Abstract
Chronic graft-versus-host disease (cGVHD) is an immune-mediated disorder and is the major long-term complication of allogeneic hematopoietic stem cell transplantation (allo-HSCT). The oral mucosa including the salivary glands is affected in the majority of cGVHD patients; however, at present there is only a limited understanding of disease pathobiology. In this study, we performed a quantitative proteomic analysis of saliva pooled from oral cGVHD(+) and oral cGVHD(-) patients using iTRAQ (isobaric Tags for Relative and Absolute Quantification) labeling, followed by tandem mass spectrometry. Among 249 salivary proteins identified by tandem mass spectrometry, 82 proteins exhibited altered expression in oral cGVHD patients compared to allo-HSCT patients without oral cGVHD. Many of the identified proteins function in innate or acquired immunity, or are associated with tissue maintenance functions such as proteolysis or the cytoskeleton. Using ELISA immunoassays, we further confirmed that two of these proteins, IL-1 receptor antagonist and Cystatin B, showed decreased expression in patients with active oral cGVHD (P < 0.003). Receiver Operator Characteristic analysis revealed that these two markers were able to distinguish oral cGVHD with a sensitivity of 85% and specificity of 60%, and showed slightly better discrimination in newly diagnosed patients studied within 12 months of allo-HSCT transplantation (sensitivity, 92%; specificity 73%). In addition to identifying novel potential salivary cGVHD biomarkers, our study demonstrates that there is coordinated regulation of protein families involved in inflammation, anti-microbial defense and tissue protection in oral cGVHD that may also reflect changes in salivary gland function and damage to the oral mucosa.
Keywords: chronic graft-versus-host disease, saliva, proteomics, iTRAQ mass spectrometry, biomarker discovery
INTRODUCTION
Chronic graft-versus-host disease (cGVHD) is a life-threatening immunological condition that occurs following allogeneic hematopoietic stem cell transplantation (allo-HSCT), affecting 30–70% of patients who survive more than 3 months [1–3]. Its presentation may be progressive, arising from acute GVHD that merges into chronic, or de novo, where there is no prior acute GVHD. cGVHD has been characterized as both an alloimmune and autoimmune disease, affecting multiple tissues of the transplant recipient including the skin, oral mucosa, liver and eyes [1]. It involves different, predominantly T cell-mediated immunologic mechanisms including donor-derived alloreactive T-lymphocytes, autoreactive T-lymphocytes and dysregulated expression of inflammatory mediators [4]. While T-lymphocytes are the primary mediators of cGVHD, a role for B-lymphocytes in cGVHD is suggested by the presence of serum autoantibodies and B-cell markers such as B-cell activating factor (BAFF), as well as recent promising results obtained using drugs that target B-cell surface antigens [5].
The oral mucosa is affected in the majority (51–63%) of cGVHD patients at initial diagnosis and is the second most commonly involved tissue after the skin [2]. Common manifestations of oral cGVHD include tissue atrophy, erythema and edema, lichenoid changes and mucoceles [6,7]. Damage to salivary glands frequently leads to xerostomia, which, together with reduced salivary immunoglobulin production increases the patient’s susceptibility to oral infections [8]. In more severely affected patients, significant pain associated with oral lesions and sclerodermatous changes can lead to fibrosis that causes trismus (limited mouth opening) and compromised oral function. In addition to the significant morbidity and mortality associated with cGVHD, the disease can mimic other autoimmune or inflammatory conditions such as scleroderma and lichen planus [1,9]. Hence, biomarkers that could distinguish cGVHD from other clinically-similar immune conditions would be very useful as diagnostic tools.
Since the oral mucosa and salivary glands are major target organs of a number of human diseases, salivary proteomics is an appropriate methodology for the molecular profiling of oral-associated diseases including cGVHD, Sjögren’s syndrome, periodontitis and oral cancer (reviewed in [10]). Saliva represents an ideal starting point for identifying potential cGVHD biomarkers since changes in the salivary proteome should be directly associated with the localized oral pathology. Saliva also has a lower proteomic complexity compared to serum, which in principle reduces the time and cost required for the analysis of mass spectrometry data [11]. Whole saliva is composed of fluid including proteins produced by major and minor salivary glands, as well as both secreted and non-secreted proteins produced by mucosal, periodontal, and immune cells that reside in the mouth. To date, several studies have used mass spectrometry or immunoassay-based approaches to identify potential oral GVHD markers [7,12,13]; however, to date there is little consensus as to whether any of the identified salivary proteins might be useful for diagnosis or predicting patient outcomes. In an effort to obtain a global profile of proteomic alterations occurring in oral cGVHD, we employed a quantitative mass spectrometry (MS) approach to identify salivary proteins displaying altered expression in patients with active oral cGVHD. We identified 82 salivary proteins that showed quantitative changes in expression in oral cGVHD, and have further validated two proteins using ELISA immunoassays.
MATERIALS AND METHODS
Study Populations
The clinical details and demographics of the allo-HSCT study population are described in Table 1. Patients who had received an allogeneic transplantation procedure at the Fred Hutchinson Cancer Research Center/Seattle Cancer Care Alliance (SCCA) were recruited for this study through the Long Term Follow-up (LTFU) Program, either at the anniversary visit, or during a later appointment required for ongoing treatment of cGVHD [14]. At these appointments a comprehensive assessment of patients was completed by an attending oncologist including computed tomography scan, complete physical examination including evaluation of cGVHD status, and profiling of serum proteins and electrolytes as needed. Global cGVHD severity was scored according to the NIH global severity scale [15] on a scale of 0–4 for each of 7 organs (mouth, skin, eye, gastrointestinal tract, liver, lung and joints/fascia; Asymptomatic=0 to severe=4). All allo-HSCT patients were in remission at the time of saliva collection.
Table 1.
Clinical characteristics of allo-HSCT patients used for proteomic studies
| Phase I | Phase II | |||||
|---|---|---|---|---|---|---|
| Characteristic a | Oral cGVHD(+)(N=10) | Oral cGVHD(−)(N=10) | P value b | Oral cGVHD(+)(N=36) | Oral cGVHD(−)(N=10) | P value b |
| Age, median (range) | 55 (38–67) | 56.5 (21-63) | 0.91 | 42 (25–76) | 38.5 (31–68) | 0.51 |
| Gender, n | ||||||
| Male | 5 (50%) | 5 (50%) | 1.0 | 23 (64%) | 3 (30%), | 0.077 |
| Female | 5 (50%) | 5 (50%) | 13 (36%) | 7 (70%) | ||
| Original disease, n | ||||||
| AML | 5 (50%) | 3 (30%) | 1.0 | 9 (25%) | 2 (20%) | 0.77 |
| ALL | 0 | 2 (20%) | 4 (11%) | 2 (20%) | ||
| CML | 0 | 2 (20%) | 2 (5.5%) | 1 (10%) | ||
| Myelofibrosis | 2 (20%) | 0 | 3 (8.3%) | 0 | ||
| NHL | 1 (10%) | 2 (20%) | 3 (8.3%) | 1 (10%) | ||
| Other | 2 (20%) | 1 (10%) | 15 (41.6%) | 4 (40%) | ||
| Conditioning regimen, n | ||||||
| Radiotherapy/chemotherapy | 6 (60%) | 6 (60%) | 1.0 | 23 (64%) | 3 (30%) | 0.77 |
| Chemotherapy only | 4 (40%) | 4 (40%) | 13 (36%) | 7 (70%) | ||
| Type of donor, n | ||||||
| Related | 4 (40%) | 4 (40%) | 1.0 | 16 (44.4%) | 5 (50%) | 1.0 |
| Unrelated | 6 (60%) | 5 (50%) | 20 (55.6%) | 5 (50%) | ||
| Haploidentical | 0 | 1 (10%) | 0 | 0 | ||
| Cell source, n | ||||||
| Bone marrow | 0 | 6 (60%) | 0.003 | 8 (22.2%) | 1 (10%) | 0.13 |
| PBSCs | 10 (100%) | 3 (30%) | 27 (75%) | 7 (70%) | ||
| Cord Blood | 0 | 1 (10%) | 1 (2.8%) | 2 (20%) | ||
| Type of transplant, n | ||||||
| Myeloablative | 5 (50%) | 4 (40%) | 1.0 | 29 (80.6%) | 7 (70%) | 0.67 |
| Non-myeloablative | 5 (50%) | 6 (60%) | 7 (19.4%) | 3 (30%) | ||
| GVHD prophylaxis, n | ||||||
| MTX + Tacrolimus | 3 (30%) | 4 (40%) | 0.76 | 18 (50%) | 4 (40%) | 0.62 |
| MMF + Tacrolimus | 2 (20%) | 1 (10%) | 1 (2.8%) | 0 | ||
| CSP + MMF | 3 (30%) | 5 (50%) | 5 (13.9%) | 1 (10%) | ||
| CSP + MTX | 1 (10%) | 0 | 3 (8.4%) | 0 | ||
| Other | 1 (10%) | 0 | 9 (25%) | 5 (50%) | ||
| Acute GVHD c, n | 7 (70%) | 9 (90%) | 0.58 | 20 (55.5%) | 6 (60%) | 1.0 |
| Months post-transplant saliva was collected, mean (range) | 16.1 (6.7–34.5) | 23.3 (12–62.7) | 0.26 | 40.8 (10–121.6) | 24.4 (12–80.9) | 0.16 |
Abbreviations used- Diseases: ALL, Acute Lymphocytic Leukemia; AML, Acute Myelogenous Leukemia; CML, Chronic Myelogenous Leukemia; NHL, Non-Hodgkin’s Lymphoma. Conditioning regimens: CSP, Cyclosporin; MMF, Mycophenolate plus Cellcept and Myfortic; MTX, Methotrexate
Age and saliva collection data were compared between oral cGVHD(+) and oral cGVHD(-) patient groups using a two-tailed Student T-test. The other patient characteristics were compared using a two-tailed exact Chi-square test.
Indicates number of patients where acute GVHD was seen in one or more tissues
Oral examinations to assess oral cGVHD were performed at the Oral Medicine clinic at SCCA. Diagnosis of oral cGVHD was based on mucosal changes including atrophy of the mucosal surfaces including loss of normal surface keratinization of gingiva and dorsal tongue; erythema, especially vascular inflammation; hyperkeratinization including lichenoid and plaque-like changes; mucoceles, especially on the soft palate and lower labial mucosa; ulcers; and erythema of the parotid gland duct [6]. The protocol was approved by the Institutional Review Board of the Fred Hutchinson Cancer Research Center, with all subjects providing informed consent.
For phase I of the study, an additional 20 healthy adults were recruited for salivary proteomic analysis through the University of Washington School of Dentistry. They were divided into 1) middle aged to elderly adults (n = 10) with an average age of 58 years (range 50–68 yrs) and 2) young adults (n = 10) with an average age of 27 years (range 21 to 34 yrs). Each group contained an equal number of males and females. The protocol was approved by the UW Institutional Review Board.
Saliva collection and processing
Unstimulated whole saliva was collected from each consented human subject essentially as described previously [16]. Subjects were asked to refrain from eating, drinking or brushing their teeth for at least 1 hour prior to collection. Immediately prior to collection, subjects rinsed their mouth with water. Whole saliva was collected from each subject over a period of 15 minutes directly into a sterile 50 ml vial which was kept on ice throughout. In cases where subjects had difficulty producing adequate saliva, a funnel was inserted on top of the vial to facilitate collection. After collection, saliva was centrifuged at 17,000g for 15 minutes at 4°C, which removes bacteria and mucosal cells. The supernatant was removed and treated with Protease Inhibitor Cocktail (50 μl/1 ml of whole saliva; Sigma Aldrich, St. Louis, MO, USA) to minimize protein degradation. Saliva samples were divided into 0.5 ml aliquots and stored at −80°C.
iTRAQ labeling of saliva samples
Proteins from four different pools of saliva, obtained from the oral cGVHD(+) and oral cGVHD(-) patients collected in Phase I (Table 1), as well as from two groups of healthy adults (to allow identification of salivary proteins that showed altered expression as a result of normal physiologic aging), were first trysinized and labeled with isobaric Tags for Relative and Absolute Quantification (iTRAQ) chemical tags.
Protein samples (240 μg) from each pooled saliva sample were prepared in a 8 M urea buffer after trichloroacetic acid precipitation and then reduced by incubation with 5 mM trifluoroacetic acid (TCEP) for 1 hour at 37°C; cysteine residues were blocked by incubation for 10 minutes in 10 mM methyl methane (MMTS) at room temperature. Proteins were digested by the addition of 5 μl of sequencing grade trypsin (Promega, Madison, WI) and incubation at 37°C for 2 hours, followed by a second incubation with an additional 5 μl of trypsin at 37°C for 16 hours. The trypsin-digested samples were labeled with iTRAQ reagents according to the manufacturer’s protocol (Applied Biosystems, Foster City, CA) as described previously [17]. Each pooled saliva sample was labeled with a different iTRAQ label, and then the 4 saliva samples were combined to create one sample for MS analysis.
Glycoprotein enrichment and analysis
Glycoproteins/peptides were separately isolated from the iTRAQ-labeled saliva pools as described previously [18]. Following trypsin digestion and iTRAQ labeling, the samples were desalted through a C18 column (Waters Corporation, Milford, MA) with 0.1% trifluoroacetic acid to remove uncoupled iTRAQ reagent. The samples were then oxidized with sodium periodate (final concentration 12.5 mM) by incubation for 1 hour at room temperature. Following a further desalting step on a C18 column, the samples were dried down and the flow-through fraction saved for tandem MS analysis. The freeze dried samples were coupled to hydrazide resin (Biorad, Hercules, CA) in 0.1 M sodium acetate, 0.15 M NaCl, pH 5.5. Following the 24 hour binding step, the supernatant was collected and the hydrazine resin was washed three times with 1.5 M NaCl, three times with 80% acetonitrile, three times with 100% methanol, three times with deionized water and finally three times with 0.1 M NH4HCO3, pH 8.3. The supernatants (non-glycopeptides) were saved for tandem MS analysis. The hydrazine resin was then treated with Peptide:N-Glycosidase F (PNGase F)(New England Biolabs, Beverly, MA) dissolved in 0.1 M NH4HCO3, pH 8.3, for a total of 48 hours. The supernatant containing the released glycopeptides was collected and the resin was further rinsed three times in 0.05 M NH4HCO3 buffer and then twice with 80% acetonitrile followed by centrifugation at 3,000g for 5 minutes after each wash. The glycopeptide-containing supernatants were then processed for tandem MS as described below.
Tandem mass spectrometry analysis and protein identification
Quantitative tandem MS analysis was performed using the 4800 Proteomics Analyzer with TOF/TOF Optics™ (AB SCIEX, Framingham, MA) and the MS reflector positive ion mode was used with automated acquisition of 800–4000 m/z range with 1000 shots per spectrum. A maximum of 15 peaks were selected per spot, with minimum signal to noise of 50 and cluster area of 500. Approximately 48,872 precursors were subjected to tandem MS in the whole study. Proteins were identified using the MASCOT algorithm (Matrix Science, Boston, MA) and searched against a human International Protein Index (IPI) database (version 3.84) with the ProteinPilot™ software (version 3.0) and the Paragon™ method [19]. The raw peptide identification results from the Paragon™ algorithm searches are further processed with the Pro Group™ algorithm within the ProteinPilot™ software before final display. Protein quantification was achieved by averaging iTRAQ ratios of all the peptides identified within each polypeptide. Normalization using a Gaussian distribution with median of 1 (when comparing all peptides between control and experimental groups) was performed after iTRAQ ratios were calculated. A protein was considered to be significantly increased in expression if the iTRAQ ratio (e.g. oral cGVHD(+)/oral cGVHD(-)) was >1.6, and to be significantly decreased if the iTRAQ ratio was <0.65 [20].
In order to exclude salivary proteins that might be markers of normal physiologic aging, all proteins showing a significantly increased iTRAQ ratio in both the allo-HSCT patient population (i.e. oral cGVHD(+)/oral cGVHD(-)) and during normal aging (i.e. elderly adults/young adults) were removed from the final MS dataset. The same exclusion criteria were applied to proteins that showed a significantly decreased iTRAQ ratio in both groups.
Protein function and pathway analysis
Gene Ontology (GO) analysis was performed using the online program Database for Annotation, Visualization and Integrated Discovery (DAVID), version 6.7 [21]. Proteins upregulated and downregulated in oral cGVHD(+) patients, relative to oral cGVHD(-) patients, were classified according to their biological process, cellular component, molecular function and cellular pathway.
ELISA assays
Proteins were chosen for validation as potential oral cGVHD biomarkers based on 1) a significant iTRAQ ratio (iTRAQ ratio < 0.65 or >1.6)[20], and 2) the availability of a high quality ELISA assay. IL-1 receptor antagonist (IL-1ra) and Cystatin B were measured using ELISA assay kits purchased from R&D Systems (Minneapolis, MN). Dilutions of saliva typically used in immunoassays were 1:1000 for IL-1Ra and 1:400 for Cystatin B. ELISA data was analyzed using parametric 4-PL curve fitting software to determine concentrations of samples based on a standard curve (Readerfit, Hitachi Solutions America). R square (R2) values for our ELISA standard curves were typically in the range of 0.98–0.995. Protein assays were carried out using a BCA protein assay kit (Pierce Biotechnology, Rockford, IL), and ELISA immunoassay data normalized to protein concentration, per milligram of salivary protein.
Statistical analysis
Protein levels determined using ELISA immunoassays were compared between groups using the Mann-Whitney U test and the correlation between marker protein levels was described by the Spearman rank correlation. Protein expression data was analyzed using Sigmaplot (Systat Software, San Jose, CA) or SPSS version 19 (IBM, Armonk, NY). A Receiver Operator Characteristic (ROC) analysis was used to assess the diagnostic potential of selected proteins, including ROC curves for single and combined protein markers, using Sigmaplot or SPSS. Differences were considered statistically significant at P < 0.05. Patient characteristics were compared using the Student t-test and exact Chi-square test.
RESULTS
Patient characteristics
Table 1 displays the clinical characteristics of the entire allo-HSCT population used in our study. The patients with or without oral cGVHD collected in phase I were used for the mass spectrometry studies, while patients collected in both phase I and phase II were used for the validation studies using immunoassays. The two patient groups in each phase of our study were generally well matched in terms of age, gender, original disease and type of transplant. Among the oral cGVHD patients, 63% exhibited a history of acute GVHD compared to 75% of the oral cGVHD(-) group (Table 1). The NIH global severity score of oral cGVHD patients varied from 1–10 (mean = 3.5, n = 42 patients), and the number of involved tissues varied from 1–4. After the oral mucosa (100% affected), skin was the most commonly involved site (n = 22, 52%), followed by the eye (n = 19, 45%). The vast majority (83%) of oral cGVHD patients showed disease involvement at two or more sites.
The mean whole saliva flow rates in oral cGVHD(+) patients was slightly lower than the mean flow rate seen in oral cGVHD(-) patients and in healthy adult controls, but the difference was not statistically significant (Table 2). Overall, patients with oral cGVHD were an average of 36 months post-transplant at the time of saliva collection, while the oral cGVHD(-) group were an average of 31.7 months post-allo-HSCT at sampling (P = 0.65).
Table 2.
Salivary flow rates of allo-HSCT patients and healthy adult subjects
| Study Group | Number of subjects | Salivary flow rate a | P value b |
|---|---|---|---|
| Oral cGVHD(+) | 37 | 0.37 ml/min ± 0.23 | - |
| Oral cGVHD(-) | 15 | 0.415 ml/min ± 0.16 | 0.49 |
| Elderly adults | 10 | 0.415 ml/min ± 0.05 | 0.53 |
| Young adults | 10 | 0.45 ml/min ± 0.1 | 0.30 |
flow rates are shown in ml/min ± 1 standard deviation (SD)
P values are shown comparing the salivary flow rate of each group of subjects to the oral cGVHD(+) patient population, using a two-tailed Student t-test
The oral cGVHD proteome
For the mass spectrometry (phase I) of the project, the four saliva samples, collected from 40 subjects split into two groups of allo-HSCT patients and two groups of healthy adults were each labeled with a different iTRAQ label, and then combined and subjected to tandem MS simultaneously. Out of a total of 249 proteins identified by tandem MS, 82 proteins were significantly changed in expression as a result of oral cGVHD, based on the iTRAQ data comparing the saliva from patients with oral cGVHD vs. no oral cGVHD. Among those, 44 proteins were significantly upregulated in oral cGVHD (Table 3) while 38 proteins were downregulated (Table 4). Of the 82 salivary proteins altered in oral cGVHD, 13 were identified by hydrazine affinity chromatography and tandem MS as being glycoproteins (Tables 3 and 4). Proteins involved in innate and acquired immunity and inflammation, as well as oral (tooth) protection and various housekeeping functions, were prominently represented in the MS dataset (see below).
Table 3.
Salivary Proteins Upregulated in Patients with Oral Chronic Graft-versus-Host Disease
| ACCESSION NUMBER | PROTEIN/GENE NAME | NO. OF PEPTIDES/% COVERAGE | ITRAQ RATIO (GVHD+/GVHD−) | ITRAQ RATIO (ADULT CONTROL)† |
|---|---|---|---|---|
| IPI00790899 | 55 kDa protein | 10/19.7% | 1.58 | 0.94 |
| IPI00300786 | alpha-amylase 1/AMY1A | 889/83.4% | 4.88 | 0.65 |
| IPI00847635 | Alpha-1-antichymotrypsin isoform 1/SERPINA3 | 8/16.3% | 2.38 | 1.23 |
| IPI00552432 | Basic salivary proline-rich protein 2/PRB2 | 12/29.7% | 12.3 | 1.3 |
| IPI00019482 | Basic salivary proline-rich protein 4/PRB4 | 12/29.7% | 10.72 | 1.14 |
| IPI00296654 | BPI fold-containing family B member 2/BPIFB2 | 17/22.7% | 2.36 | 0.50 ¶ |
| IPI00032293 | Cystatin C/CST3 | 11/45.9% | 2.1 | 0.9 |
| IPI00002851 | Cystatin D/CST5 | 23/43% | 2.85 | 1.14 |
| IPI00032294 | Cystatin S/CST4* | ND | 2.94 | 0.72 |
| 127/67.4% | 3.76 | 0.86 | ||
| IPI00305477 | Cystatin SN/CST1* | ND | 1.92 | 1.03 |
| 136/66.7% | 37.5 | 1.13 | ||
| IPI00220327 | Cytokeratin-1/KTR1 | 25/34.9% | 3.26 | 1.44 |
| IPI00021304 | Cytokeratin-2e/KRT2 | 10/14.9% | 2.25 | 1.19 |
| IPI00300725 | Cytokeratin-6A/KRT6A | 5/8.7% | 10.73 | 0.80 |
| IPI00019359 | Cytokeratin-9/KRT9 | 5/8% | 1.87 | 0.9 |
| IPI00009865 | Cytokeratin-10/KRT10 | 23/29.5% | 2.26 | 1.48 |
| IPI00873806 | FAM3B isoform A/FAM3B* | ND | 3.79 | 0.89 |
| IPI00784293 | Golgi membrane protein 1/GOLM1 | 7/17% | 2.03 | 0.64 ¶ |
| IPI00853068 | Hemoglobin alpha-2/HBA2 | 10/53.5 | 11.0 | 0.23 ¶ |
| IPI00654755 | Hemoglobin subunit beta/HBB | 13/55.1% | 15.0 | 0.35 ¶ |
| IPI00012024 | Histatin-1/HTN1 | 18/36.8% | 6.82 | 0.33 ¶ |
| IPI00012026 | Histatin-3/HTN3 | 0.56/27.5% | 5.53 | 0.395 ¶ |
| IPI00387120 | Ig kappa chain V-IV region Len | 15.8% | 2.29 | 1.02 |
| IPI00854743 | IgG heavy chain variable region/ENSP00000375034 | 2/13.5% | 2.10 | 0.99 |
| IPI00304808 | Kallikrein-1/KLK1 | 0.93/23.3% | 1.67 | 1.23 |
| IPI00383717 | Kallikrein, Intron-containing (fragment) | 6/22.2% | 1.53 | 1.09 |
| IPI00025023 | Lactoperoxidase/LPO | 20/19.5% | 3.05 | 0.47 ¶ |
| IPI00848342 | Lactotransferrin precursor/LTF* | ND | 1.53 | 0.96 |
| 61/48.3% | 3.16 | 0.39 ¶ | ||
| IPI00019038 | Lysozyme C/LYZ | 16/37.8% | 7.85 | 0.14 ¶ |
| IPI00009123 | Nucleobindin-2/NUCB2 | 12/23.3% | 1.94 | 0.87 |
| IPI00646304 | Peptidyl-prolyl cis-trans isomerase B/PPIB | 5/20.4% | 1.67 | 0.70 |
| IPI00004573 | Polymeric immunoglobulin receptor/PIGR | 87/43.9% | 1.91 | 0.26 ¶ |
| IPI00748533 | PRB3 protein/PRB3 | 18/28.5% | 2.20 | 0.94 |
| IPI00022974 | Prolactin-inducible protein/PIP | 29/65.8% | 13.0 | 0.41 ¶ |
| IPI00377025 | Proline-rich protein HaeIII subfamily 1/PRH1;PRH2 | 190/79.5% | 12.76 | 0.43 ¶ |
| IPI00856018 | Proline-rich protein 4/PRR4 | 7/26.1% | 5.40 | 0.74 |
| IPI00023011 | Proline-rich protein 3/SMR3B | 72/67.1% | 10.37 | 0.84 |
| IPI00008580 | Secretory leukocyte protease inhibitor/SLPI | 1/9.1% | 3.1 | 0.48 ¶ |
| IPI00642739 | TIMP1 metallopeptidase inhibitor 1/TIMP1 | 2/14.7% | 2.06 | 0.64 ¶ |
| IPI00299729 | Transcobalamin-1/TCN1 | 5/10.6% | 9.91 | 0.82 |
| IPI00414884 | Uncharacterized protein C4orf40/C4orf40 | 5/23.3% | 2.29 | 0.96 |
| IPI00374315 | UPF0762 protein C6orf58/C6orf58 | 11/24.6% | 1.65 | 0.45 ¶ |
| IPI00006705 | Uteroglobin/SCGB1A1 | 1.1/11% | 1.61 | 1.08 |
| IPI00784119 | V-ATPase subunit S1/ATP6AP1 | ND | 2.04 | 0.76 |
| IPI00166729 | Zinc-alpha-2-glycoprotein/AZGP1 | 61/52% | 21.57 | 0.69 |
Proteins obtained from the glycoprotein dataset. In some cases the same protein was identified in both the glycoprotein (*) and non-glycoprotein samples
iTRAQ ratios of healthy elderly adult/young adult saliva sample
Proteins (n=15) in the adult control saliva dataset that show the opposite trend in relative expression compared with the oral cGVHD(+)/oral cGVHD(−) dataset
ND, not determined
Table 4.
Salivary Proteins Downregulated in Patients with Oral Chronic Graft-versus-Host Disease
| ACCESSION NUMBER | PROTEIN/GENE NAME | NO. OF PEPTIDES/% COVERAGE | ITRAQ RATIO (GVHD+/GVHD−) | ITRAQ RATIO (ADULT CONTROL)† |
|---|---|---|---|---|
| IPI00021263 | 14-3-3 protein zeta-delta/YWHAZ | 1.43/28.6% | 0.50 | 2.0 ¶ |
| IPI00747533 | 56 kDa protein/PGD | 8/16.2% | 0.52 | 1.23 |
| IPI00021439 | Actin, cytoplasmic 1/ACTB | ND/60.8% | 0.44 | 2.09 ¶ |
| IPI00465248 | Alpha-enolase/ENO1 | 19/35.3% | 0.60 | 1.5 ¶ |
| IPI00419215 | Alpha-2-macroglobulin-like protein 1/A2ML1* | ND | 0.56 | 1.46 |
| ND/7.7% | 0.66 | 0.95 | ||
| IPI00654709 | Aldehyde dehydrogenase/ALDH3A1 | 3/5.7% | 0.59 | 1.30 |
| IPI00304557 | BPI fold-containing family A member 2 (SPLUNC2)/BPIFA2* | 0.47 | 1.42 | |
| 42/63.5% | 0.60 | 0.69 | ||
| IPI00027486 | Carcinoembryonic antigen/CEACAM5* | ND | 0.47 | 1.78 ¶ |
| IPI00292532 | Cathelicidin antimicrobial peptide/CAMP | 1/5.2% | 0.58 | 2.08 ¶ |
| IPI00021828 | Cystatin-B/CST6 | 31/79.6% | 0.45 | 1.34 |
| IPI00021885 | Fibrinogen alpha chain/FGA | 7/6.4% | 0.45 | 1.02 |
| IPI00298497 | Fibrinogen beta chain/FGB | 9/20% | 0.37 | 1.37 |
| IPI00219757 | Glutathione S-transferase P/GSTP1 | 7/43.3% | 0.63 | 1.90 ¶ |
| IPI00478493 | Haptoglobin-related protein/HRP* | ND | 0.49 | 1.66 ¶ |
| IPI00641737 | Haptoglobin/HP* | ND | 0.49 | 1.47 |
| IPI00003111 | Ig kappa chain V-I region AU/LOC652694 | 0.97/28.8% | 0.49 | 0.86 |
| IPI00430847 | IGKC protein/IGKC | 0.88/55.1% | 0.44 | 0.86 |
| IPI00829626 | IGL@ protein | 23/36.5% | 0.37 | 0.70 |
| IPI00479708 | Ig mu chain C region/IGHM* | ND/9% | 0.56 | 2.06 |
| 24/27.7% | 0.19 | 1.74 ¶ | ||
| IPI00175024 | IL-1 receptor antagonist protein/IL1RN | 9/49.7% | 0.40 | 1.24 |
| IPI00219077 | Leukotriene A-4 hydrolase, isoform 1/LTA4H | 1.10/5.2% | 0.57 | 1.54 ¶ |
| IPI00004310 | Ly6/PLAUR domain-containing 3/LYPD3* | ND | 0.56 | 1.57 ¶ |
| IPI00027409 | Myeloblastin/PRTN3 | 6/25.8% | 0.64 | 1.81 ¶ |
| IPI00236556 | Myeloperoxidase, proteinase 3/MPO* | ND | 0.54 | 0.96 |
| 8/7.3% | 0.55 | 0.72 | ||
| IPI00005721 | Neutrophil defensin 1/DEFA1 | 1.34/19.2% | 0.57 | 1.71 ¶ |
| IPI00027769 | Neutrophil elastase/ELANE | 2/2.6% | 0.61 | 1.405 |
| IPI00169383 | Phosphoglycerate kinase 1/PGK1 | 8/23.7% | 0.61 | 1.48 |
| IPI00010796 | Protein disulfide-isomerase/P4HB | 15/30.3% | 0.45 | 1.45 |
| IPI00479186 | Pyruvate kinase isozymes M1, M2/PKM2 | 9/16% | 0.60 | 1.41 |
| IPI00816799 | Rheumatoid factor D5 light chain (Fragment) | 7/35.6% | 0.41 | 1.04 |
| IPI00027462 | S100A9/MRP14 | 45/80.7% | 0.48 | 3.07 ¶ |
| IPI00216298 | Thioredoxin/TXN | 11/51.4% | 0.57 | 1.085 |
| IPI00300376 | Transglutaminase-3/TGM3 | 8/11.1% | 0.48 | 1.41 |
| IPI00788802 | Transkelotase variant (Fragment)/TKT | 1.07/8.8% | 0.62 | 1.13 |
| IPI00426051 | Uncharacterized protein DKFZp686C15213 | 11/20% | 0.55 | 1.02 |
| IPI00423462 | Uncharacterized protein DKFZp686K18196 | 88/34.7% | 0.50 | 1.07 |
| IPI00877792 | Fibrinogen gamma chain/FGG* | ND | 0.38 | 1.64 ¶ |
| 1.04/11% | 0.32 | 1.08 | ||
| IPI00887678 | Uncharacterized protein/LOC654188 | 6/21.5% | 0.60 | 1.27 |
Proteins obtained from the glycoprotein dataset. In some cases the same protein was identified in both the glycoprotein (*) and non-glycoprotein samples
iTRAQ ratios of healthy elderly adult/young adult saliva sample
Proteins (n=14) in the adult control saliva dataset that show the opposite trend in relative expression compared with the oral cGVHD(+)/oral cGVHD(−) dataset
ND, not determined
Additional analysis of the iTRAQ dataset obtained from the two healthy adult groups revealed that 29 proteins (35%) identified as part of the oral cGVHD proteome showed the opposite trend in expression, based on iTRAQ data, during normal physiologic aging (Tables 3 and 4). Specifically, 15 proteins that were upregulated in oral cGVHD saliva were downregulated during normal aging (comparing young adults to middle-aged adults); 14 proteins that were downregulated in oral cGVHD saliva were upregulated with normal physiologic aging.
Functional classification and pathway analysis
In order to functionally classify the salivary proteins that displayed altered expression in oral cGVHD, we performed Gene Ontology analysis using the DAVID Bioinformatics Knowledgebase [21]. The majority (33/59, 56%) of cGVHD-altered proteins that were identified by this database were either secreted or known to have an extracellular function (Supplementary Figure 1A). A large number of the proteins also had a protein binding activity, either as proteinase inhibitors or as enzymes such as proteases or hydrolases.
To identify specific trends in the expression data, we grouped proteins belonging to the same gene family or that were functionally similar, and determined a mean iTRAQ value (Supplementary Figure 1B). Among proteins upregulated in oral cGVHD were numerous proteins that function in innate immunity or oral tissue protection, including salivary cystatins, histatins and proline-rich proteins (Supplementary Figure 1B). Cytokeratins were also abundant in oral cGVHD saliva. Among the downregulated proteins were several proteinases including neutrophil elastase and myeloperoxidase, and as well as several glycolytic enzymes and anti-apoptotic factors (Supplementary Figure 1B). The results show that proteins within a family or that have a common function such as proteolysis or anti-microbial activity were frequently co-regulated in oral cGVHD.
Examination of IL-1ra and Cystatin B as potential oral cGVHD biomarkers
Fig. 1 summarizes the salivary expression of IL-1ra and CSTB in oral cGVHD(+) and oral cGVHD(-) patients, normalized to salivary protein concentration. Levels of IL-1ra and CSTB were both reduced ~2.3-fold in oral cGVHD patients compared to the oral cGVHD(-) group (Fig. 1, P < 0.003, Mann Whitney U test). The Spearman rank correlation between the two proteins was 0.69 (P < 0.001).
Figure 1. IL-1ra and Cystatin B are downregulated in oral cGVHD.
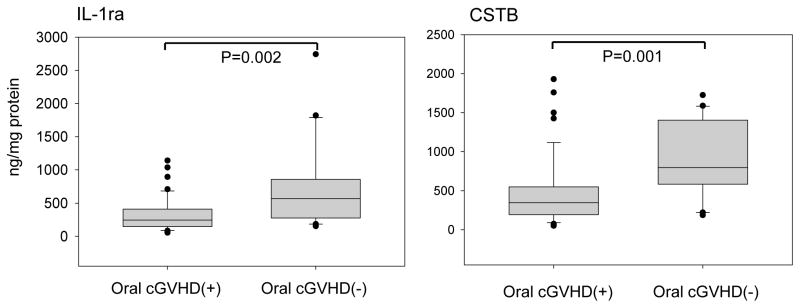
The levels of salivary IL-1Ra and CSTB were measured in individual oral cGVHD(+) patients and oral cGVHD(-) patients by ELISA immunoassay, and the results displayed as boxplots. The top and bottom of each box indicate the 75th and 25th percentile, respectively. The line inside each box indicates the median concentration. The top and the bottom ends of the whiskers represent 90th and 10th percentile, respectively. Outliers are shown as dots. Significance levels were calculated using Mann-Whitney U tests. The median values (from left to right) were: 244.7 ng/ml (oral cGVHD(+)) and 567.5 ng/ml (oral cGVHD(-))(IL-1ra); 345.8 ng/ml (oral cGVHD(+)) and 796.5 ng/ml (oral cGVHD(-))(CSTB).
Receiver Operator Characteristic (ROC) analysis was performed to further assess marker performance. The area under the curve (AUC) for the oral cGVHD group was 0.76 for the two markers, with a sensitivity of 85% and specificity of 60% (Table 5). Patients with oral cGVHD who were examined within 12 months of allogeneic transplantation exhibited slightly better sensitivity (92%) and specificity (73%) with the two-marker panel, though the number of patients was relatively small (n = 24)(Table 5).
Table 5.
Receiver Operator Characteristics for Oral cGVHD Biomarkers
| Marker | AUC c | P-value d | Sensitivity | Specificity |
|---|---|---|---|---|
| All allo-HSCT patients a | ||||
| IL-1ra | 0.74 | 0.002 | 87% | 60% |
| CSTB | 0.765 | 0.001 | 78% | 80% |
| IL-1ra + CSTB | 0.76 | 0.001 | 85% | 60% |
| Newly diagnosed patients b | ||||
| IL-1ra | 0.76 | 0.034 | 85% | 64% |
| CSTB | 0.78 | 0.019 | 92% | 64% |
| IL-1ra + CSTB | 0.8 | 0.014 | 92% | 73% |
Comparison was between 46 oral cGVHD(+) and 20 oral cGVHD(−) patients
Comparison was between 13 oral cGVHD(+) and 11 oral cGVHD(−) patients who were studied ≤ 12 months post-transplantation
Area under curve
Tests whether AUC value is significantly different from a value of 0.5
To date all proteins measured by immunoassay, including IL-1ra and CSTB, showed the same upward or downward trend in expression as was predicted from the iTRAQ analysis of pooled saliva samples. These findings support the use of the iTRAQ/tandem MS approach for proteomic discovery [17, 20].
DISCUSSION
Chronic GVHD remains the primary long term complication of allo-HSCT and is a major hurdle to the more widespread use of allo-HSCT worldwide [2,22]. Chronic inflammation, tissue damage and apoptosis are central aspects of cGVHD pathobiology; this tissue damage can be mediated directly by infiltrating T-lymphocytes and indirectly by pro-inflammatory cytokines including IL-1, TNF-α and IL-17 released by various types of immune cells [4,8,23]. In accordance with this disease model, we identified many proteins associated with oral cGVHD that function in inflammation and/or innate immunity [24], as well as proteins involved in normal cellular maintenance and survival (Tables 3 and 4). The presence of elevated cytokeratins in oral cGVHD saliva (Supplementary Figure 1B) is likely a biochemical marker of mucosal tissue damage and cell death [9]. Alterations in innate and acquired immunity is closely associated with the pathobiology of cGVHD in different target organs [13,25,26], and is clinically relevant to cGVHD-associated mortality which in most cases results from serious bacterial and/or fungal infections [1].
Saliva is largely a product of the parotid and submandibular salivary glands, and its protein and non-protein composition is a reflection of both local and systemic (serum-derived) sources. In cGVHD, damage to salivary glands is rapid and often severe, affecting both overall saliva production and composition [27–29]. Patients typically have higher concentrations of metal ions such as sodium and magnesium, as well as albumin, IgG and total protein [30,31]. Salivary flow rates are generally reduced [27], though some recent studies have found no difference in flow rates between oral cGVHD(+) and oral GVHD(-) patients ([13] and this study). Damage to salivary glands likely varies depending on the intensity of conditioning (including whether or not radiotherapy is used) and accompanying levels of T-lymphocytes and pro-inflammatory cytokines.
We examined the expression of two proteins selected from the MS dataset and confirmed in an expanded patient population that IL-1ra and cystatin B had significantly altered expression in association with oral cGVHD (Fig. 1). IL-1ra is a protein that binds to the IL-1 receptor and blocks IL-1 signaling. It is produced in both secreted (soluble) and intracellular forms by immune cells, keratinocytes and several other cell types [32]. Altered expression of IL-1Ra is associated with many chronic inflammatory diseases including rheumatoid arthritis. Donor and recipient IL-1Ra genotype have been shown to affect the incidence and/or severity of acute and chronic GVHD, respectively [33,34]. An altered balance between IL-1 and IL-1ra has been proposed to be one factor that governs the severity of several inflammatory conditions including rheumatoid arthritis and Crohn’s disease [35]. Salivary IL-1Ra levels were markedly reduced in oral cGVHD(+) patients compared to oral cGVHD(-)patients (Fig. 1), which might favor excessive IL-1 signaling in the oral mucosa [7]. Cystatin B (CSTB or Stefin B), a protease inhibitor and member of the cystatin family, functions as an inhibitor of cathepsins L, H and B and may regulate cathepsin-mediated apoptosis in neuronal cells [36]. In addition to being a protease inhibitor, CSTB has been proposed to have other functions including the regulation of glycolysis and inhibition of the interferon-β pathway in association with HIV infection [37]. One of the proteins CSTB interacts with is pyruvate kinase M2, an enzyme that is downregulated in oral cGVHD saliva (Table 4). Hence, the markedly diminished expression of CSTB in oral cGVHD could affect multiple cellular functions including cathepsin protease activity and interferon signaling. CSTB is present in human saliva primarily as several S-modified forms; however, it is unclear how these modifications might influence function [38].
An important focus of current GVHD research is to develop biomarker panels which might eventually be used as diagnostic tools and to inform patient prognosis and treatment strategies [39]. To date, a number of candidate cGVHD biomarkers have been reported, including serum B cell-activating factor (BAFF)[25], serum MHC class I-related chain A (MICA) and MICA antibodies [40], serum IL-15 [41], serum CXCL9 [42] and salivary lactotransferrin and lactoperoxidase [13]. These studies vary in terms of the biofluid examined (serum or saliva), type of population studied (pediatric vs. adult) and proteomic analysis methodology. While there is considerable overlap between the serum and salivary proteome [11], it is likely that serum and salivary biomarkers for cGVHD will be distinct, and measure disease activity in different tissues. In the case of IL-1ra and CSTB, additional studies with both independent sets of cGVHD patients as well as longitudinal studies will be necessary to determine their potential utility as oral biomarkers of cGVHD. Another recent report indicated that salivary IL-1ra levels were reduced in oral cGVHD patients, suggesting that this protein may indeed have utility as an oral cGVHD biomarker [43].
In summary, we have utilized a proteomic approach to describe the alterations in salivary protein expression that occurs in oral cGVHD patients, and have identified two potential biomarkers for this disease. These studies provide new insights into the inflammatory and anti-inflammatory pathways that may be involved in the persistence of this chronic inflammatory disease, and adds new proteins that might be used as research or diagnostic tools in the study of oral cGVHD.
Supplementary Material
A) Salivary proteins (n = 59) that exhibited altered expression in oral cGVHD were classified according to their molecular function. 37 proteins have a protein binding function, either as proteinase inhibitors or enzymes. B) Classification of upregulated (black bars) and downregulated (grey bars) proteins in oral cGVHD, grouped by common molecular function or pathway. The mean iTRAQ value for each family or functional group of proteins was determined from the data in Tables S1 and S2. The number of proteins represented in each functional grouping is shown in parentheses. In all cases proteins within a group showed the same trend in expression, with the exception of the cysteine proteinase inhibitor cystatin B, which is downregulated in the saliva of cGVHD patients.
Acknowledgments
We thank Drs. Paul Martin and Edmond Truelove for helpful discussions during the course of this project, and Dr. M. Elvira Correa (State University of Campinas, Campinas, Brazil) for help with saliva collections. We are also grateful to the many patients seen at Seattle Cancer Care Alliance who participated in this study. Finally, we wish to thank the anonymous reviewers for their helpful suggestions and comments. Financial Disclosure: Supported by grants from the National Institutes of Health (R21 DE017634) and the Douglass L. Morell Dentistry Research Fund (to R.B.P.).
Abbreviations used
- allo-HSCT
allogeneic hematopoietic stem cell transplantation
- AUC
area under the curve
- cGVHD
chronic graft-versus-host disease
- CSTB
cystatin B
- GVHD
graft-versus-host disease
- IL-1ra
interleukin-1 receptor antagonist
- iTRAQ
isobaric Tags for Relative and Absolute Quantification
- MS
mass spectrometry
- ROC
receiver operator characteristic
Footnotes
Conflict of Interest Statement: There are no conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee SJ, Vogelsang G, Flowers ME. Chronic graft-versus-host disease. Biol Blood Marrow Transplant. 2003;9:215–233. doi: 10.1053/bbmt.2003.50026. [DOI] [PubMed] [Google Scholar]
- 2.Lee SJ, Flowers ME. Recognizing and managing chronic graft-versus-host disease. Hematology Am Soc Hematol Educ Program. 2008:134–141. doi: 10.1182/asheducation-2008.1.134. [DOI] [PubMed] [Google Scholar]
- 3.Filipovich AH, Weisdorf D, Pavletic S, et al. National Institutes of Health consensus development project on criteria for clinical trials in chronic graft-versus-host disease: I. Diagnosis and staging working group report. Biol Blood Marrow Transplant. 2005;11:945–956. doi: 10.1016/j.bbmt.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Shlomchik WD. Graft-versus-host disease. Nat Rev Immunol. 2007;7:340–352. doi: 10.1038/nri2000. [DOI] [PubMed] [Google Scholar]
- 5.Shimabukuro-Vornhagen A, Hallek MJ, Storb RF, von Bergwelt-Baildon MS. The role of B cells in the pathogenesis of graft-versus-host disease. Blood. 2009;114:4919–4927. doi: 10.1182/blood-2008-10-161638. [DOI] [PubMed] [Google Scholar]
- 6.Schubert MM, Sullivan KM, Morton TH, et al. Oral manifestations of chronic graft-v-host disease. Arch Intern Med. 1984;144:1591–1595. [PubMed] [Google Scholar]
- 7.Fall-Dickson JM, Mitchell SA, Marden S, et al. Oral symptom intensity, health-related quality of life, and correlative salivary cytokines in adult survivors of hematopoietic stem cell transplantation with oral chronic graft-versus-host disease. Biol Blood Marrow Transplant. 2010;16:948–956. doi: 10.1016/j.bbmt.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagler RM, Nagler A. The molecular basis of salivary gland involvement in graft--vs.--host disease. J Dent Res. 2004;83:98–103. doi: 10.1177/154405910408300203. [DOI] [PubMed] [Google Scholar]
- 9.Pimentel VN, de Matos LS, Soares TC, et al. Perforin and granzyme B involvement in oral lesions of lichen planus and chronic GVHD. J Oral Pathol Med. 2010;39:741–746. doi: 10.1111/j.1600-0714.2010.00917.x. [DOI] [PubMed] [Google Scholar]
- 10.Al-Tarawneh SK, Border MB, Dibble CF, Bencharit S. Defining salivary biomarkers using mass spectrometry-based proteomics: a systematic review. OMICS. 2011;15:353–361. doi: 10.1089/omi.2010.0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loo JA, Yan W, Ramachandran P, Wong DT. Comparative human salivary and plasma proteomes. J Dent Res. 2010;89:1016–1023. doi: 10.1177/0022034510380414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imanguli MM, Atkinson JC, Harvey KE, et al. Changes in salivary proteome following allogeneic hematopoietic stem cell transplantation. Exp Hematol. 2007;35:184–192. doi: 10.1016/j.exphem.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bassim CW, Ambatipudi KS, Mays JW, et al. Quantitative Salivary Proteomic Differences in Oral Chronic Graft-versus-Host Disease. J Clin Immunol. 2012;32:1390–1399. doi: 10.1007/s10875-012-9738-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegenbart U, Niederwieser D, Sandmaier BM, et al. Treatment for acute myelogenous leukemia by low-dose, total-body, irradiation-based conditioning and hematopoietic cell transplantation from related and unrelated donors. J Clin Oncol. 2006;24:444–453. doi: 10.1200/JCO.2005.03.1765. [DOI] [PubMed] [Google Scholar]
- 15.Arai S, Jagasia M, Storer B, et al. Global and organ-specific chronic graft-versus-host disease severity according to the 2005 NIH Consensus Criteria. Blood. 2011;118:4242–4249. doi: 10.1182/blood-2011-03-344390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devic I, Hwang H, Edgar JS, et al. Salivary alpha-synuclein and DJ-1: potential biomarkers for Parkinson’s disease. Brain. 2011;134:e178. doi: 10.1093/brain/awr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdi F, Quinn JF, Jankovic J, et al. Detection of biomarkers with a multiplex quantitative proteomic platform in cerebrospinal fluid of patients with neurodegenerative disorders. J Alzheimers Dis. 2006;9:293–348. doi: 10.3233/jad-2006-9309. [DOI] [PubMed] [Google Scholar]
- 18.Ramachandran P, Boontheung P, Xie Y, Sondej M, Wong DT, Loo JA. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J Proteome Res. 2006;5:1493–1503. doi: 10.1021/pr050492k. [DOI] [PubMed] [Google Scholar]
- 19.Shilov IV, Seymour SL, Patel AA, et al. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Drummelsmith J, Winstall E, Bergeron MG, Poirier GG, Ouellette M. Comparative proteomics analyses reveal a potential biomarker for the detection of vancomycin-intermediate Staphylococcus aureus strains. J Proteome Res. 2007;6:4690–4702. doi: 10.1021/pr070521m. [DOI] [PubMed] [Google Scholar]
- 21.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 22.Seggewiss R, Einsele H. Immune reconstitution after allogeneic transplantation and expanding options for immunomodulation: an update. Blood. 2010;115:3861–3868. doi: 10.1182/blood-2009-12-234096. [DOI] [PubMed] [Google Scholar]
- 23.Serody JS, Hill GR. The IL-17 differentiation pathway and its role in transplant outcome. Biol Blood Marrow Transplant. 2012;18:S56–61. doi: 10.1016/j.bbmt.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujii H, Cuvelier G, She K, et al. Biomarkers in newly diagnosed pediatric-extensive chronic graft-versus-host disease: a report from the Children’s Oncology Group. Blood. 2008;111:3276–3285. doi: 10.1182/blood-2007-08-106286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shulman HM, Kleiner D, Lee SJ, et al. Histopathologic diagnosis of chronic graft-versus-host disease: National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease: II. Pathology Working Group Report. Biol Blood Marrow Transplant. 2006;12:31–47. doi: 10.1016/j.bbmt.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 27.Nagler RM, Nagler A. Salivary gland involvement in graft-versus-host disease: the underlying mechanism and implicated treatment. Isr Med Assoc J. 2004;6:167–172. [PubMed] [Google Scholar]
- 28.Soares AB, Faria PR, Magna LA, et al. Chronic GVHD in minor salivary glands and oral mucosa: histopathological and immunohistochemical evaluation of 25 patients. J Oral Pathol Med. 2005;34:368–373. doi: 10.1111/j.1600-0714.2005.00322.x. [DOI] [PubMed] [Google Scholar]
- 29.Noce CW, Gomes A, Copello A, et al. Oral involvement of chronic graft-versus-host disease in hematopoietic stem cell transplant recipients. Gen Dent. 2012;59:458–462. [PubMed] [Google Scholar]
- 30.Norhagen G, Engstrom PE, Bjorkstrand B, Hammarstrom L, Smith CI, Ringden O. Salivary and serum immunoglobulins in recipients of transplanted allogeneic and autologous bone marrow. Bone Marrow Transplant. 1994;14:229–234. [PubMed] [Google Scholar]
- 31.Nagler RM, Nagler A. The effect of pilocarpine on salivary constituents in patients with chronic graft-versus-host disease. Arch Oral Biol. 2001;46:689–695. doi: 10.1016/s0003-9969(01)00035-8. [DOI] [PubMed] [Google Scholar]
- 32.Arend WP, Malyak M, Guthridge CJ, Gabay C. Interleukin-1 receptor antagonist: role in biology. Annu Rev Immunol. 1998;16:27–55. doi: 10.1146/annurev.immunol.16.1.27. [DOI] [PubMed] [Google Scholar]
- 33.Cullup H, Dickinson AM, Jackson GH, Taylor PR, Cavet J, Middleton PG. Donor interleukin 1 receptor antagonist genotype associated with acute graft-versus-host disease in human leucocyte antigen-matched sibling allogeneic transplants. Br J Haematol. 2001;113:807–813. doi: 10.1046/j.1365-2141.2001.02811.x. [DOI] [PubMed] [Google Scholar]
- 34.Rocha V, Franco RF, Porcher R, et al. Host defense and inflammatory gene polymorphisms are associated with outcomes after HLA-identical sibling bone marrow transplantation. Blood. 2002;100:3908–3918. doi: 10.1182/blood-2002-04-1033. [DOI] [PubMed] [Google Scholar]
- 35.Arend WP. The balance between IL-1 and IL-1Ra in disease. Cytokine Growth Factor Rev. 2002;13:323–340. doi: 10.1016/s1359-6101(02)00020-5. [DOI] [PubMed] [Google Scholar]
- 36.Sun T, Turk V, Turk B, Kopitar-Jerala N. Increased expression of stefin B in the nucleus of T98G astrocytoma cells delays caspase activation. Front Mol Neurosci. 2012;5:93. doi: 10.3389/fnmol.2012.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rivera-Rivera L, Perez-Laspiur J, Colon K, Melendez LM. Inhibition of interferon response by cystatin B: implication in HIV replication of macrophage reservoirs. J Neurovirol. 2011;18:20–29. doi: 10.1007/s13365-011-0061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cabras T, Manconi B, Iavarone F, et al. RP-HPLC-ESI-MS evidenced that salivary cystatin B is detectable in adult human whole saliva mostly as S-modified derivatives: S-Glutathionyl, S-cysteinyl and S-S 2-mer. J Proteomics. 2011;75:908–913. doi: 10.1016/j.jprot.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 39.Levine JE, Paczesny S, Sarantopoulos S. Clinical applications for biomarkers of acute and chronic graft-versus-host disease. Biol Blood Marrow Transplant. 2012;18:S116–124. doi: 10.1016/j.bbmt.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boukouaci W, Busson M, Peffault de Latour R, et al. MICA-129 genotype, soluble MICA, and anti-MICA antibodies as biomarkers of chronic graft-versus-host disease. Blood. 2009;114:5216–5224. doi: 10.1182/blood-2009-04-217430. [DOI] [PubMed] [Google Scholar]
- 41.Pratt LM, Liu Y, Ugarte-Torres A, et al. IL15 levels on day 7 after hematopoietic cell transplantation predict chronic GVHD. Bone Marrow Transplant. 2013;48:722–728. doi: 10.1038/bmt.2012.210. [DOI] [PubMed] [Google Scholar]
- 42.Kitko CL, Levine JE, Storer BE, et al. Plasma CXCL9 elevations correlate with chronic GVHD diagnosis. Blood. 2014;123:786–793. doi: 10.1182/blood-2013-08-520072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mays JW, Weller ML, Bassim CW, et al. Oral graft-versus-host disease is distinguished by inflammation and impaired anti-microbial immunity. 43rd Annual Meeting of the American Association for Dental Research; 2014. URL: https://iadr.confex.com/iadr/43am/webprogram/start.html. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Salivary proteins (n = 59) that exhibited altered expression in oral cGVHD were classified according to their molecular function. 37 proteins have a protein binding function, either as proteinase inhibitors or enzymes. B) Classification of upregulated (black bars) and downregulated (grey bars) proteins in oral cGVHD, grouped by common molecular function or pathway. The mean iTRAQ value for each family or functional group of proteins was determined from the data in Tables S1 and S2. The number of proteins represented in each functional grouping is shown in parentheses. In all cases proteins within a group showed the same trend in expression, with the exception of the cysteine proteinase inhibitor cystatin B, which is downregulated in the saliva of cGVHD patients.


