem cell-derived Sca-1+ progenitors differentiate into smooth muscle cells, which is mediated by collagen IV-integrin alpha1/beta1/alphav and PDGF receptor pathways. Am. J. Physiol Cell Physiol. 2007;292:C342–C352. doi: 10.1152/ajpcell.00341.2006. [DOI] [PubMed] [Google Scholar]
- 32.Passman JN, et al. A sonic hedgehog signaling domain in the arterial adventitia supports resident Sca1+ smooth muscle progenitor cells. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9349–9354. doi: 10.1073/pnas.0711382105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J. Clin. Invest. 2006;116:36–48. doi: 10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vladykovskaya E, et al. Reductive metabolism increases the proinflammatory activity of aldehyde phospholipids. J. Lipid Res. 2011;52:2209–2225. doi: 10.1194/jlr.M013854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 36.Cherepanova OA, et al. Oxidized phospholipids induce type VIII collagen expression and vascular smooth muscle cell migration. Circ. Res. 2009;104:609–618. doi: 10.1161/CIRCRESAHA.108.186064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salmon M, Gomez D, Greene E, Shankman L, Owens GK. Cooperative binding of KLF4, pELK-1, and HDAC2 to a G/C repressor element in the SM22alpha promoter mediates transcriptional silencing during SMC phenotypic switching in vivo. Circ. Res. 2012;111:685–696. doi: 10.1161/CIRCRESAHA.112.269811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Regan CP, Adam PJ, Madsen CS, Owens GK. Molecular mechanisms of decreased smooth muscle differentiation marker expression after vascular injury. J. Clin. Invest. 2000;106:1139–1147. doi: 10.1172/JCI10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feinberg MW, et al. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. 2007;26:4138–4148. doi: 10.1038/sj.emboj.7601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao X, et al. Kruppel-like factor 4 regulates macrophage polarization. J. Clin. Invest. 2011;121:2736–2749. doi: 10.1172/JCI45444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma N, et al. Myeloid Kruppel-like factor 4 deficiency augments atherogenesis in ApoE−/− mice--brief report. Arterioscler. Thromb. Vasc. Biol. 2012;32:2836–2838. doi: 10.1161/ATVBAHA.112.300471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou G, et al. Endothelial Kruppel-like factor 4 protects against atherothrombosis in mice. J. Clin. Invest. 2012;122:4727–4731. doi: 10.1172/JCI66056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen AT, et al. Smooth muscle cell plasticity: fact or fiction? Circ. Res. 2013;112:17–22. doi: 10.1161/CIRCRESAHA.112.281048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang Z, et al. Differentiation of multipotent vascular stem cells contributes to vascular diseases. Nat. Commun. 2012;3:875. doi: 10.1038/ncomms1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foster KW, et al. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene. 2005;24:1491–1500. doi: 10.1038/sj.onc.1208307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jaubert J, Cheng J, Segre JA. Ectopic expression of kruppel like factor 4 (Klf4) accelerates formation of the epidermal permeability barrier. Development. 2003;130:2767–2777. doi: 10.1242/dev.00477. [DOI] [PubMed] [Google Scholar]
- 47.Katz JP, et al. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katz JP, et al. Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology. 2005;128:935–945. doi: 10.1053/j.gastro.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 49.Dandre F, Owens GK. Platelet-derived growth factor-BB and Ets-1 transcription factor negatively regulate transcription of multiple smooth muscle cell differentiation marker genes. Am. J. Physiol Heart Circ. Physiol. 2004;286:H2042–H2051. doi: 10.1152/ajpheart.00625.2003. [DOI] [PubMed] [Google Scholar]
- 50.Clement N, et al. Notch3 and IL-1beta exert opposing effects on a vascular smooth muscle cell inflammatory pathway in which NF-kappaB drives crosstalk. J. Cell Sci. 2007;120:3352–3361. doi: 10.1242/jcs.007872. [DOI] [PubMed] [Google Scholar]
- 51.Vengrenyuk Y, et al. Cholesterol Loading Reprograms the MicroRNA-143/145-Myocardin Axis to Convert Aortic Smooth Muscle Cells to a Dysfunctional Macrophage-Like Phenotype. Arterioscler Thromb Vasc Biol. 2015;35:535–546. doi: 10.1161/ATVBAHA.114.304029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Owens GK. Regulation of differentiation of vascular smooth muscle cells. Physiol Rev. 1995;75:487–517. doi: 10.1152/physrev.1995.75.3.487. [DOI] [PubMed] [Google Scholar]
- 53.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 54.Wirth A, et al. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat. Med. 2008;14:64–68. doi: 10.1038/nm1666. [DOI] [PubMed] [Google Scholar]
- 55.Vooijs M, Jonkers J, Berns A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2001;2:292–297. doi: 10.1093/embo-reports/kve064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wirth A, et al. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat. Med. 2008;14:64–68. doi: 10.1038/nm1666. [DOI] [PubMed] [Google Scholar]
- 57.Gomez D, Shankman LS, Nguyen AT, Owens GK. Detection of histone modifications at specific gene loci in single cells in histological sections. Nat. Methods. 2013;10:171–177. doi: 10.1038/nmeth.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katz JP, et al. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vooijs M, Jonkers J, Berns A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2001;2:292–297. doi: 10.1093/embo-reports/kve064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wamhoff BR, et al. A G/C element mediates repression of the SM22alpha promoter within phenotypically modulated smooth muscle cells in experimental atherosclerosis. Circ. Res. 2004;95:981–988. doi: 10.1161/01.RES.0000147961.09840.fb. [DOI] [PubMed] [Google Scholar]
- 61.Alexander MR, et al. Genetic inactivation of IL-1 signaling enhances atherosclerotic plaque instability and reduces outward vessel remodeling in advanced atherosclerosis in mice. J. Clin. Invest. 2012;122:70–79. doi: 10.1172/JCI43713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Caplice NM, et al. Smooth muscle cells in human coronary atherosclerosis can originate from cells administered at marrow transplantation. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4754–4759. doi: 10.1073/pnas.0730743100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McDonald OG, Owens GK. Programming smooth muscle plasticity with chromatin dynamics. Circ. Res. 2007;100:1428–1441. doi: 10.1161/01.RES.0000266448.30370.a0. [DOI] [PubMed] [Google Scholar]
- 64.Liao X, et al. Kruppel-like factor 4 regulates macrophage polarization. J. Clin. Invest. 2011;121:2736–2749. doi: 10.1172/JCI45444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deaton RA, Gan Q, Owens GK. Sp1-dependent activation of KLF4 is required for PDGF-BB-induced phenotypic modulation of smooth muscle. Am. J. Physiol Heart Circ. Physiol. 2009;296:H1027–H1037. doi: 10.1152/ajpheart.01230.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salmon M, Gomez D, Greene E, Shankman L, Owens GK. Cooperative binding of KLF4, pELK-1, and HDAC2 to a G/C repressor element in the SM22alpha promoter mediates transcriptional silencing during SMC phenotypic switching in vivo. Circ. Res. 2012;111:685–696. doi: 10.1161/CIRCRESAHA.112.269811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Geisterfer AA, Peach MJ, Owens GK. Angiotensin II induces hypertrophy, not hyperplasia, of cultured rat aortic smooth muscle cells. Circ. Res. 1988;62:749–756. doi: 10.1161/01.res.62.4.749. [DOI] [PubMed] [Google Scholar]
- 73.Rong JX, Shapiro M, Trogan E, Fisher EA. Transdifferentiation of mouse aortic smooth muscle cells to a macrophage-like state after cholesterol loading. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13531–13536. doi: 10.1073/pnas.1735526100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.