Abstract
There is considerable support for the view that aerobic exercise may confer cognitive benefits to mild cognitively impaired elderly persons. However, the biological mechanisms mediating these effects are not entirely clear. As a preliminary step towards informing this gap in knowledge, we enrolled older adults confirmed to have mild cognitive impairment (MCI) in a 6-month exercise program. Male and female subjects were randomized into a 6-month program of either aerobic or stretch (control) exercise. Data collected from the first 10 completers, aerobic exercise (n=5) or stretch (control) exercise (n=5), were used to determine intervention-induced changes in the global gene expression profiles of the aerobic and stretch groups. Using microarray, we identified genes with altered expression (relative to baseline values) in response to the 6-month exercise intervention. Genes whose expression were altered by at least two-fold, and met the p-value cutoff of 0.01 were inputted into the Ingenuity Pathway Knowledge Base library to generate gene-interaction networks. After a 6-month aerobic exercise-training, genes promoting inflammation became down-regulated, whereas genes having anti-inflammatory properties and those modulating immune function or promoting neuron survival and axon growth, became up-regulated (all fold change ≥ ± 2.0, p < 0.01). These changes were not observed in the stretch group. Importantly, the differences in the expression profiles correlated with significant improvement in maximal oxygen uptake (VO2max) in the aerobic program as opposed to the stretch group. We conclude that three distinct cellular pathways may collectively influence the training effects of aerobic exercise in MCI subjects. We plan to confirm these effects using rt-PCR and correlate such changes with the cognitive phenotype.
Keywords: mild cognitive impairment, microarray, gene expression, aerobic exercise
1.1. INTRODUCTION
Neuroinflammation, a prevalent feature of Alzheimer’s disease (AD), is heralded by insoluble protein deposition, reactive astrocytes, activated microglia, dystrophic neurites, and intracellular neurofibrillary tangles (NFTs) (1–3). Abundant evidence suggest that inflammatory mechanisms within the central nervous system (CNS) likely promote cognitive deterioration (4). AD progression is marked by uncontrolled glial cell activation, neuroinflammation and consequent neuronal/synaptic dysfunction, inciting a vicious cycle of increased glial activation and neuronal damage (5). Therefore, strategies to reduce neuroinflammation may attenuate the risk of sporadic AD (6).
Physical activity can reduce inflammation (7), and has been demonstrated to improve memory in MCI and possibly AD subjects (8). In support of this view, but through a yet undiscerned mechanism, moderate exercise reduced amyloid plaque burden in transgenic mouse models of AD, and improved cognitive function (9). Independently, aerobic exercise can modulate immune response, increase the population of regulatory T cells, decrease the population of inflammatory monocytes, and down-regulate inflammation (10–15). However, whether pro-inflammatory proteins facilitating cell death or anti-inflammatory genes promoting neuronal survival and axonal growth, are involved in this regulatory mechanism have not been systematically examined in MCI subjects. Importantly, data are lacking on the mechanisms underlying the beneficial effects of exercise in African American (AA) MCI subjects. As a preliminary step to this investigative exercise, we examined the training-induced changes in the overall gene expression profile from blood cells of MCI volunteers randomized into a 6-month aerobic exercise-training program versus a stretch-exercise control group. We hypothesized that aerobic training adaptation would promote favorable changes in the expression of pro-inflammatory or anti-inflammatory genes, and of genes modulating immune response or promoting neuronal survival. Since MCI is prodromal for AD, insight into these mechanisms may inform the development of interventions and treatment regimens for AD patients.
1.2. MATERIAL and METHODS
The protocols used in this investigation were approved by the Howard University Institutional Review Board (IRB). As required for studies involving human subjects, all participants completed a signed informed consent form prior to enrollment in the study.
1.2.1. Level I Screening
Main eligibility criteria used for inclusion consisted of the following: age > 55 years; ability to exercise vigorously without causing harm to self; diagnostic designation as MCI according to Petersen criteria (16) using education adjusted scores; have a committed study partner; be in good general health; and willing to exercise for the entire study duration; and undergo required medical and study-related assessments. After obtaining informed consent from volunteers, we collected data on demographics and general medical history. Based on this initial evaluation, we excluded volunteers with the following history: head trauma, uncontrolled diabetes mellitus and hypertension; current chronic renal, liver, respiratory or neurologic disorders; recent myocardial infarction (within the previous 6 months), and unstable angina. Also excluded were persons using hormone replacement therapy (HRT) or medications that may affect memory (e.g. anticholinergics, sedative hypnotics, narcotics, and antiparkinsonian agents). Volunteers having unstable medical conditions, evidenced by starting new medications within 6 weeks of enrollment, and those having history of chronic alcohol and drug abuse, were also excluded from the study. Subjects were allowed to continue using medications to treat AD (Reminyl, Aricept, Exelon, Namenda, and Ginkgo Biloba).
Diagnosis of MCI was made using the following criteria: memory complaints, education adjusted Mini-Mental State Examination (MMSE) scores, where the Adjusted MMSE=Raw MMSE--(0.471 x [education-12]) + (0.131 x [age – 70])) with a score of 24–30 being inclusive (17), objective memory loss ascertained by performance on education adjusted scores on the Wechsler Memory Scale Logical Memory II, Clinical Dementia Rating Scale (CDR) of ≤0.5, modified Hachinski score < 4, and Geriatrics Depression Scale (GDS) < 6. We also excluded persons with probable dementia according to National Institute of Neurological and Communicative Disorders and Stroke/Alzheimer’s Disease and Related Disorders Association (MINCDS/ADRDA) criteria, and those having memory loss from medical, neurological, or psychiatric conditions, medication effects or delirium.
1.2.2. Level II Screening
From the remaining eligible volunteers, we acquired vital signs and anthropometric measurements, and then performed detailed general physical and neurological examinations. We collected fasting blood samples to assess plasma lipoprotein/lipids, hemoglobin and hematocrit, basic metabolic panel (BMP), rapid plasma reagin (RPR) and plasma levels of thyroxine (T4), and thyroid stimulating hormone from all eligible volunteers.
The remaining qualified and willing volunteers underwent a maximal treadmill exercise test using the Bruce protocol (18) to screen for cardiovascular disease (CVD). The test was terminated when the subject could no longer continue, or CVD signs and symptoms occurred (19). CVD indicators including blood pressure, heart rate, and ECG were recorded before the test, at the end of every exercise stage, and every 2 minutes for 6 minutes after discontinuing the test. The test was terminated if the subject had >2-mV ST-segment depression, extra systole, chest pain, arrhythmias, hypotension or dizziness, or significant ST segment elevation during the treadmill tests (19, 20). The maximal oxygen consumption (VO2max) was determined using a validated customized on-line system (K4b2 by Cosmed, Chicago Illinois). As part of the initial evaluation and prior to the treadmill screening tests, each participant had a brain MRI to exclude significant intracranial pathology (such as clinically significant cerebrovascular disease including cortical infarcts, strategically located subcortical gray matter or extensive white matter abnormalities). Prior to randomization, study partners and subjects were instructed to maintain regular caloric intake during the study period, and maintain an American Heart Association Step 1 diet: <30% of energy from fat, ~55% from carbohydrate, ~15% from protein, and cholesterol intake <300 mg/day.
1.2.3. Randomization and Blinding Procedures
Randomization of subjects occurred prior to baseline tests. All staff, except those directly monitoring exercise-training, were blinded to group assignments. The data were de-identified using assigned unique identifiers for labeling and tracking.
1.2.4. Baseline Testing
At the completion of dietary stabilization and randomization, subjects were familiarized with treadmill screening tests, before undergoing the actual treadmill exercise test. We then determined baseline VO2max and endurance capacity, using a modified Bruce protocol. Discontinuation criteria for this test were similar to those used for the level II screening test.
During the 24-hour period prior to blood drawing for baseline tests, participants were instructed not to have anything by mouth, consume no alcohol or smoke cigarettes, and to not use any anti-inflammatory medications. Additionally, subjects were told to refrain from exercise for 72 hours prior to testing, and to confirm that they had no infection in the week prior to testing. For the gene expression analyses, and prior to the initiation of exercise-training, we collected overnight fasting blood samples using sterile techniques by personnel trained in phlebotomy, and stored samples in heparinized collection tubes. Subsequently, the samples were centrifuged at 500g after which the buffy coat layer containing leukocyte population was removed and stored in aliquots at −80°C.
1.2.5. Aerobic Exercise-Training Protocol
We inferred each subject’s maximum heart rate from baseline VO2max tests. Both the intervention (aerobic exercise) and control (stretch exercise) groups underwent supervised training 3 times/week.
The aerobic exercise protocol complied with the American College of Sports Medicine Guidelines (ACSM) (21), and included treadmill walking or jogging, stair-stepping, and elliptical climbing. Subjects underwent a warm up period, followed by exercise-training and an appropriate cool-down period. Initial training sessions lasted 20 min at 50% VO2max, while study staff used records of exercise heart rate and duration to monitor protocol compliance. During each session, training duration increased by 5 min/week until subjects completed 40 min of exercise at 50% VO2max. Subsequently, training intensity was increased by 5% VO2max weekly until achieving 70% VO2max while taking necessary safety precautions. Participants were asked to add an unsupervised 45–60 min lower intensity walk on the weekend after the first 4–6 weeks of exercise. Overall exercise-training lasted 6 months, and subjects continued all interventions until the completion of final testing.
1.2.6. Stretch Exercise Protocol
Static rather than dynamic stretch of joints increases flexibility, joint range, and prevents motion loss, but has minimal impact on VO2max(22). As such, training of the stretch group consisted of maintaining exercise positions for 15–30 seconds to produce a slight pull on the muscle but not to the point of triggering the sensation of pain. Using different positions for a total of about 40 minutes, each stretch was directed at often tight muscles (e.g. hamstrings, hip flexors, calves and chest), and repeated slowly, 3–5 times on each body side 3 days/week (22). To more closely match the increasing intensity and duration of aerobic-exercise, stretch training targeted hamstrings in week-1; hamstring + hip flexors in week-2; hamstring + hip flexors + calves in week-3; and hamstring + hip flexors + calves + chest muscle in week-4, and continued until study completion and final testing.
1.2.7. Follow-up Test at 6 Months
After subjects completed the 6-month aerobic or stretch exercise-training, all baseline tests (e.g. VO2max, blood tests for microarray analyses) were repeated using methods analogous to baseline testing.
1.2.8. Microarray Analyses
The microarray analysis included data from 10 MCI subjects randomly assigned to either an aerobic exercise (n=5) or a stretch exercise control (n=5) group. To determine changes in global gene expression profiles, we performed microarray analysis on blood samples collected before (baseline) and after a 6-month exercise-training in the 10 MCI volunteers who completed the protocol. Total RNA was isolated from buffy coat samples and extracted using the RNeasy Mini Kit (QIAGEN, Valencia, CA) according to the manufacturer’s instructions. After the extraction, RNA concentration and purity (OD260/280) were measured using the NanoDrop ND-1000 spectrophotometer (Thermo Fischer Scientific, Waltham, MA, USA), and the RNA integrity number (RIN) was determined using an Agilent 2100 Bioanalyzer Instrument (Agilent, Santa Clara, CA, USA). We profiled gene expression using the Illumina HumanHT-12 v4 Expression BeadChip platform containing 47,000 probes covering RefSeq and Unigene annotated genes (Illumina Inc., San Diego, CA, USA). 500 ng total RNA was labeled using the Illumina TotalPrep-96 RNA Amplification Kit (Ambion, Inc., Austin, TX). Briefly, the protocol features first- and second-strand reverse transcription steps, followed by a single in vitro transcription (IVT) amplification that incorporates biotin-labeled nucleotides to generate biotinylated cRNA. The purified cRNA was quantified and the fragment size was determined using an Agilent 2100 Bioanalyzer Instrument (Agilent, Santa Clara, CA, USA). Then, 750 ng of labeled biotinylated cRNA probe was hybridized overnight to the Illumina HumanHT-12 v4 Expression BeadChip. The hybridization, washing, and scanning were performed according to the manufacturer’s instructions. The BeadChips were scanned using a HiScanSQ System (Illumina Inc., San Diego, CA, USA). The microarray images were registered and extracted automatically during the scan using the manufacturer’s default settings.
Ingenuity pathway analysis (IPA) was used to analyze conduits of relevant genes. Genes with significant changes in expression compared to baseline values were determined by applying a paired t-test on each individual gene, and using the cutoff combination of the Benjamini-Hochberg multi-test with an adjusted p-value < 0.01 and absolute fold change ≥ 2.0. Qualified genes were provided to the IPA program for core analysis. Based on the context of molecular mechanisms stored in the Ingenuity Database, networks were generated for up-regulated and down-regulated genes that met the threshold. Using the Ingenuity system for core analysis, corresponding networks were determined from all genes with significantly altered expression.
1.3. RESULTS
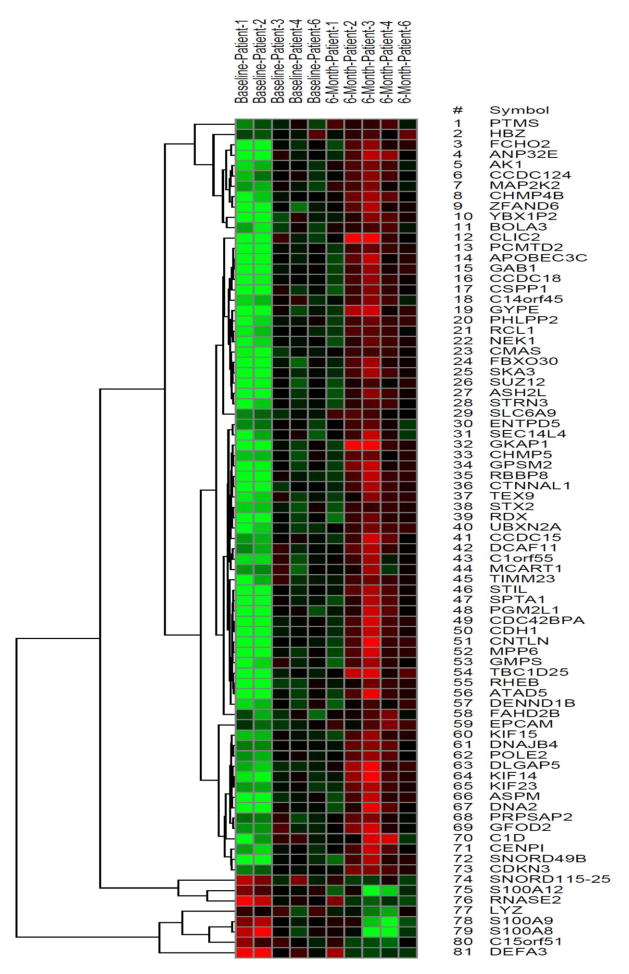
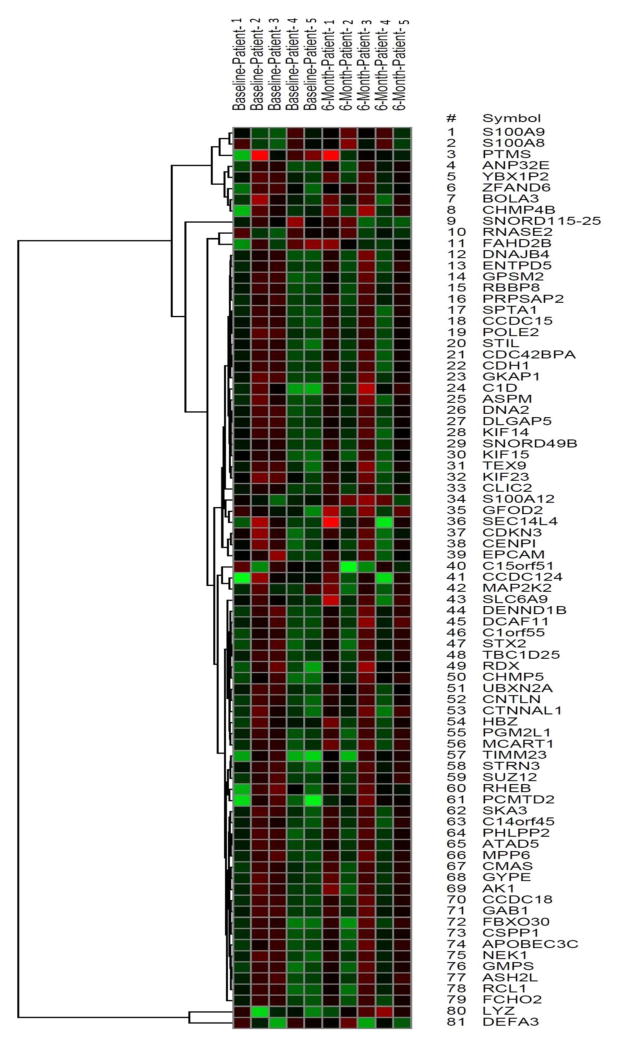
We used microarrays to obtain gene expression profiles of human blood cells that were derived from MCI subjects at baseline and after a 6-month intervention of aerobic or stretch exercise. The samples consisted of AAs (3 males and 7 females) with ages as follows: aerobic=73±5 years and stretch=70±9 years (mean ± standard deviation). In total, 81 genes in our sample were differentially expressed in response to aerobic exercise-training (Figure 1: aerobic exercise heat map) compared to only 6 unique and non-overlapping genes in the stretch-training group, using a cut-off p-value < 0.01 and fold change ≥ 2.0. In analyses restricted to the 81 differentially expressed genes obtained from the aerobic exercise group, the stretch group showed no statistically significant alteration in training-induced gene expression profiles (fold change ≤ 2 at p ≥ 0.01) (Figure 4: stretch exercise heat map). Many of the 81 genes with altered expression in the exercise group (Table 1) play important roles in immune function, cell survival and inflammation.
Figure 1.
Heat map of the 81 genes whose expression were altered by a 6-month aerobic exercise training
Figure 4.
Heat map of the 81 genes in the stretch control group, whose expression were altered by a 6-month aerobic exercise training.
Table 1.
Notable Genes Involved in Immune Function, Promotion of Neuronal Survival and Axonal Growth with ≥ 2 Fold Change (p < 0.01) after a 6-Month Aerobic Exercise-Training Program
| Symbol | Name | p-Value | Fold-change | Function |
|---|---|---|---|---|
| Genes involved in immunological function | ||||
| DEFA3 | Defensin, alpha-3, neutrophil-specific | 0.009578 | −3.3 | Kill microbes by permeabilizing their plasma membrane (Nature Reviews Immunology (2003) 3, 710–720) |
| C15orf51 | Dynamin-1 pseudogene | 0.00554 | −3.3 | Unknown |
| RNASE2 | Ribonuclease, RNase A family-2 | 0.001291 | −3.1 | Selectively chemotactic for dendritic cells (Blood (2003) 102, 3396–3403) |
| S100A8 | S100 calcium binding protein A8 | 0.000489 | −2.8 | Potent amplifier of inflammation in autoimmunity and in cancer development/spread (Refs (33, 34); see Discussion) |
| LYZ | Lysozyme | 0.00392 | −2.6 | Innate immunity, monocyte-macrophage system and enhanced activity of immune agents (Ric Clin Lab (1978) 8, 211–231) |
| S100A9 | S100 calcium binding protein A9 | 0.001877 | −2.3 | Potent amplifier of inflammation in autoimmunity and in cancer development/spread (Refs (33, 34); see Discussion) |
| SNORD115-25 | Small nucleolar RNA, C/D box 115-25 | 0.007939 | −2.1 | RNA methylation (Nature (1998) 383, 732–735) |
| S100A12 | S100 calcium binding protein A12 | 0.002473 | −2.0 | Regulation of inflammatory processes and immune response (Ref (24); see Discussion) |
| MAP2K2 | Mitogen-activated protein kinase kinase-2 | 0.007647 | 2.2 | Role in mitogen growth factor signal transduction (Refs (59, 60, 62); see Discussion) |
| PTMS | Parathymosin | 0.004324 | 2.5 | Mediate immune function by blocking the effect of prothymosin alpha (Refs (51, 52, 54); see Discussion) |
| CMAS | Cytidine monophosphate Neu5Ac synthetase | 0.005699 | 2.9 | Sialylation of glycoproteins (Proc Natl Acad Sci USA (1998) 95, 9140–9145; see Discussion) |
| Genes promoting neuronal survival and axon growth | ||||
| GAB1 | GRB2-associated binding protein-1 | 0.007016 | 3.1 | Involved in cell growth and enhancement of neuronal survival through EGFR (Ref (71, 72); see Discussion) |
| CDH1 | Cadherin-1, type-1, E-cadherin (epithelial) | 0.007616 | 2.2 | Regulates cell-cell adhesions, mobility, axonal growth, synaptic control (Refs (68, 70); see Discussion) |
| RHEB | Ras homolog enriched in brain | 0.006289 | 2.1 | Promotes neuronal survival and axon regeneration through mTOR pathway (Ref (80, 81); see Discussion) |
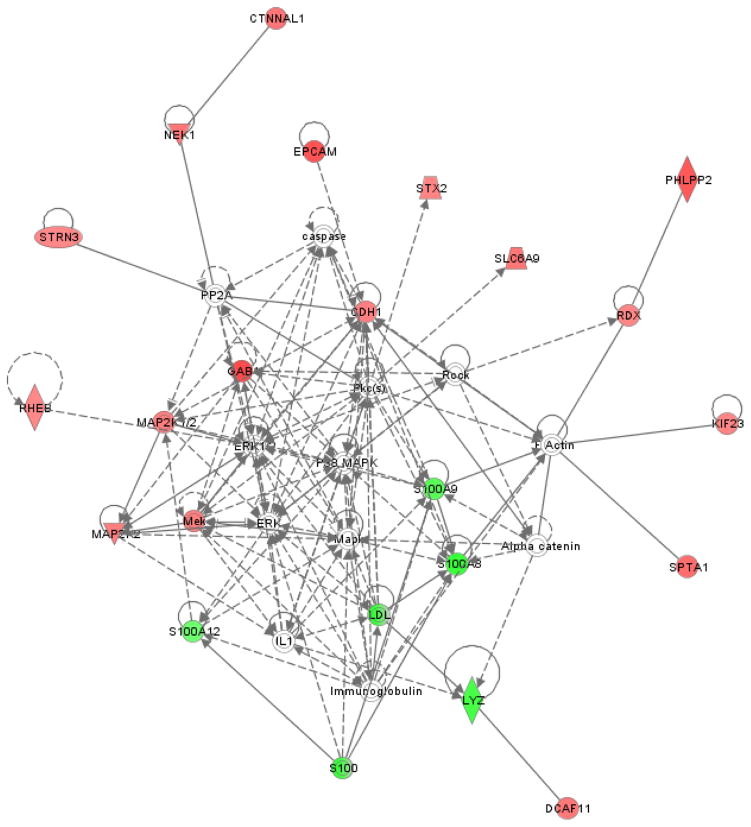
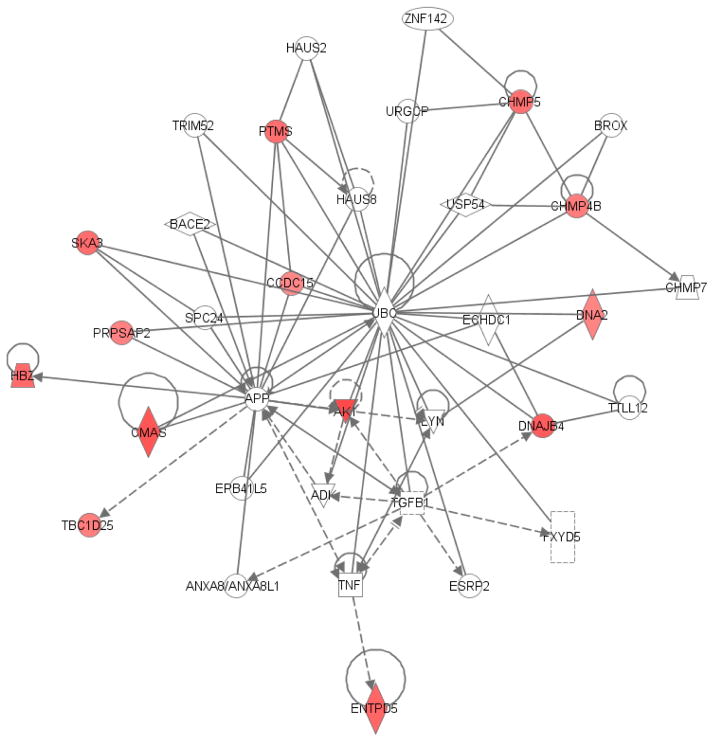
Using the Ingenuity Knowledge Base, we determined biologically relevant networks of genes using all the aerobic-exercise regulated genes in this study (Figure 2 and Figure 3). Networks identified by the Ingenuity Knowledge Base library are premised on known interactions amongst various genes. Specifically, we identified pathways constituting ribonuclease, RNase A family-2 (RNASE2), and interleukin-1 (IL-1), with RNASE2 significantly down-regulated in this study (Table 1). Similarly, the S100 calcium-binding protein family (S100A8, S100A9, and S100A12), defensin (alpha-3, neutrophil-specific) (DEFA1) and lysozyme (LYZ) (Figures 1 and 2 and Table 1) were also down-regulated. Contrary to the overall increased expression of anti-inflammatory genes in the aerobic exercise group, the expression of S100A12 and LYZ increased 1.5 and 2.2 fold, respectively, in the stretch-exercise control group, but at p > 0.01 (Table 2). The down-regulated genes in the aerobic exercise group (Table 1) have important immunological and pro-inflammatory properties. Three of the up-regulated genes (Table 1) in the expression data (parathymosin [PTMS] (Figure 3), cytidine monophosphate Neu5Ac synthetase [CMAS] and mitogen-activated protein kinase kinase-2 [MAP2K2] (Figure 2) have also been reported to exert significant immunological and anti-inflammatory properties, easing the immune response when up-regulated (see Discussion). Cadherin-1 (type-1, E-cadherin, epithelial) (CDH1), GRB2-associated binding protein-1 (GAB1), and Ras homolog enriched in brain (Rheb) are other up-regulated genes that play important roles in neuronal survival and axon growth. Training-induced up-regulation of these latter genes may promote neuronal survival in AD.
Figure 2. A network constituting some genes with altered expression after a 6-month aerobic exercise program (inflammation and cancer).
Genes are represented by nodes. Nodes colored in “red” represent up-regulated genes and nodes colored in “green” indicate down-regulated genes (p-value < 0.01, fold change ≥ 2), and the intensity of the color demonstrates the degree of up/down-regulation when compared to normal levels. Uncolored nodes are genes not identified in this study but have been integrated into the Ingenuity Pathways system/memory. Nodes are displayed using various shapes that represent the functional class of the gene product. Straight lines with arrows indicate the biological relationship between two nodes, and the nature of the relationship between two different nodes is displayed by the edges. Edges are supported by scientific publications that are stored within the Ingenuity Pathways Knowledge Base. This network was updated using the 2014 Ingenuity Pathways software.
Figure 3. A network constituting some genes with altered expression after a 6-month aerobic exercise program (cancer, cell cycle, and cellular signaling).
Genes are represented by nodes. Nodes colored in “red” represent up-regulated genes and nodes colored in “green” indicate down-regulated genes (p-value < 0.01, fold change ≥ 2), and the intensity of the color demonstrates the degree of up/down-regulation when compared to normal levels. Uncolored nodes are genes not identified in this study but have been integrated into the Ingenuity Pathways system/memory. Nodes are displayed using various shapes that represent the functional class of the gene product. Straight lines with arrows indicate the biological relationship between two nodes, and the nature of the relationship between two different nodes is displayed by the edges. Edges are supported by scientific publications that are stored within the Ingenuity Pathways Knowledge Base. This network was updated using the 2014 Ingenuity Pathways software.
Table 2.
Notable Genes with ≥ 1.5 Fold Change in the 6-Month Stretch Control Group*
| Symbol | Name | P-Value | Fold Change | Function |
|---|---|---|---|---|
| TIMM23 | Translocase of inner mitochondrial membrane 23 homolog (yeast) | 0.2468806 | 1.5 | Mediates the transport of transit peptide-containing proteins across the membrane |
| APOBEC3C | Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3C | 0.2501128 | 1.5 | Have roles in growth or cell cycle control |
| S100A12 | S100 calcium binding protein A12 | 0.0490725 | 1.5 | Regulation of inflammatory processes and immune response |
| C1D | C1D nuclear receptor corepressor | 0.0963231 | 1.5 | Localized in the nucleus and encodes DNA binding and apoptosis-inducing protein |
| CHMP5 | Charged multivesicular body protein 5 | 0.0581199 | 1.6 | Its complex is involved in degradation of surface receptor proteins and formation of endocytic multivesicular bodies (MVBs) |
| PCMTD2 | Protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 | 0.039969 | 1.9 | Protein coding gene |
| LYZ | Lysozyme | 0.0460297 | 2.2 | Enhances the activity of immune agents (Ric Clin Lab (1978) 8, 211–231) |
| SNORD115-25 | Small nucleolar RNA, C/D box 115-25 | 0.1892712 | −1.6 | RNA methylation (Nature (1996) 383, 732–735) |
Note that the p-values are > 0.01
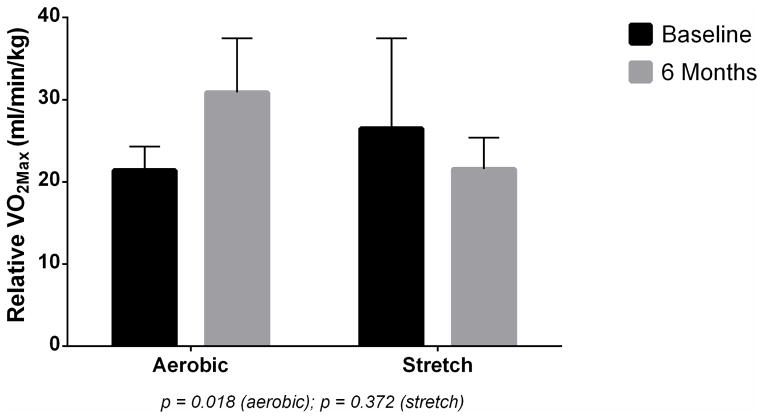
At baseline, the aerobic exercise group had lower relative VO2max (21.39 ml/kg/min) compared to the stretch group (26.48 ml/kg/min), respectively. However, after the 6-month training intervention, the aerobic group had significantly greater training-induced changes in relative VO2max (+9.72 ml/kg/min) compared to a reduction in the stretch exercise (−4.92 ml/kg/min) (see Figure 5). The training-induced changes in VO2max of the aerobic exercise group correlates with favorable changes in gene expression after the intervention.
Figure 5.
Baseline and 6-month VO2max (ml/kg/min) measurements in aerobic and stretch exercise groups.
1.4. DISCUSSION
We observed that a 6-month standardized aerobic exercise-training regimen significantly altered the regulation of several genes. Particularly, robust training-induced effects were noted on the regulation of genes involved in inflammation, cell survival and axonal growth. Our analysis revealed the down-regulation of three pro-inflammatory genes (S100A8, S100A9 and S100A12) in the S100 family. By promoting inflammation at their steady state, these genes likely play an important role in AD pathology (23–25). On the other hand, 3 genes exerting anti-inflammatory properties and others involved in neuronal survival and axonal growth were up-regulated (see below). Because of the centrality of inflammation to AD pathology, these results may, in part, mechanistically explain the beneficial effects of exercise on cognition. If confirmed in future studies, directed modulation of the expression of pro- and anti-inflammatory genes, and the enhancement of genes promoting neuronal survival, may offer useful interventions, and potentially become targets for the development of drugs to delay AD development in those at risk.
1.4.1. Down-Regulation of Pro-inflammatory Genes
It is widely accepted that neuroinflammation accompanied by excessive expression of immune cytokines is a “principal culprit” of sporadic AD (26). Pro-inflammatory proteins such as S100a8/a9 have been demonstrated to be markedly increased in response to cytokine stimuli (27, 28). In addition, S100a8/a9 may act as leukocyte chemo-attractants (29) and induce the expression of pro-inflammatory cytokines (30), causing a positive feedback in inflammation. In aged mice and humans, increases in the mRNA levels of S100a8 and S100a9 have been demonstrated in multiple tissues including the central nervous system (31). Recently, S100a9 (calgranulin B), an important pro-inflammatory mediator in acute and chronic inflammation (32–34), was identified as a contributor to amyloid plaque accumulation (35). In support of its role in AD, knockdown of S100a9 expression reduced amyloid plaque abundance and improved cognition in a mouse model (25). It appears that during AD pathogenesis, normal aging causes an increase in the population of amyloid precursor protein and/or Aβ production (36), inducing the production of pro-inflammatory proteins such as S100a9 (25). These pro-inflammatory proteins may drive AD neuropathology by promoting amyloid plaque formation. Therefore, aerobic exercise training-induced down-regulation of the S100 family of genes may ameliorate the destructive effects of inflammatory molecules and preserve cognition.
S100A12 is another pro-inflammatory protein that shares sequence homology with S100A8 and S100A9 (37). It is a potent chemo-attractant for monocytoid cells (38) and induces the expression of adhesion molecules and pro-inflammatory cytokines (39). It activates various cell types by binding the Receptor for Advanced Glycosylation End products (RAGE) associated with AD-characteristic lesions (40). Secretion of S100A9 during inflammation promotes the formation of amyloid plaques (35) and the co-existence of both S100A9 and S100A12 in pathological lesions of AD support their roles in AD pathogenesis (24).
Given the enhancing effects of exercise on memory (41–45), significant exercise-training-induced down-regulation of pro-inflammatory genes in older MCI subjects, may mechanistically relate fitness effects to reduced neuro-inflammation and AD pathogenesis. Indeed, growing independent evidence now suggest that moderate exercise can modulate inflammation, decrease pro-inflammatory cytokines (46, 47), and therefore, preserve neuronal integrity, and potentially, preserve cognition (48, 49).
1.4.2. Up-regulation of Anti-inflammatory Genes
Our results also show that aerobic-exercise training up-regulated three genes (PTMS, CMAS, MAP2K2) involved in reduced immunological response.
PTMS likely mediates immune function by blocking the effects of prothymosin alpha (50), and reduces the production of IL-2 and IL-2R (IL-2 receptor) required for proliferation of CD4+ T cells (51, 52). As a pro-inflammatory cytokine, IL-2 stimulates lymphocytes, monocytes, lymphokine-activated killer cells, and natural killer cells critical to immune function (53). Consistent with the idea that anti-inflammatory genes may be down-regulated in AD, down-regulation of parathymosin was noted in amyloid beta-stimulated microglia from postmortem cases (54). Given the pivotal role of inflammation in AD, exercise-induced up-regulation of parathymosin and consequent modulation of the immune response may reduce inflammation and benefit cognitive processes.
CMAS, the second up-regulated anti-inflammatory gene in our expression data, encodes the enzyme N-acetylneuraminate cytidylyltransferase responsible for adding sialic acid residues to glycoproteins and glycolipids in humans (55). Neuronal membranes possess sialic acid which promotes neural transmission, maintains ganglioside structure in synaptogenesis (56), and enhances brain development and learning (57). Removal of sialic acid residues from the neuronal glycocalyx activates the classical complement pathway and phagocytosis by microglia (58). Therefore, training-induced up-regulation of CMAS expression, may enhance sialic acid-dependent glycosylation of neuronal membranes, protect neurons against complement-induced loss, and potentially reduce brain inflammation, neuronal degeneration and cognitive loss.
MAP2K2, the third up-regulated anti-inflammatory gene in our data, encodes a dual specificity protein kinase belonging to the MAP kinase family. This protein is essential for mitogen growth factor signal transduction and cell cycle progression (59, 60). Although activated MAP2K2 may promote the production of some pro-inflammatory cytokines (61), its enhancement of IL-10 production may inhibit the syntheses of IFN-gamma and TNF-alpha (62, 63). Therefore, as a differential immune modulator, increased expression of MAP2K2 by a 6-month aerobic exercise-training may ease amyloid-induced immunologic responses, and perhaps preserve cognition. Though we observed no significant training-induced fold change in the expression of the amyloid precursor protein (APP) gene in our samples, the direct interaction of both CMAS and PTMS with APP (Figure 3) may indicate their potential roles in AD pathogenesis.
1.4.3. Genes Promoting Neuronal Survival
In our data, a 6-month aerobic-exercise program also up-regulated the expression of three genes (CDH1, GAB1, Rheb) that can promote neuronal survival and axon growth.
CDH1 is implicated in fundamental nervous system processes, such as axonal growth, patterning (64, 65), and synaptic control (66, 67). In addition, the degradative action of the CDH1-anaphase-promoting complex (APC) is an essential survival mechanism in post-mitotic neurons (68). Given that the inactivation of CDH1 contributes to neuronal excitotoxic death from accumulation of cyclin B1 in degenerating brain areas in Alzheimer’s disease (69, 70), aerobic-exercise training-induced up-regulation of CDH1 may be vital to neuronal survival and reduced neurodegeneration.
GAB1 is a docking/scaffolding molecule which transduces signals from a variety of tyrosine kinases, including epidermal growth factor receptor (EGFR)-induced signaling (71, 72). Given previously reported impairment of learning and memory in EGF-like growth factor knockout mice (73), it is possible that EGFR acts as a mitogen for neural stem and progenitor cells (NS/NPCs) in the central nervous system (CNS), integrating EGF/TGF-α signaling in a way that affects neuronal differentiation (74), survival (75–77), and glial proliferation (78, 79). It is possible that fitness-induced up-regulation of these genes may enhance neuronal plasticity and stem the tide of neurodegeneration.
Rheb is a small GTPase that activates mTOR (80–82). The mammalian target of rapamycin (mTOR) is a serine/threonine protein kinase which regulates cellular growth, and helps to integrate growth factor activity with other external signals (83). Because Rheb-mTOR signaling confers axon protection (80), its activation may promote neuronal survival and axon regeneration (84). Given that axonal loss is central to AD and other neurodegenerative disorders, the role of Rheb-mTOR in neurodegenerative pathogenesis must be considered. In select brain regions, it is believed that axons and their synaptic terminals but not neuronal cell bodies, are the initial loci of some neurodegeneration, a cause of early disease manifestations and clinical progression (85, 86). Therefore, exercise-induced up-regulation of expression of Rheb might promote Rheb-mTor signaling, enhance the survival of neurons and regeneration of axons, and potentially improve cognitive impairment.
An important finding from this study is the concomitant training-induced increase in relative VO2max in the aerobic but not the stretch exercise group. Though the exact molecular mechanisms which underlie the changes in expression are not completely understood, the concomitant training-induced changes in VO2max suggest that training adaptation and cardiovascular fitness, can promote advantageous effects at the molecular level.
1.4.4. Limitations and Summary
One limitation of this study is that gene expression data were derived from blood cells (and not the brain or cerebrospinal fluid), and are yet to be correlated with the AD clinical phenotype. However, it is likely that such work will yield positive results, given that exercise has been demonstrated to promote neurogenesis in the dentate gyrus of exercising mice and in humans (87). Similarly, the intensity rating of physical activity and reduced TNF-alpha, were associated with increased brain volume on magnetic resonance imaging (7). These findings are consistent with our observation of increased expression of neuronal and axonal survival genes in aerobic but not in stretch exercise training. Recently, Swindell and colleagues showed significantly increased expression of S100 genes in both blood and the hippocampus of aging human tissues (31), suggesting that molecular aspects of the underlying pathology may have a systemic component, making blood cell-derived data relevant. These observations are consistent with existing evidence arguing that exercise adaptation can decrease the levels of pro-inflammatory cytokines (46, 47).
A second limitation of this study is the relatively small sample size. A recent microarray study by Stretch et al. examining sex-based differences in gene expression concluded that a larger sample size would be helpful (88). However, compared to the cross-sectional nature of the Stretch et al. study, the randomized, controlled, and supervised clinical trial approach of this rigorous exercise study in AA MCIs, limit the impact of the small sample size. To analyze the training-induced changes in gene expression, aggregate fold changes for the same set of individuals in each group were independently calculated, analyzed and normalized within one “average” background of the stretch or aerobic exercise group. The physiological significance of the identified changes is plausible since the differentially expressed genes could be grouped into functional pathways, with notable conformity in the direction of their regulation (increase or decrease) in each pathway. Moreover, the extent of overlap of such changes likely indicates whether they are due to chance or the intervention. Had significant overlap in observed differential expression occurred between the 2 groups (aerobic versus stretch), the overall results may have inspired less confidence. Nonetheless, because a larger sample size would be helpful, caution is warranted in interpreting the results.
In summary, our data show that aerobic-exercise can down-regulate the expression of pro-inflammatory genes, concomitantly up-regulate anti-inflammatory genes, and collectively reduce inflammation. Simultaneously, aerobic exercise promotes the expression of genes involved in axonal growth and neuronal survival. Given the harmful attributes of inflammation on neuronal survival and AD pathology, it appears likely that the enhancing effects of aerobic-exercise on cognition may be mediated through reduction in inflammation. Overall, our results support the role of gene-environment interactions on health. Future RT-polymerase chain reaction studies should consider the use of cerebrospinal fluid to validate our semi-quantitative observations.
Highlights.
6-Month aerobic exercise promoted training-induced alteration in global gene expression.
Aerobic exercise-training promoted significant training-induced changes in VO2max.
Training-induced changes in VO2max paralleled changes in global gene expression.
Physiologic adaptation from aerobic exercise is likely mediated at the molecular level.
Acknowledgments
This work was supported by the National Institute on Aging at the National Institutes of Health (NIH) grant R01 5R01AG31517-2 to Obisesan TO and in part by Grant #UL1TR000101 from National Center for Advancing Translational Sciences/NIH through the Clinical and Translational Science Award Program (CTSA). The funders had no role in the design, data collection, and interpretation of this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Selkoe DJ. Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 2.Lue LF, Brachova L, Civin WH, Rogers J. Inflammation, A beta deposition, and neurofibrillary tangle formation as correlates of Alzheimer’s disease neurodegeneration. J Neuropathol Exp Neurol. 1996;55:1083–1088. [PubMed] [Google Scholar]
- 3.Hoozemans JJ, Veerhuis R, Rozemuller JM, Eikelenboom P. Neuroinflammation and regeneration in the early stages of Alzheimer’s disease pathology. Int J Dev Neurosci. 2006;24:157–165. doi: 10.1016/j.ijdevneu.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Lee KS, Chung JH, Choi TK, Suh SY, Oh BH, Hong CH. Peripheral cytokines and chemokines in Alzheimer’s disease. Dement Geriatr Cogn Disord. 2009;28:281–287. doi: 10.1159/000245156. [DOI] [PubMed] [Google Scholar]
- 5.Mrak RE, Griffin WS. Glia and their cytokines in progression of neurodegeneration. Neurobiol Aging. 2005;26:349–354. doi: 10.1016/j.neurobiolaging.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 6.Vlad SC, Miller DR, Kowall NW, Felson DT. Protective effects of NSAIDs on the development of Alzheimer disease. Neurology. 2008;70:1672–1677. doi: 10.1212/01.wnl.0000311269.57716.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braskie MN, Boyle CP, Rajagopalan P, Gutman BA, Toga AW, Raji CA, et al. Physical activity, inflammation, and volume of the aging brain. Neuroscience. 2014;273:199–209. doi: 10.1016/j.neuroscience.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchman AS, Boyle PA, Yu L, Shah RC, Wilson RS, Bennett DA. Total daily physical activity and the risk of AD and cognitive decline in older adults. Neurology. 2012;78:1323–1329. doi: 10.1212/WNL.0b013e3182535d35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adlard PA, Perreau VM, Pop V, Cotman CW. Voluntary exercise decreases amyloid load in a transgenic model of Alzheimer’s disease. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2005;25:4217–4221. doi: 10.1523/JNEUROSCI.0496-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boas SR, Joswiak ML, Nixon PA, Kurland G, O’Connor MJ, Bufalino K, et al. Effects of anaerobic exercise on the immune system in eight- to seventeen-year-old trained and untrained boys. J Pediatr. 1996;129:846–855. doi: 10.1016/s0022-3476(96)70028-8. [DOI] [PubMed] [Google Scholar]
- 11.Gleeson M, Bishop NC, Stensel DJ, Lindley MR, Mastana SS, Nimmo MA. The anti-inflammatory effects of exercise: mechanisms and implications for the prevention and treatment of disease. Nat Rev Immunol. 2011;11:607–615. doi: 10.1038/nri3041. [DOI] [PubMed] [Google Scholar]
- 12.McTiernan A. Mechanisms linking physical activity with cancer. Nat Rev Cancer. 2008;8:205–211. doi: 10.1038/nrc2325. [DOI] [PubMed] [Google Scholar]
- 13.McTiernan A. Physical activity after cancer: physiologic outcomes. Cancer Invest. 2004;22:68–81. doi: 10.1081/cnv-120027581. [DOI] [PubMed] [Google Scholar]
- 14.Ouchi N, Parker JL, Lugus JJ, Walsh K. Adipokines in inflammation and metabolic disease. Nat Rev Immunol. 2011;11:85–97. doi: 10.1038/nri2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spence RR, Heesch KC, Brown WJ. Exercise and cancer rehabilitation: a systematic review. Cancer Treat Rev. 2010;36:185–194. doi: 10.1016/j.ctrv.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Petersen RC. Mild cognitive impairment as a diagnostic entity. Journal of internal medicine. 2004;256:183–194. doi: 10.1111/j.1365-2796.2004.01388.x. [DOI] [PubMed] [Google Scholar]
- 17.Mungas D, Marshall SC, Weldon M, Haan M, Reed BR. Age and education correction of Mini-Mental State Examination for English and Spanish-speaking elderly. Neurology. 1996;46:700–706. doi: 10.1212/wnl.46.3.700. [DOI] [PubMed] [Google Scholar]
- 18.Bruce RA, Hornsten TR. Exercise stress testing in evaluation of patients with ischemic heart disease. Prog Cardiovasc Dis. 1969;11:371–390. doi: 10.1016/0033-0620(69)90027-9. [DOI] [PubMed] [Google Scholar]
- 19.American College of Sports Medicine Position Stand. . The recommended quantity and quality of exercise for developing and maintaining cardiorespiratory and muscular fitness, and flexibility in healthy adults. Med Sci Sports Exerc. 1998;30:975–991. doi: 10.1097/00005768-199806000-00032. [DOI] [PubMed] [Google Scholar]
- 20.White RD, Evans CH. Performing the exercise test. Prim Care. 2001;28:29–53. vi. doi: 10.1016/s0095-4543(05)70006-3. [DOI] [PubMed] [Google Scholar]
- 21.American College of Sports Medicine position stand. . The recommended quantity and quality of exercise for developing and maintaining cardiorespiratory and muscular fitness in healthy adults. Med Sci Sports Exerc. 1990;22:265–274. [PubMed] [Google Scholar]
- 22.American College of Sports Medicine Position Stand. . Exercise and physical activity for older adults. Med Sci Sports Exerc. 1998;30:992–1008. [PubMed] [Google Scholar]
- 23.Sen J, Belli A. S100B in neuropathologic states: the CRP of the brain? J Neurosci Res. 2007;85:1373–1380. doi: 10.1002/jnr.21211. [DOI] [PubMed] [Google Scholar]
- 24.Shepherd CE, Goyette J, Utter V, Rahimi F, Yang Z, Geczy CL, et al. Inflammatory S100A9 and S100A12 proteins in Alzheimer’s disease. Neurobiol Aging. 2006;27:1554–1563. doi: 10.1016/j.neurobiolaging.2005.09.033. [DOI] [PubMed] [Google Scholar]
- 25.Ha TY, Chang KA, Kim J, Kim HS, Kim S, Chong YH, et al. S100a9 knockdown decreases the memory impairment and the neuropathology in Tg2576 mice, AD animal model. PLoS One. 2010;5:e8840. doi: 10.1371/journal.pone.0008840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Streit WJ, Mrak RE, Griffin WS. Microglia and neuroinflammation: a pathological perspective. Journal of neuroinflammation. 2004;1:14. doi: 10.1186/1742-2094-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu K, Champaiboon C, Guenther BD, Sorenson BS, Khammanivong A, Ross KF, et al. Anti-Infective Protective Properties of S100 Calgranulins. Antiinflamm Antiallergy Agents Med Chem. 2009;8:290–305. doi: 10.2174/187152309789838975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lackmann M, Rajasekariah P, Iismaa SE, Jones G, Cornish CJ, Hu S, et al. Identification of a chemotactic domain of the pro-inflammatory S100 protein CP-10. J Immunol. 1993;150:2981–2991. [PubMed] [Google Scholar]
- 30.Benedyk M, Sopalla C, Nacken W, Bode G, Melkonyan H, Banfi B, et al. HaCaT keratinocytes overexpressing the S100 proteins S100A8 and S100A9 show increased NADPH oxidase and NF-kappaB activities. J Invest Dermatol. 2007;127:2001–2011. doi: 10.1038/sj.jid.5700820. [DOI] [PubMed] [Google Scholar]
- 31.Swindell WR, Johnston A, Xing X, Little A, Robichaud P, Voorhees JJ, et al. Robust shifts in S100a9 expression with aging: a novel mechanism for chronic inflammation. Sci Rep. 2013;3:1215. doi: 10.1038/srep01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foell D, Frosch M, Sorg C, Roth J. Phagocyte-specific calcium-binding S100 proteins as clinical laboratory markers of inflammation. Clin Chim Acta. 2004;344:37–51. doi: 10.1016/j.cccn.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 33.Gebhardt C, Nemeth J, Angel P, Hess J. S100A8 and S100A9 in inflammation and cancer. Biochem Pharmacol. 2006;72:1622–1631. doi: 10.1016/j.bcp.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 34.Gebhardt C, Breitenbach U, Tuckermann JP, Dittrich BT, Richter KH, Angel P. Calgranulins S100A8 and S100A9 are negatively regulated by glucocorticoids in a c-Fos-dependent manner and overexpressed throughout skin carcinogenesis. Oncogene. 2002;21:4266–4276. doi: 10.1038/sj.onc.1205521. [DOI] [PubMed] [Google Scholar]
- 35.Zhang C, Liu Y, Gilthorpe J, van der Maarel JR. MRP14 (S100A9) protein interacts with Alzheimer beta-amyloid peptide and induces its fibrillization. PLoS One. 2012;7:e32953. doi: 10.1371/journal.pone.0032953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maarouf CL, Daugs ID, Kokjohn TA, Walker DG, Hunter JM, Kruchowsky JC, et al. Alzheimer’s disease and non-demented high pathology control nonagenarians: comparing and contrasting the biochemistry of cognitively successful aging. PLoS One. 2011;6:e27291. doi: 10.1371/journal.pone.0027291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Z, Tao T, Raftery MJ, Youssef P, Di Girolamo N, Geczy CL. Proinflammatory properties of the human S100 protein S100A12. J Leukoc Biol. 2001;69:986–994. [PubMed] [Google Scholar]
- 38.Miranda LP, Tao T, Jones A, Chernushevich I, Standing KG, Geczy CL, et al. Total chemical synthesis and chemotactic activity of human S100A12 (EN-RAGE) FEBS Lett. 2001;488:85–90. doi: 10.1016/s0014-5793(00)02392-9. [DOI] [PubMed] [Google Scholar]
- 39.Hofmann MA, Drury S, Fu C, Qu W, Taguchi A, Lu Y, et al. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97:889–901. doi: 10.1016/s0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- 40.Sasaki N, Toki S, Chowei H, Saito T, Nakano N, Hayashi Y, et al. Immunohistochemical distribution of the receptor for advanced glycation end products in neurons and astrocytes in Alzheimer’s disease. Brain Res. 2001;888:256–262. doi: 10.1016/s0006-8993(00)03075-4. [DOI] [PubMed] [Google Scholar]
- 41.Barnes DE, Blackwell T, Stone KL, Goldman SE, Hillier T, Yaffe K. Cognition in older women: the importance of daytime movement. J Am Geriatr Soc. 2008;56:1658–1664. doi: 10.1111/j.1532-5415.2008.01841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yaffe K, Barnes D, Nevitt M, Lui LY, Covinsky K. A prospective study of physical activity and cognitive decline in elderly women: women who walk. Arch Intern Med. 2001;161:1703–1708. doi: 10.1001/archinte.161.14.1703. [DOI] [PubMed] [Google Scholar]
- 43.Podewils LJ, Guallar E, Kuller LH, Fried LP, Lopez OL, Carlson M, et al. Physical activity, APOE genotype, and dementia risk: findings from the Cardiovascular Health Cognition Study. Am J Epidemiol. 2005;161:639–651. doi: 10.1093/aje/kwi092. [DOI] [PubMed] [Google Scholar]
- 44.Laurin D, Verreault R, Lindsay J, MacPherson K, Rockwood K. Physical activity and risk of cognitive impairment and dementia in elderly persons. Arch Neurol. 2001;58:498–504. doi: 10.1001/archneur.58.3.498. [DOI] [PubMed] [Google Scholar]
- 45.Middleton LE, Mitnitski A, Fallah N, Kirkland SA, Rockwood K. Changes in cognition and mortality in relation to exercise in late life: a population based study. PLoS One. 2008;3:e3124. doi: 10.1371/journal.pone.0003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.You T, Arsenis NC, Disanzo BL, Lamonte MJ. Effects of exercise training on chronic inflammation in obesity: current evidence and potential mechanisms. Sports Med. 2013;43:243–256. doi: 10.1007/s40279-013-0023-3. [DOI] [PubMed] [Google Scholar]
- 47.Nascimento CM, Pereira JR, de Andrade LP, Garuffi M, Talib LL, Forlenza OV, et al. Physical exercise in MCI elderly promotes reduction of pro-inflammatory cytokines and improvements on cognition and BDNF peripheral levels. Current Alzheimer research. 2014;11:799–805. doi: 10.2174/156720501108140910122849. [DOI] [PubMed] [Google Scholar]
- 48.Porto FH, Coutinho AM, Pinto AL, Gualano B, Duran FL, Prando S, et al. Effects of Aerobic Training on Cognition and Brain Glucose Metabolism in Subjects with Mild Cognitive Impairment. Journal of Alzheimer’s disease: JAD. 2015 doi: 10.3233/JAD-150033. [DOI] [PubMed] [Google Scholar]
- 49.Erickson KI, Voss MW, Prakash RS, Basak C, Szabo A, Chaddock L, et al. Exercise training increases size of hippocampus and improves memory. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3017–3022. doi: 10.1073/pnas.1015950108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abiko T, Sekino H. Synthesis of rat parathymosin alpha fragment 1-28 and examination of its inhibitory activity towards the restoring activity of thymosin alpha 1 on the impaired T-lymphocytes of uremic patients. Chem Pharm Bull (Tokyo) 1991;39:2647–2652. doi: 10.1248/cpb.39.2647. [DOI] [PubMed] [Google Scholar]
- 51.Baxevanis CN, Reclos GJ, Economou M, Arsenis P, Katsiyiannis A, Seferiades K, et al. Mechanism of action of prothymosin alpha in the human autologous mixed lymphocyte response. Immunopharmacol Immunotoxicol. 1988;10:443–461. doi: 10.3109/08923978809006448. [DOI] [PubMed] [Google Scholar]
- 52.Baxevanis CN, Frillingos S, Seferiadis K, Reclos GJ, Arsenis P, Katsiyiannis A, et al. Enhancement of human T lymphocyte function by prothymosin alpha: increased production of interleukin-2 and expression of interleukin-2 receptors in normal human peripheral blood T lymphocytes. Immunopharmacol Immunotoxicol. 1990;12:595–617. doi: 10.3109/08923979009019679. [DOI] [PubMed] [Google Scholar]
- 53.Liao W, Lin JX, Leonard WJ. IL-2 family cytokines: new insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr Opin Immunol. 2011;23:598–604. doi: 10.1016/j.coi.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker DG, Lue LF, Beach TG. Gene expression profiling of amyloid beta peptide-stimulated human post-mortem brain microglia. Neurobiol Aging. 2001;22:957–966. doi: 10.1016/s0197-4580(01)00306-2. [DOI] [PubMed] [Google Scholar]
- 55.Varki A, Gagneux P. Multifarious roles of sialic acids in immunity. Ann N Y Acad Sci. 2012;1253:16–36. doi: 10.1111/j.1749-6632.2012.06517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang B, Brand-Miller J. The role and potential of sialic acid in human nutrition. Eur J Clin Nutr. 2003;57:1351–1369. doi: 10.1038/sj.ejcn.1601704. [DOI] [PubMed] [Google Scholar]
- 57.Wang B. Molecular mechanism underlying sialic acid as an essential nutrient for brain development and cognition. Adv Nutr. 2012;3:465S–472S. doi: 10.3945/an.112.001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Linnartz B, Kopatz J, Tenner AJ, Neumann H. Sialic acid on the neuronal glycocalyx prevents complement C1 binding and complement receptor-3-mediated removal by microglia. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32:946–952. doi: 10.1523/JNEUROSCI.3830-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pages G, Lenormand P, L’Allemain G, Chambard JC, Meloche S, Pouyssegur J. Mitogen-activated protein kinases p42mapk and p44mapk are required for fibroblast proliferation. Proc Natl Acad Sci U S A. 1993;90:8319–8323. doi: 10.1073/pnas.90.18.8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roovers K, Assoian RK. Integrating the MAP kinase signal into the G1 phase cell cycle machinery. Bioessays. 2000;22:818–826. doi: 10.1002/1521-1878(200009)22:9<818::AID-BIES7>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 61.Thalhamer T, McGrath MA, Harnett MM. MAPKs and their relevance to arthritis and inflammation. Rheumatology (Oxford) 2008;47:409–414. doi: 10.1093/rheumatology/kem297. [DOI] [PubMed] [Google Scholar]
- 62.Yi AK, Yoon JG, Yeo SJ, Hong SC, English BK, Krieg AM. Role of mitogen-activated protein kinases in CpG DNA-mediated IL-10 and IL-12 production: central role of extracellular signal-regulated kinase in the negative feedback loop of the CpG DNA-mediated Th1 response. J Immunol. 2002;168:4711–4720. doi: 10.4049/jimmunol.168.9.4711. [DOI] [PubMed] [Google Scholar]
- 63.Spagnoli GC, Juretic A, Schultz-Thater E, Dellabona P, Filgueira L, Horig H, et al. On the relative roles of interleukin-2 and interleukin-10 in the generation of lymphokine-activated killer cell activity. Cell Immunol. 1993;146:391–405. doi: 10.1006/cimm.1993.1035. [DOI] [PubMed] [Google Scholar]
- 64.Konishi Y, Stegmuller J, Matsuda T, Bonni S, Bonni A. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science. 2004;303:1026–1030. doi: 10.1126/science.1093712. [DOI] [PubMed] [Google Scholar]
- 65.Lasorella A, Stegmuller J, Guardavaccaro D, Liu G, Carro MS, Rothschild G, et al. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature. 2006;442:471–474. doi: 10.1038/nature04895. [DOI] [PubMed] [Google Scholar]
- 66.Juo P, Kaplan JM. The anaphase-promoting complex regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Curr Biol. 2004;14:2057–2062. doi: 10.1016/j.cub.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 67.van Roessel P, Elliott DA, Robinson IM, Prokop A, Brand AH. Independent regulation of synaptic size and activity by the anaphase-promoting complex. Cell. 2004;119:707–718. doi: 10.1016/j.cell.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 68.Almeida A, Bolanos JP, Moreno S. Cdh1/Hct1-APC is essential for the survival of postmitotic neurons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2005;25:8115–8121. doi: 10.1523/JNEUROSCI.1143-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bossy-Wetzel E, Schwarzenbacher R, Lipton SA. Molecular pathways to neurodegeneration. Nat Med. 2004;10 (Suppl):S2–9. doi: 10.1038/nm1067. [DOI] [PubMed] [Google Scholar]
- 70.Maestre C, Delgado-Esteban M, Gomez-Sanchez JC, Bolanos JP, Almeida A. Cdk5 phosphorylates Cdh1 and modulates cyclin B1 stability in excitotoxicity. EMBO J. 2008;27:2736–2745. doi: 10.1038/emboj.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yart A, Mayeux P, Raynal P. Gab1, SHP-2 and other novel regulators of Ras: targets for anticancer drug discovery? Curr Cancer Drug Targets. 2003;3:177–192. doi: 10.2174/1568009033481976. [DOI] [PubMed] [Google Scholar]
- 72.Hoeben A, Martin D, Clement PM, Cools J, Gutkind JS. Role of GRB2-associated binder 1 in epidermal growth factor receptor-induced signaling in head and neck squamous cell carcinoma. Int J Cancer. 2013;132:1042–1050. doi: 10.1002/ijc.27763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oyagi A, Moriguchi S, Nitta A, Murata K, Oida Y, Tsuruma K, et al. Heparin-binding EGF-like growth factor is required for synaptic plasticity and memory formation. Brain Res. 2011;1419:97–104. doi: 10.1016/j.brainres.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 74.Anchan RM, Reh TA, Angello J, Balliet A, Walker M. EGF and TGF-alpha stimulate retinal neuroepithelial cell proliferation in vitro. Neuron. 1991;6:923–936. doi: 10.1016/0896-6273(91)90233-p. [DOI] [PubMed] [Google Scholar]
- 75.Morrison RS, Kornblum HI, Leslie FM, Bradshaw RA. Trophic stimulation of cultured neurons from neonatal rat brain by epidermal growth factor. Science. 1987;238:72–75. doi: 10.1126/science.3498986. [DOI] [PubMed] [Google Scholar]
- 76.Morrison RS, Keating RF, Moskal JR. Basic fibroblast growth factor and epidermal growth factor exert differential trophic effects on CNS neurons. J Neurosci Res. 1988;21:71–79. doi: 10.1002/jnr.490210111. [DOI] [PubMed] [Google Scholar]
- 77.Alexi T, Hefti F. Trophic actions of transforming growth factor alpha on mesencephalic dopaminergic neurons developing in culture. Neuroscience. 1993;55:903–918. doi: 10.1016/0306-4522(93)90307-2. [DOI] [PubMed] [Google Scholar]
- 78.Leutz A, Schachner M. Epidermal growth factor stimulates DNA-synthesis of astrocytes in primary cerebellar cultures. Cell Tissue Res. 1981;220:393–404. doi: 10.1007/BF00210517. [DOI] [PubMed] [Google Scholar]
- 79.Simpson DL, Morrison R, de Vellis J, Herschman HR. Epidermal growth factor binding and mitogenic activity on purified populations of cells from the central nervous system. J Neurosci Res. 1982;8:453–462. doi: 10.1002/jnr.490080233. [DOI] [PubMed] [Google Scholar]
- 80.Cheng HC, Kim SR, Oo TF, Kareva T, Yarygina O, Rzhetskaya M, et al. Akt suppresses retrograde degeneration of dopaminergic axons by inhibition of macroautophagy. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2011;31:2125–2135. doi: 10.1523/JNEUROSCI.5519-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang Q, Inoki K, Kim E, Guan KL. TSC1/TSC2 and Rheb have different effects on TORC1 and TORC2 activity. Proc Natl Acad Sci U S A. 2006;103:6811–6816. doi: 10.1073/pnas.0602282103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sato T, Nakashima A, Guo L, Tamanoi F. Specific activation of mTORC1 by Rheb G-protein in vitro involves enhanced recruitment of its substrate protein. J Biol Chem. 2009;284:12783–12791. doi: 10.1074/jbc.M809207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 84.Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cheng HC, Ulane CM, Burke RE. Clinical progression in Parkinson disease and the neurobiology of axons. Ann Neurol. 2010;67:715–725. doi: 10.1002/ana.21995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Selkoe DJ. Alzheimer’s disease is a synaptic failure. Science. 2002;298:789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- 87.Pereira AC, Huddleston DE, Brickman AM, Sosunov AA, Hen R, McKhann GM, et al. An in vivo correlate of exercise-induced neurogenesis in the adult dentate gyrus. Proc Natl Acad Sci U S A. 2007;104:5638–5643. doi: 10.1073/pnas.0611721104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stretch C, Khan S, Asgarian N, Eisner R, Vaisipour S, Damaraju S, et al. Effects of sample size on differential gene expression, rank order and prediction accuracy of a gene signature. PLoS One. 2013;8:e65380. doi: 10.1371/journal.pone.0065380. [DOI] [PMC free article] [PubMed] [Google Scholar]