Abstract
Aims: Hydrogen sulfide (H2S) exerts a wide range of actions in the body, especially in the modulation of mitochondrial functions. The normal replication of mitochondrial DNA (mtDNA) is critical for cellular energy metabolism and mitochondrial biogenesis. The aim of this study was to investigate whether H2S affects mtDNA replication and the underlying mechanisms. We hypothesize that H2S maintains mtDNA copy number via inhibition of Dnmt3a transcription and TFAM promoter methylation. Results: Here, we demonstrated that deficiency of cystathionine gamma-lyase (CSE), a major H2S-producing enzyme, reduces mtDNA copy number and mitochondrial contents, and it inhibits the expressions of mitochondrial transcription factor A (TFAM) and mitochondrial marker genes in both smooth muscle cells and aorta tissues from mice. Supply of exogenous H2S stimulated mtDNA copy number and strengthened the expressions of TFAM and mitochondrial marker genes. TFAM knockdown diminished H2S-enhanced mtDNA copy number. In addition, CSE deficiency induced the expression of DNA methyltransferase 3a (Dnmt3a) and TFAM promoter DNA methylation, and H2S repressed Dnmt3a expression, resulting in TFAM promoter demethylation. We further found that H2S S-sulfhydrates transcription repressor interferon regulatory factor 1 (IRF-1) and enhances the binding of IRF-1 with Dnmt3a promoter after reduced Dnmt3a transcription. H2S had little effects on the expression of Dnmt1 and Dnmt3b as well as on ten-eleven translocation methylcytosine dioxygenase 1, 2, and 3. Innovation: A sufficient level of H2S is able to inhibit TFAM promoter methylation and maintain mtDNA copy number. Conclusion: CSE/H2S system contributes to mtDNA replication and cellular bioenergetics and provides a novel therapeutic avenue for cardiovascular diseases. Antioxid. Redox Signal. 23, 630–642.
Introduction
Hydrogen sulfide (H2S) can be endogenously produced in the body, and cystathionine gamma-lyase (CSE) is a main H2S-producing enzyme in the vascular system (43, 47). Cystathionine beta-synthase (CBS) and 3-mercaptopyruvate sulfurtransferase (3MST) also have been reported to contribute to the endogenous generation of H2S in vascular cells (34, 35). The effects of H2S in the vascular system are complex and permit H2S to exert a wide range of actions, including vasodilatation, angiogenesis, cellular bioenergetics, and energy production (13, 38, 39). The signaling mechanisms underlying the multifaceted vascular effects of H2S have been attributed to activating KATP channels, inducing cell membrane hyperpolarization, increasing intracellular calcium, altering cellular cGMP and cAMP levels, attenuating inflammation and oxidative stress, and so on (4, 26, 43). In many cases, nitric oxide (NO) mediates the cytoprotective signaling of H2S in cardiovascular functions, and H2S and NO cooperatively regulate vascular tone by activating a neuroendocrine HNO-TRPA1-CGRP signaling pathway or AKT/p38-MAPK pathway (3, 12, 20). Deficiency of CSE/H2S system leads to various vascular disorders, such as hypertension and atherosclerosis (25, 46, 47).
Altered mitochondrial biogenesis and mitochondrial dysfunction are often involved in vascular disorders. Many factors contribute to the regulation of mitochondrial biogenesis, and PGC-1α is a key transcriptional regulation factor that induces mitochondrial biogenesis by activating different transcription factors, including nuclear respiratory factor 1 and nuclear respiratory factor 2, which activate mitochondrial transcription factor A (TFAM) (7, 22). Mammalian mitochondrial DNA (mtDNA), a ∼16.6 Kb circular double-stranded genome, encodes 13 proteins constituting the essential subunits of the mitochondrial respiratory chain (6). The normal replication of mtDNA is critical for cellular energy metabolism and mitochondrial biogenesis (44). TFAM, a nuclear coding gene, directly regulates mitochondrial DNA transcription and replication, and it maintains mitochondrial biogenesis and mass (9, 40). TFAM displays sequence-specific binding to both heavy strand promoter and light strand promoter in the mitochondrial genome, and it initiates the DNA replication (29, 46). Disruption of the TFAM gene in mice results in depletion of mtDNA and reduction of mtDNA-encoded polypeptides (21, 28). On the other hand, overexpression of TFAM ameliorates the decline in mtDNA copy number of transgenic (Tg) mice (15).
Innovation.
Hydrogen sulfide (H2S) as a novel gasotransmitter induces cellular bioenergetics and energy production, and H2S post-translational modification of proteins contributes to most of its actions in the body. In this study, we highlight a novel function for H2S in maintenance of mitochondrial DNA (mtDNA) replication and mitochondrial marker gene expression. H2S, which by S-sulfhydration of interferon regulatory factor 1 (IRF-1), enhances IRF-1 binding with Dnmt3a promoter, inhibits Dnmt3a expression and mitochondrial transcription factor A (TFAM) promoter methylation after increased expression of TFAM and mitochondrial marker genes. These findings point to an important function of H2S in improving mtDNA replication and cellular bioenergetics via epigenetic modification of TFAM.
DNA methylation is a specific postsynthetic modification of DNA, which is the transfer of methyl groups from S-adenosyl methionine (SAM) to the 5′ position of cytosine forming 5-methylcytosine (5). SAM can also be transported into the mitochondria with the mitochondrial SAM transporter, where it is converted into S-adenosyl homocysteine in methylation reactions of DNA (1). DNA methylation plays an important role in the epigenetic modulation of gene expression and gene silencing as well as in normal organismal development (19). The DNA methylation is mostly established by the de novo DNA methyltransferase 3a (Dnmt3a) and Dnmt3b and maintained by Dnmt1 during replication (19, 30, 31). The dynamic regulation of Dnmt expression substantially contributes to DNA methylation profiles on both a genome-wide and a gene-specific scale. Maintaining DNA methylation status by balancing methylation and demethylation processes impacts various processes from normal development to diseases (23).
H2S regulation of mitochondrial functions has been reported. By sensing the oxygen levels in mitochondria, CSE translocates into mitochondria, leading to the production of H2S inside mitochondria and conferring resistance to hypoxia by increasing ATP synthesis in smooth muscle cells (SMCs) (13). Lack of H2S suppresses respiratory reserve capacity and ATP turnover as well as glycolysis in cancer cells (38). H2S therapy increased mitochondrial biogenesis in the cortex and protected brain damage after cardiac arrest and resuscitation in rats (33). Nonetheless, H2S epigenetic regulation of mtDNA replication and the underlying mechanisms in vascular cells have been unknown.
In this study, we investigated the epigenetic regulation of H2S on mtDNA replication and mitochondrial biogenesis by using cultured SMCs and aorta tissues from both CSE knockout (KO) mice and wild-type (WT) littermates. Our findings demonstrated that H2S inhibits Dnmt3a transcription by S-sulfhydrating transcription repressor interferon regulatory factor 1 (IRF-1), which leads to TFAM promoter demethylation and enhanced TFAM expression as well as to higher mtDNA copy number and mitochondrial marker gene expression.
Results
H2S induces an increase in mtDNA copy number and mitochondrial maker gene expression
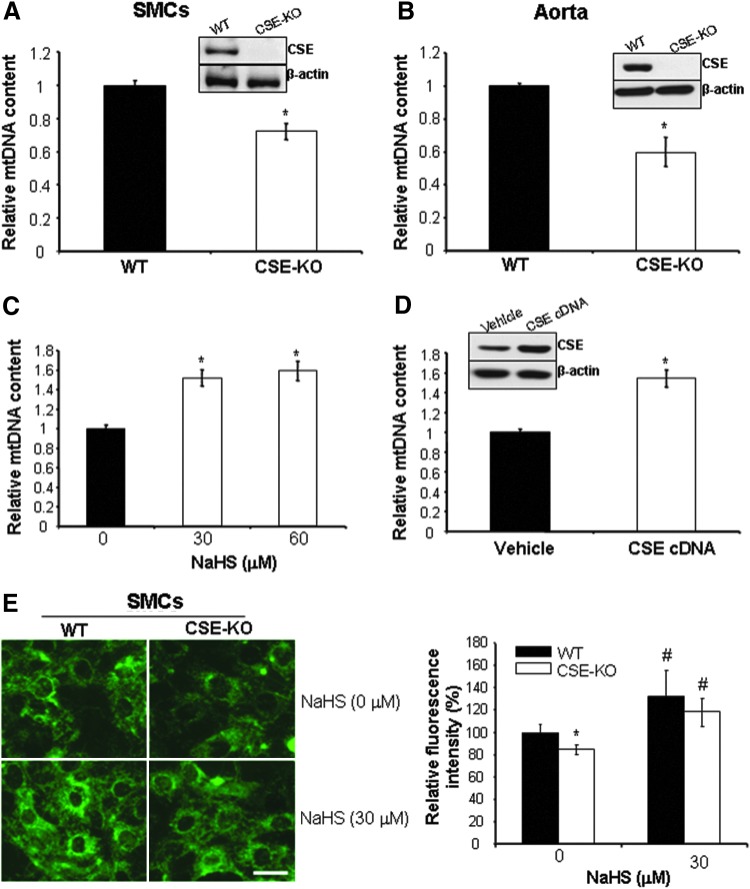
Using real-time PCR, we found that mtDNA copy number is significantly lower in SMCs and aorta tissues from CSE-KO mice in comparison with those from WT littermates (Fig. 1A, B). Exogenously applied NaHS (30 and 60 μM, Fig. 1C) or CSE overexpression (Fig. 1D) significantly enhanced mtDNA copy number in WT-SMCs. Moreover, we examined the effects of CSE/H2S system on mitochondrial contents by using mitotracker green probe. Fluorescent microscope observations and fluorimetric measurements showed that mitochondrial mass is slightly but significantly decreased in CSE-KO SMCs, and incubation of both WT-SMCs and CSE-KO SMCs with NaHS (30 μM) increased mitochondrial contents (Fig. 1E). Neither CSE deficiency nor exogenously applied NaHS had an effect on 3MST expression (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/ars), and CBS mRNA and protein was not found in both WT and CSE-KO SMCs.
FIG. 1.
Hydrogen sulfide (H2S) enhances mitochondrial DNA (mtDNA) copy number. Cystathionine gamma-lyase (CSE) deficiency leads to decreased mtDNA copy number in smooth muscle cells (SMCs) (A) and aorta tissues (B) isolated from mice. mtDNA copy number was determined using real-time PCR assay and normalized to β-actin gene. *p<0.05. (C) Exogenously applied H2S induces mtDNA copy number. Wild-type (WT)-SMCs were incubated with NaHS (30 and 60 μM) for 48 h after detection of mtDNA copy number. *p<0.05. (D) CSE overexpression increases mtDNA copy number. WT-SMCs were transfected with CSE cDNA for 48 h after detection of CSE expression and mtDNA copy number. *p<0.05. (E) CSE/H2S system stimulates mitochondrial contents. WT-SMCs and CSE-KO-SMCs were incubated with or without NaHS (30 μM) for 48 h after detection of mitochondrial contents by mitotracker greed probe. *p<0.05 versus all other groups; #p<0.05 versus WT-SMCs without NaHS treatment. Scale bar: 20 μm. The experiments in (A, C–E) were repeated at least thrice, and three to four mice were used for each group in (B). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
To further elucidate the effect of H2S on mtDNA replication, quantitative PCR was performed to detect the variation in the expression of genes related to mitochondrial function and biogenesis. As shown in Figure 2A and B, CSE deficiency significantly decreased the expression of Cytochrome C oxidase subunit 1 (MT-CO1), Cytochrome b (CytB), and ATP synthase subunit 6 (Atp6) in both SMCs and aorta tissues, all of which are mitochondrial proteins encoded by mtDNA. Twinkle and citrate synthase (CS), two nuclear-coding mitochondrial proteins, were not changed by either CSE deficiency or exogenously applied NaHS. The expression of MT-CO1, CytB, and Atp6 was significantly enhanced after incubation of WT-SMCs with 30 μM NaHS for 24 h and further increased at 48 h (Fig. 2C).
FIG. 2.
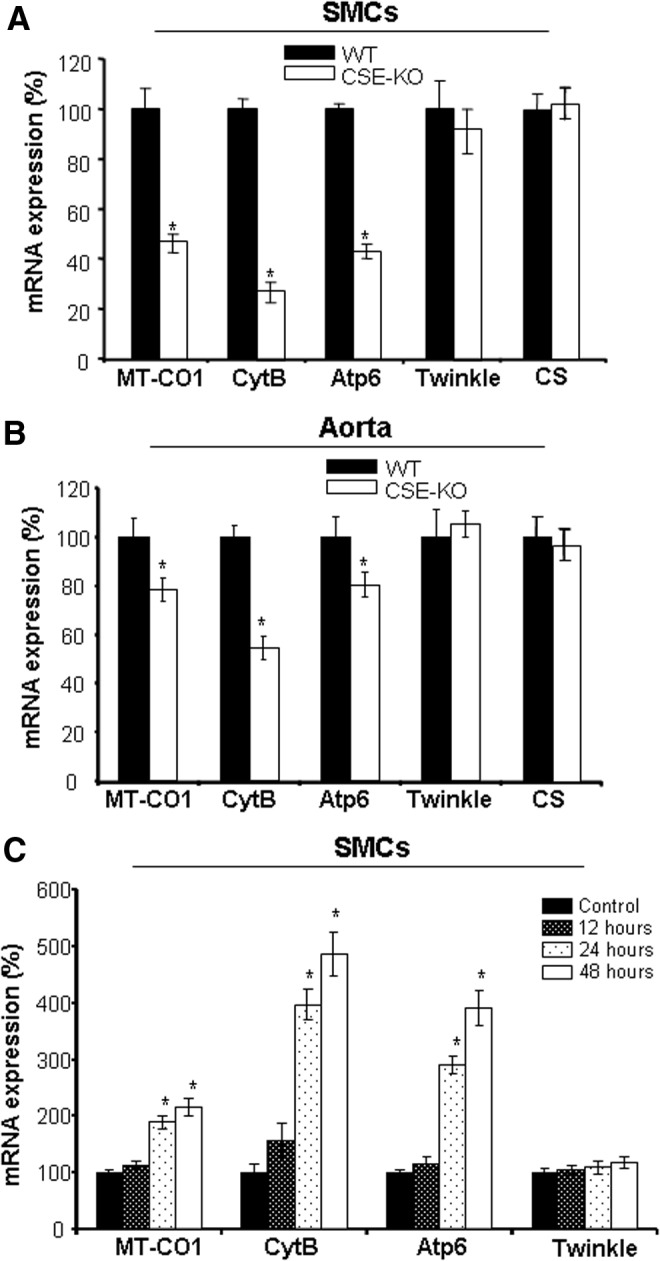
H2S stimulates the expression of mitochondrial marker genes. (A, B) The mRNA expression of MT-CO1, CytB, and Atp6 is lower in CSE-deficient aorta tissues and SMCs compared with that from WT littermates. *p<0.05. (C) Exogenously applied H2S induces the mRNA expression of MT-CO1, CytB, and Atp6 in SMCs. WT-SMCs were incubated with NaHS (30 μM) for the indicated time (12–48 h) after detection of mRNA expression by real-time PCR. *p<0.05 versus the control. The experiments in (A, C) were repeated at least thrice, and four mice were used for each group in (B).
TFAM mediates H2S-induced mtDNA copy number
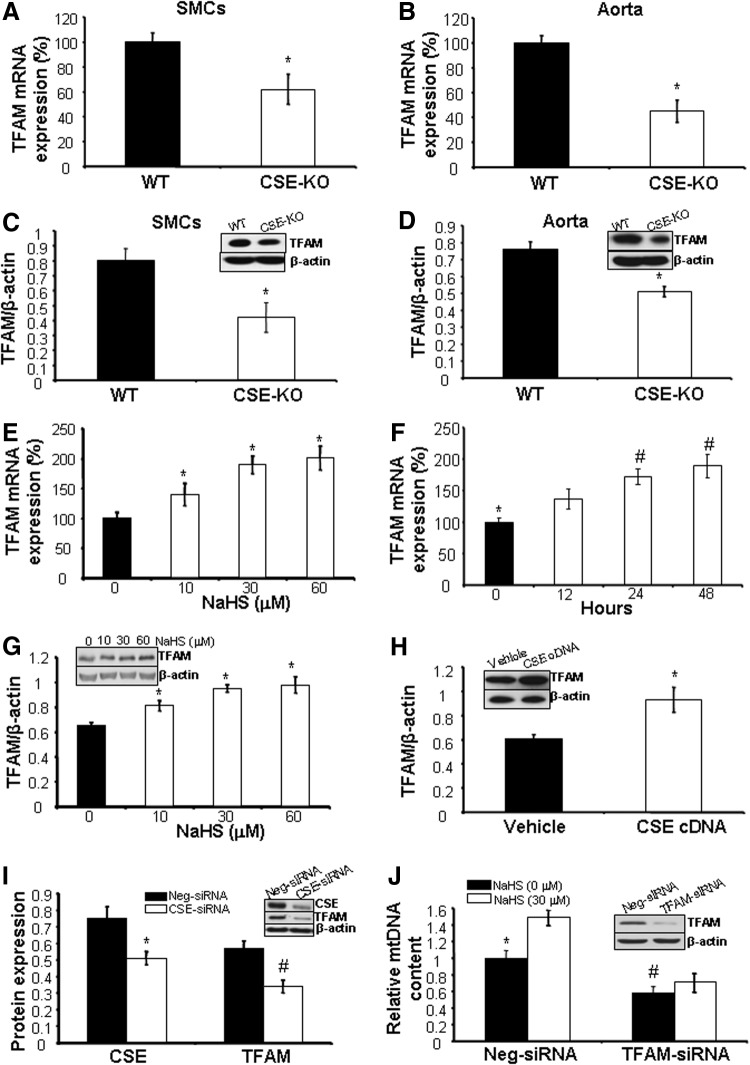
TFAM functions as a master transcription activator for mtDNA replication. We found that both mRNA and protein expression of TFAM was significantly lower in SMCs and aorta tissues from CSE-KO mice compared with WT mice (Fig. 3A–D). Incubation of WT-SMCs with NaHS stimulated the expression of both mRNA and protein of TFAM at a concentration as low as 10 μM (Fig. 3E, G). The stimulatory role of NaHS in TFAM expression was time dependent, which reaches significance at the first 12 h of incubation with NaHS and further increases at 24 and 48 h, respectively (Fig. 3F). Transfection of WT-SMCs with CSE cDNA induced but short interfering RNA (siRNA)-mediated CSE knockdown reduced TFAM protein expression (Fig. 3H, I), suggesting the stimulatory role of CSE/H2S system in TFAM expression.
FIG. 3.
H2S induces mitochondrial transcription factor A (TFAM) expression. (A, B) TFAM mRNA expression is lower in CSE-deficient SMCs and aorta. *p<0.05. (C, D) TFAM protein expression is lower in CSE-deficient SMCs and aorta. *p<0.05. (E) H2S stimulates the mRNA expression of TFAM in a dose-dependent manner. WT-SMCs were incubated with NaHS (10, 30, and 60 μM) for 48 h after detection of TFAM mRNA by real-time PCR. *p<0.05. (F) H2S stimulates the mRNA expression of TFAM in a time-dependent manner. WT-SMCs were incubated with 30 μM NaHS for the indicated time (12–48 h) after detection of TFAM mRNA by real-time PCR. *p<0.05 versus all other groups; #p<0.05 versus the cells treated with NaHS for 12 h. (G) H2S increases TFAM protein expression. WT-SMCs were incubated with NaHS (10, 30, and 60 μM) for 48 h after detection of TFAM protein expression by Western blotting. *p<0.05. (H) CSE overexpression increases TFAM expression. WT-SMCs were transfected with CSE cDNA for 48 h after detection of TFAM protein expression. *p<0.05. (I) siRNA-mediated CSE knockdown reduces TFAM expression. WT-SMCs were transfected with CSE-specific siRNA (CSE-siRNA) or negative siRNA (Neg-siRNA) for 72 h after detection of CSE and TFAM protein expression. * and #p<0.05 versus Neg-siRNA transfected cells. (J) siRNA-mediated TFAM knockdown abolishes H2S-induced mtDNA copy number. After WT-SMCs were transfected with TFAM-specific siRNA (TFAM-siRNA) or Neg-siRNA for 48 h, NaHS (30 μM) was added for additional 24 h after detection of TFAM protein expression and mtDNA copy number. *p<0.05 versus all groups. #p<0.05 versus Neg-siRNA-transfected cells without NaHS treatment. All experiments were repeated at least thrice, and four to six mice were used in each group to isolate aorta tissue.
To explore the involvement of TFAM in H2S-induced mtDNA copy number, we knocked down TFAM expression with gene-specific siRNA (Fig. 3J). We then found that the stimulatory role of H2S in mtDNA copy number is diminished (Fig. 3J), indicative of the mediation of TFAM in H2S-induced mtDNA copy number.
H2S inhibits TFAM promoter methylation by suppressing Dnmt3a expression
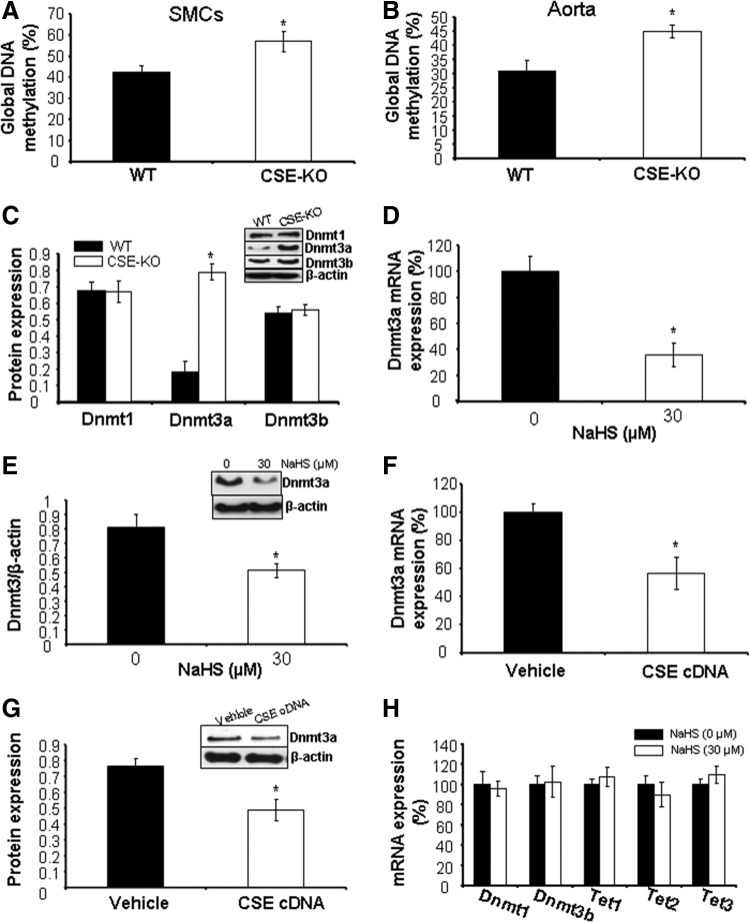
A growing body of evidence suggests that epigenetic modification, in particular hypermethylation, is involved in gene silencing. We first measured the global methylation ratio between WT and CSE-KO mice, and found that DNA methylation is significantly higher in aorta and SMCs from CSE-KO mice (Fig. 4A, B). The regulation of Dnmts substantially contributes to DNA methylation. We next assessed the effects of H2S on the expression of Dnmts. CSE deficiency significantly induced the expression of Dnmt3a but not Dnmt1 and Dnmt3b in the aorta (Fig. 4C). We further found that exogenously applied NaHS or CSE overexpression decreases the mRNA and protein expression of Dnmt3a (Fig. 4D–G). Two other Dnmts (Dnmt1 and Dnmt3b) and three DNA demethylases, Ten-eleven translocation methylcytosine dioxygenase 1 (Tet1), 2, and 3, were not changed by NaHS (Fig. 4H), suggesting that the higher global DNA methylation is attributed to the lower level of H2S and the higher expression of Dnmt3a.
FIG. 4.
H2S lessens Dnmt3a expression. (A, B) Global DNA methylation is higher in CSE-deficient SMCs and aorta in mice. *p<0.05. (C) CSE deficiency induces the expression of Dnmt3a but not Dnmt1 and Dnmt3b in mouse aorta. *p<0.05. (D, E) Exogenously applied H2S reduces the mRNA and protein expression of Dnmt3a. WT-SMCs were incubated with NaHS (30 μM) for 48 h after detection of Dnmt3a mRNA and protein expression. *p<0.05. (F, G) CSE overexpression decreases Dnmt3a mRNA and protein expression. WT-SMCs were transfected with CSE cDNA for 48 h after detection of Dnmt3a mRNA and protein expression. *p<0.05. (H) H2S has no effect on the mRNA expression of Dnmt1, Dnmt3b, and Tet 1–3. WT-SMCs were incubated with NaHS (30 μM) for 48 h after detection of mRNA expression of Dnmt1, Dnmt3b, and Tet 1–3. All experiments were repeated at least thrice, and four to six mice were used in each group to isolate aorta tissue.
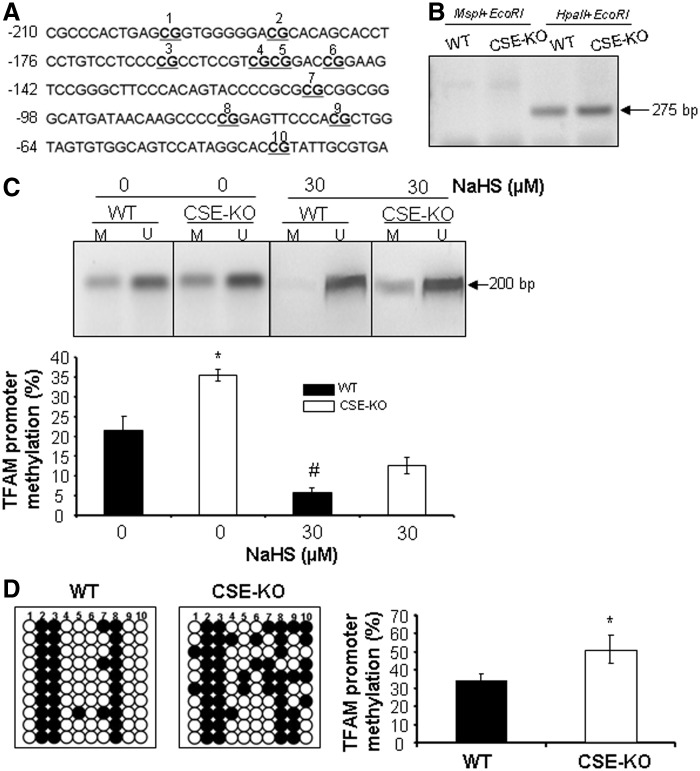
To investigate the crosstalk between H2S and TFAM in mitochondrial biogenesis, we analyzed TFAM promoter via Methprimer (www.urogene.org/methprimer/) and found one CpG island in the promoter region −210 to −30, in which there are 10 potential CpG methylation loci (Fig. 5A). By using methylation-sensitive restriction endonuclease (MS-RE)-PCR, we first found that the CpG island in the TFAM promoter is methylated (Fig. 5B). Then, methylation-special PCR (MSP) and bisulfite genomic sequencing were performed to quantify the TFAM promoter methylation levels in both WT-SMCs and CSE-KO SMCs, respectively. The results showed that CSE deficiency induces substantial levels of DNA methylation in SMCs (Fig. 5C, D). On average, 5.1 of the 10 potential CpG methylation loci in the CpG island of TFAM promoter were methylated in CSE-KO SMCs but only 3.4 in WT-SMCs (Fig. 5D). Incubation of both WT-SMCs and CSE-KO SMCs with NaHS significantly decreased TFAM promoter methylation (Fig. 5C).
FIG. 5.
H2S attenuates TFAM promoter methylation. (A) There are 10 potential CpG methylation loci in the predicted CpG islands between −210 bp and −30 bp of the mouse TFAM promoter. (B) MS-RE-PCR data show that the CpG islands in the TFAM promoter are methylated in both WT and CSE-KO SMCs. (C) MSP analysis validates that CSE deficiency induces but exogenously applied H2S reduces the methylation ratio in the CpG islands of the TFAM promoter. Both WT and CSE-KO SMCs were incubated with NaHS (30 μM) for 48 h. M, methylation-specific primers; U, unmethylation-specific primers. *p<0.05 versus all other groups; #p<0.05 versus WT-SMCs without NaHS treatment. (D) Higher DNA methylation in CSE-deficient SMCs evidenced by bisulfite genomic sequencing. *p<0.05. The experiments in (B, C) were repeated thrice, and 10 clones were randomly chosen for sequencing in (D).
We then determined whether TFAM expression and mitochondrial biogenesis can be modified by 5-aza-2′-deoxycytidine (AZA), a DNA methyltransferase inhibitor. We found that the expression of TFAM, MT-CO1, and CytB is increased at 24 h after AZA incubation in WT-SMCs (Fig. 6A). siRNA-mediated knockdown of Dnmt3a also enhanced TFAM expression (Fig. 6B, C), pointing to the critical role of Dnmt3a in epigenetic regulation of TFAM gene expression.
FIG. 6.
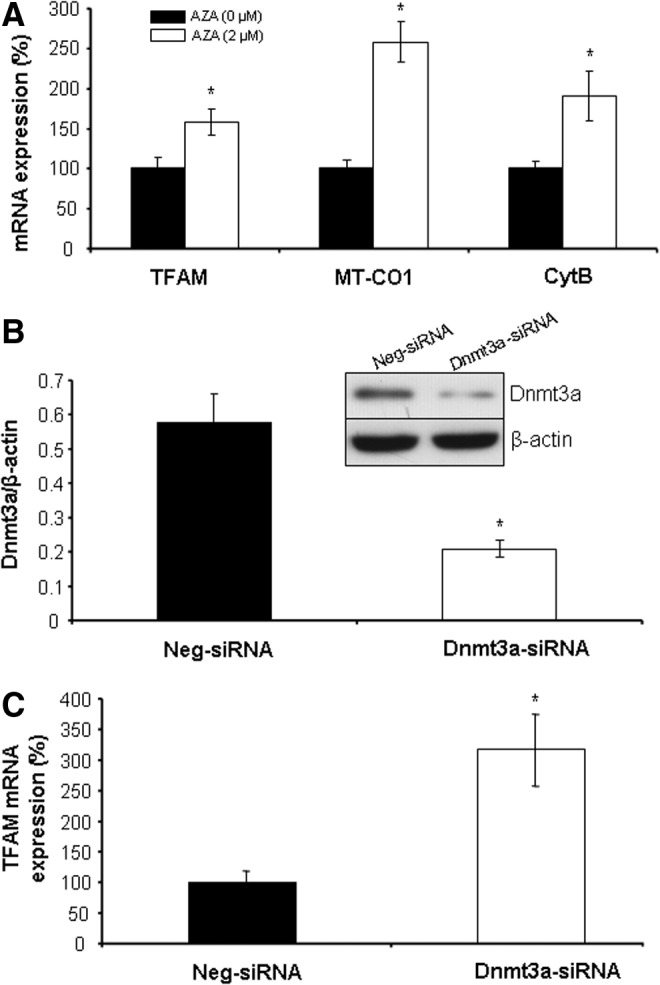
DNA demethylation enhances the expression of TFAM. (A) AZA stimulates the expression of TFAM, MT-CO1, and CytB. WT-SMCs were incubated with AZA (2 μM) for 96 h. *p<0.05. (B) siRNA-mediated knockdown of Dnmt3a. WT-SMCs were transfected with Dnmt3a-specific siRNA (Dnmt3a-siRNA) or Neg-siRNA for 72 h after detection of Dnmt3a protein expression. *p<0.05. (C) Dnmt3a knockdown induces TFAM expression. WT-SMCs were transfected with Dnmt3a-siRNA or Neg-siRNA for 72 h after detection of TFAM mRNA expression. *p<0.05. All experiments were repeated at least thrice.
H2S S-sulfhydrates IRF-1 after increased binding with Dnmt3a promoter
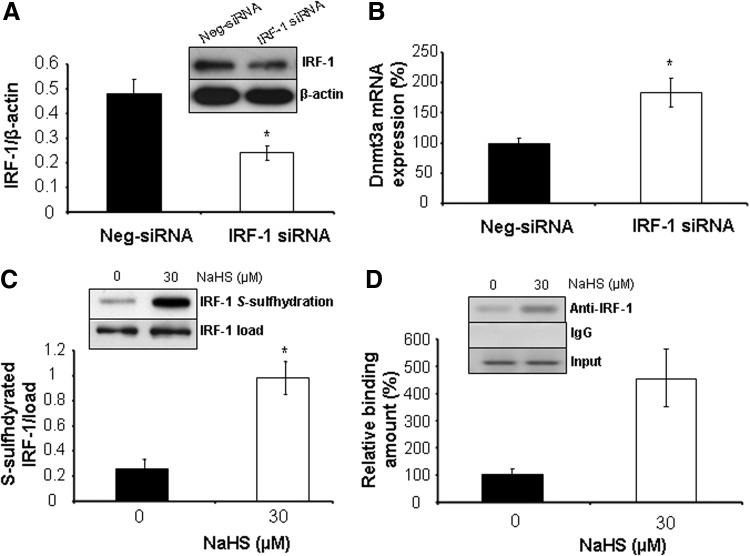
We next investigated how H2S affects Dnmt3a expression. H2S inhibition of Dnmt3a mRNA expression suggests that H2S may act on Dnmt3a transcription. IRF-1 is a transcription activator or repressor that has been predicted to target at the Dnmt3a promoter. Here, we first found that knockdown of IRF-1 induces Dnmt3a expression (Fig. 7A, B). NaHS had little effect on IRF-1 expression but significantly S-sulfhydrated IRF-1 (Fig. 7C). We further validated that NaHS stimulates the binding of IRF-1 with Dnmt3a promoter, as evidenced by chromatin immunoprecipitation (ChIP) assay (Fig. 7D). No immunoprecipitation and amplification were seen with nonspecific IgG antibody. All these data suggest that H2S may strengthen the inhibitory activity of IRF-1 on Dnmt3a transcription.
FIG. 7.
H2S S-sulfhydrates IRF-1 and stimulates the binding of IRF-1 with Dnmt3a promoter. (A) siRNA-mediated knockdown of IRF-1. WT-SMCs were transfected with IRF-1-specific siRNA (IRF-1-siRNA) or Neg-siRNA for 72 h after detection of IRF-1 protein expression. *p<0.05. (B) IRF-1 knockdown enhances Dnmt3a mRNA expression. WT-SMCs were transfected with IRF-1-siRNA or Neg-siRNA for 72 h after detection of Dnmt3a mRNA expression. *p<0.05. (C) H2S S-sulfhydrates IRF-1. WT-SMCs were incubated with NaHS (30 μM) for 2 h after detection of IRF-1 S-sulfhydration by biotin switch assay. *p<0.05. (D) H2S stimulates the binding of IRF-1 with Dnmt3a promoter. WT-SMCs were incubated with NaHS (30 μM) for 2 h after detection of IRF-1 binding with Dnmt3a promoter by ChIP assay. *p<0.05. All experiments were repeated at least thrice.
Discussion
The gasotransmitter's roles of H2S in biology and medicine have been widely studied, which demonstrate that H2S is involved in most cellular functions and physiologic activities (27, 38, 43, 47). It is well known that H2S level is significantly lower in cardiovascular diseases and supply of exogenous H2S improves cardiovascular functions (25, 46, 47). The pathogenesis of cardiovascular diseases is multifactorial and complex, resulting from the tight interactions between genetic and environmental factors (11, 16, 36). Several lines of evidence pointed to the altered mitochondrial structure and function in various cardiovascular disorders. In this study, we provided evidence showing that CSE deficiency in both SMCs and mouse aorta tissues decreases mitochondrial contents and mtDNA copy number, and incubation of SMCs with exogenous H2S stimulates mitochondrial contents and mtDNA copy number. Abnormal mtDNA replication would cause impaired mitochondrial function and structure, which may be directly linked to the vascular dysfunction, such as hypertension and atherosclerosis. Transcriptions of MT-CO1, CytB, and Atp6, three mtDNA-encoded genes, were significantly lower in CSE-deficient SMCs. In contrast, exogenously applied H2S enhanced the expression of these three genes. These data suggest that H2S may enhance mtDNA replication by directly stimulating the transcription of mtDNA-encoded genes.
TFAM is a key activator of mitochondrial transcription and mitochondrial genome replication. TFAM has been shown to be modified by phosphorylation, microRNAs, and cAMP phosphodiesterase Prune (17, 24, 49). Our data presented here clearly demonstrated that H2S induces the expression of TFAM mRNA and protein. Either complete KO of CSE or partial knockdown of CSE significantly suppressed TFAM expression. In addition, H2S-induced mtDNA copy number was abolished when the expression of TFAM was suppressed. These data implicated that TFAM definitely contributes to CSE/H2S-improved mtDNA replication and mitochondrial homeostasis.
Our main focus was to understand H2S regulation of TFAM expression. CSE/H2S system strengthening TFAM mRNA expression points to the possibility of H2S regulation of TFAM at the transcription level. DNA methylation in the form of 5-methylcytosine in promoter CpG islands is known as the most common epigenetic modification of gene expression (37). CpG island is a short interspersed DNA sequence in which GC-rich or CpG-rich nucleotides are predominantly methylated, in comparison with the average genomic pattern. Our data first found that the global DNA methylation rate is significantly higher in CSE-deficient SMCs and aorta tissues. Interestedly, the transcription of Dnmt3a but not Dnmt1 and Dnmt3b was induced by CSE overexpression or H2S treatment. On the other hand, only the expression of Dnmt3a was suppressed by CSE KO, pointing to the mediation of Dnmt3a in CSE-deficiency-induced global DNA methylation. We further excluded the involvement of DNA demethylases in CSE/H2S system-regulated DNA methylation, because H2S had little effect on the expression of Tet1, Tet2, and Tet3. All of these 3 Tets can oxidize 5-hydroxymethylcytosine to 5-Formylcytosine and 5-Carboxylcytosine and lead to DNA demethylation (10, 18). Dnmts use SAM as the methyl donor and catalyze the transfer of a methyl group to DNA, and SAM is generated and maintained via the methionine cycle and transsulfuration pathway (8). Methionine is first converted to SAM. In losing its methyl group, SAM becomes S–adenosyl homocysteine, which is then converted to homocysteine. Homocysteine is either converted back to methionine, or it enters the transsulfuration pathway. CSE is a critical enzyme in the transsulfuration pathway to produce H2S, and deficiency of CSE may generate feedback inhibition, leading to the accumulation of SAM and higher DNA methylation, which deserves to be further investigated.
We next explored whether H2S-regulated TFAM transcription correlates with the DNA methylation status of its CpG island promoter region. Sequence analysis first revealed that there are 10 potential CpG methylation loci in the predicted CpG island region between −221 bp and −30 bp of the TFAM promoter, suggesting that cytosine methylation may play a role in the regulation of TFAM expression. We then used three gold-standard methods to identify and quantify TFAM DNA methylation, including MS-RE-PCR, MS-PCR, and bisulfite genomic sequencing. All these assays proved that TFAM promoter methylation level is significantly higher in CSE-deficient SMCs and aorta tissues. Furthermore, exogenously applied H2S attenuated TFAM promoter methylation. Broad inhibition of Dnmt activity by AZA or specific knockdown of Dnmt3a induced the expression of TFAM as well as MT-CO1 and CytB, two mtDNA-encoded genes. It has been reported that methylation of the TFAM gene promoter is associated with insulin resistance and chronic obstructive pulmonary disease (14). Here, we further validated that TFAM gene methylation is also linked to H2S-related vascular dysfunction.
The mechanism underlying the change of Dnmt3a expression by CSE/H2S system was subsequently elucidated. It is proposed that the major signaling mechanism of H2S is through the S-sulfhdyration of reactive cysteine residues on target proteins by yielding a hydropersulfide moiety (-SSH), with the potential to alter protein conformation and the final function and activity of target proteins (27, 48). So far, there have been a dozen of proteins observed to be modified by H2S through S-sulfhydration (41, 51). To investigate whether H2S may directly S-sulfhydrate the transcriptional activators or repressors of Dnmt3a gene, we focused on IRF-1, which has been predicted to target at Dnmt3a promoter and regulate Dnmt3a transcription (32). IRF-1 alters expression of target genes by binding to an interferon-stimulated response (ISR) element in their promoters (50). Knockdown of IRF-1 in SMCs markedly stimulated Dnmt3a expression, and ChIP assay further proved that IRF-1 binds with Dnmt3a promoter at the basal condition. These results suggest that IRF-1 may act as a transcription repressor of Dnmt3a. In the presence of exogenous H2S, the binding of IRF-1 with Dnmt3a promoter containing ISR element was increased by more than thrice, consistent with the inhibitory role of H2S in Dnmt3a transcription. In addition, IRF-1 was basally S-sulfhydrated and further strengthened by exogenous applied H2S, as proved by biotin switch assay with anti-IRF-1 antibody. H2S S-sulfhydration of IRF-1 may enhance its binding capacity with Dnmt3a promoter after reduced Dnmt3a transcription. IRF-1 protein binds to the ISR element via an N-terminal helix-turn-helix DNA-binding domain, and amino-acid sequence analysis showed there are six cysteine residues in IRF-1 protein and only one cysteine (cysteine-53) is located in its DNA-binding domain. Further studies need to be performed to determine the responsible cysteine residue(s) for H2S S-sulfhydration modification of IRF-1.
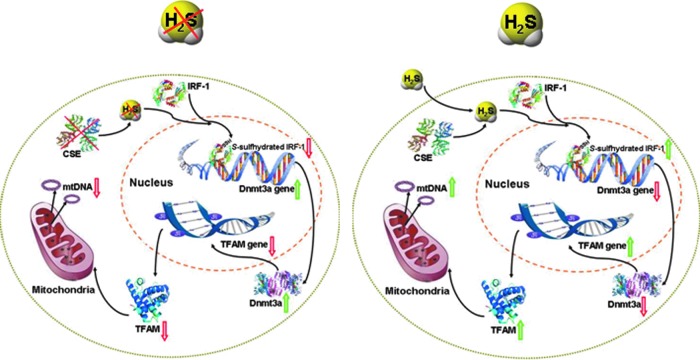
H2S acts as a biologically active mediator in the cardiovascular system by regulating vascular tones. Here, we provided novel data that H2S maintains mtDNA copy number, mitochondrial contents, and the transcriptions of mtDNA-encoded genes, which is mediated by S-sulfhydration of IRF-1, inhibition of Dnmt3a transcription, and TFAM promoter methylation after enhanced TFAM expression, at least in the vascular system (Fig. 8). Deficiency of CSE caused reduced H2S generation and diminished the level of S-sulfhydrated IRF-1, which strengthens Dnmt3a expression and TFAM methylation, leading to lower mitochondrial contents and mtDNA replication. These findings will help advance our understanding of H2S biology and physiology and develop novel therapeutic avenues for treatment of cardiovascular disease linked to abnormal mitochondrial mass and functions.
FIG. 8.
Putative signal pathways underlying the effects of H2S on TFAM/Dnmt3a and mtDNA copy number. H2S induces IRF-1 activity by S-sulfhydration and leads to IRF-1 binding with Dnmt3a promoter, which decreases Dnmt3a transcription after reduced TFAM promoter methylation and increased TFAM expression and mtDNA replication. When endogenous H2S level is lower (left panel), lesser IRF-1 proteins are S-sulfhydrated, which leads to IRF-1 dissociation from Dnmt3a and promotes Dnmt3a expression, global DNA methylation, as well as TFAM promoter methylation after reduced expression of TFAM, lower mtDNA copy number, and transcriptions of mtDNA-encoded genes. In contrast, sufficient H2S maintains mtDNA copy number and mitochondrial contents by S-sulfhydrating IRF-1, inhibiting Dnmt3a transcription and TFAM promoter methylation, and stimulating the transcriptions of TFAM and mtDNA-encoded genes (right panel). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Materials and Methods
Cell culture and collection of mouse aorta tissues
CSE-KO mice were generated as previously described (46). All animals were maintained on standard rodent chow, and had free access to food and water. SMCs isolated from CSE-KO mice and WT littermates were cultured in phenol red-free Dulbecco's modified Eagle's medium containing 10% (v/v) fetal bovine serum and 100 U/ml penicillin, and 100 μg/ml streptomycin (Sigma, Oakville, ON) at 37°C in an atmosphere of 95% O2 and 5% CO2 (40). The experiments were performed when the cells reached 70–80% confluence between passages 10 and 16. In all studies, cells were first incubated in the serum-free medium for 12 h and 10% serum added together with different treatments, and the media were changed every 2 days. In most studies, the cells were incubated with NaHS at 30 μM for 48 h but only 2 h for S-sulfhydration experiments.
After the mice (18–20 week and male) were anaesthetized, the tissues of aorta were dissected and cleaned for protein and nucleic acid extraction. The animal procedures described in this study were approved by the Lakehead University Animal Care Committee, and animals' experiments were conducted in compliance with the NIH Guide for the Care and Use of Laboratory Animals (NIH Publication No. 85–23, revised 1996).
CSE overexpression and siRNA transfection
The plasmid CSE cDNA/pIRES2-EGFP was constructed in our previous study and used to transfect SMCs with Lipofectamine 2000 (Invitrogen, Burlington, ON) in an OptiMEM medium to overexpress CSE (45). The vector pIRES2-EGFP without CSE gene acted as a control. siRNA transfection was performed as previously described (36). SMCs were transfected with gene-specific siRNA targeting CSE, Dnmt3a, or TFAM (Santa Cruz Biotechnology, Santa Cruz, CA) for 48 h using Lipofectamine™ 2000 transfection agent in serum-free medium following the manufacturer's protocol. The cells transfected with scrambled siRNA acted as a nonsilencing control (36).
mtDNA copy number
Total genomic DNA was isolated using GenElute™ Mammalian Genomic DNA Miniprep kit (Sigma). The ratio mtDNA versus nuclear DNA (nDNA) was determined using a real-time PCR assay as described elsewhere (2, 42), with the following adaptations: Briefly, 50 ng purified DNA was amplified in a 25 μl PCR reaction containing SYBR Green PCR Master Mix (Bio-Rad, Mississauga, ON) and 40 nM of each primer. Real-time PCR was performed in an iCycler iQ5apparatus (Bio-Rad) associated with the iCycler optical system software (version 3.1). The primers were designed to target nDNA (β-actin, 5′-ATGGTGGGAATGGGTCAGAA-3′ and 5′-CTTTTCACGGTTGGCCTTAG-3′) or mtDNA (ND1, 5′-AAACGCCCTAACAACCAT-3′ and 5′-GGATAGGATGCTCGGATT-3′). The cycling reaction conditions were as follows: initial activation step for 10 min at 95°C, followed by 38 cycles of 15 s at 95°C, 1 min at 54 or 60°C, 20 s at 95°C, and 1 min at 55°C. The mtDNA content was normalized against that of β-actin gene to calculate the relative mtDNA copy number. Each measurement was repeated in triplicate, and a nontemplate control was included in each experiment.
Determination of mRNA level
Total RNA was isolated using TriReagent (Invitrogen). First-strand cDNA was synthesized according to the manufacturer's protocol (New England Biolabs, Ipswich, MA). Real-time PCR was performed in an iCycler iQ5 apparatus (Bio-Rad) associated with the iCycler optical system software (version 3.1) using SYBR Green PCR Master Mix, as previously described (45). The primers used were shown next: CBS (5′-GTGGCATGGCGACTGAAGAACGAA-3′ and 5′-ATGCGGGCAAAGGCGAAGGAAT-3′), 3MST (5′-CCGCCCCGCAACTTTTCCGAG-3′ and 5′-GAAGCCGCCGTCCAGCAATGA-3′), MT-CO1 (5′-TGTTCATTAATCGTTGATTAT-3′ and 5′- GTTGACCTAATTCTGCTCGA-3′); CytB (5′-ATCTGCCTTATAGTCCAAATCCTC-3′ and 5′- TTACTGTAGCGCCTCAAAATG-3′); Atp6 (5′-CTCAAAACGCCTAATCAACAAC-3′ and 5′- TACGGCTCCAGCTCATAGTG-3′); Twinkle (5′-GGATGGGGCCAAAGAGGGTGTC-3′ and 5′- AGGCCGGGGGATGGTGGTC-3′); CS (5′-GCAGCAACATGGGAAGACAGTGGT-3′ and 5-ATATGCCCGGGCGAAGTTGC-3′) TFAM (5′-GGAGGCAAAGGATGATTCGG-3′ and 5′-TCGTCCAACTTCAGCCATCT-3′), Dnmt3a (5′-CGGCAGAATAGCCAAGTTCA-3′ and 5′-AAGCCAAACACCCTTTCCAT-3′), Dnmt1 (5′-ACTGCGTCTCGGTCATTC-3′ and 5′-GTCTGTGCCTCCCTCCAT-3′), Dnmt3b (5′-CAGAAGTGATGGCTAAGATA-3′ and 5′-TCCTGGGAAATGCTACAA-3′), Tet1 (5′-TCTCGGGTCAGCATCACT-3′ and 5′-GAGCACCTCGCATTTCCT-3′), Tet2 (5′-TGGCAAATGTGAAGGATG-3′ and 5′-GATTAGGTCGCACTCGTA-3′), Tet3 5′-CACCAAGGCTGAGAACCC-3′ and 5′-TTGCCCGTGTAGATGACC-3′), and β-actin (5′-ATGGTGGGAATGGGTCAGAA-3′ and 5′-CTTTTCACGGTTGGCCTTAG-3′). β-actin was used as an internal reference. The PCR conditions were as follows: denaturation at 95°C for 90 s, followed by 38 cycles of denaturation at 95°C for 10 s, and annealing at 60°C for 20 s. Relative mRNA quantification was calculated by using the arithmetic formula “2-ΔΔCT,” where ΔCT is the difference between the CT (threshold cycle) of a given target cDNA and an endogenous β-actin cDNA.
MSP, bisulfite genomic sequencing, and global DNA methylation analysis
MS-RE-PCR was used to detect TFAM promoter methylation. Total genomic DNA was digested by MspI/EcoRI or HpaII/EcoRI. MspI and HpaII have the same recognize site, MspI is sensitive to methylated locus, and HpaII can cut methylated locus. The modified DNA was amplified with the primers (5′-GGCACCGTATTGCGTGAGAC-3′ and 5′-CAGGAAGTGGGGTCGGGTA-3′). We further carried out bisulfite conversion of fragmented purified DNA with the EZ DNA Methylation-Gold™ Kit (ZYMO Research, Irvine, CA) following the manufacturer's recommendation. For methylation analysis of TFAM promoter, genomic DNA (100 ng) was treated with sodium bisulfate following PCR with methylation-specific primers (5′-CGTATAGTATTTTTTGTTTTTTTCGT-3′ and 5′-TATAAACTACCACACTACCAACGTA-3′) or unmethylation-specific primers (5′-GGATGTATAGTATTTTTTGTTTTTTTTG-3′ and 5′-TATAAACTACCACACTACCAACATA-3′). PCR products were loaded onto 2% agarose gels, stained with ethidium bromide, and visualized under ultraviolet light.
Genomic sequencing analysis of TFAM promoter region spanning from −210 to −30 was performed on bisulfite-modified genomic DNA. The bisulfite-modified promoter of TFAM was amplified using the following primers: 5′-GTTTGTGTGTGTGTGTATG-3′ and 5′-CATCCAACACCTCCACCTA-3′. The amplified PCR products were gel-purified with the GenEluteTmGel Extraction Kit (Sigma) and cloned into pGEM-T Easy vector (Promega, Madison, WI). At least 10 independent clones for each sample were randomly chosen for sequencing (MOBIX Lab, McMaster University). By comparing the bisulfite-converted sequence data against the unconverted DNA sequence, we quantified the number of methylation sites for the region of interest to determine the percentage of methylation.
The global methylation analysis was used with Imprint® Methylated DNA Quantification Kit (Sigma) according to the manufacturer's instructions.
Western blotting
All samples were mixed with loading buffer and subjected to 10% SDS–polyacrylamide gel electrophoresis. Each lane was loaded with 50–100 μg of equal amounts of protein. All samples were then transferred onto polyvinylidenedifluoride membranes and blocked with 5% milk in Tris-buffered saline–Tween 20 (TBST) buffer for 1 h at room temperature. The membranes were subsequently exposed to the first antibody against 3MST, CBS, Dnmt1, Dnmt3a, Dnmt3b, TFAM, IRF-1, or CSE (Abcam Inc, Toronto, ON) at the dilution of 1:1,000 in 5% milk in TBST overnight at 4°C; then, membranes were washed and incubated with the secondary antibody. Antibody binding was detected using enhanced chemiluminescence (BioRad) and exposed to X-ray film. The results were quantified by densitometry using Image J software.
ChIP assay
After treatment with NaHS, SMCs in 100-mm dishes were fixed directly by adding formaldehyde to culture medium for 10 min at 37°C to cross-link protein to DNA. The fixed cells were harvested and prepared for ChIP assay following the manufacturer's manual (Sigma) (48). The sonicated supernatant was incubated with an antibody against IRF-1 antibody or a nonspecific IgG antibody overnight at 4°C with rotation. A fraction of the protein-DNA was not precipitated but set aside for the total chromatin examination (termed input). The aimed sequence containing potential IRF-1 binding sites in the Dnmt3a promoter was amplified by PCR using primers 5′-AGCCCCAGCCTGCCGCCTACTG-3′ and 5′-TTGGGGTTCACTCCGCTTCTCCA-3′. A single 320 bp band was generated and electrophoresed on a 2% agarose, stained with ethidium bromide. The input was used as a positive control. The PCR bands produced were sequenced to confirm their identity. Quantitative analysis of IRF-1 and the Dnmt3a promoter interaction was determined by real-time PCR, and the binding intensity of IRF-1 with the Dnmt3a promoter was normalized to the level of input by using the same primers.
Biotin switch assay
S-sulfhydration assay was performed as previously described (27, 48). Briefly, cells were homogenized in HEN buffer (250 mM Hepes-NaOH (pH 7.7), 1 mM EDTA, and 0.1 mM neocuproine) supplemented with 100 μM deferoxamine and centrifuged at 13,000 g for 30 min at 4°C. Cell lysates were added to blocking buffer (HEN buffer with 2.5% SDS and 20 mM MMTS) at 50°C for 20 min with frequent vortexing. MMTS was then removed by acetone, and the proteins were precipitated at −20°C for 20 min. After acetone removal, the proteins were resuspended in HENS buffer (HEN buffer with 1% SDS). To the suspension, 4 mM biotin-HPDP in dimethyl sulfoxide was added without ascorbic acid. After incubation for 3 h at 25°C, biotinylated proteins were precipitated by streptavidin-agarose beads, which were then washed with HENS buffer. The biotinylated proteins were eluted by SDS-PAGE gel and subjected to Western blotting analysis using anti-IRF-1 antibody.
Detection of mitochondrial content
Mitochondria were labeled using the mitochondria-specific dye mitotracker green (Molecular Probes, Life Technologies Ltd., Burlington, ON) according to the manufacturer's protocol (33). Briefly, after incubation with NaHS (30 μM) for 48 h, SMCs were incubated with 100 nM mitotracker green for 30 min at 37°C. Cells were washed twice with phosphate-buffered saline after observation under an inverted Olympus IX70 fluorescent microscope (Tokyo, Japan). Fluorescence intensity was quantified with Image-J software 1.48.
Statistical analysis
All data are expressed as means±SE and represent at least three independent experiments. Statistical comparisons were made using Student's t-test or one-way ANOVA followed by a post hoc analysis (Tukey test) where applicable. p<0.05 was considered statistically significant.
Supplementary Material
Abbreviations Used
- 3MST
3-mercaptopyruvate sulfurtransferase
- Atp6
ATP synthase subunit 6
- AZA
5-aza-2′-deoxycytidine
- CBS
cystathionine beta-synthase
- ChIP
chromatin immunoprecipitation
- CS
citrate synthase
- CSE
cystathionine gamma-lyase
- CytB
cytochrome b
- Dnmt
DNA methyltransferase
- H2S
hydrogen sulfide
- IRF-1
interferon regulatory factor 1
- ISR
interferon stimulated response
- KO
knockout
- MSP
methylation-specific PCR
- MS-RE
methylation-sensitive restriction endonuclease
- MT-CO1
cytochrome C oxidase subunit 1
- mtDNA
mitochondrial DNA
- ND1
NADH dehydrogenase 1
- nDNA
nuclear DNA
- NO
nitric oxide
- SAM
S-adenosyl methionine
- siRNA
short interfering RNA
- SMCs
smooth muscle cell
- Tet
ten-eleven translocation methylcytosine dioxygenase
- TFAM
mitochondrial transcription factor A
- WT
wild type.
Acknowledgments
This study was supported by a grant-in-aid from the Heart and Stroke Foundation of Canada. G.Y. was supported by a New Investigator award from the Heart and Stroke Foundation of Canada.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Agrimi G, Di Noia MA, Marobbio CM, Fiermonte G, Lasorsa FM, and Palmieri F. Identification of the human mitochondrial S-adenosylmethionine transporter: bacterial expression, reconstitution, functional characterization and tissue distribution. Biochem J 379: 183–190, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso A, Martín P, Albarrán C, García P, García O, de Simón LF, García-Hirschfeld J, Sancho M, de la Rúa C, and Fernández-Piqueras J. Real-time PCR designs to estimate nuclear and mitochondrial DNA copy number in forensic and ancient DNA studies. Forensic Sci Int 139: 141–149, 2004 [DOI] [PubMed] [Google Scholar]
- 3.Altaany Z, Yang G, and Wang R. Crosstalk between hydrogen sulfide and nitric oxide in endothelial cells. J Cell Mol Med 17: 879–888, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bibli SI, Yang G, Zhou Z, Wang R, Topouzis S, and Papapetropoulos A. Role of cGMP in hydrogen sulfide signaling. Nitric Oxide 46: 7–13, 2015 [DOI] [PubMed] [Google Scholar]
- 5.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21, 2002 [DOI] [PubMed] [Google Scholar]
- 6.Byun HM, and Baccarelli AA. Environmental exposure and mitochondrial epigenetics: study design and analytical challenges. Hum Genet 133: 247–257, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell CT, Kolesar JE, and Kaufman BA. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim Biophys Acta 1819: 921–929, 2012 [DOI] [PubMed] [Google Scholar]
- 8.Chu J, Qian J, Zhuang Y, Zhang S, and Li Y. Progress in the research of S-adenosyl-L-methionine production. Appl Microbiol Biotechnol 97: 41–49, 2013 [DOI] [PubMed] [Google Scholar]
- 9.Clayton DA. Replication and transcription of vertebrate mitochondrial DNA. Annu Rev Cell Biol 7: 453–478, 1991 [DOI] [PubMed] [Google Scholar]
- 10.Dhliwayo N, Sarras MP, Jr, Luczkowski E, Mason SM, and Intine RV. Parp inhibition prevents ten-eleven translocase enzyme activation and hyperglycemia-induced DNA demethylation. Diabetes 63: 3069–3076, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding Y, Xia B, Yu J, Leng J, and Huang J. Mitochondrial DNA mutations and essential hypertension (Review). Int J Mol Med 32: 768–774, 2013 [DOI] [PubMed] [Google Scholar]
- 12.Eberhardt M, Dux M, Namer B, Miljkovic J, Cordasic N, Will C, Kichko TI, de la Roche J, Fischer M, Suárez SA, Bikiel D, Dorsch K, Leffler A, Babes A, Lampert A, Lennerz JK, Jacobi J, Martí MA, Doctorovich F, Högestätt ED, Zygmunt PM, Ivanovic-Burmazovic I, Messlinger K, Reeh P, and Filipovic MR. H2S and NO cooperatively regulate vascular tone by activating a neuroendocrine HNO-TRPA1-CGRP signalling pathway. Nat Commun 5: 4381, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu M, Zhang W, Wu L, Yang G, Li H, and Wang R. H2S metabolism in mitochondria and its regulatory role in energy production. Proc Natl Acad Sci U S A 109: 2943–2948, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gemma C, Sookoian S, Dieuzeide G, García SI, Gianotti TF, González CD, and Pirola CJ. Methylation of TFAM gene promoter in peripheral white blood cells is associated with insulin resistance in adolescents. Mol Genet Metab 100: 83–87, 2010 [DOI] [PubMed] [Google Scholar]
- 15.Hokari M, Kuroda S, Kinugawa S, Ide T, Tsutsui H, and Iwasaki Y. Overexpression of mitochondrial transcription factor A (TFAM) ameliorates delayed neuronal death due to transient forebrain ischemia in mice. Neuropathology 30: 401–407, 2010 [DOI] [PubMed] [Google Scholar]
- 16.Ikeuchi M, Matsusaka H, Kang D, Matsushima S, Ide T, Kubota T, Fujiwara T, Hamasaki N, Takeshita A, Sunagawa K, and Tsutsui H. Overexpression of mitochondrial transcription factor a ameliorates mitochondrial deficiencies and cardiac failure after myocardial infarction. Circulation 112: 683–690, 2005 [DOI] [PubMed] [Google Scholar]
- 17.Jiang J, Yang J, Wang Z, Wu G, and Liu F. TFAM is directly regulated by miR-23b in glioma. Oncol Rep 30: 2105–2110, 2013 [DOI] [PubMed] [Google Scholar]
- 18.Kang KA, Piao MJ, Kim KC, Kang HK, Chang WY, Park IC, Keum YS, Surh YJ, and Hyun JW. Epigenetic modification of Nrf2 in 5-fluorouracil-resistant colon cancer cells: involvement of TET-dependent DNA demethylation. Cell Death Dis 17: e1183, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, and Sasaki H. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet 16: 2272–2280, 2007 [DOI] [PubMed] [Google Scholar]
- 20.King AL, Polhemus DJ, Bhushan S, Otsuka H, Kondo K, Nicholson CK, Bradley JM, Islam KN, Calvert JW, Tao YX, Dugas TR, Kelley EE, Elrod JW, Huang PL, Wang R, and Lefer DJ. Hydrogen sulfide cytoprotective signaling is endothelial nitric oxide synthase-nitric oxide dependent. Proc Natl Acad Sci U S A 111: 3182–3187, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, and Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintance and embryogenesis in mice. Nat Genet 18: 231–236, 1998 [DOI] [PubMed] [Google Scholar]
- 22.LeBleu VS, O'Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM, Asara JM, and Kalluri R. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol 16: 992–1003, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li P, Demirci F, Mahalingam G, Demirci C, Nakano M, and Meyers BC. An integrated workflow for DNA methylation analysis. J Genet Genomics 40: 249–260, 2013 [DOI] [PubMed] [Google Scholar]
- 24.Lu B, Lee J, Nie X, Li M, Morozov YI, Venkatesh S, Bogenhagen DF, Temiakov D, and Suzuki CK. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol Cell 49: 121–132, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mani S, Li H, Austin RC, Dickhout JG, Lhoták S, Meng QH, Yang G, Wu L, and Wang R. Decreased endogenous production of hydrogen sulfide accelerates atherosclerosis. Circulation 127: 2523–2534, 2013 [DOI] [PubMed] [Google Scholar]
- 26.Mustafa AK, Sikka G, Gazi SK, Steppan J, Jung SM, Bhunia AK, Barodka VM, Gazi FK, Barrow RK, Wang R, Amzel LM, Berkowitz DE, and Snyder SH. Hydrogen sulfide as endothelium-derived hyperpolarizing factor sulfhydrates potassium channels. Circ Res 109: 1259–1268, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, and Snyder SH. H2S signals through protein S-sulfhydration. Sci Signal 2: ra72, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ngo HB, Kaiser JT, and Chan DC. The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat Struct Mol Biol 18: 1290–1296, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ngo HB, Lovely GA, Phillips R, and Chan DC. Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat Commun 5: 3077, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okano M, Xie S, and Li E. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res 26: 2536–2540, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pacaud R, Sery Q, Oliver L, Vallette FM, Tost J, and Cartron PF. DNMT3L interacts with transcription factors to target DNMT3L/DNMT3B to specific DNA sequences: role of the DNMT3L/DNMT3B/p65-NFκB complex in the (de-)methylation of TRAF1. Biochimie 104: 36–49, 2014 [DOI] [PubMed] [Google Scholar]
- 32.Pan H, Xie X, Chen D, Zhang J, Zhou Y, and Yang G. Protective and biogenesis effects of sodium hydrosulfide on brain mitochondria after cardiac arrest and resuscitation. Eur J Pharmacol 741: 74–82, 2014 [DOI] [PubMed] [Google Scholar]
- 33.Pohjoismäki JL, Wanrooij S, Hyvärinen AK, Goffart S, Holt IJ, Spelbrink JN, and Jacobs HT. Alterations to the expression level of mitochondrial transcription factor A, TFAM, modify the mode of mitochondrial DNA replication in cultured human cells. Nucleic Acids Res 34: 5815–5828, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sen U, Sathnur PB, Kundu S, Givvimani S, Coley DM, Mishra PK, Qipshidze N, Tyagi N, Metreveli N, and Tyagi SC. Increased endogenous H2S generation by CBS, CSE, and 3MST gene therapy improves ex vivo renovascular relaxation in hyperhomocysteinemia. Am J Physiol Cell Physiol 303: C41–C51, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sobenin IA, Zhelankin AV, Mitrofanov KY, Sinyov VV, Sazonova MA, Postnov AY, and Orekhov AN. Mutations of mitochondrial DNA in atherosclerosis and atherosclerosis-related diseases. Curr Pharm Des 21: 1158–1163, 2015 [DOI] [PubMed] [Google Scholar]
- 36.Song CX. and He C. Potential functional roles of DNA demethylation intermediates. Trends Biochem Sci 38: 480–484, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szabo C, Coletta C, Chao C, Módis K, Szczesny B, Papapetropoulos A, and Hellmich MR. Tumor-derived hydrogen sulfide, produced by cystathionine-β-synthase, stimulates bioenergetics, cell proliferation, and angiogenesis in colon cancer. Proc Natl Acad Sci U S A 110: 12474–12479, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang G, Yang G, Jiang B, Ju Y, Wu L, and Wang R. H2S is an endothelium-derived hyperpolarizing factor. Antioxid Redox Signal 19: 1634–1646, 2013 [DOI] [PubMed] [Google Scholar]
- 39.Tian M, Bao H, Martin FL, Zhang J, Liu L, Huang Q, and Shen H. Association of DNA methylation and mitochondrial DNA copy number with human semen quality. Biol Reprod 91: 101, 2014 [DOI] [PubMed] [Google Scholar]
- 40.Vandiver MS, Paul BD, Xu R, Karuppagounder S, Rao F, Snowman AM, Ko HS, Lee YI, Dawson VL, Dawson TM, Sen N, and Snyder SH. Sulfhydration mediates neuroprotective actions of parkin. Nat Commun 4: 1626, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker JA, Hedges DJ, Perodeau BP, Landry KE, Stoilova N, Laborde ME, Shewale J, Sinha SK, and Batzer MA. Multiplex polymerase chain reaction for simultaneous quantitation of human nuclear, mitochondrial, and male Y-chromosome DNA: application in human identification. Anal Biochem 337: 89–97, 2005 [DOI] [PubMed] [Google Scholar]
- 42.Wang R. Gasotransmitters: growing pains and joys. Trends Biochem Sci 39: 227–232, 2014 [DOI] [PubMed] [Google Scholar]
- 43.Wang R. Signaling pathways for the vascular effects of hydrogen sulfide. Curr Opin Nephrol Hypertens 20: 107–112, 2011 [DOI] [PubMed] [Google Scholar]
- 44.Wen SL, Zhang F, and Feng S. Decreased copy number of mitochondrial DNA: a potential diagnostic criterion for gastric cancer. Oncol Lett 6: 1098–1102, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang G, Cao K, Wu L, and Wang R. Cystathionine gamma-lyase overexpression inhibits cell proliferation via a H2S-dependent modulation of ERK1/2 phosphorylation and p21Cip/WAK-1. J Biol Chem 279: 49199–49205, 2004 [DOI] [PubMed] [Google Scholar]
- 46.Yang G, Li H, Tang G, Wu L, Zhao K, Cao Q, Xu C, and Wang R. Increased neointimal formation in cystathionine gamma-lyase deficient mice: role of hydrogen sulfide in α5β1-integrin and matrix metalloproteinase-2 expression in smooth muscle cells. J Mol Cell Cardiol 52: 677–688, 2012 [DOI] [PubMed] [Google Scholar]
- 47.Yang G, Wu L, Jiang B, Yang W, Qi J, Cao K, Meng Q, Mustafa AK, Mu W, Zhang S, Snyder SH, and Wang R. H2S as a physiologic vasorelaxant: hypertension in mice with deletion of cystathionine gamma-lyase. Science 322: 587–590, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang G, Zhao K, Ju Y, Mani S, Cao Q, Puukila S, Khaper N, Wu L, and Wang R. Hydrogen sulfide protects against cellular senescence via S-sulfhydration of Keap1 and activation of Nrf2. Antioxid Redox Signal 18: 1906–1919, 2013 [DOI] [PubMed] [Google Scholar]
- 49.Zhang F, Qi Y, Zhou K, Zhang G, Linask K, and Xu H. The cAMP phosphodiesterase Prune localizes to the mitochondrial matrix and promotes mtDNA replication by stabilizing TFAM. EMBO Rep 2015. [Epub ahead of print]; DOI: 10.15252/embr.201439636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L, Cardinal JS, Bahar R, Evankovich J, Huang H, Nace G, Billiar TR, Rosengart MR, Pan P, and Tsung A. Interferon regulatory factor-1 regulates the autophagic response in LPS-stimulated macrophages through nitric oxide. Mol Med 18: 201–208, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao K, Ju Y, Li S, Altaany Z, Wang R, and Yang G. S-sulfhydration of MEK1 leads to PARP-1 activation and DNA damage repair. EMBO Rep 15: 792–800, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.