Abstract
We present a mathematical model for ionotropic glutamate receptors (iGluRs) that is built on mechanistic understanding and yields a number of thermodynamic and kinetic properties of channel gating. iGluRs are ligand-gated ion channels responsible for the vast majority of fast excitatory neurotransmission in the central nervous system. The effects of agonist-induced closure of the ligand-binding domain (LBD) are transmitted to the transmembrane channel (TMC) via inter-domain linkers. Our model shows that, relative to full agonists, partial agonists may reduce either the degree of LBD closure or the curvature of the LBD free energy basin, leading to less stabilization of the channel open state and hence lower channel open probability. A rigorous relation is derived between the channel closed-to-open free energy difference and the tension within the linker. Finally by treating LBD closure and TMC opening as diffusive motions, we observe gating trajectories that resemble stochastic current traces from single-channel recordings and are able to calculate the rate constants for transitions between the channel open and closed states. Our model can be implemented by molecular dynamics simulations to realistically depict iGluR gating and may guide functional experiments in gaining deeper insight into this essential family of channel proteins.
Keywords: channel gating, ligand-gated ion channels, partial agonism, single channel kinetics
Introduction
Transmembrane proteins that form ion channels receive stimuli to open a pore and allow the passage of ions. This process, termed gating, is frequently modeled at three different levels. Kinetic models derived from single-channel or whole-cell recordings provide phenomenological descriptions of channel activity but cannot capture molecular details underlying the kinetic steps. Structural models of gating, either from structure determination (e.g., X-ray crystallography) at putatively different functional states or from mutagenesis-based (e.g., cysteine substitution) functional studies, provide qualitative descriptions of the crucial movements involved in gating, but cannot inform how the energetics and dynamics of the relevant structural elements quantitatively define the gating process. Computational models, e.g., from molecular dynamics simulations, can capture some of the energetics and dynamics, but are inevitably limited in the temporal and conformational scales that can be explored. A fourth type of model 1–2, termed mathematical here, allows thermodynamic and kinetic properties of the gating process to be calculated from the energetics and dynamics of the conformational changes involved, thus bridging some of the gaps in the preceding three types of models. The present study aims to present such a mathematical model for ionotropic glutamate receptors (iGluRs).
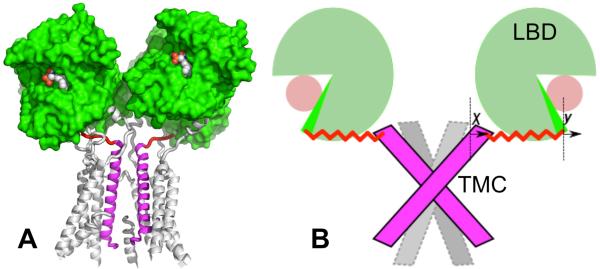
iGluRs are ligand-gated ion channels responsible for the vast majority of fast excitatory neurotransmission in the central nervous system. The two main iGluR subtypes, AMPA and NMDA receptors, are essential to all aspects of brain function including higher order processes such as learning and memory. These tetrameric assemblies convert agonist (glutamate or glycine) binding to the ligand-binding domain (LBD) into currents through the transmembrane channel (TMC; Fig. 1A) 3–5. iGluR subunits are composed of discrete, highly modular domains that are separated from each other by flexible linkers 6–7. The extracellular LBD is composed of two lobes termed D1 and D2. The LBDs from various iGluR subunits have been genetically isolated and crystallized in an assortment of ligand-bound forms 5. The transmembrane domain consists of three transmembrane helices (termed M1, M3, and M4) and a re-entrant helix (termed M2). The TMC is formed by the tetrameric assembly of the transmembrane domains of the receptor's four subunits, with M3 as the major pore-lining helix and forming the activation gate at the C-terminus 3, 6. The M2–M3 region has some similarity in sequence and structure with the counterpart in potassium channels 6, 8–9.
Figure 1.
Structure of an AMPA receptor and our mechanism-based mathematical model. (A) Crystal structure of an AMPA receptor (Protein Data Bank entry 3KG2). The LBD is rendered as green surface; bound ligand is shown as spheres; the TMC is shown as cartoon, with one subunit undisplayed for clarity. The M3 helices from two diagonal subunits are highlighted in magenta and the corresponding M3-D2 linkers highlighted in red. (B) Model for channel gating. The agonist-bound LBD can be closed (dark green) or transiently open (light green); y denotes the degree of LBD closure (the 0 value of y, where the LBD free energy is at minimum, is indicated by a vertical line next to the symbol “y”). The C-termini of the M3 helices move outward (inward) to open (close) the channel; x denotes the degree of channel opening (the 0 value of x is indicated by a vertical line next to the symbol “x”). The distance between the two vertical lines is L0. When x and y are displaced from their 0 values, L0 + y – x is the length of the linker.
The gating mechanisms of iGluRs have been studied extensively. Kinetic modeling of single-channel and whole-cell currents reveals complex behavior, including multiple open and closed substates at saturating agonist concentrations 10–16. Crystal structures of isolated LBDs and near-full-length receptors 6, 17–20 have clearly defined the agonist-induced lobe closure of the LBD and also suggested a crucial role for the LBD-TMC linkers, especially the M3-D2 linker, in transmitting the effect of agonist binding further to the TMC, but have revealed little about the conformations adopted by the TMC in the open state. On the other hand, functional data based on cysteine substitution have shown that the M3 helix is an essential component of iGluR pore opening and appears to be largely rigid 21–23, but structural elements around M3 also significantly contribute to the activation process 24–25. In addition, molecular dynamics simulations 26–27 have presented a likely scenario for the coupling between LBD lobe closure and TMC pore opening: the closure of the D2 lobe toward D1 produces outward pulling of the M3-D2 linker, which in turn leads to channel opening (Fig. 1). This coupling mechanism was further addressed in a combined functional and computational study 16, which showed that glycine insertions in the M3-D2 linker designed to increase the linker length and hence weaken the LBD-TMC coupling significantly reduced the channel open probability. Also contributing to our understanding of channel gating are free energy calculations for the LBD lobe closure through molecular dynamics simulations 28–30.
Here, building on the significant mechanistic understanding achieved so far on iGluR gating, we present a mathematical model for calculating thermodynamic and kinetic properties of the gating process from the energetics and dynamics of intra- and inter-domain motions. We show illustrative results on partial agonism, effects of linker insertions, and transition rates between channel open and closed states.
The Model
We model the energetics for the lobe closure of the agonist-bound LBD and the pore opening of the TMC, as well as their coupling by the M3-D2 linker (Fig. 1B). The free energy surface for LBD closure has been calculated through molecular dynamics simulations 28–30. To model the single well of this free energy surface, we use a harmonic potential:
| (1) |
where y is the degree of LBD closure, with y = 0 corresponding to the free energy minimum and negative y signifying opening of the agonist-bound LBD; and kb is the LBD spring constant. Similarly, we model the small-amplitude extension and contraction of the M3-D2 linker from its optimal length Lm by a harmonic potential
| (2) |
where x is the degree of TMC opening; Δ = L0 – Lm, with L0 denoting the length of the linker at x = y = 0; and k1 is the linker spring constant.
We assume that the TMC has a global minimum corresponding to the closed state and a shallow minimum corresponding to the open state; the quiescent channel predominantly stays in the closed state but may make rare excursions to the open state. This is captured by the potential (Fig. 2A)
| (3) |
which has the global and shallow minima located at x = −1 and x = 1, respectively, with a free energy difference of 4ε/3. The free energy for the receptor as a whole is then
| (4) |
The 2-dimensional free energy surface at ε = 20, kb = k1 = 30, and Δ = 1 is shown in Fig. 2B. All energies are measured in units of the product of Boltzmann's constant and absolute temperature, and lengths are in units of Å. The chosen value of Δ is such that the linker is in its relaxed state (i.e., neither compressed nor extended) when the LBD is at its free energy minimum (i.e., y = 0) and the TMC is at its open state minimum (i.e., x = 1). If the TMC were to move toward the closed state, the linker would be extended, resulting in an energetic penalty. The LBD-TMC coupling via the linker therefore leads to an overall free energy surface that has two nearly evenly matched minima, one at (x, y) = (1, 0) for the channel open state and the other at (−0.83, −0.91) for the channel closed state. The stabilization effect on the channel open state by the coupling to the LBD can be seen by comparing the free energy function Wc(x) of the isolated TMC with the potential of mean force, Wpmf(x), in x for the full receptor (Fig. 2A), defined through
| (5) |
With an appropriate choice of the arbitrary normalization constant C, the resulting potential of mean force is
| (6) |
The second term on the right-hand side of Eq. (6) arises from the coupling to the LBD. Its value is 0 at x = Δ = 1 and positive at x = −1.
Figure 2.
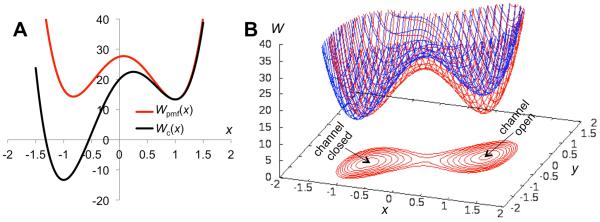
The free energy surface of the receptor. (A) The free energy function of the isolated TMC and the potential of mean force for TMC opening when coupled to the LBD via the M3-D2 linker. (B) The free energy surface for the receptor. Shown in red is the free energy surface (also displayed as contours) when the LBD is bound with a full agonist. In blue is the free energy surface when the LBD is bound with an NMDAR-type partial agonist, which reduces the curvature (i.e., kb) of the free energy function for LBD closure (from 30 to 20).
The two stable states are separated by a saddle-shaped barrier. The lowest barrier heights, located at the saddle point (0.076, −0.46), are 14.4 and 13.4 when measured from the channel open and closed minima, respectively. These will be denoted as ΔWo→sp and ΔWc→sp, respectively.
Results
Our model predicts a number of important functional properties of iGluR gating, allowing for validation and refinement by future electrophysiological measurements.
Partial agonism
Full agonists evoke maximal currents through the TMC, whereas partial agonists have submaximal efficacy. Partial agonism manifests itself first through the LBD, and this manifestation has been the subject of many experimental and computational studies 29, 31–43. For AMPA receptors, a correlation between the degree of LBD closure and agonist efficacy was observed in crystal structures 33, 37. This correlation is absent in NMDA receptors, as their LBDs bound with full and partial agonists have very similar structures 34–36, 42. Recently we found that the GluN1 LBD responds to partial agonist binding by reducing the curvature of the free energy surface while largely preserving the minimum position 28.
How is partial agonism transmitted to the TMC to produce submaximal currents? We can now directly address this question with our model. Agonist efficacy can be measured by the channel open probability 15, 44, which in our model is given by
| (7) |
where x‡ denotes the barrier of the potential of mean force separating the channel open and closed states. For AMPA receptors, reduced closure of the LBD by a partial agonist would lead to compression of the linker if the TMC is in the open state; the strain in the linker would be relieved if the TMC retracts to the closed state (Fig. 1B). This effect can be illustrated by reducing L0 and hence Δ by 0.2. The potentials of mean force for the full and AMPAR-type partial agonists are compared in Fig. 3A. Correspondingly the channel open probability is reduced from 0.71 to 0.011. The dependence of the relative efficacy, defined as Po′/Po, on Δ′/Δ is shown in Fig. 3B; the unprimed and primed symbols denote quantities related to the full and partial agonists, respectively.
Figure 3.
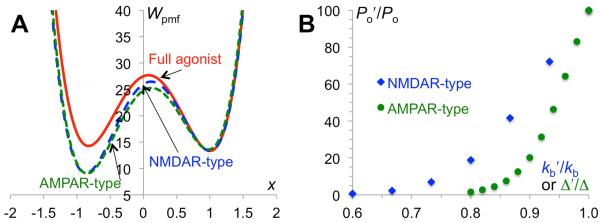
Partial agonism. (A) Potentials of mean force for full and AMPAR-type (reduced degree of cleft closure) and NMDAR-type (reduced curvature of LBD free energy basin) partial agonists. (B) Relative efficacies of the two types of partial agonists.
For the NMDAR-type partial agonists, following our earlier study 28, we assume that the curvature of the LBD free energy function is reduced. As a result, the penalty for moving away from the LBD-closed conformation (i.e., y = 0) is lowered; when coupled to the TMC via the linker, the stabilization of the channel open state is correspondingly less. The 2-dimensional free energy surface for an NMDAR-type partial agonist, with kb reduced from 30 to 20, is illustrated in Fig. 2B. The resulting potential of mean force in x is compared to that for the full agonist in Fig. 3A. This partial agonist reduces the channel open probability from 0.71 to 0.016. The dependence of the relative efficacy on kb′/kb is shown in Fig. 3B.
Linker insertions
Molecular dynamics simulations have shown that the M3-D2 linker is critical for transmitting the effect of the agonist-induced LBD conformational change to the TMC 26–27. To directly probe the role of the M3-D2 linker, Kazi et al. 16 introduced glycine insertions. The resulting lengthening of the linker was found to reduce the channel open probability. We can explain this observation with our model by noting that a glycine insertion would increase Lm, the optimal linker length. As a result, Δ is decreased. The net effect of the glycine insertion is thus identical to that of an AMPAR-type partial agonist.
Kazi et al. 16 empirically interpreted the change in the closed-to-open free energy difference, ΔGc→o, by a glycine insertion in terms of the tension in the linker. With our model, we can derive a rigorous relation between insertion-induced change in ΔGc→o and linker tension. We first note that a spring (with spring constant kl) when extended by δ generates a tension klδ within. Hence to find the tension within the linker we need to calculate the average extension of the linker. When the channel is in the closed state, the average extension is
| (8) |
where the subscript “c” in the integrals signifies that the area of integration is restricted to the channel closed state. Similarly, the average extension of the linker in the channel open state is
| (9) |
In our model with kb = 30 and Δ = 1, the average linker extensions are 0.01 and 0.91, respectively, in the channel open and closed states. That is, the linker is more extended and hence there is greater tension in the channel closed state.
The closed-to-open free energy difference is given by
| (10) |
or
| (11) |
We now take the derivative of ΔGc→o with respect to the linker optimal length Lm:
| (12) |
Using Eqs. (8) and (9), we can simplify the last result as
| (13) |
Note that klδo and klδc can be recognized as the linker tensions in the channel open and closed states, respectively, to be denoted as Fo Fc. For a small change in Lm, denoted as δLm, the preceding equation allows us to find the corresponding change in ΔGc→o as
| (14) |
That is, the insertion-induced change in ΔGc→o, when divided by the change in linker optimal length, is equal to the difference in linker tension between the channel closed and open states in the wild-type receptor. Since our model predicts Fc > Fo, an linker insertion leads to an increase in ΔGc→o, i.e., a decrease in channel open probability. This is just what was observed by Kazi et al.16.
Single-channel gating kinetics
So far we have only dealt with equilibrium properties. By assuming appropriate dynamics for the two coordinates x and y, our model can also predict kinetic properties of channel gating. Here we assume that x and y follow diffusive dynamics, with diffusion constant Dx and Dy, respectively. A trajectory with many transitions between the channel closed and open states then resembles the stochastic current trace of a single-channel recording (Fig. 4A). The probabilities, Ho(τ) and Hc(τ), that the residence times in the channel open and closed states are longer than τ compare well with single exponentials, , and , respectively (Fig. 4B), as can be expected by the high energy barriers that must be crossed in order to leave the energy wells. With Dx and Dy set to 2 × 104 (in units of Å2/ms), the mean residence times, and , are 6.6 and 2.7 (in units of ms), in the channel open and closed states, respectively. Note that the ratio agrees well with Po, confirming that the trajectory represents an equilibrium sampling of the free energy surface.
Figure 4.
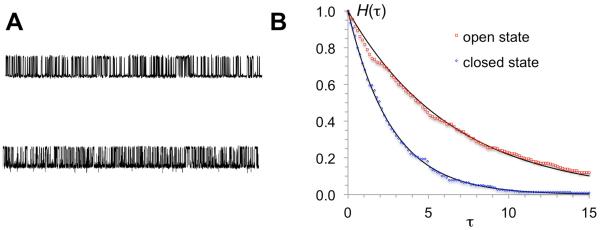
“Current” traces and channel open and closed times. (A) Upper trace: values of x from a Brownian dynamics simulation of the model at Dx = Dy = 2 × 104, using the Ermak-McCammon algorithm 45 with a time step of 5 × 10−8. Lower trace: currents from a single-channel recording of the GluN1/GluN2A NMDA receptor under steady-state conditions at pH 8 in the on-cell attached configuration. (B) Probabilities for residence times in channel open and closed states to be longer than τ. Symbols are from binning the residence times from the simulation; curves are single exponentials with exponents given by the mean residence times in the channel open and closed states.
The inverses of and can be recognized as the rate constants for the transitions between the channel open and closed states. The rate constants for the transitions between the stable states of a 2-dimensional free energy surface like the one described here can be predicted by a recent theory of Berezhkovskii et al.46. Either rate constant, e.g., ko→c for the channel open-to-closed transition, can be written in the form
| (15) |
where kL is the rate constant obtained by Langer 47 by assuming that the inter-well transition is rate-limited by passage through the saddle point region, and k0 is the rate constant for the transition on the potential of mean force, U0(y), in y. The latter is defined via
| (16) |
where C, like in Eq. (5), denotes an arbitrary constant. The Langer result is given by
| (17) |
where K is the matrix of second derivatives of W(x, y):
| (18) |
which is to be evaluated either at the channel open minimum (where x = 1) or at the saddle point (where x = 0.076). The remaining parameter γ in Eq. (17) is the absolute value of the only negative eigenvalue of the matrix Ksp · D, where D is the diffusion matrix, assumed here to be diagonal (with diagonal elements Dx and Dy).
As noted by Berezhkovskii et al. 46, when Dy → 0, motion along y, i.e., on the potential of mean force U0(y) becomes rate-limiting. This potential has a double-well shape, with one minimum at y = 0 corresponding to the channel open state and one minimum at y = −0.91 corresponding to the channel closed state, separated by a barrier at y = −0.48, to be denoted as y‡. The rate constant for the transition on U0(y) is given by
| (19) |
At Dx = Dy = 2 × 104, ko→c is rate-limited by kL and the predicted value for its inverse is 6.4, which is close to the value of obtained in the Brownian dynamics simulation. Similarly, the predicted value of 2.6 for the inverse of kc→o is close to the simulation value of . Partial agonists will change both ko→c and kc→o. For the blue free energy surface in Fig. 2B modeling an NMDAR-type partial agonist, ΔWo→sp decreases while ΔWc→sp increases. Therefore one should expect an increase in ko→c and a decrease in kc→o.
Discussion
We have presented a mechanism-based mathematical model that yields a number of thermodynamic and kinetic properties of iGluR gating. The model demonstrates that, by reducing either the degree of LBD closure (as in AMPA receptors) or the curvature of the LBD free energy basin (as in NMDA receptors), partial agonists can decrease the stabilization of the channel open state provided by agonist-induced LBD closure, thereby decreasing the channel open probability and thus agonist efficacy. With the model we also derive a rigorous relation between the channel closed-to-open free energy difference and the tension within the LBD-TMC linker. Finally by treating LBD closure and TMC opening as diffusive motions, we observe gating trajectories that resemble single-channel recordings and predict the rate constants for the transitions between the channel open and closed states.
In its present form, our model provides a conceptual framework for predicting functional observables from the energetics and dynamics of intra- and inter-domain motions. The results presented are illustrative only, though the orders of magnitude for the channel open probability (~0.7), the energy barrier for channel opening or closing (~8 kcal/mol), and the mean residence time in the channel open or closed state (~5 ms) are in line with observed iGluR single-channel activity under certain conditions 13, 16, 48–49. The diffusion constants assumed here (~2 × 104 Å2/ms) for intra-domain motions are approximately 500-fold smaller than the translational diffusion constant of a single-domain protein 50. Kinetic studies on residue-residue contact formation in peptides have found slower intra-chain diffusion relative to translational diffusion of free amino acids 51.
It is worth noting that the free energy functions and the diffusion constants in the model can be obtained from molecular dynamics simulations. In fact, the free energy function for LBD closure has already been computed in several studies 28–30. Similar approaches can be used to obtain the free energy functions for TMC opening 52–54 and for the linker extension. A number of important details will have to be accounted for. For example, the receptor contains four subunits. Here we treated the tetrameric TMC as a single unit but included only a single LBD monomer and the associated LBD-TMC linker. All the four LBD monomers (perhaps with inter-subunit coupling) and the associated linkers will have to be included. While we used a single coordinate here to represent the conformational freedom of the LBD or TMC, a more realistic representation may require more than one coordinate. Indeed, two coordinates were used for LBD closure in the recent free energy calculations 28–30. The TMC will likewise require at least two coordinates, e.g., lateral displacements of the A/C and B/D M3 helix C-termini. On the other hand, each LBD-TMC linker may still be modeled well by a single coordinate, i.e., the end-to-end extension, though the relation between this quantity and the LBD and TMC coordinates will not be as simple as the one presented here. In the end one will be able to construct a high-dimensional free energy surface for the full receptor, from which thermodynamic properties like channel open probability and linker tension can be predicted. With diffusion constants of conformational coordinates calculated from additional simulations, prediction of kinetic properties of channel gating will also be possible.
Another direction for future development is to treat additional functional properties. Histograms of residence times from single-channel recordings on iGluRs at saturating agonist concentrations deviate from single exponentials, indicating multiple open and closed substates 11–16. The molecular basis for these multiple substates remains unclear. One possibility worth pursuing is that the channel-closed (or open) free energy basin encompasses multiple minima. In addition to the open and closed states, the receptor can enter a desensitized state in the continual presence of agonists. Channel desensitization can be accounted for by introducing a new free energy minimum for the LBD dimer 17, 19, 27, 55–56 or the TMC (LPW, unpublished data). It is also of both experimental and physiological interest to study channel gating evoked by a brief application of agonists, replicating what occurs at synapses. Treating this condition will require the consideration of agonist association and dissociation kinetics. Our modeling approach will facilitate mechanistic interpretation of functional data and guide the design of functional experiments, on iGluRs as well as on other families of ligand-gated ion channels 57 and channel proteins in general.
ACKNOWLEDGMENT
This work was supported in part by NIH Grant GM58187.
ABBREVIATIONS
- iGluR
ionotropic glutamate receptor
- LBD
ligand-binding domain
- TMC
transmembrane channel.
REFERENCES
- 1.Sigg D, Qian H, Bezanilla F. Kramers' Diffusion Theory Applied to Gating Kinetics of Voltage-Dependent Ion Channels. Biophys. J. 1999;76:782–803. doi: 10.1016/S0006-3495(99)77243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou HX. A Theory for the Proton Transport of the Influenza Virus M2 Protein: Extensive Test against Conductance Data. Biophys. J. 2011;100:912–921. doi: 10.1016/j.bpj.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate Receptor Ion Channels: Structure, Regulation, and Function. Pharmacol. Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paoletti P, Bellone C, Zhou Q. NMDA Receptor Subunit Diversity: Impact on Receptor Properties, Synaptic Plasticity and Disease. Nat. Rev. Neurosci. 2013;14:383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- 5.Kumar J, Mayer ML. Functional Insights from Glutamate Receptor Ion Channel Structures. Annu. Rev. Physiol. 2012;75:313–337. doi: 10.1146/annurev-physiol-030212-183711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sobolevsky AI, Rosconi MP, Gouaux E. X-Ray Structure, Symmetry and Mechanism of an AMPA-Subtype Glutamate Receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karakas E, Furukawa H. Crystal Structure of a Heterotetrameric NMDA Receptor Ion Channel. Science. 2014;344:992–997. doi: 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wo ZG, Oswald RE. Unraveling the Modular Design of Glutamate-Gated Ion Channels. Trends Neurosci. 1995;18:161–168. doi: 10.1016/0166-2236(95)93895-5. [DOI] [PubMed] [Google Scholar]
- 9.Wood MW, VanDongen HMA, VanDongen AMJ. Structural Conservation of Ion Conduction Pathways in K Channels and Glutamate Receptors. Proc. Natl. Acad. Sci. USA. 1995;92:4882–4886. doi: 10.1073/pnas.92.11.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lester RA, Jahr CE. NMDA Channel Behavior Depends on Agonist Affinity. J. Neurosci. 1992;12:635–643. doi: 10.1523/JNEUROSCI.12-02-00635.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyllie DJ, Behe P, Colquhoun D. Single-Channel Activations and Concentration Jumps: Comparison of Recombinant NR1a/NR2a and NR1a/NR2d NMDA Receptors. J. Physiol. 1998;510:1–18. doi: 10.1111/j.1469-7793.1998.001bz.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banke TG, Traynelis SF. Activation of NR1/NR2b NMDA Receptors. Nat. Neurosci. 2003;6:144–152. doi: 10.1038/nn1000. [DOI] [PubMed] [Google Scholar]
- 13.Popescu G, Auerbach A. Modal Gating of NMDA Receptors and the Shape of Their Synaptic Response. Nat. Neurosci. 2003;6:476–483. doi: 10.1038/nn1044. [DOI] [PubMed] [Google Scholar]
- 14.Auerbach A, Zhou Y. Gating Reaction Mechanisms for Nmda Receptor Channels. J. Neurosci. 2005;25:7914–7923. doi: 10.1523/JNEUROSCI.1471-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kussius CL, Popescu GK. Kinetic Basis of Partial Agonism at NMDA Receptors. Nat. Neurosci. 2009;12:1114–1120. doi: 10.1038/nn.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazi R, Dai J, Sweeney C, Zhou HX, Wollmuth LP. Mechanical Coupling Maintains the Fidelity of NMDA Receptor-Mediated Currents. Nat. Neurosci. 2014;17:914–922. doi: 10.1038/nn.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of Glutamate Receptor Desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- 18.Chen L, Durr KL, Gouaux E. X-Ray Structures of AMPA Receptor-Cone Snail Toxin Complexes Illuminate Activation Mechanism. Science. 2014;345:1021–1026. doi: 10.1126/science.1258409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durr KL, Chen L, Stein RA, De Zorzi R, Folea IM, Walz T, McHaourab HS, Gouaux E. Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States. Cell. 2014;158:778–792. doi: 10.1016/j.cell.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yelshanskaya MV, Li M, Sobolevsky AI. Structure of an Agonist-Bound Ionotropic Glutamate Receptor. Science. 2014;345:1070–1074. doi: 10.1126/science.1256508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobolevsky AI, Beck C, Wollmuth LP. Molecular Rearrangements of the Extracellular Vestibule in NMDAR Channels During Gating. Neuron. 2002;33:75–85. doi: 10.1016/s0896-6273(01)00560-8. [DOI] [PubMed] [Google Scholar]
- 22.Sobolevsky AI, Prodromou ML, Yelshansky MV, Wollmuth LP. Subunit-Specific Contribution of Pore-Forming Domains to NMDA Receptor Channel Structure and Gating. J. Gen. Physiol. 2007;129:509–525. doi: 10.1085/jgp.200609718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sobolevsky AI, Yelshansky MV, Wollmuth LP. Different Gating Mechanisms in Glutamate Receptor and K+ Channels. J. Neurosci. 2003;23:7559–7568. doi: 10.1523/JNEUROSCI.23-20-07559.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kazi R, Gan Q, Talukder I, Markowitz M, Salussolia CL, Wollmuth LP. Asynchronous Movements Prior to Pore Opening in NMDA Receptors. J. Neurosci. 2013;33:12052–12066. doi: 10.1523/JNEUROSCI.5780-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talukder I, Borker P, Wollmuth LP. Specific Sites within the Ligand-Binding Domain and Ion Channel Linkers Modulate NMDA Receptor Gating. J. Neurosci. 2010;30:11792–11804. doi: 10.1523/JNEUROSCI.5382-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai J, Zhou HX. An NMDA Receptor Gating Mechanism Developed from MD Simulations Reveals Molecular Details Underlying Subunit-Specific Contributions. Biophys. J. 2013;104:2170–2181. doi: 10.1016/j.bpj.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong H, Zhou HX. Atomistic Mechanism for the Activation and Desensitization of an AMPA-Subtype Glutamate Receptor. Nat. Commun. 2011;2:354. doi: 10.1038/ncomms1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai J, Zhou HX. Reduced Curvature of Ligand-Binding Domain Free Energy Surface Underlies Partial Agonism at NMDA Receptors. Structure. 2014;23:237–247. doi: 10.1016/j.str.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau AY, Roux B. The Free Energy Landscapes Governing Conformational Changes in a Glutamate Receptor Ligand-Binding Domain. Structure. 2007;15:1203–1214. doi: 10.1016/j.str.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao Y, Belcher J, Berger AJ, Mayer ML, Lau AY. Conformational Analysis of Nmda Receptor GluN1, GluN2, and GluN3 Ligand-Binding Domains Reveals Subtype-Specific Characteristics. Structure. 2013;21:1788–1799. doi: 10.1016/j.str.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmed AH, Ptak CP, Fenwick MK, Hsieh CL, Weiland GA, Oswald RE. Dynamics of Cleft Closure of the GluA2 Ligand-Binding Domain in the Presence of Full and Partial Agonists Revealed by Hydrogen-Deuterium Exchange. J. Biol. Chem. 2013;288:27658–27666. doi: 10.1074/jbc.M113.495564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arinaminpathy Y, Sansom MS, Biggin PC. Binding Site Flexibility: Molecular Simulation of Partial and Full Agonists within a Glutamate Receptor. Mol. Pharmacol. 2006;69:11–18. doi: 10.1124/mol.105.016691. [DOI] [PubMed] [Google Scholar]
- 33.Armstrong N, Gouaux E. Mechanisms for Activation and Antagonism of an AMPA-Sensitive Glutamate Receptor: Crystal Structures of the GluR2 Ligand Binding Core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 34.Furukawa H, Gouaux E. Mechanisms of Activation, Inhibition and Specificity: Crystal Structures of the NMDA Receptor NR1 Ligand-Binding Core. EMBO J. 2003;22:2873–2885. doi: 10.1093/emboj/cdg303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen KB, Tajima N, Risgaard R, Perszyk RE, Jorgensen L, Vance KM, Ogden KK, Clausen RP, Furukawa H, Traynelis SF. Structural Determinants of Agonist Efficacy at the Glutamate Binding Site of N-Methyl-D-Aspartate Receptors. Mol. Pharmacol. 2013;84:114–127. doi: 10.1124/mol.113.085803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inanobe A, Furukawa H, Gouaux E. Mechanism of Partial Agonist Action at the NR1 Subunit of NMDA Receptors. Neuron. 2005;47:71–84. doi: 10.1016/j.neuron.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 37.Jin RS, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Structural Basis for Partial Agonist Action at Ionotropic Glutamate Receptors. Nat. Neurosci. 2003;6:803–810. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- 38.Lau AY, Roux B. The Hidden Energetics of Ligand Binding and Activation in a Glutamate Receptor. Nat. Struct. Mol. Biol. 2011;18:283–7. doi: 10.1038/nsmb.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maltsev AS, Ahmed AH, Fenwick MK, Jane DE, Oswald RE. Mechanism of Partial Agonism at the GluR2 AMPA Receptor: Measurements of Lobe Orientation in Solution. Biochemistry. 2008;47:10600–10610. doi: 10.1021/bi800843c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Postila PA, Ylilauri M, Pentikainen OT. Full and Partial Agonism of Ionotropic Glutamate Receptors Indicated by Molecular Dynamics Simulations. J. Chem. Inf. Model. 2011;51:1037–1047. doi: 10.1021/ci2000055. [DOI] [PubMed] [Google Scholar]
- 41.Ramaswamy S, Cooper D, Poddar N, MacLean DM, Rambhadran A, Taylor JN, Uhm H, Landes CF, Jayaraman V. Role of Conformational Dynamics in Alpha-Amino-3-Hydroxy-5-Methylisoxazole-4-Propionic Acid (AMPA) Receptor Partial Agonism. J. Biol. Chem. 2012;287:43557–43564. doi: 10.1074/jbc.M112.371815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao Y, Harrison CB, Freddolino PL, Schulten K, Mayer ML. Molecular Mechanism of Ligand Recognition by NR3 Subtype Glutamate Receptors. EMBO J. 2008;27:2158–2170. doi: 10.1038/emboj.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ylilauri M, Pentikainen OT. Structural Mechanism of N-Methyl-D-Aspartate Receptor Type 1 Partial Agonism. PLoS One. 2012;7:e47604. doi: 10.1371/journal.pone.0047604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erreger K, Geballe MT, Dravid SM, Snyder JP, Wyllie DJ, Traynelis SF. Mechanism of Partial Agonism at NMDA Receptors for a Conformationally Restricted Glutamate Analog. J. Neurosci. 2005;25:7858–7866. doi: 10.1523/JNEUROSCI.1613-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ermak DL, Mccammon JA. Brownian Dynamics with Hydrodynamic Interactions. J. Chem. Phys. 1978;69:1352–1360. [Google Scholar]
- 46.Berezhkovskii AM, Szabo A, Greives N, Zhou HX. Multidimensional Reaction Rate Theory with Anisotropic Diffusion. J. Chem. Phys. 2014;141:204106. doi: 10.1063/1.4902243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langer JS. Statistical Theory of Decay of Metastable States. Ann. Phys. 1969;54:258–275. [Google Scholar]
- 48.Prieto ML, Wollmuth LP. Gating Modes in AMPA Receptors. J. Neurosci. 2010;30:4449–4459. doi: 10.1523/JNEUROSCI.5613-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Talukder I, Wollmuth LP. Local Constraints in Either the GluN1 or GluN2 Subunit Equally Impair NMDA Receptor Pore Opening. J. Gen. Physiol. 2011;138:179–94. doi: 10.1085/jgp.201110623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou HX. Calculation of Translational Friction and Intrinsic Viscosity. II. Application to Globular Proteins. Biophys. J. 1995;69:2298–2303. doi: 10.1016/S0006-3495(95)80100-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lapidus LJ, Eaton WA, Hofrichter J. Measuring the Rate of Intramolecular Contact Formation in Polypeptides. Proc. Natl. Acad. Sci. USA. 2000;97:7220–7225. doi: 10.1073/pnas.97.13.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fowler PW, Sansom MS. The Pore of Voltage-Gated Potassium Ion Channels Is Strained When Closed. Nat. Commun. 2013;4:1872. doi: 10.1038/ncomms2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park S, Im W. Two Dimensional Window Exchange Umbrella Sampling for Transmembrane Helix Assembly. J. Chem. Theory Comput. 2013;9:13–17. doi: 10.1021/ct3008556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu F, Hummer G. Pore Opening and Closing of a Pentameric Ligand-Gated Ion Channel. Proc. Natl. Acad. Sci. USA. 2010;107:19814–19819. doi: 10.1073/pnas.1009313107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carbone AL, Plested AJ. Coupled Control of Desensitization and Gating by the Ligand Binding Domain of Glutamate Receptors. Neuron. 2012;74:845–857. doi: 10.1016/j.neuron.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 56.Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S. Structural Mechanism of Glutamate Receptor Activation and Desensitization. Nature. 2014;514:328–334. doi: 10.1038/nature13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Du J, Dong H, Zhou HX. Size Matters in Activation/Inhibition of Ligand-Gated Ion Channels. Trends Pharmacol. Sci. 2012;33:482–493. doi: 10.1016/j.tips.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]