Summary
We developed radioactivity-based and fluorescence-based assays for the DNA repair enzyme APE1 and showed that its decreased activity is associated with increased lung cancer risk. This suggests that ‘bad DNA repair’ rather than ‘bad luck’ is involved in cancer etiology.
Abstract
The key role of DNA repair in removing DNA damage and minimizing mutations makes it an attractive target for cancer risk assessment and prevention. Here we describe the development of a robust assay for apurinic/apyrimidinic (AP) endonuclease 1 (APE1; APEX1), an essential enzyme involved in the repair of oxidative DNA damage. APE1 DNA repair enzymatic activity was measured in peripheral blood mononuclear cell protein extracts using a radioactivity-based assay, and its association with lung cancer was determined using conditional logistic regression with specimens from a population-based case–control study with 96 lung cancer cases and 96 matched control subjects. The mean APE1 enzyme activity in case patients was 691 [95% confidence interval (CI) = 655–727] units/ng protein, significantly lower than in control subjects (mean = 793, 95% CI = 751–834 units/ng protein, P = 0.0006). The adjusted odds ratio for lung cancer associated with 1 SD (211 units) decrease in APE1 activity was 2.0 (95% CI = 1.3–3.1; P = 0.002). Comparison of radioactivity- and fluorescence-based assays showed that the two are equivalent, indicating no interference by the fluorescent tag. The APE1Asp148Glu SNP was associated neither with APE1 enzyme activity nor with lung cancer risk. Taken together, our results indicate that low APE1 activity is associated with lung cancer risk, consistent with the hypothesis that ‘bad DNA repair’, rather than ‘bad luck’, is involved in cancer etiology. Such assays may be useful, along with additional DNA repair biomarkers, for risk assessment of lung cancer and perhaps other cancers, and for selecting individuals to undergo early detection techniques such as low-dose CT.
Introduction
The rate at which mutations accumulate in DNA is a key element in the development of cancer. It is governed by the extent of exposure to DNA-damaging agents, both internal and external, as well as the ability to repair DNA once damaged (1–3). Indeed, germ-line deficiencies in DNA repair have been clearly shown to cause a number of hereditary cancer-prone diseases such as BRCA1- and BRCA2-associated breast cancer, mismatch repair-associated hereditary non-polyposis colorectal cancer, the nucleotide excision repair deficient xeroderma pigmentosum, and MutYH-related attenuated familial adenomatous polyposis (4–8). Moreover, epidemiological studies have suggested that imbalances in DNA repair are associated with increased risk of sporadic cancers (9–19). The complexity of DNA repair mechanisms suggests that the contribution of specific DNA repair activities to cancer risk needs to be evaluated, in an effort to generate a panel of DNA repair risk factors for a particular cancer. To this end we, and others have shown that imbalances in enzymatic activity of 8-oxoguanine DNA glcosylase (OGG1) (10,11) and N-methylpurine DNA glycosylase (MPG) (12,16,17), as measured in peripheral blood mononuclear cells (PBMC), are associated with increased lung cancer risk. These enzymes, like the other known human DNA glycosylases, remove damaged bases as the first step in base excision repair. Their action generates an abasic site (also termed AP site; apurinic/apyrimidimic site), an intermediate, which is in fact a secondary DNA damage. The repair of this abasic site is initiated by AP endonuclease 1 (APE1, APEX1) which nicks the DNA 5′ to the abasic site, followed by the activities of a DNA polymerase and a DNA ligase to complete this accurate repair process (3,20). Thus, the function of APE1 is critical in completing the repair initiated by DNA glycosylases.
APE1 has several additional important functions (i): initiation of the repair of spontaneously formed abasic sites that account for ~10000 DNA damages/cell/day, making them the most abundant type of spontaneous DNA damage (21); (ii) alkylation of certain DNA bases drastically weakens the glycosylic bond linking the deoxyribose to the alkylated base. This causes facilitated spontaneous release of the alkylated base from DNA, thereby creating additional abasic sites that need to be dealt with by APE1 (3); (iii)stimulation of the activity of at least certain DNA glycosylases, by facilitating their turnover (22,23); (iv)involvement in the repair of single strand breaks, another abundant type of DNA damage (24); (v) involvement in nucleotide incision repair (25); and (vi) acting as a redox factor via its REF domain (26). These multiple functions may explain the high levels of this enzyme, estimated to be 350000–7000000 molecules/cell (27), and are reflected in APE1’s specific activity in protein extracts prepared from peripheral blood mononuclear cells, which was found to be 4–5 orders of magnitude higher than the activity of OGG1 and MPG (18). In addition, APE1 is essential in mice (28). These features of APE1 make inter-individual variations in its activity a potential risk factor in cancer.
Here, we describe the development of a robust assay for the enzymatic activity of APE1 in protein extracts prepared from PBMC, and demonstrate that assays based on 32P-labeled (APE1-P) or fluorescence-tagged DNA substrates (APE1-F) are equivalent. We also show that the APE1 Asp149Glu SNP is neither associated with APE1 enzyme activity level, nor with lung cancer risk. However, reduced APE1 enzymatic activity is significantly associated with increased lung cancer risk.
Materials and methods
Study participants
The study participants have been described previously (16,18). Case patients (100) were recruited from the Rambam Medical Center (Haifa, Israel). Control subjects were enrollees of Clalit Health Services identified from the same geographical area. Blood samples were drawn prior to any treatment. One hundred control subjects were individually matched to the case patients by gender, year of birth (±1 year), place of residence and ethnic group (Jews versus non-Jews). The only exclusion criterion was former diagnosis of lung cancer. Participants provided written informed consent at time of recruitment, and were interviewed in-person to obtain information about their personal and family history of cancer, and smoking history. Diagnoses of lung cancer, made at the diagnosing hospital, included information on histological type, TNM staging and tumor grade. The institutional review board at Carmel Medical Center, Haifa, approved all procedures.
Blood samples, isolation of PBMC and preparation of protein extracts
Each study participant provided a sample (17ml) of peripheral blood collected in two ACD blood collection tubes (BD vacutainers, Catalog No. 364606). Blood samples were processed 18–24h after collection to isolate PBMC, as described previously (16). Protein extracts were prepared by freeze-thaw lysis in hypotonic buffer, followed by salt extraction as described previously (16). Both the isolated PBMC and protein extracts can be stored frozen for at least 1.5 years at −80°C with essentially no loss of DNA repair activity. For the development stage, 500ml bags of whole blood with citrate-phosphate-dextrose-adenine (CPDA-1; Teva medical, Ashdod, Israel) as anticoagulant, obtained from healthy donors, or leukocytes bags (buffy coats, ~100ml) were purchased from the National Blood Bank at the Sheba Medical Center (Tel Hashomer, Israel). Overall, blood from 20 different donors was used.
DNA substrates
APE1-P assay
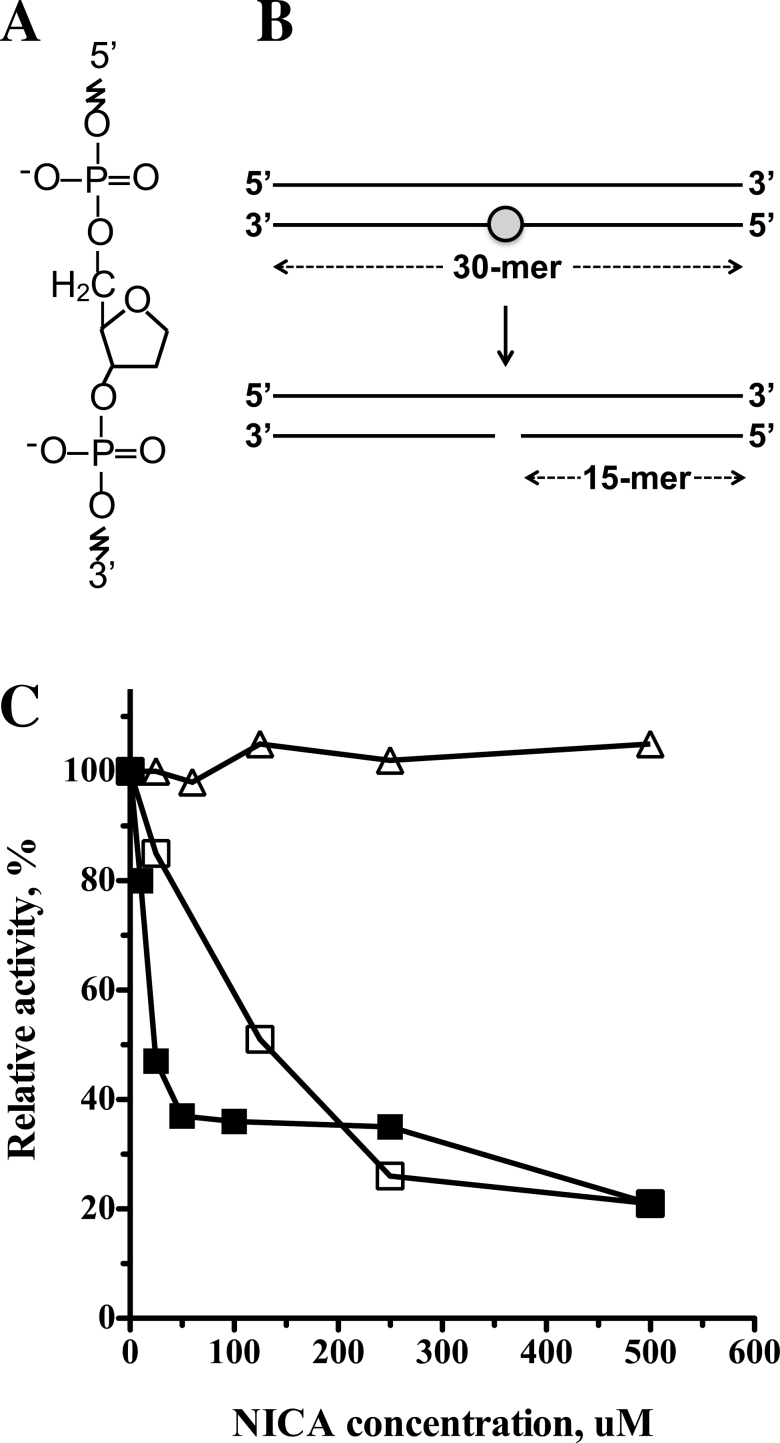
30mer oligonucleotide containing furanyl abasic site (Figure 1A) had the sequence 5′- G GTG CAT GAC ACT GTF ACC TAT CCT CAG CG -3′ (F = furanyl abasic site). The complementary oligonucleotide had the sequence 5′- CG CTG AGG ATA GGT CAC AGT GTC ATG CAC C -3′.
Figure 1.
APE1 DNA repair assay. (A) The structure of a furanyl abasic site. (B) Outline of the APE1 DNA repair assay. A radiolabeled synthetic short double-stranded DNA carrying a site-specific furanyl abasic site (marked by a circle) was incubated with a protein extract. The action of APE1 caused a nick at the lesion site, which enabled subsequent quantification of reaction products. (C) Inhibition of DNA repair activities by the APE1 inhibitor NICA. Closed squares, purified APE1 enzyme (0.1 unit per in the reaction); Open squares, APE1 activity in PBMC extract (0.025ng/µl protein extract in the reaction); Open triangles, OGG1 activity in PBMC extract (0.4 µg/µl protein extract in reaction).
APE1-F assay
30mer oligonucleotide containing furanyl abasic site (Figure 1A) and 3′ Yakima yellow fluorescent tag had the sequence 5′- G GTG CAT GAC ACT GTF ACC TAT CCT CAG CG Y-3′ (F = furanyl abasic site; Y = Yakima yellow tag). The complementary oligonucleotide had the sequence 5′- CG CTG AGG ATA GGT CAC AGT GTC ATG CAC C -3′.
APE1 activity assay
The assays were adapted to a robotic platform, in which liquid handling of the nicking reactions were performed automatically by a Freedom EVO 200 robot (Tecan) and Freedom EVOware software (Tecan). Denatured radioactive DNA products were analyzed by electrophoresis on a 15% polyacrylamide gel containing 8M urea, in 89mM Tris.borate, 2.5mM ethylenediaminetetraacetic acid (EDTA) pH 8.0, at 1500V for 2h at 45°C–50°C. The distribution of radiolabeled DNA products was visualized and quantified using a Fuji BAS 2500 phosphorimager. Fluorescent denatured DNA products were analyzed by capillary gel electrophoresis, using the ABI3130XL genetic analyzer (Applied Biosystems), and the GeneMapper (Applied Biosystems) and PeakAnalyzer (Robiotec, Rehovot, Israel) softwares. The optimized final APE1-F and APE1-P reaction conditions are presented below.
The reaction mixture (20 µl) contained 75 mmol/l Tris (pH 7.8), 0.1 mmol/l EDTA, 9 mmol/l magnesium chloride (MgCl2), 42.4 mmol/l potassium chloride (KCl), 0.25 µg/µl bovine γ-globulin, 0.25% polyvinyl alcohol (PVA), 0.25 mmol/l Spermidine; 0.05 mmol/l Spermine; 35 nmol/l substrate, and 0.015ng/µl protein extract for APE1-F or 0.02ng/µl protein extract for APE1-P. The reaction was carried out at 37°C for 15min, after which it was stopped by heat inactivation at 95°C for 2min. One unit of APE1 activity is defined to cleave 1fmol of DNA substrate in 1h at 37°C, under these conditions.
OGG1 assay
The fluorescence-based OGG1 assay conditions have been described previously (16).
Analysis of the APE1 Asp148Glu SNP
For APE1 (APEX1) genotyping analysis, genomic DNA was extracted from blood using a commercially available kit according to the manufacturer’s recommendations (PUREGENE).
Genotyping of APEX1 (SNP Assay-on-Demand C_8921503_10) was performed by allelic discrimination using a Taqman-based SNP genotyping assay on the ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). The assay was done in 20 μl reaction volume containing 1 μl genomic DNA, 0.15 μl primer/probe mix, 5 μl TaqMan genotyping master mix (Applied Biosystems), and 14 μl of double distilled water. The thermocycling included a pre-run of 2min at 50°C followed by 10min at 95°C, then 40 cycles of 10 s at 95°C followed by 60 s at 60°C. For APEX1 Assay-on-Demand, the context sequence is AATTCTGTTTCATTTCTATAGGCGA[G/T]GAGGAGCATGATCAGGAAGGCCGGG. Approximately 5% of duplicated samples were used for internal quality control. No discrepancies were observed.
Optimization of APE1 assay reaction conditions
APE1 assay conditions were rigorously optimized, including buffer type and pH (Tris pH 7.0–8.0; Na-Phosphate pH 7.0–8.0; 3-(N-morpholino)propanesulfonic acid (MOPS) pH 7.0–7.8; and Tricin pH 7.4–8.0), salts [KCl and sodium chloride (NaCl)] at concentration of 10–200mM, metal ions at 0.1 or 1mM [Ni2+, Co2+, Ca2+, Mn2+, Fe3+, Fe2+, Zn2+ and Mg2+ (0 to 15mM)], stabilizing agents including bovine serum albumin (1 or 5mg/ml), bovine γ-globulin (0–10mg/ml), polyethylene glycol 6000 (1% or 5%), Ficol (1% or 5%), Dextran (1% or 5%), and PVA (0–5%). Using these stabilizing agents, optimization was also performed to stabilize the diluted extract added to the APE1 reaction. In addition, the effect of the following compounds on the APE1 reaction was examined: polydA-polydT (0.3–3 pmol), poly dT (0.1–1 pmol), tRNA (0.5 or 1mg), EDTA (0–2mM), ethyleneglycol-bis(aminoethylether)-tetraacetic acid (0.1 and 1mM), spermine (0.1–1mM), spermidine (0.1–1mM), dithiothreitol (0–10mM), adenosine triphosphate (0.2 and 1mM), the amount of DNA substrate (0.125–8 pmol), and the amount of protein extract (5–200ng/ml protein). Reaction kinetics was performed for 10–120min.
Protein and inhibitor
Purified APE1 was purchased from NEB (cat# M0282), and 7-nitro-1H-indole-2-carboxylic acid was bought from Acros Organics (cat# KM09181DA).
Statistical analysis
Statistical analysis was performed essentially as described previously (16). The odds ratio of lung cancer was estimated for the APE1 test using a conditional logistic regression model adjusting for smoking status, automatically adjusting for the matching variables, sex, age, place of residence and ethnicity. Odds ratios were estimated for the APE1 test as a continuous variable (assuming a linear relation with the log odds), and also categorized into three groups according to the tertiles of the controls. In the latter case, a test for a linear trend across tertiles was conducted using scores of 1, 2 and 3 for the three tertile groups. The same methods were used for evaluating the relationship between APE1 Asp148Glu SNP and lung cancer. Models evaluating the three combinations of alleles (2 degrees of freedom test) and the trend (1 degree of freedom test) were examined.
Significance tests for odds ratios and for comparing models combining the APE1 assay and APE1 Asp148Glu SNP with models including only the APE1 assay were performed using the likelihood ratio test.
The fluorescent assay results were compared with the radioactive assay by calculating Pearson correlations within the case patients and control subjects separately and overall, and by Altman–Bland analysis examining the standard deviation of pairwise differences in test results. Relationships between APE1 Asp148Glu SNP and APE1 assay results were evaluated by multiple linear regression with APE1 as the dependent variable.
All the statistical analyses were performed using S-Plus (TIBCO Software) and/or SAS software (version 9.2; SAS Institute).
Results
Characterization of APE1 activity in PBMC protein extracts
To determine whether inter-individual variations in APE1 activity are associated with increased risk of lung cancer, we developed an assay for measuring the enzymatic activity of APE1 in protein extracts prepared from PBMC, based on an assay routinely used in the DNA repair field (29). The assay measures the incision activity of APE1 at a synthetic AP site (Figure 1A) in a 32P end-labeled 30-base pair synthetic DNA duplex, which leads to the conversion of the labeled 30mer to a 15mer oligonucleotide (Figure 1B). The reaction can be monitored by electrophoresis in a denaturing urea-polyacrylamide gel, followed by phosphorimaging visualization, and quantification (Supplementary Figure 1, available at Carcinogenesis Online).
Human cells contain two AP endonucleases, APE1 and APE2. However, APE1 is the main AP endonuclease in human cells, accounting for >95% of the total cellular AP site incision activity (30,31). To examine whether the incision of the substrate at the AP site is indeed due to the activity of APE1 in the PBMC extract we used the specific APE1 inhibitor 7-nitro-1H-indole-2-carboxylic acid (CRT0044876) (32). As can be seen in Figure 1C, 7-nitro-1H-indole-2-carboxylic acid (NICA) strongly inhibited the incision activity by purified APE1, as expected. It also strongly inhibited the incision of the AP site in PBMC extracts, although at higher concentrations. In contrast, OGG1 DNA glycosylase activity was unaffected (Figure 1C).
Development of the APE1 assay
The APE1 assay was rigorously optimized to yield a reproducible and robust assay, suitable for measuring inter-individual variations in the population. This required optimizing protein extract preparation, extract stability, and incision reaction conditions.
Protein extract preparation
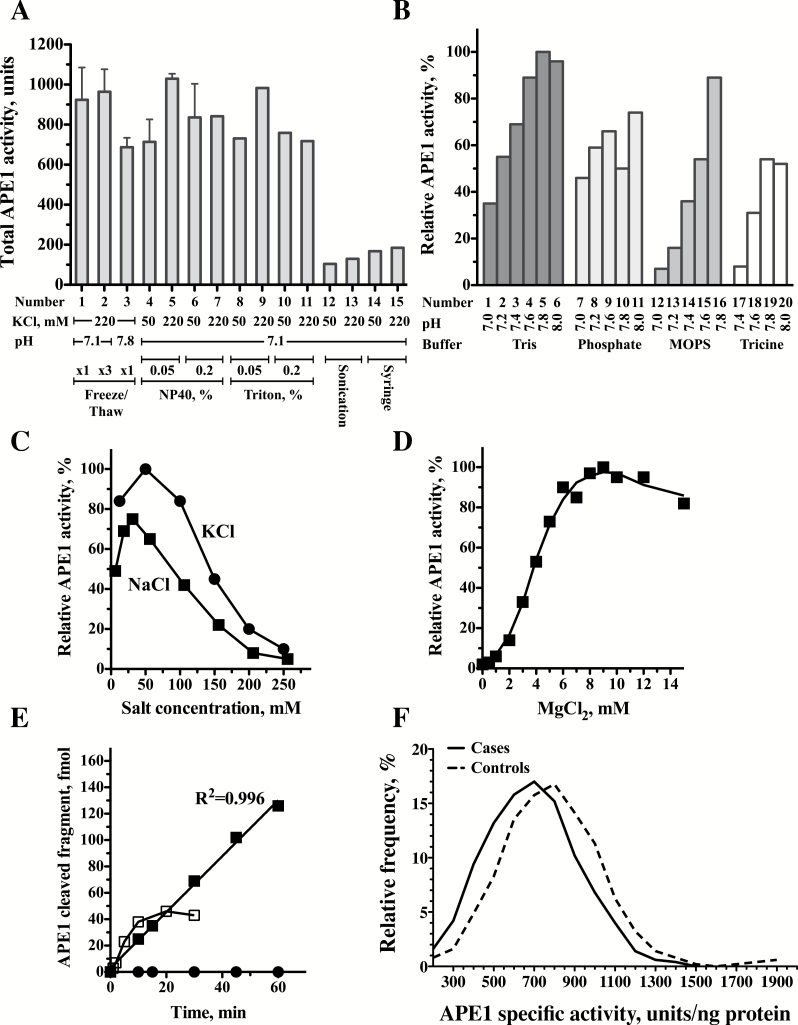
Preparing the whole PBMC protein extract is critical to ensure that maximal APE1 activity is retrieved. The two critical stages in such a procedure are lysis of the cells, and salt extraction of the APE1 from the nucleus. We tested five methods of cell lysis: freeze-thaw in hypotonic buffer, treatment with the detergents NP40 or Triton X-100, sonication, and sheering by passage through a syringe needle. Salt extraction was performed with 50 or 220mM KCl. As can be seen in Figure 2A, there were big differences in the total APE1 activity extracted by the various methods. Notably, sonication or sheering followed by salt extraction gave low APE1 activity yields, whereas the other three methods gave high yields that were comparable with each other. The freeze-thaw method was selected, because it was used also to prepare the extracts for assaying the DNA glycosylases OGG1 and MPG, and therefore allowed measurement of multiple enzyme activities in the same extract.
Figure 2.
Optimizations of the radioactivity-based APE1 DNA repair assay. (A) Optimization of the preparation of protein extracts. Protein extracts were prepared under various conditions and assayed for APE1 activity. Lanes 1–3 Freeze-Thaw extraction; Lanes 4–7 extraction with NP40; Lanes 8–11 extraction with Triton; Lanes 12–13 Extraction by sonication; Lanes 14–15 Extraction by Syringe. (B) Effects of buffers and pH on APE1 activity. APE1 enzyme activity is presented relative to the activity in Tris pH 7.8 (set as 100%). Lanes 1–6 Tris buffer; Lanes 7–11 Phosphate buffer; Lanes 12–16 MOPS buffer; Lanes 17–20 Tricine buffer. (C) Effect of different salt concentrations on APE1 activity. APE1 enzyme activity is presented relative to the activity in 50mM KCl (set as 100%) Closed circles, KCl; Closed squares, NaCl. (D) Effect of MgCl2 concentrations on APE1 activity. APE1 enzyme activity is presented relative to the activity in 9mM MgCl2 (set as 100%). (E) Time course of APE1 DNA repair activity in protein extracts prepared from peripheral blood mononuclear cells. Closed squares, reaction under optimized conditions; Open squares, reaction before optimization; Closed circles, control DNA without the abasic site. (F) Relative frequency plots for APE1 activities were determined in 99 case patients (continuous line) and 99 matched controls subjects (dashed line). The relative frequencies as percent were plotted using GraphPad Prism version 5.00, with bin width of 100 units that was automatically chosen by the software. The relative frequency plots were smoothed by two neighbors on each size, zero order of polynomial smoothing. Case patients exhibit a shift to lower values of APE1 enzyme activity.
Stabilizing the protein extract
When very low amounts of protein extract are used in enzymatic reaction mixtures, it is often difficult to obtain reproducible activities due to non-specific losses from adherence to the tube, or dissociation of complexes involved in the reaction under study. This was clearly the case for the APE1 assay, as indicated by the decrease in the reaction rate with time (Supplementary Figure 2A, available at Carcinogenesis Online). To overcome this problem we tried various stabilizing additives including protein carriers (bovine γ-globulin and bovine serum albumin), polycationic polyamines (spermine and spermidine), and different macromolecular crowding agents (Ficoll, dextran, polyethylene glycol and PVA). Of these, addition of 1% PVA to the reaction mixture dramatically improved the linearity of the APE1 activity with time (Supplementary Figure 2A, available at Carcinogenesis Online). Stabilization by PVA was also needed for the diluted extract, to avoid loss of activity during preparation of the reaction mixtures. Thus, while pre-incubating the diluted extract for 30min prior to its addition to the reaction mixture caused an ~50% reduction in the APE1 activity (Supplementary Figure 2B, available at Carcinogenesis Online), including 1% PVA in the extract dilution buffer maintained full activity (Supplementary Figure 2B, available at Carcinogenesis Online). After further optimization, the extract dilution buffer for the APE1 reaction contained 50mM Tris (pH 7.8), 0.5% PVA, 50mM KCl, 0.5mM spermidine, 0.1mM spermine and 0.5mg/ml bovine γ-globulin.
Optimization of APE1 assay conditions
To select the buffer to be used in the APE1 reaction, we examined the Tris–HCl, Na-phosphate, MOPS–KOH and Tricine, at a pH range of 7.0–8.0 (Figure 2B). Tris–HCl pH = 7.8 was chosen for the APE1 reaction because it exhibited the highest activity. Titration of KCl and NaCl concentrations showed higher APE1 activity with KCl than with NaCl, with optimal APE1 activity observed at 50mM KCl (Figure 2C). The incision by APE1 requires Mg2+ as a cofactor. Titration of MgCl2 concentrations showed an optimal APE1 activity at 9mM (Figure 2D). The final standard reaction mixture contained 75mM Tris (pH 7.8), 0.1mM EDTA, 9mM MgCl2, 42.4mM KCl, 0.25 μg/μl bovine γ-globulin, 0.25% PVA, 0.25mM spermidine, 0.05mM spermine, 35nM substrate and 0.02ng/μl protein extract. Under these conditions, the reaction was greatly improved over the reaction under non-optimized conditions (Figure 2E). APE1 incision was not observed with a control substrate without an AP site, and it was linear with time up to 60min and with extract protein concentration up to 20ng/μl (Supplementary Figure 1, available at Carcinogenesis Online).
Low APE1 activity is associated with increased lung cancer risk
The association between APE1 enzymatic activity and lung cancer risk was examined in 99 lung cancer patients and 99 matched control subjects. Blood specimens were collected from each participant, after which PBMC were isolated, and protein extracts prepared and assayed for APE1 enzymatic activity under the optimized conditions described above. Figure 2F shows the distribution of APE1 activity spanning 348–1235 and 413–1885 units/ng protein for case patients and control subjects, respectively. A shift to lower APE1 values is observed in case patients compared with control subjects (Figure 2F). The mean value of APE1 in cases was 691 [95% confidence interval (CI) = 655–727] units/ng protein, significantly lower than in control subjects, who had a mean of 793 (95% CI = 751–834) units/ng protein (P = 0.0006). The APE1 activity in the control subjects appeared lower in males (749 units/ng protein) than in females (857 units/ng protein), but the difference between the genders among patients was much smaller (684 units/ng protein in males versus 702 in females) (test for gender-disease interaction: P = 0.055; Table 1). The APE1 activity was lower in those aged >65 years than in those aged ≤65 years (P = 0.009) (Table 1). There was no significant difference in APE1 activity between patients with adenocarcinoma and patients with squamous cell carcinoma (Table 1).
Table 1.
Distribution of selected characteristics and APE1 activity value in lung cancer patients and control subjectsa
| Variable | Control subjects (n = 100) | Case patients (n = 100) | P | ||
|---|---|---|---|---|---|
| No. | APE1 mean (95% CI) | No. | APE1 mean (95% CI) | ||
| Allb | 99 | 793 (751–834) | 99 | 691 (655–727) | 0.0006c |
| SQCC | 30 | 656 (598–713) | |||
| Adenocarcinoma | 45 | 689 (643–735) | |||
| P comparing subgroupsd | 0.43 | ||||
| P interactione | 0.99 | ||||
| Age, y | |||||
| ≤65 | 40 | 844 (781–907) | 40 | 713 (655–771) | |
| >65 | 59 | 758 (705–812) | 59 | 676 (631–722) | |
| P comparing subgroupsf | 0.009 | ||||
| P interactione | 0.54 | ||||
| Sex | |||||
| Male | 59 | 749 (711–788) | 59 | 684 (642–725) | |
| Female | 40 | 857 (774–940) | 40 | 702 (637–767) | |
| P comparing subgroupsf | 0.041 | ||||
| P interactione | 0.055 | ||||
| Smoking status | |||||
| Never smoked | 50 | 819 (753–885) | 24 | 704 (634–775) | |
| Past smoker | 27 | 768 (700–837) | 36 | 702 (637–767) | |
| Current smoker | 22 | 765 (691–839) | 36 | 664 (618–711) | |
| P comparing subgroupsf | 0.16 | ||||
| P interactione | 0.72 | ||||
aAPE1 activity was measured as described in the ‘Materials and methods’. One participant did not have a known APE1 value. This participant and the matched controls were excluded from the analysis. Three case participants did not have a known smoking status.
bOf the 100 lung cancer cases, 30 had squamous cell carcinoma (SQCC), 46 had adenocarcinoma, 14 BAC, 4 adenosquamous carcinoma, 4 adenoBAC; 1 small cell carcinoma and 1 unknown histology.
cAnalysis of covariance comparing cases with controls, with matched pair and smoking status as a covariate.
dAnalysis of covariance comparing histological type within cases, with smoking status, age (continuous) or gender as covariates.
eTest for interaction between case–control status and the variable of interest.
fAnalysis of covariance comparing subsets defined by the variable of interest and stratified by cases and controls, with smoking status, age (continuous) or gender as covariates, as appropriate.
To determine whether smoking status is associated with APE1 activity level we calculated the mean APE1 activity level for each smoking status in case patients and control subjects. As can be seen in Table 1, the mean levels of APE1 activity among current, past and never smokers were similar, indicating that smoking did not materially affect the level of APE1 enzyme activity in PBMC. In addition, no interaction between smoking status and APE1 activity was found when a test for interaction was performed.
The association between the level of activity of APE1 and the probability of having lung cancer, adjusted for smoking status, was explored using conditional logistic regression. When APE1 activity was used as a continuous variable the adjusted odds ratio for lung cancer associated with 1 SD (211 units) decrease in APE1 activity was 2.0 (95% CI = 1.3–3.1; P = 0.002) (Table 2). When APE1 activity values were divided into tertiles using the controls’ values, the adjusted odds ratio for the lowest tertile versus the highest was 3.3 (95% CI = 1.4–8.1; P = 0.008). A test for trend over the three tertiles was also significant (P = 0.004, Table 2). Thus, low APE1 activity is associated with lung cancer risk.
Table 2.
Conditional logistic regression analysis of APE1 activity value in lung cancer patients and control subjects
| Variable | No. of control subjects (%) | No. of case patients (%) | Adjusteda OR (95% CI) |
|---|---|---|---|
| APE1 (per 211U decrease)b,c | 96 (100.0) | 96 (100.0) | 2.0 (1.3–3.1) P = 0.002 |
| APE1 (by tertiles)d | 1.0 (referent) | ||
| >847 U | 32 (33.3) | 19 (19.8) | 0.9 (0.3–2.2) P = 0.77 |
| 718–847 U | 32 (33.3) | 17 (17.7) | 3.3 (1.4–8.1) P = 0.008 |
| ≤718 U | 32 (33.3) | 60 (62.5) | Trend testd P = 0.004 |
aConditional logistic regression for matched sets adjusted for smoking status (smoker, ex-smoker, never smoker).
bAPE activity was measured as described in the Materials and methods and was first fitted in the conditional logistic regression model as a continuous variable and with adjustment for smoking status. The odds ratio for smoking that was obtained with this model, was: ex-smoker v never smoker: 3.4 (95% CI= 1.4–8.2); current smoker v never smoker: 2.6 (1.1–6.1).
c211U represents 1 SD in the control group.
dTertiles of the control subjects’ values. The upper tertile was chosen as the referent.
Comparison of radioactivity- and fluorescence-based APE1 assays
The 32P-based assay has the advantage that the radioactive label has no effect on the enzymatic reaction. However, the special safety measures needed for handling radioactive material and the rather short half-life of the 32P label limit its broad usage. End labeling with a fluorescent tag offers an attractive alternative; however, these tags are generally big chemical moieties, and once attached to the DNA substrate might be recognized as ‘DNA damage’ and interfere with the target repair reaction. Having developed the 32P-based APE1 assay, we developed a parallel fluorescence-based version, and examined whether the two are equivalent.
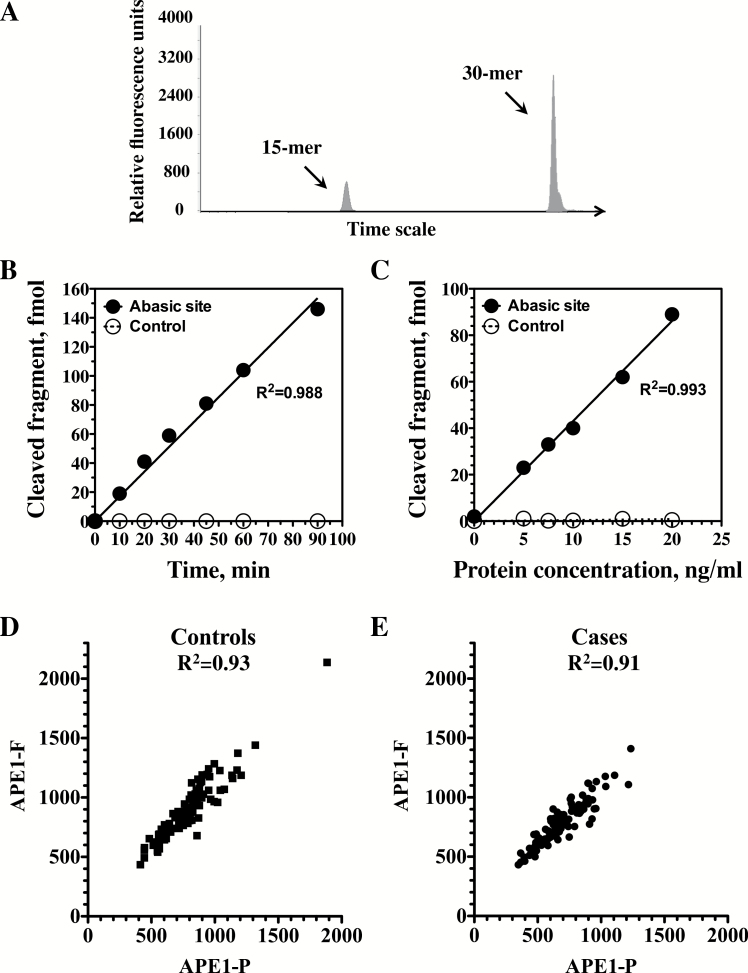
The fluorescence-based assay is similar to the radioactivity-based assay, except for the substitution of the fluorescent tag, Yakima yellow, as a 3′-end-label for the substrate. Detection was performed by capillary gel electrophoresis, using a commercial DNA sequencing machine (Figure 3A). The fluorescence-based APE1 assay was optimized, yielding essentially the same optimal reaction conditions, except for different protein extract concentrations. Under these conditions APE1 activity was linear with time for up to 90min of reaction time (Figure 3B; R 2 = 0.988), and with increasing protein concentrations up to 20ng/ml (Figure 3C; R 2 = 0.993). Cleavage was observed with the substrate carrying the AP site, but not with the control oligonucleotide, which carried a G instead of the lesion (Figure 3B and C). Overall, the assay was developed to be highly reproducible and robust, with coefficient of variation of 15%.
Figure 3.
Fluorescence-based APE1 DNA repair assay. (A) Example of a fluorescent plot of the APE1 reaction products analyzed by capillary gel electrophoresis, using the ABI3130XL genetic analyzer and the GeneMapper software. (B and C) Time course and protein extract titration, respectively, of APE1 DNA repair activity under optimized reaction conditions in protein extracts prepared from peripheral blood mononuclear cells. Quantification was done by quantifying fluorescent plots such as the one presented in A. Closed circles, substrate with an abasic site; Open circles, control DNAs without the lesion. (D and E) Correlation between the radioactivity-based APE1 assay (APE1-P) and the fluorescence-based APE1 assay (APE1-F) in control subjects (closed squares; D) and case patients (closed circles; E).
In order to determine whether the fluorescent version of the APE1 assay could be an adequate replacement for the radioactive version, an Altman–Bland analysis comparing the different versions of test was performed assuming the radioactive test to be the gold standard. The standard deviation (SD = 81.3) of the differences between the fluorescent and radioactive versions was less than half of the standard deviation (SD = 202) of the radioactive version. The standard deviation of the residual for the regression of the radioactive assay on the fluorescent assay (SD = 75.8) that represents the error in predicting the radioactive assay result from the fluorescent result was also less than half of the likely difference between two individuals having the radioactive test. Likewise, the R 2 for regression of the radioactive assay on the fluorescent assay result shows that the fluorescent test explains 86% of the variance of the radioactive test. Furthermore, a comparison of the radioactivity-based and fluorescent-based assay revealed a very high correlation of 0.93 in control subjects and 0.91 in case patients (Figure 3D and E). These analyses suggest that the fluorescent assay is an adequate replacement for the radioactive version.
The APE1 Asp148Glu SNP is not associated with lung cancer
There are conflicting reports on the association of the APE1 Asp148Glu polymorphic variant with lung cancer risk. We determined the occurrence of this polymorphism for all the case–control subjects and tabulated the mean APE1 test value for each SNP allele (Table 3A). Using conditional logistic regression, we found no association between the Asp148Glu SNP and lung cancer when taken as a stand-alone factor (Chi-squared = 0.51 on 2 df, P = 0.77; test for trend: Chi-squared = 0.51 on 1 df, P = 0.48). In addition, no association was found between the Asp148Glu SNP and APE1 activity. This was true for both the radioactivity-based APE1 assay (Table 3A: F-test = 1.93 on 2, 97 df, P = 0.15; test for trend: F-test = 0.39 on 1, 98 df, P = 0.53); and the fluorescence-based APE1 assay (Supplementary Table 1A, available at Carcinogenesis Online: F-test = 2.62 on 2, 97 df, P = 0.08; test for trend: F-test = 0.82 on 1, 98 df, P = 0.37). In order to test whether Asp148Glu SNP affects the association between the APE1 activity and lung cancer, we compared the odds ratio (OR) for this association in three logistic regression models in which the Asp138Glu SNP was either included or not included in the model (Radioactive APE1—Table 3B, Fluorescent APE1—Supplementary Table 1B, available at Carcinogenesis Online). We found that the ORs did not change substantially on entry of Asp148Glu SNP to the model (Radioactive APE1—Table 3B: OR = 1.40 (95% CI = 1.13–1.73), P = 0.002 versus OR = 1.44 (95% CI = 1.15–1.81), P = 0.001 or 1.41 (95% CI = 1.14–1.76) P = 0.002; Fluorescent APE1– Supplementary Table 1B, available at Carcinogenesis Online: OR = 1.34 (95% CI, 1.11–1.61) P = 0.002 versus OR = 1.38 (95% CI = 1.14–1.69), P = 0.001 or 1.36 (95% CI = 1.12–1.65), P = 0.002) suggesting that the Asp148Glu SNP does not explain any of the association between the APE1 activity and lung cancer.
Table 3.
Analysis of the relationships between APE1 Asp148Glu, APE1 activity and lung cancer risk
A. Association of APE1 Asp148Glu with lung cancer, and with APE1 activity (radioactivity-based)
| Allele | Controls | Cases | Overall | χ 2 for association with lung cancer a | F-test for association with APE1 activity b | |
| All | n | 99 | 99 | 198 (100%) | 0.51 (2 df) | 1.93 (2, 97 df) |
| Mean APE1 (SE) | 793 (21) | 691 (18) | 742 (14) | P = 0.77 | P = 0.15 | |
| T/T | n | 42 | 34 | 76 (38.4%) | ||
| Mean APE1 (SE) | 736 (29) | 673 (33) | 708 (22) | |||
| T/G | n | 46 | 50 | 96 (48.5%) | ||
| Mean APE1 (SE) | 843 (34) | 712 (26) | 774 (22) | |||
| G/G | n | 11 | 15 | 26 (13.1%) | ||
| Mean APE1 (SE) | 804 (53) | 663 (33) | 723 (32) | |||
| Trend | 0.51 (1 df) | 0.39 (1, 98 df) | ||||
| P = 0.48 | P = 0.53 |
B. Logistic regression analysis of APE1 Asp148Glu and APE1 activity (radioactivity-based) in lung cancer
| Model | OR c APE1 (95%CI) | OR SNP1 (95%CI) | OR SNP2 (95%CI) | OR SNP trend (95%CI) |
| Smoking + APE1 | 1.40 (1.13, 1.73), P = 0.002 | — | — | — |
| Smoking + APE1 + SNP1 (wt v htrz) + SNP2 (wt v homz) | 1.44 (1.15, 1.81), P = 0.001 | 1.67 (0.81, 3.44), P = 0.37 | 1.42 (0.48, 4.24), P = 0.37 | — |
| Smoking + APE1 + SNP trend | 1.41 (1.14, 1.76), P = 0.002 | — | — | 1.32 (0.79, 2.19), P = 0.29 |
aTest for association between SNP and lung cancer using conditional logistic regression adjusted for smoking: 2 degree of freedom test and test for trend.
bTest for association between SNP and APE1 activity (radioactivity-based assay) using multiple linear regression, controlling for smoking and matched pairs: 2 degree of freedom test and test for trend.
cOdds ratios are expressed per 100 APE1 units.
Discussion
Effective methods to measure DNA repair in the population are expected to provide effective tools for cancer risk assessment (33). In this context, functional DNA repair enzyme assays offer the advantage of measuring the actual DNA repair capability, because they combine the outcome of multiple processes that regulate gene expression and enzyme activity, including gene polymorphism, epigenetic status, transcription, splicing, messenger RNA stability, translation, protein stability, post-translational modification, as well as lifestyle and environmental effects (34). Although many efforts have been made to use genomic markers, such as gene polymorphism and messenger RNA levels, such approaches have generally yielded small and often conflicting effects (35,36). For example, there are conflicting reports on whether the APE1 Asp148Glu polymorphism is or is not associated with lung cancer risk (37–39). In our study, we found this SNP to be associated neither with APE1 enzyme activity, nor with lung cancer risk; however, we found that low activity of APE1 enzyme was strongly associated with lung cancer risk.
The need to use labeled substrates for assaying DNA repair enzymatic activity raised the question of suitability of radioactive versus fluorescent labels. Fluorescently tagged DNA substrates are more user-friendly than the classical 32P-labeled DNA substrates. Although the 32P-label offers superior sensitivity and does not interfere with enzymatic measurements, its employment necessitates taking special safety precautions in the laboratory, thus limiting its use. On the other hand, while the sensitivity of fluorescence-based DNA repair assays is sufficient for most applications, fluorescent tags are foreign moieties, and when covalently linked to a DNA substrate may be recognized as ‘DNA damage’ by the DNA repair machinery, and interfere with its measurement. In this study, we examined this issue by separately developing APE1 assays using as substrates 32P-labeled and fluorescently tagged DNA substrates, and using each to measure APE1 activity in the protein extracts prepared from all case patients and control subjects. Our results clearly show that the two substrates yield equivalent results; therefore the fluorescence-based DNA substrate can be used as a valid substrate for measuring APE1 enzyme activity in PBMC extracts. This paves the way to broad use of the assay, and enables increased throughput via automated assays, readout and analysis, as we have done.
The association of low APE1 enzymatic activity with increased lung cancer risk can be explained by the multiple DNA repair functions of APE1, which cover both spontaneously generated and oxidation-induced abundant lesions, such as abasic sites and single-strand breaks. If not repaired, these lesions can cause mutations during trans-lesion DNA synthesis due to the lack of coding information, and moreover, can easily deteriorate into double-strand breaks, leading to deletion, translocations and other chromosomal aberrations, thereby facilitating carcinogenesis. Interestingly, increased expression of APE1 was reported in lung cancer tissues (40), and was correlated with poor prognosis following chemotherapy (41,42). Thus, while low expression of APE1 is a risk factor for lung cancer, the tumor itself often exhibits increased APE1 expression and activity. This apparently contradictory behavior of APE1 activity can be explained by a dual role of APE1 in carcinogenesis. Under normal conditions, low activity of APE1 allows an increased level of mutations and genome instability, which facilitates carcinogenesis. However, as the cells progress along the path towards cancer, those with increased APE1 activity are selected because better repair endows an advantage during proliferation. Such increased APE1 expression was also reported to cause drug resistance in lung cancer patients (42).
We have recently reported that a personalized integrated DNA repair score composed of a weighted combination of three DNA repair activities, OGG1, MPG and APE1, is strongly associated with lung cancer risk (18). APE1, as well as OGG1 and MPG, act to repair oxidative DNA damage, which is caused not only by agents such as tobacco smoke and heavy metals, but also by internal processes, primarily inflammation (43–45), and therefore inter-individual differences in DNA repair activities may play an important role in other cancer types. It was recently suggested that the majority of cancers are due to ‘bad luck’ (46), implying that they are unpreventable. Somewhat surprisingly, that report did not take into account inter-individual variations in DNA repair. Based on our results, and results from other labs, we suggest that certain individuals’ deficiency in DNA repair capacity rather than ‘bad luck’ is a major cause of cancer.
Supplementary material
Supplementary Table 1 and Figures 1–2 can be found at http://carcin.oxfordjournals.org/
Funding
NIH/NCI/EDRN (#1 U01 CA111219 to Z.L., T.P.E. and G.R.); Flight Attendant Medical Research Institute, Florida (#032001_CoE to Z.L. and T.P.E.).
Conflict of interest: T.P.E. and Z.L. have a patent on the OGG1 risk factor for lung cancer, and a patent application pending for the OGG1, MPG and APE1 panel of DNA repair biomarkers.
Supplementary Material
Glossary
Abbreviations
- AP
apurinic/apyrimidinic
- APE1
apurinic/apyrimidinic endonuclease 1
- CI
confidence interval
- EDTA
ethylenediaminetetraacetic acid
- KCl
potassium chloride
- MPG
N-methylpurine DNA glycosylase
- NaCl
sodium chloride
- OGG1
8-oxoguanine DNA glycosylase
- OR
odds ratio
- PBMC
peripheral blood mononuclear cell
- PVA
polyvinyl alcohol
References
- 1. Vogelstein B., et al. (2004) Cancer genes and the pathways they control. Nat. Med., 10, 789–799. [DOI] [PubMed] [Google Scholar]
- 2. Hanahan D., et al. (2011) Hallmarks of cancer: the next generation. Cell, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- 3. Friedberg E.C., et al. (2006) DNA Repair and Mutagenesis. 2nd edn. ASM Press, Washington, DC. [Google Scholar]
- 4. Hoeijmakers J.H. (2001) Genome maintenance mechanisms for preventing cancer. Nature, 411, 366–374. [DOI] [PubMed] [Google Scholar]
- 5. Welcsh P.L., et al. (2001) BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum. Mol. Genet., 10, 705–713. [DOI] [PubMed] [Google Scholar]
- 6. Markowitz S.D., et al. (2009) Molecular origins of cancer: molecular basis of colorectal cancer. N. Engl. J. Med., 361, 2449–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheadle J.P., et al. (2007) MUTYH-associated polyposis—from defect in base excision repair to clinical genetic testing. DNA Repair (Amst)., 6, 274–279. [DOI] [PubMed] [Google Scholar]
- 8. Modrich P. (1994) Mismatch repair, genetic stability, and cancer. Science, 266, 1959–1960. [DOI] [PubMed] [Google Scholar]
- 9. Wei Q., et al. (2000) Repair of tobacco carcinogen-induced DNA adducts and lung cancer risk: a molecular epidemiologic study. J. Natl. Cancer Inst., 92, 1764–1772. [DOI] [PubMed] [Google Scholar]
- 10. Paz-Elizur T., et al. (2003) DNA repair activity for oxidative damage and risk of lung cancer. J. Natl. Cancer Inst., 95, 1312–1319. [DOI] [PubMed] [Google Scholar]
- 11. Gackowski D., et al. (2003) Products of oxidative DNA damage and repair as possible biomarkers of susceptibility to lung cancer. Cancer Res., 63, 4899–4902. [PubMed] [Google Scholar]
- 12. Speina E., et al. (2003) Decreased repair activities of 1,N(6)-ethenoadenine and 3,N(4)-ethenocytosine in lung adenocarcinoma patients. Cancer Res., 63, 4351–4357. [PubMed] [Google Scholar]
- 13. Kennedy D.O., et al. (2005) DNA repair capacity of lymphoblastoid cell lines from sisters discordant for breast cancer. J. Natl. Cancer Inst., 97, 127–132. [DOI] [PubMed] [Google Scholar]
- 14. Paz-Elizur T., et al. (2006) Reduced repair of the oxidative 8-oxoguanine DNA damage and risk of head and neck cancer. Cancer Res., 66, 11683–11689. [DOI] [PubMed] [Google Scholar]
- 15. Wu X., et al. (2007) Mutagen sensitivity: a genetic predisposition factor for cancer. Cancer Res., 67, 3493–3495. [DOI] [PubMed] [Google Scholar]
- 16. Leitner-Dagan Y., et al. (2012) N-methylpurine DNA glycosylase and OGG1 DNA repair activities: opposite associations with lung cancer risk. J. Natl. Cancer Inst., 104, 1765–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crosbie P.A., et al. (2012) Elevated N3-methylpurine-DNA glycosylase DNA repair activity is associated with lung cancer. Mutat. Res, 732, 43–46. [DOI] [PubMed] [Google Scholar]
- 18. Sevilya Z., et al. (2014) Low integrated DNA repair score and lung cancer risk. Cancer Prev. Res. (Phila)., 7, 398–406. [DOI] [PubMed] [Google Scholar]
- 19. Leitner-Dagan Y., et al. (2014) Enzymatic MPG DNA repair assays for two different oxidative DNA lesions reveal associations with increased lung cancer risk. Carcinogenesis, 35, 2763–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krokan H.E., et al. (2013) Base excision repair. Cold Spring Harb. Perspect. Biol., 5, a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- 22. Hill J.W., et al. (2001) Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res., 29, 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pope M.A., et al. (2002) Escherichia coli apurinic-apyrimidinic endonucleases enhance the turnover of the adenine glycosylase MutY with G:A substrates. J. Biol. Chem., 277, 22605–22615. [DOI] [PubMed] [Google Scholar]
- 24. Parsons J.L., et al. (2005) APE1-dependent repair of DNA single-strand breaks containing 3′-end 8-oxoguanine. Nucleic Acids Res., 33, 2204–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ishchenko A.A., et al. (2002) Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature, 415, 183–187. [DOI] [PubMed] [Google Scholar]
- 26. Tell G., et al. (2009) The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxid. Redox Signal., 11, 601–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Al-Safi R.I., et al. (2012) Small-molecule inhibitors of APE1 DNA repair function: an overview. Curr. Mol. Pharmacol., 5, 14–35. [PubMed] [Google Scholar]
- 28. Li M., et al. (2014) Human apurinic/apyrimidinic endonuclease 1. Antioxid. Redox Signal., 20, 678–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wilson D.M., III, et al. (1995) Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J. Biol. Chem., 270, 16002–16007. [DOI] [PubMed] [Google Scholar]
- 30. Hadi M.Z., et al. (2000) Second human protein with homology to the Escherichia coli abasic endonuclease exonuclease III. Environ. Mol. Mutagen., 36, 312–324. [PubMed] [Google Scholar]
- 31. Kim Y.J., et al. (2012) Overview of base excision repair biochemistry. Curr. Mol. Pharmacol., 5, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Madhusudan S., et al. (2005) Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res., 33, 4711–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paz-Elizur T., et al. (2008) DNA repair of oxidative DNA damage in human carcinogenesis: potential application for cancer risk assessment and prevention. Cancer Lett., 266, 60–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Paz-Elizur T., et al. (2005) Interrogating DNA repair in cancer risk assessment. Cancer Epidemiol. Biomarkers Prev., 14, 1585–1587. [DOI] [PubMed] [Google Scholar]
- 35. Hung R.J., et al. (2008) International Lung Cancer Consortium: pooled analysis of sequence variants in DNA repair and cell cycle pathways. Cancer Epidemiol. Biomarkers Prev., 17, 3081–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vineis P., et al. (2009) A field synopsis on low-penetrance variants in DNA repair genes and cancer susceptibility. J. Natl. Cancer Inst., 101, 24–36. [DOI] [PubMed] [Google Scholar]
- 37. Popanda O., et al. (2004) Specific combinations of DNA repair gene variants and increased risk for non-small cell lung cancer. Carcinogenesis, 25, 2433–2441. [DOI] [PubMed] [Google Scholar]
- 38. Chen W., et al. (2014) The association of APE1 Asp148Glu gene polymorphisms and lung cancer risk: an updated meta-analysis. Tumour Biol., 35, 3597–3603. [DOI] [PubMed] [Google Scholar]
- 39. Li X., et al. (2014) The interaction of APEX1 variant with polycyclic aromatic hydrocarbons on increasing chromosome damage and lung cancer risk among male Chinese. Molecular carcinogenesis. [DOI] [PubMed] [Google Scholar]
- 40. Yoo D.G., et al. (2008) Alteration of APE1/ref-1 expression in non-small cell lung cancer: the implications of impaired extracellular superoxide dismutase and catalase antioxidant systems. Lung Cancer, 60, 277–284. [DOI] [PubMed] [Google Scholar]
- 41. Wang D., et al. (2009) APE1 overexpression is associated with cisplatin resistance in non-small cell lung cancer and targeted inhibition of APE1 enhances the activity of cisplatin in A549 cells. Lung Cancer, 66, 298–304. [DOI] [PubMed] [Google Scholar]
- 42. Li Z., et al. (2014) Predictive value of APE1, BRCA1, ERCC1 and TUBB3 expression in patients with advanced non-small cell lung cancer (NSCLC) receiving first-line platinum-paclitaxel chemotherapy. Cancer Chemother. Pharmacol., 74, 777–786. [DOI] [PubMed] [Google Scholar]
- 43. Lonkar P., et al. (2011) Reactive species and DNA damage in chronic inflammation: reconciling chemical mechanisms and biological fates. Int. J. Cancer, 128, 1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ohnishi S., et al. (2013) DNA damage in inflammation-related carcinogenesis and cancer stem cells. Oxid. Med. Cell. Longev., 2013, 387014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ferguson L.R. (2010) Chronic inflammation and mutagenesis. Mutat. Res., 690, 3–11. [DOI] [PubMed] [Google Scholar]
- 46. Tomasetti C., et al. (2015) Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science, 347, 78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.