Significance
This work shows that active DNA demethylation governs ripening, an important plant developmental process. Our work defines a molecular mechanism, which has until now been missing, to explain the correlation between genomic DNA demethylation and fruit ripening. It demonstrates a direct cause-and-effect relationship between active DNA demethylation and induction of gene expression in fruits. The importance of these findings goes far beyond understanding the developmental biology of ripening and provides an innovative strategy for its fine control through fine modulation of epimarks in the promoters of ripening related genes. Our results have significant application for plant breeding especially in species with limited available genetic variation.
Keywords: active DNA demethylation, DNA glycosylase lyase, epigenetic, tomato, fruit ripening
Abstract
In plants, genomic DNA methylation which contributes to development and stress responses can be actively removed by DEMETER-like DNA demethylases (DMLs). Indeed, in Arabidopsis DMLs are important for maternal imprinting and endosperm demethylation, but only a few studies demonstrate the developmental roles of active DNA demethylation conclusively in this plant. Here, we show a direct cause and effect relationship between active DNA demethylation mainly mediated by the tomato DML, SlDML2, and fruit ripening— an important developmental process unique to plants. RNAi SlDML2 knockdown results in ripening inhibition via hypermethylation and repression of the expression of genes encoding ripening transcription factors and rate-limiting enzymes of key biochemical processes such as carotenoid synthesis. Our data demonstrate that active DNA demethylation is central to the control of ripening in tomato.
Genomic DNA methylation is a major epigenetic mark that is instrumental to many aspects of chromatin function, including gene expression, transposon silencing, or DNA recombination (1–4). In plants, DNA methylation can occur at cytosine both in symmetrical (CG or CHG) and nonsymmetrical (CHH) contexts and is controlled by three classes of DNA methyltransferases, namely, the DNA Methyltransferase 1, Chromomethylases, and the Domain Rearranged Methyltransferases (5–7). Indeed, in all organisms, cytosine methylation can be passively lost after DNA replication in the absence of methyltransferase activity (1). However, plants can also actively demethylate DNA via the action of DNA Glycosylase-Lyases, the so-called DEMETER-Like DNA demethylases (DMLs), that remove methylated cytosine, which is then replaced by a nonmethylated cytosine (8–11). Initially identified as enzymes necessary for maternal imprinting in Arabidopsis thaliana (12), the role of DMLs has since been established in various processes such as limiting extensive DNA methylation at gene promoters (13), determining the global demethylation of seed endosperm (8, 14) and promoting plant responses to pathogens (15). Of note, Arabidopsis ros1, dml2, and dml3 single, double, or triple mutants showed little or no developmental alterations (9, 16, 17), suggesting that active DNA demethylation is not critical for development in this species. However, as mentioned above, genomic DNA methylation is an important mechanism that influences gene expression, and methylation at promoters is known to inhibit gene transcription (5, 18). Hence, it is likely that the active removal of methylation marks is an important mechanism during plant development and plant cell fate reprogramming, leading to the hypomethylation of sites important for DNA–protein interaction and gene expression, as already observed in human cells (19).
Indeed, accumulating evidence suggests that active DNA demethylation might play a greater role in controlling gene expression in tomato. In support of this idea, recent work describing the methylome dynamics in tomato fruit pericarp revealed substantial changes in the distribution of DNA methylation over the tomato genome during fruit development, and demethylation during ripening at specific promoters such as the NON RIPENING (NOR) and COLORLESS NON RIPENING (CNR) promoters (20, 21). This observation is consistent with previous studies indicating that genome cytosine methylation levels decrease by 30% in pericarp of fruits during ripening, although DNA replication is very limited at this stage (22).
Here, we investigated active DNA demethylation as a possible mechanism governing the reprogramming of gene expression in fruit pericarp cells at the onset of fruit ripening.
Results
The Tomato Genome Contains Four DNA Glycosylase Genes with Specific Expression Patterns.
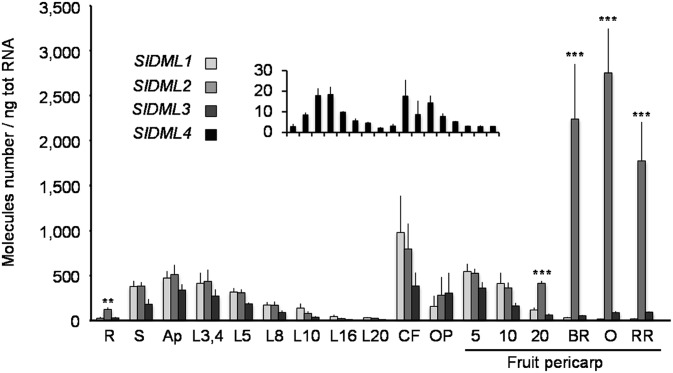
The tomato genome contains four putative DML genes encoding proteins with characteristic domains of functional DNA glycosylase-lyases (23) (SI Appendix, Fig. S1 A and C and Table S1). SlDML1 and -2 are orthologous to the Arabidopsis AtROS1 (Repressor of Silencing 1) gene and SlDML3 to AtDME (DEMETER), whereas SlDML4 has no closely related Arabidopsis ortholog (SI Appendix, Fig. S1B). All four SlDML genes are ubiquitously expressed in tomato plants, although SlDML4 is expressed at a very low level in all organs analyzed. In leaves, flowers, and young developing fruits, the four genes present coordinated expression patterns characterized by high expression levels in young organs that decrease when organs develop. However, unlike SlDML1, SlDML3, and SlDML4, which are barely expressed during fruit ripening, SlDML2 mRNA abundance increases dramatically in ripening fruits, suggesting an important function at this developmental phase (Fig. 1).
Fig. 1.
Differential expression of SlDML genes in tomato organs. Absolute quantification of SlDML1, SlDML2, SlDML3, and SlDML4 mRNA; SlDML4 gene expression is presented in a separate diagram because of its very low expression level. Fruit pericarp is at 5, 10, 20 dpa and at Breaker (BR, 39 dpa), orange (O), and red ripe (RR). Asterisks indicate significant difference [Student’s t test (n = 3)] between SlDML2 and all other SlDML genes: *P < 0.05; **P < 0.01; ***P < 0.001. Error bars indicate means ± SD. Ap, stem apex; CF, closed flowers; L, leaves at positions 3, 4, 5, 8, 10, 16, and 20 from apex; OP, open flowers 5, 10, and 20; R, roots; S, stem from whole seedlings.
Transgenic Plants with Reduced DML Gene Expression Present Various Fruit and Plant Phenotypes.
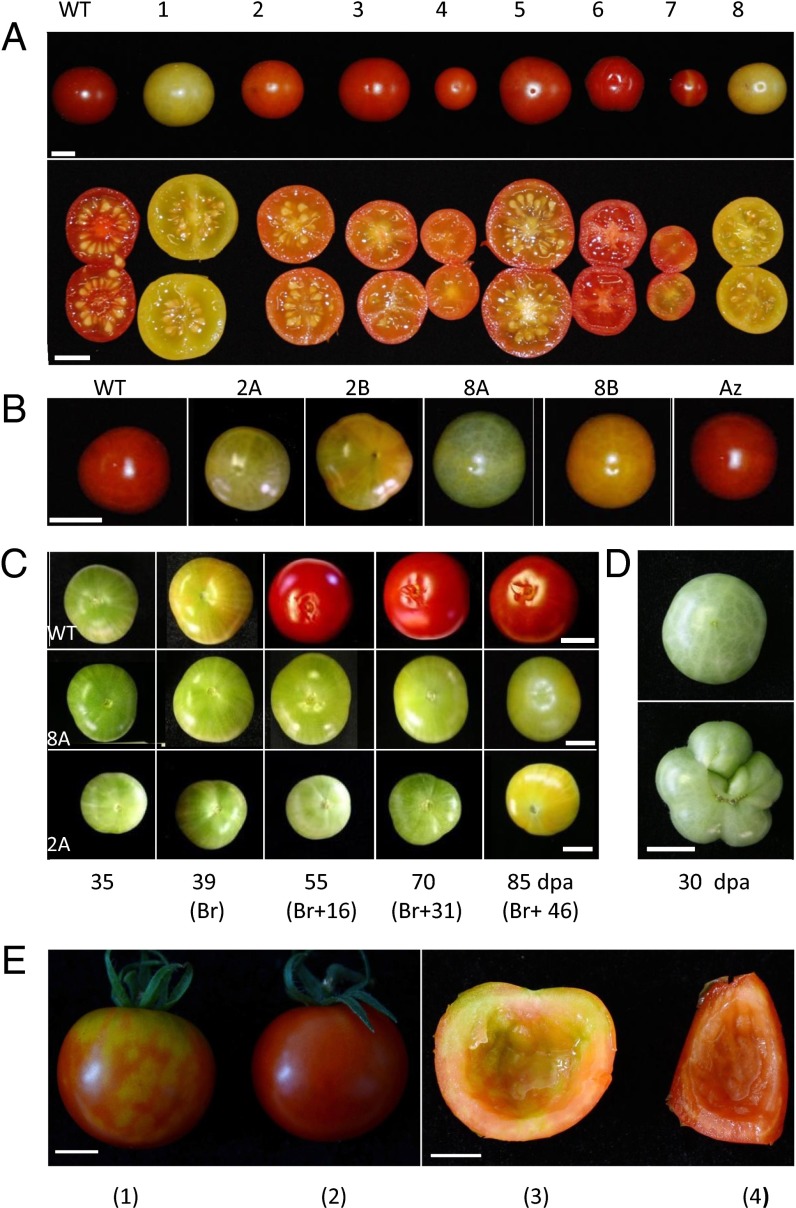
The physiological significance of tomato DMLs was addressed through RNAi-mediated gene repression using the highly conserved Helix–hairpin–Helix-Gly/Pro rich domain (HhH-GPD) specific to DML proteins as a target sequence (SI Appendix, Fig. S2A). Our goal was to repress simultaneously all tomato SlDML genes, anticipating potential functional redundancy among these four genes; 23 independent T0 transgenic lines were generated and 22 showed alterations of fruit development, including delayed ripening, modified fruit shape, altered color, shiny appearance, parthenocarpy, or combinations of these phenotypes (Fig. 2A).
Fig. 2.
Phenotypes of tomato DML RNAi fruits. (A) Fruits (70 dpa) (upper lane) or fruit sections (lower lane) from eight independent representative T0 RNAi plants. (B) Fruits (85 dpa) from T2 plants (left to right); WT plants, line 2 plants (DML2A and DML2B), line 8 plants (DML8A and DML8B), and an azygous plant (AZ). (C) Ripening kinetics of WT (Top), DML8A (Middle), and DML2A (Bottom). (D) WT bicarpel (Upper) DML2B multicarpel fruits (Lower). (E) VIGS experiment on 47-dpa (Br + 5) fruits injected with PVX/SlDML2 [fruits (1) and (3)] or PVX [fruits (2) and (4)] at 12 dpa [fruits (3) and (4)] inside of fruits (1) and (2), respectively. (Scale bars: 1 cm.)
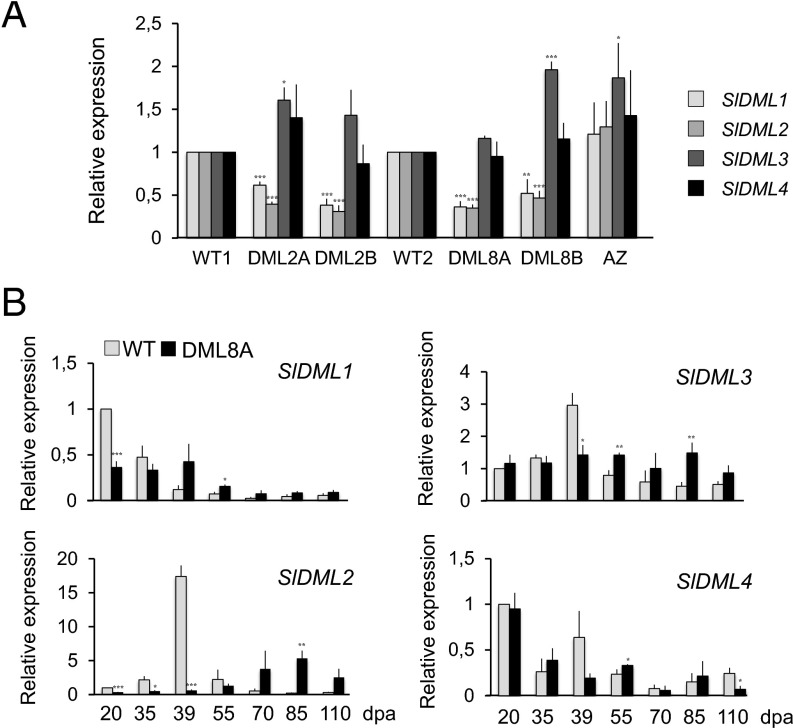
Lines 2 and 8, which showed delayed and inhibited ripening phenotypes, were chosen to investigate the possible link between ripening and DNA demethylation. In both cases, 10–25 T1 and T2 plants were grown that showed maintenance and strengthening of the nonripening phenotypes in subsequent generations coincident with the presence of the transgene. The loss of the RNAi transgene in segregating lines led to reversion to a wild-type (WT) phenotype, indicating a lack of memory effect across generations when fruit ripening is considered (Fig. 2 A and B and SI Appendix, Fig. S3A). In plants of both RNAi lines, analysis of SlDML gene residual expression in 20 days postanthesis (dpa) fruits indicates that only SlDML1 and SlDML2 are repressed to 40–60% of the WT level, whereas SlDML3 and SlDML4 are either unaffected or induced compared with WT (Fig. 3A). This is most likely attributable to the lower homology level of these two genes, with SlDML1 in the part of the gene used for the RNAi construct (SI Appendix, Fig. S2A). During ripening, SlDML2 expression is reduced to 10% of WT at the Breaker (Br) stage and remains low at 55 dpa (Br + 16) but increases slightly at 70 dpa (Br + 31) (Fig. 3B and SI Appendix, Fig. S2B), coincident with the partial ripening observed in transgenic RNAi fruits (Fig. 2C and SI Appendix, Fig. S3B). Whether the increase in SlDML2 expression at late ripening stages is attributable to a weaker effect of the RNAi remains unclear. None of the three remaining genes, SlDML1, SlDML3, and SlDML4, which are weakly expressed during ripening, displayed significantly reduced expression compared with WT fruits of the same age, indicating that observed ripening phenotypes are likely attributable to SlDML2 gene repression. This hypothesis was further confirmed using virus induced gene silencing (VIGS) to specifically repress the SlDML2 gene; 17.5% of the fruits injected with a PVX/SlDML2 vector presented non ripening sectors contrary to those injected with a control PVX virus that all ripened normally (Fig. 2E and SI Appendix, Fig. S4B). Indeed, SlDML2 was down-regulated in nonripening sectors of fruits injected with the PVX/SlDML2 vector, whereas none of the three other SlDML genes was repressed (SI Appendix, Fig. S4C), demonstrating that the specific knock down of SlDML2 is sufficient to inhibit ripening.
Fig. 3.
Residual expression of SlDML genes in fruits of transgenic DML RNAi plants. Normalized expression of the SlDML genes (A) in 20-dpa transgenic fruits of plants from line 2 (DML2A and -2B), line 8 (DML8A and -8B), an azygous plant (AZ), and the respective WT1 and WT2 controls (B) in WT2 and DML8A fruits at seven developmental stages. Expression of the SlDML genes was normalized to EF1α and to the corresponding WT fruits at 20 dpa. For each SlDML gene, asterisks indicate significant difference [Student’s t test (n = 3)] between transgenic plants and WT controls, respectively, at 20 dpa (A) or at the same age during fruit development (B). *P < 0.05; **P < 0.01; ***P < 0.001). Error bars indicate mean ± SD.
It was noteworthy that some plants from line 2 developed additional phenotypes affecting plant growth, leaf shape, flower development, and fruit carpel number that were not observed in T0 and T1 generations (Fig. 2D and SI Appendix, Fig. S3 B and C). The screening of additional lines revealed other independent transgenic lines that presented flower, fruit, and plant phenotypes similar to line 2 (SI Appendix, Fig. S3D). These observations indicate that the severity of the phenotypes increases over generations and suggest that DMLs may also be involved in other aspects of tomato plant development beyond fruit ripening.
All Aspects of Fruit Ripening Are Delayed and Limited in RNAi Transgenic Lines.
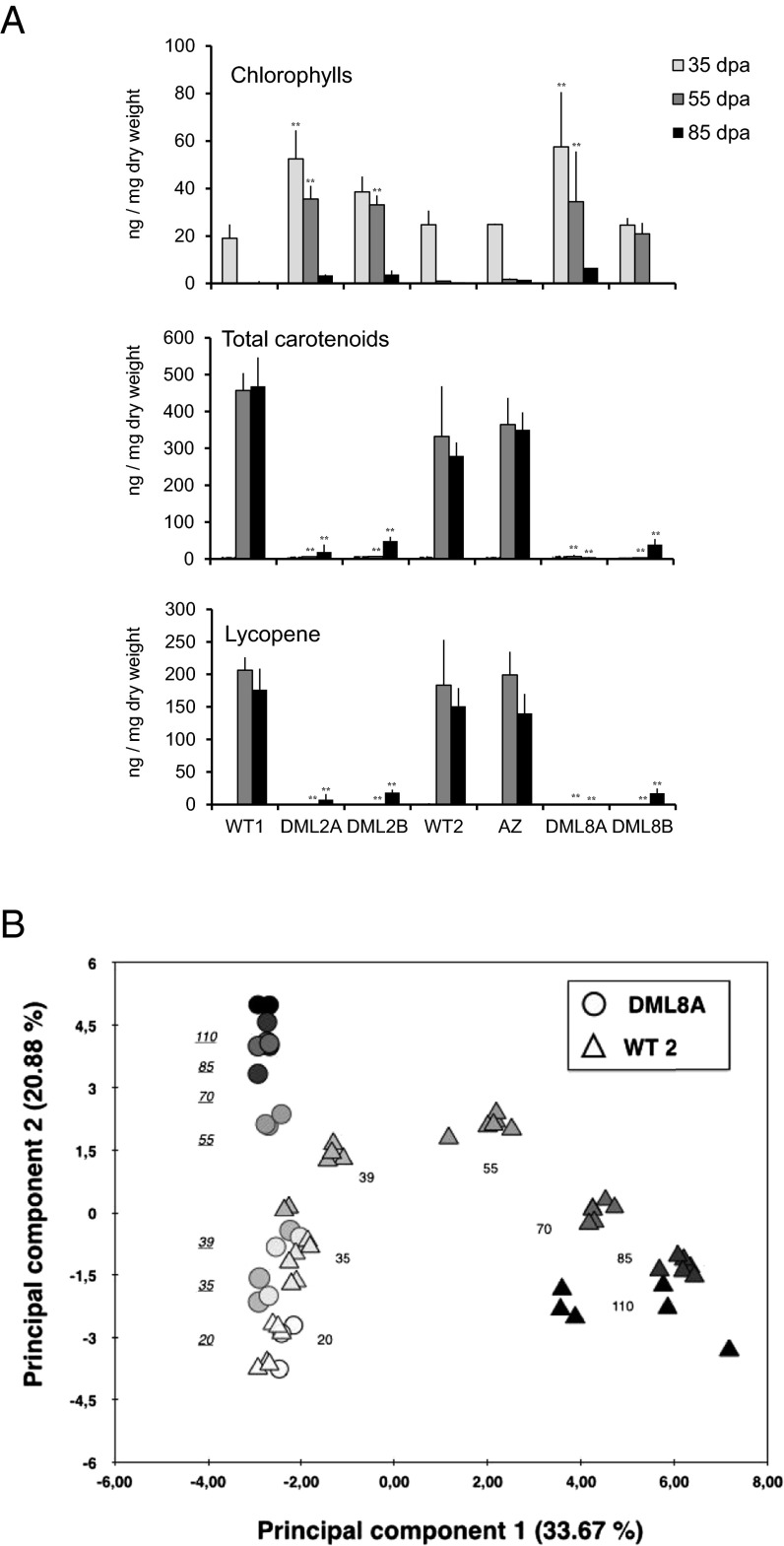
Fruits of transgenic lines 2 and 8 were further analyzed to investigate the consequences of DNA demethylation on the ripening process. Indeed, in fruits of both transgenic lines, the onset of fruit ripening was delayed from 10 to 20 d compared with WT or Azygous revertant fruits, and ripening of transgenic fruits was never completed even after 45 d or longer maturation times (Fig. 2 B and C and SI Appendix, Fig. S3B). The ripening defect is further demonstrated by the late and extremely reduced total carotenoids and lycopene accumulation and the delayed chlorophyll degradation (Fig. 4A). Primary metabolite composition was also modified, as visualized by principal component analysis (PCA) using the absolute concentration of 31 primary metabolites issued from 1H-NMR analysis (Fig. 4B and SI Appendix, Fig. S5A). The first two principal components (PCs), explain more than 54% of total variability. During early development (20, 35, and 39 dpa), WT and transgenic samples follow parallel trajectories as highlighted by the PCA in which the second PC (PC2) explains 21% of the total variability. However, at 55-dpa and later ripening stages, PC1, which accounts for 33.67% of the global variability, separates WT fruits from all other samples. Hence, WT fruit samples harvested at 55-dpa and older stages are clearly distinct from transgenic fruit samples of the same age. Metabolic differences between ripening WT and transgenic fruits are mainly attributable to overaccumulation of malate and reduction or delayed accumulation of compounds typical of ripening fruits, including glucose, fructose, glutamate, rhamnose, and galactose (SI Appendix, Fig. S5 B–D). Climacteric rise of ethylene production was also dramatically reduced in fruits of both DML RNAi lines, although low ethylene accumulation occurred to a degree and timing consistent with the late and limited ripening process of RNAi fruits (SI Appendix, Fig. S6).
Fig. 4.
Metabolic profiling of carotenoids and primary metabolites in transgenic DML RNAi fruits. (A) Chlorophylls (Top), total carotenoids (Middle), and lycopene (Bottom) content. Asterisks indicate significant difference [Student’s t test (n = 3)] between DML2A and -2B, DML8A and -8B, and WT1 and WT2, respectively, at the same age: *P < 0.05; **P < 0.01; ***P < 0.001. Error bars indicate means ± SD. (B) PCA using primary metabolites in WT2 (△) and DML8A (○) fruits at seven developmental stages. Color indicates the fruit developmental stages: white is 20 dpa and from light gray to black are 35, 39 (Br), 55, 70, 85, and 110 dpa.
Fruit-Ripening Defects Are Correlated with the Repression and Hypermethylation of Genes Necessary for This Developmental Process.
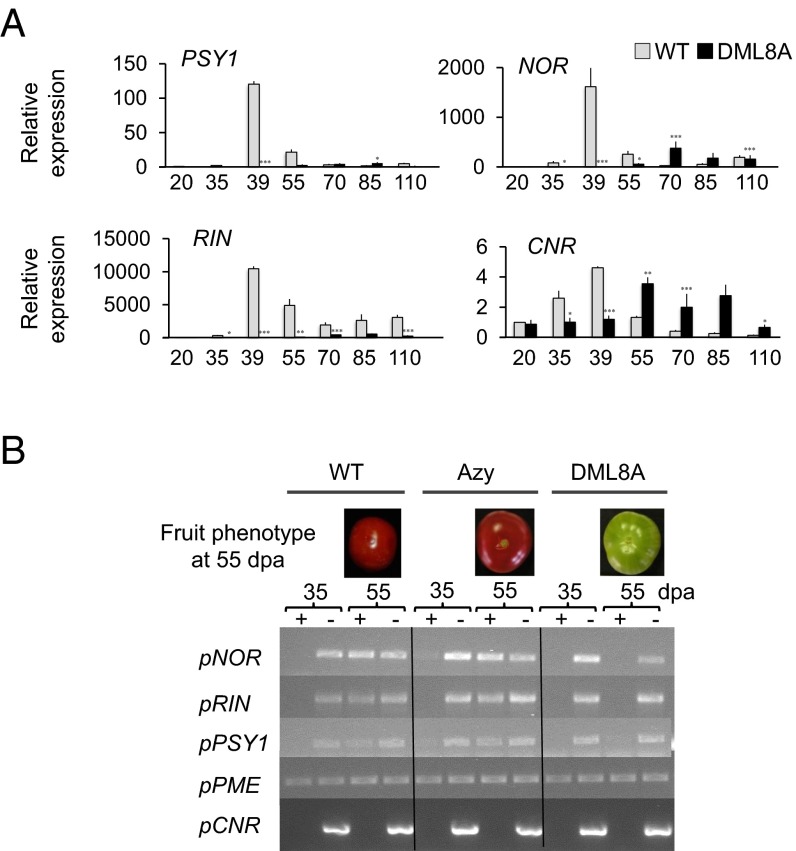
To demonstrate a causal relationship between fruit ripening defects of transgenic lines and the impairment of active DNA demethylation, the expression of CNR (21), RIPENING INHIBITOR (RIN) (24), NOR (25), and PHYTOENE SYNTHASE 1 (PSY1) (26, 27) genes was assessed in RNAi transgenic plants. These genes were selected among others because they are necessary for the overall ripening process (CNR, RIN, NOR), or specifically govern carotenoid accumulation (PSY1), an important quality trait of mature tomato fruit. Moreover, the promoter regions of these genes showed reduced methylation levels during fruit ripening in WT tomato (20, 21). It is noteworthy that CNR gene induction was delayed 15 d in transgenic fruits, and all three other genes showed a dramatic reduction in expression level consistent with the ripening defect of the transgenic lines (Fig. 5A and SI Appendix, Fig. S7). To assess whether repression of CNR, RIN, NOR, and PSY1 gene expression in ripening fruits results from the maintenance of a high cytosine methylation status of their promoter upon down-regulation of SlDML2, methylsensitive-PCR (McrBC-PCR) analysis of the corresponding promoters was performed. This approach revealed a ripening-associated demethylation of the RIN, NOR, and PSY1 promoters in WT and Azygous revertant fruits but not in SlDML RNAi fruits (Fig. 5B). No detectable variations of methylation in the CNR promoter during ripening of WT fruits were revealed with this method. The putative differentially methylated regions (DMRs) in the NOR and PSY1 promoter regions were subsequently analyzed by gene specific bisulfite pyrosequencing (28). Methylation analysis of the CNR promoter was targeted to a region known to be methylated at all stages (CNR1) (SI Appendix, Fig. S9C), used here as a control for methylation and to a previously identified DMR (CNR2) (SI Appendix, Fig. S9C) (20, 21). For all three promoters, cytosines that became demethylated in ripening WT fruits but not in transgenic fruits of the same age were identified (Fig. 6A and SI Appendix, Fig. S9). Two distinct situations were observed: (i) sequences corresponding to putative RIN binding sites (RIN BS) in the CNR and NOR promoters (20), where methylation is high at 20 and 35 dpa in all plants analyzed and drops to very low levels during ripening of WT fruits but is maintained to high levels in RNAi fruits of the same age; and (ii) sequences that are hypermethylated in transgenic fruits at all stages analyzed compared with WT fruits. These latter sequences include a newly identified DMR in the PSY1 promoter and cytosines upstream and downstream to the RIN BS in the NOR and CNR promoters. These data demonstrate the absolute requirement of promoter demethylation in critical genes for ripening to occur. The data also suggest multiple patterns of cytosine demethylation occurring either specifically during ripening or at earlier stages.
Fig. 5.
Expression and demethylation at key genes controlling ripening are inhibited in DML RNAi plants. (A) Expression of the RIN, NOR, CNR, and PSY1 genes in transgenic DML8A and WT fruits normalized to EF1α and to WT fruits at 20 dpa. Asterisks indicate significant difference [Student’s t test (n = 3)] between WT and DML8A samples at a given stage: *P < 0.05; **P < 0.01; ***P < 0.001. Error bars indicate means ± SD. (B) McrBC-PCR analysis of selected promoter fragments in fruits of WT, azygous (Azy), and DML8A plants; 1 µg of genomic DNA was digested with McrBC (NEB) during 5h (+); (–) indicates negative control for the digestion reaction that was performed without GTP. In the WT and azygous plants, the part of NOR, RIN, and PSY1 promoter regions analyzed are methylated at 35 dpa (no amplification) but are demethylated at 55 dpa (amplification). In DML8A plants, the three promoter regions behave similarly to WT at 35 dpa but remained methylated at 55 dpa (no amplification in both cases). The pectin-methyl esterase (PME) promoter is used as an unmethylated control, and the CNR promoter fragment used here was found to be sufficiently methylated at all stages for complete digestion by McrBC.
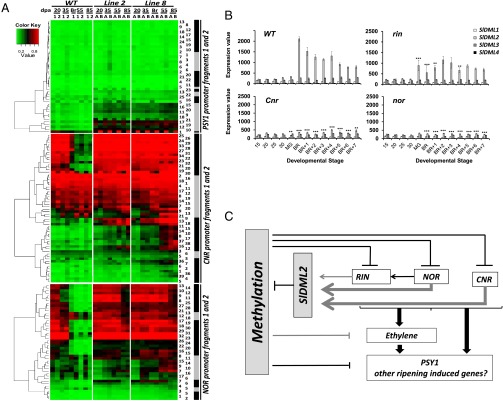
Fig. 6.
Bisulfite-sequencing analysis at the NOR, CNR, and PSY1 promoter fragments in WT and transgenic DML RNAi plants. (A) Heat-map representation of DNA methylation at selected NOR, CNR, and PSY1 promoter regions (SI Appendix, Fig. S8) in fruits of control (WT1 and WT2) and transgenic (DML2A, -2B, -8A, and -8B) plants at five (WT and line 8) or four (line 2) developmental stages. For each promoter, two fragments have been analyzed (fragment 1, gray box; fragment 2, black box), the positions of which are shown in SI Appendix, Fig. S8 and Fig. S9. The position of the Cs within each promoter fragment is also shown (number in the columns on the right side), as defined in SI Appendix, Fig. S8. For each promoter, Cs have been clustered considering the two PCR fragments analyzed together. (B) Changes in expression of SlDML genes in fruits of Ailsa Craig (WT) and near-isogenic mutant lines rin, Cnr, and nor, as determined by microarrays analysis. For fruit development, days postanthesis are shown. Mature green is 40 dpa in Ailsa Craig and then Br is 49 dpa. For nonripening mutants, Br onward are 49 dpa + 1–7 d. Asterisks indicate significant difference (variance ratio, F tests) between WT and mutant lines for the SlDML2 gene only to avoid overloading the figure: *P < 0.05; **P < 0.01; ***P < 0.001). Details of expression results and statistical analyses for all four genes are provided in Dataset S1. Error bars indicate means ± SD. (C) Proposed function of DNA demethylation in the control of fruit ripening; SlDML2 is necessary for the active demethylation of the NOR, CNR, RIN, and PSY1 promoter region, thereby allowing these gene expressions. SlDML2 gene expression is reduced in the rin, nor, and Cnr background, suggesting a regulatory loop. There is at this time no evidence of direct regulation of the SlDML2 gene by the RIN, NOR, or CNR protein. SlDML2 may control the expression of additional ripening induced gene, as shown in this study for the PSY1 gene and suggested by the demethylation of several promoters during fruit ripening (20). Arrows indicate activation. Lines indicate repression: black, direct effects; gray, direct or indirect effects.
Discussion
Previously reported analysis of DNA cytosine methylation and RIN binding during fruit development in WT and in the rin and Cnr tomato-ripening mutants suggested a significant role for DNA methylation during ripening and a feedback loop between methylation and ripening transcription factors (20, 21, 29). Here, we demonstrate for the first time to our knowledge that active DNA demethylation is an absolute requirement for fruit ripening to occur and show a direct cause and effect relationship between hypermethylation at specific promoters and repression of gene expression. In this context, SlDML2 appears to be the main regulator of the ripening associated DNA demethylation process. (i) SlDML2 is the only SlDML gene induced concomitantly to the demethylation and induction of genes that control fruit ripening; (ii) the specific knockdown of SlDML2 in VIGS-treated fruits leads to inhibition of fruit ripening similar to DML-RNAi fruits; and (iii) the hypermethylated phenotype described in the Cnr and rin mutants (20) is associated with the specific repression of SlDML2, with none of the other SlDML genes being down-regulated (Fig. 6B and Dataset S1).
Indeed, we cannot formally rule out that SlDML1, which is repressed in the transgenic RNAi lines, also participates in the genomic DNA demethylation in fruits. However, SlDML1 is mainly expressed at early stages of fruit development and only at very low levels during fruit ripening. Hence, this protein may also be involved in demethylation events but mainly those occurring at the early stages of fruit development.
In addition to genes encoding major fruit ripening regulators, those encoding enzymes involved in various aspects of fruit ripening are also likely to be demethylated, as suggested by the observation that PSY1 gene expression also requires demethylation. Combined transcriptomic, methylomic, and metabolomic analysis of the transgenic lines described here will now be required to determine the network of genes and metabolic processes primarily targeted by demethylation in tomato fruit.
SlDML2 is the likely focal point of a feedback regulation on ripening-associated DNA demethylation, because this gene is clearly down-regulated in fruits of the rin, nor, and Cnr mutants, contrary to the other SlDML genes that are normally expressed (Fig. 6 B and C and Dataset S1). It is plausible that timing and extent of demethylation may represent an important source of variation in the diversity of kinetics and intensity of ripening found among tomato varieties, thus presenting a frontier for further investigation. Controlling the timing and kinetics of active DNA demethylation in fruits may therefore provide new strategies to enhance fruit shelf life. In addition, engineering DNA demethylation in tomato fruits would be an innovative and novel strategy for the improvement of traits of agronomical relevance in a species with little genetic diversity (30). Finally, the recent demonstration that hypermethylation of a Myb promoter blocks anthocyanin accumulation during pear and apple ripening (31, 32) supports the notion of a more general role for demethylation in fruits. However, whether this mechanism occurs similarly during the ripening of all fleshy fruit species now requires further investigation.
Materials Methods
Plant Material and Experimental Plan.
All experiments were performed using a cherry tomato variety (Solanum lycopsersicum, cv WVA106) that was grown in greenhouse conditions, except for VIGS experiments, which were performed on Solanum lycopsersicum, cv Ailsa Craig grown in growth chambers as described (21). For the array experiments, fruit pericarp of Ailsa Craig and near-isogenic mutants rin, nor, and Cnr were collected at 13 stages of fruit development and ripening with three independent biological replicates per line and immediately frozen in liquid nitrogen for RNA extraction and array analysis. Details of tomato transformation, selection of line 2 and 8 used in this study, and of VIGS experiments are provided in SI Appendix, Materials and Methods.
For all analysis, two independent transgenic T2 plants (DML2A and -B and DML8A and -B for lines 2 and 8, respectively) and an azygous plant obtained from line 8 were used. Additional T2 plants were eventually used as controls for the phenotypes of these four plants. T2 plants from line 2 presented dramatic alterations of flower development, not visible in previous generations, and were backcrossed to allow fruit development. This resulted in a limited number of fruits (see below). For this reason, not all developmental stages could be analyzed for this line.
The experimental plan was designed to span tomato fruit development and ripening in cv West Virginia 106 (WVA106) and transgenic DML RNAi plants over a period of 85 d from fruit set to account for the strongly delayed ripening phenotype of the transgenic fruits. At stages following mature green, the DML RNAi fruits diverge from the WT, because they are significantly delayed in ripening induction and almost completely ripening inhibited. Because it was not possible to select stages equivalent to the Br (39 dpa) or red ripe stages in the transgenic lines, we have chosen to analyze fruits identically staged, which allows comparing changes in the context of a developmental parameter (days postanthesis) that can be precisely measured. Two independent cultures were performed. (i) Plants from line 2 and the relevant WT control (WT1), fruits were harvested at 20, 35, 55 (Br + 16), 70 (Br + 31), and 85 (Br + 46) dpa. Because the fruit yield was reduced in line 2, a sufficient number of fruits at the Br stage could not be harvested and older fruits were preferentially selected to allow the analysis of late effects of demethylation inhibition. (ii) Line 8 was grown together with its own WT control (WT2) and an azygous plant. Because there were more fruits available for this line, the Br stage (39 dpa) was harvested in addition of the stages used for line 2.
For all fruit samples, two individual T2 plants were used, and for each sample, a minimum of six fruits separated in three biological replicates were processed and stored at −80 °C until used.
Molecular and Metabolite Analysis.
Details of molecular (gene expression, microarrays, McrBC-PCR analysis of gene DNA methylation, and gene-targeted bisulfite sequencing) and metabolite (Carotenoid, ethylene, and 1H-NMR) analysis are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Marie Mirouze for critical reading of the manuscript and Antoine Daunay and Nicolas Mazaleyrat for technical support in bisulfite pyrosequencing analysis. We acknowledge Syngenta and specifically Dr. Charles Baxter for help with the tomato GeneChip studies and Alex Marshall for help with the array analysis, as well as Cécile Cabasson and Jim Craigon for help in statistical analysis. Metabolomic profiling was performed on the Metabolome Facility of Bordeaux Functional Genomics Center and supported by the French National Infrastructure for Metabolomics and Fluxomics (MetaboHUB) funded by the Agence Nationale de Recherche (ANR, Project ANR-11-INBS-0010). R.L. was the recipient of a grant from the Chinese Scholarship Council and P.G. of a Fulbright grant. G.B.S., T.C.H., and N.H.C. acknowledge financial support from the Biotechnology and Biological Sciences Research Council, UK (Grants BB/F005458/1 and BB/J015598/1), and G.B.S. acknowledges support from the European Cooperation in Science and Technology (COST) Action FA1106. Y.H., J.K., and C.W. were supported by Hangzhou Normal University and the National Natural Science Foundation of China (NFSC, Grant 31370180). M.B. and M.L. received support from the Laboratoire d’Excellence (LABEX) entitled “Towards a Unified theory of biotic Interactions; the roLe of environmental Perturbations” (TULIP, ANR-10-LABX-41) and from the networking activities within the European COST Action FA1106. J.J.G. was supported by National Science Foundation Grant IOS-1322714.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Full array datasets of WT (Ailsa craig) and of the rin, nor, and Cnr mutants have been deposited with the Sol Genomics Network and are accessible under the following link: ftp://ftp.solgenomics.net/microarray/, file name rinnorcnr microarrays Liuetal PNAS2015. These array datasets have been used to analyze the expression of the four DML genes in the tomato rin, nor, and Cnr mutants and in WT fruits. The results are shown in Fig. 6B and Dataset S1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1503362112/-/DCSupplemental.
References
- 1.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saze H, Tsugane K, Kanno T, Nishimura T. DNA methylation in plants: Relationship to small RNAs and histone modifications, and functions in transposon inactivation. Plant Cell Physiol. 2012;53(5):766–784. doi: 10.1093/pcp/pcs008. [DOI] [PubMed] [Google Scholar]
- 3.Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6(5):351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 4.Mirouze M, et al. Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc Natl Acad Sci USA. 2012;109(15):5880–5885. doi: 10.1073/pnas.1120841109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang M, Kimatu JN, Xu K, Liu B. DNA cytosine methylation in plant development. J Genet Genomics. 2010;37(1):1–12. doi: 10.1016/S1673-8527(09)60020-5. [DOI] [PubMed] [Google Scholar]
- 6.Bender J. DNA methylation and epigenetics. Annu Rev Plant Biol. 2004;55(1):41–68. doi: 10.1146/annurev.arplant.55.031903.141641. [DOI] [PubMed] [Google Scholar]
- 7.Finnegan EJ, Kovac KA. Plant DNA methyltransferases. Plant Mol Biol. 2000;43(2-3):189–201. doi: 10.1023/a:1006427226972. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh T-F, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324(5933):1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong Z, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111(6):803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J-K. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43(1):143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu SC, Zhang Y. Active DNA demethylation: Many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11(9):607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi Y, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell. 2002;110(1):33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 13.Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu J-K. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol. 2007;17(1):54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 14.Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324(5933):1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu A, et al. Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc Natl Acad Sci USA. 2013;110(6):2389–2394. doi: 10.1073/pnas.1211757110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamuro C, et al. Overproduction of stomatal lineage cells in Arabidopsis mutants defective in active DNA demethylation. Nat Commun. 2014;5:4062. doi: 10.1038/ncomms5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Penterman J, et al. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA. 2007;104(16):6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gehring M, Henikoff S. DNA methylation dynamics in plant genomes. Biochim Biophys Acta. 2007;1769(5-6):276–286. doi: 10.1016/j.bbaexp.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Bhutani N, Burns DM, Blau HM. DNA Demethylation Dynamics. Cell. 2011;146(6):866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong S, et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol. 2013;31(2):154–159. doi: 10.1038/nbt.2462. [DOI] [PubMed] [Google Scholar]
- 21.Manning K, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38(8):948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 22.Teyssier E, et al. Tissue dependent variations of DNA methylation and endoreduplication levels during tomato fruit development and ripening. Planta. 2008;228(3):391–399. doi: 10.1007/s00425-008-0743-z. [DOI] [PubMed] [Google Scholar]
- 23.Mok YG, et al. Domain structure of the DEMETER 5-methylcytosine DNA glycosylase. Proc Natl Acad Sci USA. 2010;107(45):19225–19230. doi: 10.1073/pnas.1014348107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vrebalov J, et al. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science. 2002;296(5566):343–346. doi: 10.1126/science.1068181. [DOI] [PubMed] [Google Scholar]
- 25.Giovannoni JJ. Genetic regulation of fruit development and ripening. Plant Cell. 2004;16(Suppl):S170–S180. doi: 10.1105/tpc.019158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartley GE, Viitanen PV, Bacot KO, Scolnik PA. A tomato gene expressed during fruit ripening encodes an enzyme of the carotenoid biosynthesis pathway. J Biol Chem. 1992;267(8):5036–5039. [PubMed] [Google Scholar]
- 27.Ray J, et al. Cloning and characterization of a gene involved in phytoene synthesis from tomato. Plant Mol Biol. 1992;19(3):401–404. doi: 10.1007/BF00023387. [DOI] [PubMed] [Google Scholar]
- 28.How-Kit A, et al. Accurate CpG and non-CpG cytosine methylation analysis by high-throughput locus-specific pyrosequencing in plants. Plant Mol Biol. 2015;88(4-5):471–485. doi: 10.1007/s11103-015-0336-8. [DOI] [PubMed] [Google Scholar]
- 29.Chen W, et al. Requirement of CHROMOMETHYLASE3 for somatic inheritance of the spontaneous tomato epimutation Colourless non-ripening. Sci Rep. 2015;5:9192. doi: 10.1038/srep09192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin T, et al. Genomic analyses provide insights into the history of tomato breeding. Nat Genet. 2014;46(11):1220–1226. doi: 10.1038/ng.3117. [DOI] [PubMed] [Google Scholar]
- 31.Telias A, et al. Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol. 2011;11(1):93. doi: 10.1186/1471-2229-11-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, et al. The methylation of the PcMYB10 promoter is associated with green-skinned sport in Max Red Bartlett pear. Plant Physiol. 2013;162(2):885–896. doi: 10.1104/pp.113.214700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.