Abstract
R-loops, nucleic acid structures consisting of an RNA-DNA hybrid and displaced single-stranded DNA, are ubiquitous in organisms from bacteria to mammals. First described in bacteria where they initiate DNA replication, it now appears that R-loops regulate diverse cellular processes such as gene expression, immunoglobulin class switching and DNA repair. Changes in R-loop regulation induce DNA damage and genome instability, and recently it was shown that R-loops are associated with neurodegenerative disorders. Here, we discuss recent developments in the field; in particular, the regulation and effects of R-loops in cells, their effect on genomic and epigenomic stability and their potential contribution to the origin of certain diseases, including cancer and neurodegenerative disorders.
Keywords: R-loop, gene expression, genomic instability, epigenomic stability
R-loops: guardian and threat to genome stability
R-loops are three-stranded nucleic acid structures, composed of an RNA-DNA hybrid and the displaced single-stranded DNA [1]. They are formed as the nascent transcript emerges from the transcription machinery and hybridizes with the complementary DNA template. Since their original identification in bacteria [2], R-loops have been found in many organisms, from yeast to humans, and genome-wide mapping of R-loops in yeast or mammalians cells showed that R-loops can arise throughout the genome under normal conditions [3–5]. R-loops play a multitude of physiological roles in cells, regulating gene expression, DNA replication, immunoglobulin class-switch recombination (see Glossary), and DNA repair [6–8]. R-loops are also formed in the CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) system, which provides immunity for bacteria and archaea, and became popular when adapted as a genome editing tool [9,10]. However, R-loops can also induce DNA damage and genome instability [11–18], and recently, they have been associated with several neurodegenerative diseases [19–22]. On the surface, it seems puzzling that the same structure has both beneficial and detrimental effects in cells. In this review, we discuss recent advances in our understanding of the physiological roles of R-loops in cells, particularly their role in regulating gene expression and DNA repair [5,8,23,24]. We also describe the recent progress made in understanding the molecular mechanisms that underlie R-loop-induced DNA damage and genome instability. Finally, we review findings linking R-loops to epigenetic marks and to the development of diseases such as cancer and neurodegenerative disorders.
Active regulation of cellular phenotypes by R-loops
R-loops arise naturally in organisms from bacteria to mammals, and extensive research has established that they have a multitude of specialized effects in cells. R-loops play a role in bacterial DNA replication, as well as mitochondrial DNA replication in humans, and they are also involved in immunoglobulin class-switch recombination (CSR) in human B-cells [6]. These effects are the subject of extensive reviews, and are therefore not discussed in depth here [1,6,7,25].
Recent experiments suggest that R-loops can also dynamically affect fundamental cellular functions such as the regulation of gene expression or DNA repair. R-loops form preferentially both at the promoters and terminators of genes [5,23,24,26], and regulate gene expression through multiple mechanisms [5,23,24,27]. In particular, they form at CpG-island containing promoters characterized by an asymmetric distribution of guanines and cytosines (GC-skew) [5,26]. This skew is such that C-rich sequences are on the template strand and G-rich sequences are on the non-template strand, an arrangement that suggests stable secondary structures known as G4 quadruplexes may coincide with R-loops. Consistent with this idea, transcription of plasmids with a G-rich non-template strand leads to formation of R-loops with G4 DNA on the G-rich strand and an RNA-DNA hybrid on the template strand [28,29]. When generated on an episomal system, R-loops protect their underlying template from de novo DNA methylation, an epigenetic mark associated with transcriptional silencing. This finding suggests that R-loops might positively regulate gene expression [5]. By contrast, the formation of R-loops at the termination regions of certain genes allows the recruitment of the RNA/DNA helicase Senataxin (SETX), which subsequently resolves the R-loop to promote transcriptional termination [23,30]. Thus, R-loops may both promote and terminate gene expression.
R-loops are also involved in the silencing of long noncoding RNAs (lnRNAs). In Arabidopsis, R-loops inhibit the expression of the lncRNA COOLAIR, an antisense transcript needed to silence the floral repressor gene FLC [27], and in human cells, R-loops inhibit the expression of the Ube3a antisense transcript, which is associated with Prader-Willi syndrome [31]. These two findings raise the question of whether the transcriptional repression of lncRNAs by R-loops occurs at other loci, and as such is a general mechanism for regulating gene expression.
Moreover, recent studies suggest that R-loops, or more specifically RNA-DNA hybrids, might be involved in DNA repair [8]. In yeast cells lacking a homologous template for repair, a transcript RNA can be used as a template for error-free double-strand break repair. It appears that the transcript can hybridize with its complementary DNA (in cis) to make a “broken” R-loop-like structure. This finding raises some important mechanistic questions. For example, could RNA-mediated repair be a mechanism for error-free DNA repair in cells that are in G1 or not replicating and thus lack the sister chromatid required for homologous recombination (HR)? Is this an alternative to HR in S/G2 cells that involves an RNA molecule?
R-loops are clearly emerging as nucleic-acid structures that can modulate complex cellular phenotypes (Figure 1). Understanding their regulation might therefore have a profound impact on our understanding of cellular biology.
Figure 1. Physiological roles of R-loops in cells.
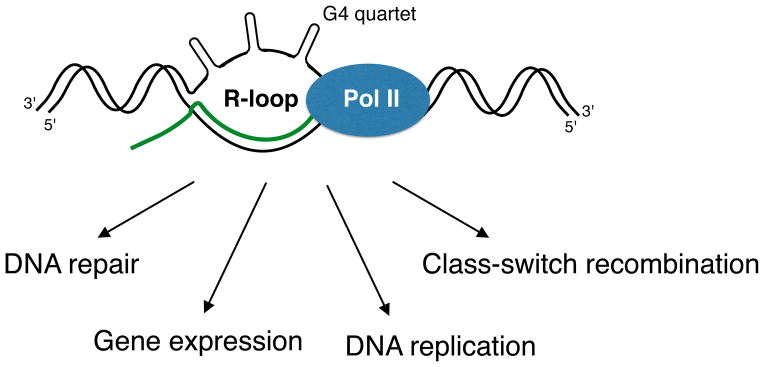
R-loops are involved in bacterial and mitochondrial DNA replication, class-switch recombination, gene expression, and DNA repair. G4 quadruplexes are proposed to form on the non-template strand.
R-loop formation and resolution within cells
R-loops are thought to form co-transcriptionally when nascent messenger RNA hybridizes with the DNA template (in cis), and because their formation is induced by the loss of many splicing factors it is possible that at least some form from unspliced messages (reviewed in [1,6]). Experiments in yeast suggest that R-loops can also form when transcripts hybridize to complementary or partially complementary sequences distant from their original site of transcription (in trans) [32]. Although R-loops are constantly produced in cells, surprisingly little is known about the factors that assist in R-loop formation. Recently, Rad51, a protein that promotes strand exchange between homologous sequences during HR, and Rad52, a protein with strand annealing properties and which can stimulates Rad51’s activity [33,34], were shown to favor hybridization of the RNA molecule to the template DNA during trans R-loop formation in yeast [8]. Moreover, bacterial RecA, a homolog of the human Rad51 protein, and human Rad52, can promote the annealing of single-stranded RNA to duplex DNA in vitro [8,35]. The presence of factors that promote R-loop formation in trans also makes it tempting to speculate that some lncRNAs or other regulatory RNAs could act through R-loop formation. As R-loops can induce DNA damage, this finding could also indicate that R-loop-induced DNA damage can occur both in cis and in trans. Trans R-loop formation is also interesting when considered in the context of RNA-directed repair, as loss of heterozygosity (LOH) could arise if a transcript from one allele is used to promote the repair of the other allele.
By contrast, there are a number of mechanisms known to resolve R-loops or prevent their formation [1]. R-loops can be resolved by RNase H, which specifically degrades the RNA moiety in RNA-DNA hybrids [16]. They can also be resolved by SETX or Aquarius (AQR) [18,23,30], which belong to a subfamily of proteins containing a conserved DEAxQ-like domain with putative RNA/DNA helicase activity [36]. Although little is known about the function of this protein family, a yeast mutant in the helicase domain of Sen1, the yeast homolog of SETX, accumulates R-loops [30]. In mammals, SETX has been suggested to prevent the accumulation of R-loops at gene terminators [23], and at least at the β-actin locus an increase in RNA-DNA hybrids was detected when SETX was depleted [37]. It was recently shown that R-loops accumulate in the nucleus of cells depleted of the putative RNA/DNA helicase Aquarius (AQR) as well, although where they accumulate in the genome is not known [18]. It has also been suggested that R-loop formation is suppressed by topoisomerase I, which resolves the negative torsional stress behind RNA polymerase II to prevent annealing of the nascent RNA with the DNA template [13,38]. This has been directly measured in yeast where R-loops accumulate at the rDNA locus in the absence of topoisomerase I [39]. Interestingly, histone 3 arginine 3 dimethylation (H3R3) prevents the formation of R-loops at the C-MYC locus by recruiting topoisomerase IIIB, which also relaxes torsional stress behind the RNA polymerase [40]. Moreover, RNA processing and RNA export factors preclude the formation of R-loops, presumably by binding to RNA as it emerges from RNA polymerase [11,41]. R-loop formation and resolution are thus highly regulated processes, and may have important effects on cellular fitness. Furthermore, the diversity of factors regulating the formation and resolution of R-loops suggests that different types of R-loops exist in the genome that may be regulated by different factors.
Multiple pathways connect R-loops to DNA damage
Interestingly, defects in factors that resolve R-loops or prevent their formation can lead to DNA damage and genome instability. The first experimental evidence for this came from yeast mutants in THO/TREX, a complex involved in transcription and RNA export, which exhibit a hyper-recombination phenotype. This phenotype can be suppressed by overexpression of RNase H1, an observation that implicates R-loops as the source of DNA damage [11]. Critical studies in yeast and human cells confirmed that R-loops induced by knocking-out factors involved in mRNA processing are a source of DNA damage [12,14–16]. Genome-wide screens identified numerous transcription, splicing or mRNA processing factors that suppress DNA damage accumulation or genome instability in an RNAse H-dependent manner [14–16]. Factors shown to prevent R-loop-induced double-strand break (DSB) formation in human cells include ASF, AQR, SETX, BRCA1, BRCA2 and topoisomerase I [12,13,17,18,37,38], suggesting that there are multiple connections between R-loops and DNA damage.
Although there are strong links between R-loops and DNA damage, one outstanding question is where in the genome this damage is formed. It is presumed that the damage occurs at the site of R-loop formation but evidence for this is limited to a few studies [11,12]. Another outstanding question concerns the mechanism by which this DNA damage arises. Diverse studies suggest that many structural elements make R-loops vulnerable to nucleases and other enzymes, and that R-loops may represent a physical barrier to other events taking place on chromatin such as replication. Below, we discuss three possible mechanisms linking R-loops to DNA damage.
The AID/APOBEC enzymes target single-stranded DNA in the R-loop
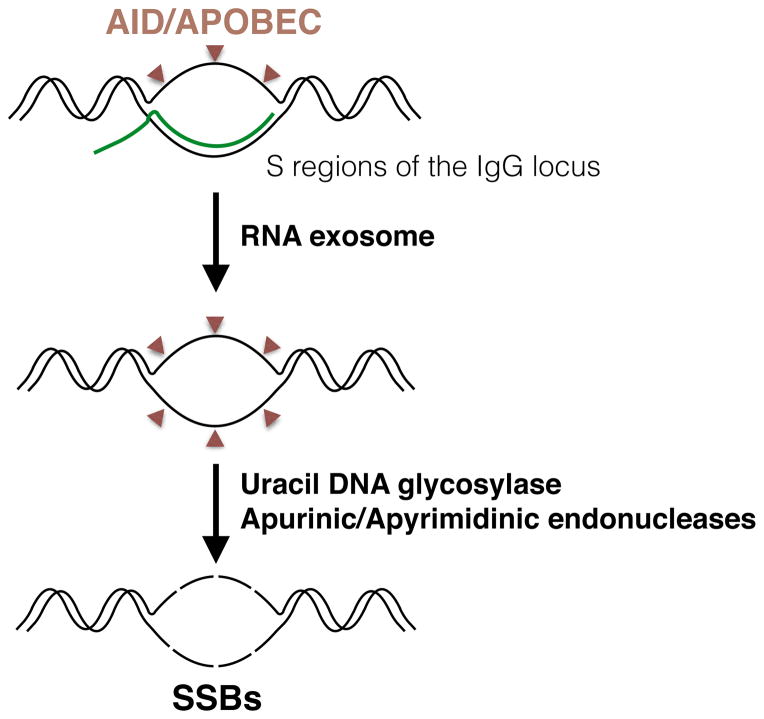
The single-stranded DNA (ssDNA) that is exposed during R-loop formation is one element of this structure that is particularly vulnerable to DNA damage. Specific nucleases target ssDNA, which in R-loops can extend up to several hundred base pairs [5,42]. One candidate is activation-induced cytidine deaminase (AID), which promotes the conversion of dC to dU residues specifically on ssDNA (Figure 2). This modification makes the DNA susceptible to the base excision repair enzyme uracil DNA glycosylase, which in turn excises the uracil base to create an abasic site and generates a DNA lesion. AID acts in this way at the immunoglobulin locus in B-cells where it targets both the non-template and template strands of R-loops formed over the S regions [43]. This mechanism might also underlie DNA damage at other R-loops in cells, because AID can initiate DSBs in a small subset of non-Ig genes [44]. Interestingly, analysis of somatic mutations in cancer identified a mutational signature associated with the APOBEC family that correlates with many cancers [45]. As a result, it has been proposed that members of the APOBEC family, of which AID is a member, might fulfill a similar function in cells that do not express AID [46]. It will be important to determine whether there is a correlation between these APOBEC mutational signatures and the presence of R-loops.
Figure 2. Formation of R-loop-mediated single-strand breaks.
AID/APOBEC enzymes can deaminate cytosine residues on the non-template DNA, but also on the template DNA once the RNA molecule is displaced by the RNA exosome. The base excision repair enzyme uracil DNA glycosylase then excises the uracil base to create an abasic site, which can be processed by apurinic-apyrimidinic endonucleases to create a single-strand break (SSB).
Transcription-coupled nucleotide excision repair factors process R-loops
Other structural elements that might be susceptible to DNA damage are the flaps that form at either end of the R-loop. Recent work indicates that these structures might be cleaved by structure-specific endonucleases within the cell [18]. In human cells, R-loops induced by the absence of diverse RNA processing factors, or by the inhibition of topoisomerase I, are actively processed into DSBs by XPF and XPG, two flap endonucleases involved in nucleotide excision repair (NER). Importantly, DSB formation also requires factors specifically involved in repair of the transcribed strand, as opposed to those involved in global genomic repair [18]. These findings suggest that the transcription-coupled nucleotide excision repair pathway (TC-NER) drives R-loop-induced DSBs and genome instability. Whether the R-loop is mistaken for DNA damage and thus aberrantly processed, or whether the processing is actually a productive event, is not yet clear.
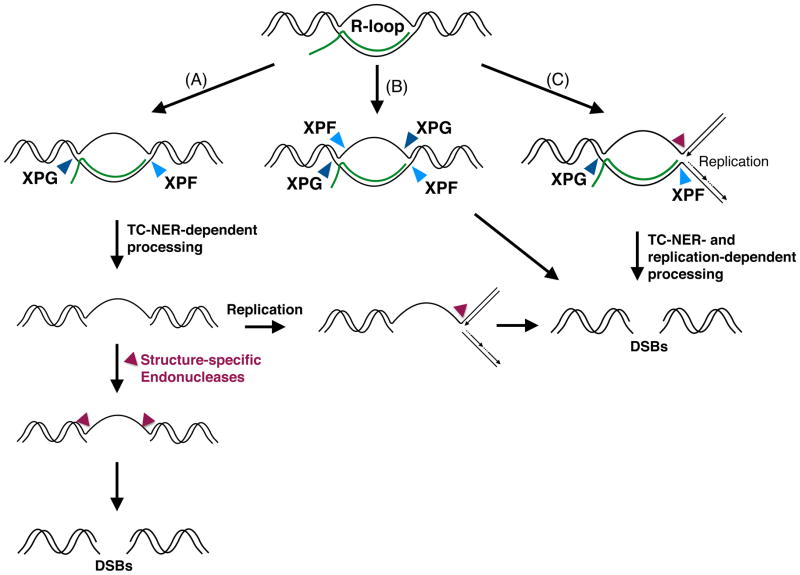
It is also unclear how the TC-NER machinery recognizes and processes an R-loop into a DSB, although at least two non-exclusive hypotheses can be proposed (Figure 3). Because XPF and XPG are flap endonucleases, they may recognize both strands of an R-loop and directly generate a DSB. Indeed in vitro studies have shown that synthetic R-loops can be cleaved on both strands by purified XPF and XPG [47]. Another possibility is that the RNA-DNA hybrid is cleaved by XPF and XPG in a manner similar to what occurs during classical NER, generating a single-stranded gap that is converted into a DSB by the action of another nuclease or upon encounter with a DNA replication fork.
Figure 3. Model for the processing of R-loops into DSBs.
(A) The endonucleases XPF and XPG can excise the RNA-DNA hybrid of an R-loop. The single-stranded gap may be converted into a DSB either by the action of structure-specific endonucleases, or by encounter with the replication fork, which can also involve structure-specific endonucleases. (B) The endonucleases XPF and XPG can recognize both strands of an R-loop and generate a DSB. (C) The TC-NER machinery may be recruited to R-loops during replication, and the concerted action of the endonucleases XPF and XPG with other structure-specific endonucleases may generate a DSB.
The collision between an R-loop and the replication machinery can lead to DSBs
A final possibility is that DSBs are generated through a physical collision between an R-loop and the replication machinery (Figure 3) [13,16,48–50]. Several lines of evidence suggest that inhibiting DNA replication prevents R-loop-induced DSB formation. First, the transcription-dependent hyper-recombination phenotype observed in the yeast THO mutant, hpr1Δ, requires transcription during S-phase [49]. Second, in human cells where ASF or TOP1 has been depleted, R-loop-induced DSBs depend on replication, and direct effects on replication fork progression can be observed [13,48]. R-loops may represent a formidable physical barrier to the replication fork, as they are relatively stable DNA secondary structures that are associated with the transcriptional machinery and other RNA-processing enzymes. Thus, R-loops might directly cause fork collapse and DSB formation, even without intervening processing by nucleases like XPF and XPG. Interestingly, the DSBs induced by ASF knockdown are suppressed by XPG knockdown [18]. As these breaks are also dependent upon S-phase, a final intriguing possibility is that the TC-NER machinery is recruited to the R-loop when the RNA polymerase collides with the replication machinery, and thus, the TC-NER pathway for R-loop processing may be directly coupled to replication.
Further work is required to understand the respective contributions of the replication fork, AID, and TC-NER factors to R-loop-induced DSB formation and genome instability. Current observations also raise the possibility that R-loops can promote genome instability throughout all of the phases of the cell cycle, since TC-NER factors and AID could create DSBs at R-loops in non-dividing cells. This is intriguing because such processing may underlie the links between R-loops and neuronal diseases [51,52]. Thus, deciphering when each of these processes act and whether some act in concert will provide a better understanding of the mechanisms underlying R-loop-induced genome instability and further our understanding of their potential contributions to human disease.
R-loops can affect genome stability
Since R-loops are a source of DSBs, they might trigger chromosomal translocations, a hallmark of numerous cancers [53], and a number of laboratories are studying the relationship between R-loops, DSBs and cancer. R-loops were found to form at a subset of common fragile sites (CFSs), specific regions in the human genome that are difficult to replicate during S phase. CFSs are prone to form DSBs, and they are frequently rearranged in cancer cells [54]. Therefore, under conditions of replication stress, collision of the replication machinery with the transcription apparatus may lead to the stabilization of R-loops, inducing the formation of DSBs and chromosomal translocations. Interestingly, ERCC1, the binding partner of XPF, as well as the endonuclease MUS81, promote sister chromatid separation during mitosis by processing the late replication intermediates formed at CFSs in the presence of aphidicolin. The processing of these late replication intermediates prevents defects in chromosome segregation, mitotic catastrophe and cell death [55]. Because aphidicolin induces both CFS and R-loops at the FRA3B region and the aphidicolin-induced instability of the FRA3B region is prevented by RNase H expression, it will be interesting to test whether the R-loops found at this site and possibly other CFS are processed by ERCC1 and MUS81.
Other regions of the genome where rearrangements have been observed in cancer cells also form R-loops. For example, R-loops are found both at the c-MYC locus [28] and the S-regions of the immunoglobulin loci [42], and it is known that translocations of c-MYC to the immunoglobulin switch regions are typical of sporadic Burkitt’s lymphomas [56] and multiple myelomas [57]. Supporting the idea that the R-loops play a causative role in these translocations, defects in H3R3 dimethylation, which drive R-loop formation by preventing topoisomerase IIIB recruitment, increase the frequency of translocations between the C-MYC and immunoglobulin loci [40]. It will be interesting to determine if other translocation sites characteristic of some cancers are also found in genomic locations where R-loops form.
Furthermore, genes involved in mRNA processing and export are often mutated in certain cancers. The FIP1L1-PDGFRA fusion gene has been identified in patients with eosinophilic leukemia [58]. FIP1L1 is a splicing factor, and mutations in its yeast ortholog lead to accumulation of R-loops. Moreover, FIP1L1-depleted human cells show an increase in DSBs and chromosome breaks [15]. Other mutations in splicing machinery and RNA export components including SF3B1, U2AF1, SRSF2 or ZRSR2 are associated with myelodysplastic syndromes (MDS) and chronic lymphocytic leukemia (CLL) [59–61]. It is still unclear whether mutations in these splicing factors simply affect the expression of essential genes; or whether defects in these factors have a more direct effect on cancer progression. Considering the role of the splicing machinery in preventing R-loop-induced DNA damage, an exciting possibility is that mutations in these splicing factors lead to increased formation of R-loops, DSBs, and genomic instability. Such a link could lead to a greater interest in the poorly understood relationship between splicing and genome stability.
Another recent study showed that R-loops accumulate at certain loci in BRCA1- and BRCA2-depleted cells, again connecting R-loops to genome maintenance pathways [17]. BRCA1 and BRCA2 are human tumor suppressor genes, and mutations in BRCA1 or BRCA2 genes increase the risk for breast cancer [62]. One of the well-characterized functions of these genes is to repair damaged DNA through HR [34], but depletion of another HR factor, Rad51, does not lead to R-loop accumulation. This finding suggests that the function of BRCA2 in preventing the accumulation of R-loops may be independent of its function in HR. It is possible that these tumor suppressor genes might help maintain genome integrity in two ways: by preventing changes in R-loops dynamics, as well as by promoting HR. Indeed, BRCA2 interacts with mRNA export factors from the TREX2 complex [17]. This interaction may help to recruit BRCA2 to co-transcriptional R-loops, where it could limit R-loop formation or restart replication forks that stall at R-loops. Further expanding the role of BRCA1 in R-loop biology, another study has shown that BRCA1 recruits SETX to the termination region of the β-actin gene where R-loops are formed, and it is suggested that BRCA1 and SETX suppress R-loop-induced DNA damage at this site [37]. Understanding the mechanism by which BRCA1 and BRCA2 dysfunction affects R-loop accumulation will further clarify the contribution of R-loops to breast cancer.
In conclusion, these data support the idea that R-loops may drive chromosomal translocations early in tumorigenesis, as well as contribute to the replication stress found in many cancer cells. If R-loops are indeed a hallmark of some cancers, it will be important to determine whether these will be useful biomarkers and whether they might be targeted to treat
Regulatory R-loops escape DNA damage
As discussed above, defects in mRNA processing factors, topoisomerase I, or RNA/DNA helicases can lead to DSBs and genome instability in an RNase H-reversible manner, suggesting R-loops are the source of damage. However, R-loops that are involved in bacterial and mitochondrial replication, gene expression, or repair do not normally lead to DSBs. Why certain R-loops are processed into DSBs while others are somehow protected remains an intriguing but unanswered question. It is unknown if regulatory R-loops and R-loops leading to DSBs are different in size, number, location, or their persistence, although several hypotheses can be proposed to explain their different fates.
In the absence of some factors resolving R-loops or preventing their formation, it has been shown that the amount of R-loops increases, either at specific loci [12,17,37] or globally as detected by immunofluorescence in fixed cells [16–18]. Thus, one possibility is that the excess of R-loops observed in these cells saturates the pathways that normally act to resolve these structures or prevent their formation, thereby allowing their aberrant processing by nucleases.
Another possibility is that the induced R-loops exist in different chromosomal contexts that may affect their processing. Early replicating fragile sites that break spontaneously during replication have been identified in human cells. These fragile sites are located in actively transcribed gene clusters and are enriched in CpG dinucleotides, where R-loops form [63]. It is therefore conceivable that R-loop fragility may be linked to their proximity to a replication origin. Data in yeast have also shown that R-loops arise at new genomic loci in mutants of RNase H or Senataxin [3,4]. It will be interesting to determine if the genome instability and DNA damage associated with these mutants results from the formation of new R-loops in proximity to replication origins.
Lastly, the persistence of R-loops that are processed into DSBs may differ from those of regulatory R-loops. Both the formation and resolution of R-loops is needed for efficient transcriptional termination, indicating these structures can be quickly turned over [23]. Thus, R-loops that are processed into DSBs may be distinguished by a longer lifetime, and consequently, a higher probability of being recognized by nucleases or hit by a replication fork.
R-loops are associated with epigenetic marks
R-loops regulate transcription through epigenetic changes
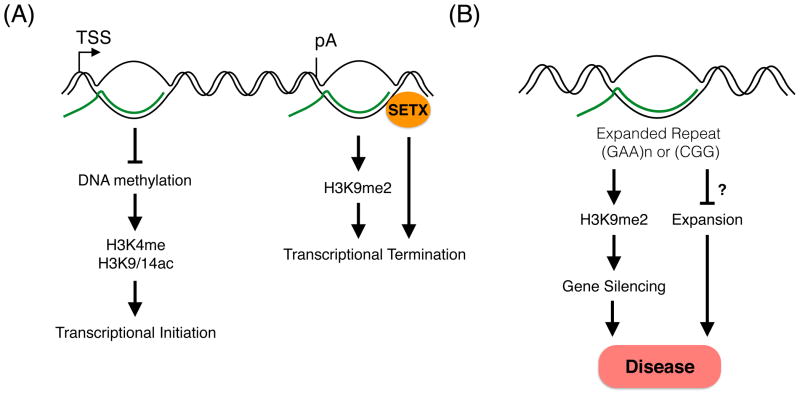
R-loops regulate numerous aspects of gene expression, from transcriptional initiation to termination and the regulation of lncRNAs [5,23,24,27], and this regulation may be intimately tied to their effects on chromatin structure and the epigenome. Strikingly, R-loops involved in gene expression appear to regulate the epigenetic marks found at promoter and termination regions. For instance, efficient transcriptional termination is mediated by R-loop-dependent heterochromatin formation (Figure 4). R-loops formed at the 3′ end of genes induce antisense transcription and the generation of double-stranded RNA (dsRNA), which then recruits the RNAi machinery. This recruitment leads to histone H3 K9 dimethylation (H3K9me2) and the formation of heterochromatin, which is proposed to pause RNA polymerase II and consequently promote efficient termination [24]. This finding is intriguing in light of others studies that have shown the 3′ end of genes are regions of open chromatin [64]. One possible explanation is that this heterochromatic state is transient and only required during the termination process.
Figure 4. R-loops are associated with chromatin marks.
(A) R-loops influence both transcriptional initiation and termination. R-loops located at promoters prevent DNA methylation, which leads to the establishment of active chromatin marks and transcriptional initiation. By contrast, R-loops located at termination regions induce the formation of heterochromatin promoting transcriptional termination. In addition, the RNA/DNA helicase Senataxin resolve R-loops at termination regions to promote transcriptional termination. It is still unknown if these two mechanisms are interconnected. (B) R-loops can form at genes with trinucleotide repeats and induce the formation of chromatin marks that silence the gene of interest. R-loops formed at these genes might also prevent further trinucleotide expansions. Gene silencing and expansion can lead to disease.
R-loops found at GC-enriched promoters provide another interesting connection to chromatin. These R-loops may prevent DNA methylation [5], although how this might occur remains obscure. Recent work showed that the endonucleases XPF and XPG are enriched at the promoters of certain genes, where they induce DNA breaks, DNA demethylation, and the establishment of active chromatin marks required for gene expression [65]. Therefore, R-loops formed at promoters could be processed by XPF and XPG, leading to DNA demethylation and consequently gene expression (Figure 4). If true, the processing of R-loops by NER factors might not be detrimental to the cell, but it might promote transcription in unperturbed cells and thus be beneficial.
As R-loops are emerging as regulators of gene expression and epigenetic modifications, one can speculate about other scenarios in which they might act. Stem cell differentiation is driven by changes in epigenetic marks that lead to the establishment of specific gene transcription programs that drive cell fate [66]. Thus, one exciting possibility is that changes in R-loops dynamics drive stem cell differentiation through their effects on epigenetic modifications.
Changes in R-loop formation and histone modifications are associated with neurodegenerative diseases and cancer
While R-loop-induced epigenetic changes might be critical to regulating gene expression, there are others scenarios in which R-loop-induced epigenetic changes may be detrimental for the cell and might lead to disease. Recently, it was shown that R-loops are formed over both the GAA and CGG expanded alleles of the FMR1 and FXN genes, respectively associated with Friedreich ataxia (FRDA) and Fragile X syndrome (FXS) (Figure 4) [21,22,67]. These stable R-loops colocalize with the repressive chromatin mark H3K9me2, which causes the silencing of the FMR1 and FXN genes in patient’s cells. Thus, R-loops, by triggering the silencing of critical genes through the establishment of repressive chromatin marks, might contribute to neurodegeneration. It is also possible that R-loops, by inhibiting the transcriptional activity of these genes, abrogate the expansion of trinucleotide repeats located in the non-coding regions of the FMR1 and FXN genes. In this scenario, the silencing of the FMR1 and FXN genes could protect the cell by preventing the unrestrained expansion of trinucleotide repeats at these loci, an event that might have lethal consequences.
R-loops are also linked to histone H3 S10 phosphorylation (H3S10P), a mark of chromatin condensation. Accumulation of H3S10P is observed in S. cerevisiae, C. elegans and human cells in which mRNA processing factors are lost. Moreover, deregulation of H3S10P, either by increasing or decreasing H3S10 phosphorylation, results in genome instability [68]. Although the molecular mechanism behind these observations is unclear, it is possible that condensed chromatin regions not only trigger the silencing of essential genes, but also impede DNA replication and/or transcription, leading ultimately to genome instability and cancer.
The relationship between R-loops and several different histone modifications raises some important questions. For instance, do these histone modifications and R-loops coexist in the same molecule? It seems unlikely that the RNA-DNA hybrid of an R-loop could wrap around a nucleosome given the properties of A- vs B-form DNA [69]. Moreover, R-loops are more likely to exist in actively transcribed genes, which are in decondensed and accessible chromatin. This would be incompatible with the formation of condensed chromatin. A more likely possibility is that the formation of the R-loop precedes the establishment of chromatin marks. R-loops might attract specific factors that allow the modification of histones in the restored B-DNA form once the R-loop is resolved; however, the histone modifications (for example H3S10P) could also be located in the chromatin adjacent to the R-loop. The application of single-molecule approaches, such as electron microscopy, to R-loop biology could provide some clarity to these questions. As we are just starting to understand the factors that regulate R-loop formation and their functions, deciphering their effects on chromatin promises to be a rich area for further work that is likely to impact our understanding of many cellular processes and diseases.
Concluding remarks
Our knowledge about R-loops has exploded over the last few years, raising new, unanswered questions. One of the challenges is to understand how R-loop processing can be both beneficial and deleterious for the cell. Although R-loops are important for regulating diverse cellular events, they can also induce DNA damage and genome instability. The processing of R-loops by TC-NER factors might remove R-loops that impede transcription, allowing the cell to restore gene expression. Alternatively, TC-NER factors may simply mistake R-loops for lesions, consequently processing them into DSBs.
Furthermore, it is unclear what distinguishes the beneficial R-loops from the deleterious ones. Genome-wide mapping of R-loops and DSBs in mutants with changes in R-loop processing and turnover might provide some answers. If R-loops located at promoters or termination regions, and R-loops induced by defects in mRNA processing, both co-localize with DSBs, it suggests that the location of R-loops is not a factor involved in their processing, and that saturation of the clearance mechanisms is more likely responsible for DSB formation.
Another exciting area to explore is the contribution of R-loops to cancer. Are R-loops promoting genome and epigenomic instability in cancer through DSB formation and/or modification of gene expression? R-loops are known to induce DSB formation and chromosomal translocations [11,54], but R-loops are also an obstacle to the transcription machinery, and thus, can alter the expression of essential genes, such as proto-oncogenes, tumor suppressor genes or DNA repair genes for example. Their effects on epigenetic marks may also affect transcription. It will be interesting to sequence the transcriptome as well as the epigenome of mutants affected in R-loop formation. R-loops are also a barrier to the replication machinery, and may be a significant source of the replication stress found in many cancer cells. Because R-loops have physiological roles throughout the genome, it will be important to determine how cells tolerate these structures during transcription and replication, and why under some conditions they lead to replication stress.
Finally, considering the link between R-loops and neurodegenerative diseases, a long-term goal for the field should be to target R-loops therapeutically. Powell and coworkers have made a first step in this direction. They observe that silencing of the paternal allele of the UBE3A gene by expressing an antisense transcript leads to the development of the rare genetic disorder, Prader-Willi syndrome. By stabilizing R-loops with topotecan, an inhibitor of topoisomerase I, the transcription of the antisense transcript is inhibited, and consequently the expression of the UBE3A gene is restored [31]. A better understanding of the molecular connection between R-loops and certain diseases, such as Friedreich ataxia and Fragile X syndrome, may ultimately allow the development of therapeutic approaches to specifically target harmful R-loops that are associated with these diseases.
R-loops have a multitude of roles in the cell and affect the genome and epigenome, both in positive and negative ways. A better molecular understanding of the mechanisms governing the formation, resolution and processing of R-loops, as well as their downstream effects will illuminate our understanding of fundamental cellular processes such as transcription and replication. It will also promise to clarify the molecular origin of diseases such as cancer and neurodegeneration.
Highlights.
R-loops are dynamic structures that regulate diverse cellular processes
R-loops threaten genomic and epigenomic stability
R-loops are associated with cancer and neurodegenerative diseases
Acknowledgments
We would like to thank all the scientists whose results we discussed in this review, and we apologize to those whose work we could not cite due to space limitations. We would also like to thank members of the Cimprich laboratory, as well as Dr. William Stork, for thoughtful discussion and helpful comments on the manuscript. This work was supported by awards from the NIH (GM100489) and the Komen Foundation (IIR 12222368).
Glossary
- AID
Activation-induced cytosine deaminase is an enzyme that deaminates cytosine bases, turning them into uracil. AID is involved in immunoglobulin diversification processes such as somatic hypermutation and class switch recombination
- APOBEC
Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like, is a family of proteins that deaminate cytosine bases into uracil, and are involved in adaptive and innate immunity. AID is a member of the APOBEC family
- Common Fragile Site
chromosomal region prone to breakage upon replication stress, and a hot spot for chromosomal rearrangements
- Class-Switch Recombination
biological process during which B cells switch from making one class of antibody to making another. The antibody retains its affinity for an antigen, but can interact with different effector molecules. This process is initiated by the AID enzyme, and involves non-homologous end joining of DNA at switch regions located in vicinity from gene segments that encode the constant regions of antibody heavy chains
- Epigenetics
the study of heritable DNA and histone modifications that affect a gene’s expression without a change in its coding sequence
- Homologous Recombination
mechanism of DNA repair that involves recombination between identical or nearby identical sequences
- Nucleotide Excision Repair
mechanism that repairs bulky lesions in the genome arising from a multitude of DNA damaging agents. There are two types of NER: GGR (Global Genome Repair) detects DNA lesions throughout the genome, while TC-NER (Transcription-coupled Nucleotide Excision Repair) is functional on the transcribed strand of expressed genes and activated when the RNA polymerase encounters a DNA lesion on the transcribed strand
- Oncogenesis
process by which normal cells become cancer cells
- Proto-oncogene
a normal gene that encodes proteins regulating cell growth and differentiation and a precursor to an oncogene. Conversion from the proto-oncogene to the oncogene can occur as a result of mutation or increased expression
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hamperl S, Cimprich KA. The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair. 2014;19:84–94. doi: 10.1016/j.dnarep.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drolet M, et al. Hypernegative supercoiling of the DNA template during transcription elongation in vitro. J Biol Chem. 1994;269:2068–2074. [PubMed] [Google Scholar]
- 3.Chan YA, et al. Genome-Wide Profiling of Yeast DNA:RNA Hybrid Prone Sites with DRIP-Chip. PLoS Genet. 2014;10:e1004288. doi: 10.1371/journal.pgen.1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Hage A, et al. Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria. PLoS Genet. 2014;10:e1004716. doi: 10.1371/journal.pgen.1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ginno PA, et al. R-Loop Formation Is a Distinctive Characteristic of Unmethylated Human CpG Island Promoters. Mol Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aguilera A, García-Muse T. R Loops: From Transcription Byproducts to Threats to Genome Stability. Mol Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes & Development. 2014;28:1384–1396. doi: 10.1101/gad.242990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keskin H, et al. Transcript-RNA-templated DNA recombination and repair. Nature. 2015;515:436–439. doi: 10.1038/nature13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szczelkun MD, et al. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci USA. 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horvath P, Barrangou R. RNA-guided genome editing. Cell Res. 2013;23:733–734. doi: 10.1038/cr.2013.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Manley JL. Inactivation of the SR Protein Splicing Factor ASF/SF2 Results in Genomic Instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Tuduri S, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paulsen RD, et al. A Genome-wide siRNA Screen Reveals Diverse Cellular Processes and Pathways that Mediate Genome Stability. Mol Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stirling PC, et al. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes & Development. 2012;26:163–175. doi: 10.1101/gad.179721.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wahba L, et al. RNase H and Multiple RNA Biogenesis Factors Cooperate to Prevent RNA:DNA Hybrids from Generating Genome Instability. MOLCEL. 2011;44:978–988. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhatia V, et al. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362–365. doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- 18.Sollier J, et al. Transcription-Coupled Nucleotide Excision Repair Factors Promote R-Loop-Induced Genome Instability. Mol Cell. 2014;56:777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haeusler AR, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groh M, et al. Mechanisms of transcriptional dysregulation in repeat expansion disorders. Biochem Soc Trans. 2014;42:1123–1128. doi: 10.1042/BST20140049. [DOI] [PubMed] [Google Scholar]
- 21.Loomis EW, et al. Transcription-Associated R-Loop Formation across the Human FMR1 CGG-Repeat Region. PLoS Genet. 2014;10:e1004294. doi: 10.1371/journal.pgen.1004294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colak D, et al. Promoter-Bound Trinucleotide Repeat mRNA Drives Epigenetic Silencing in Fragile X Syndrome. Science. 2014;343:1002–1005. doi: 10.1126/science.1245831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skourti-Stathaki K, et al. Human Senataxin Resolves RNA/DNA Hybrids Formed at Transcriptional Pause Sitesto Promote Xrn2-Dependent Termination. Mol Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skourti-Stathaki K, et al. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature. 2014;516:436–439. doi: 10.1038/nature13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helmrich A, et al. Transcription-replication encounters, consequences and genomic instability. Nat Struct Mol Biol. 2013;20:412–418. doi: 10.1038/nsmb.2543. [DOI] [PubMed] [Google Scholar]
- 26.Ginno PA, et al. GC skew at the 5″ and 3″ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Research. 2013;23:1590–1600. doi: 10.1101/gr.158436.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Q, et al. R-Loop Stabilization Represses Antisense Transcription at the Arabidopsis FLC Locus. Science. 2013;340:619–621. doi: 10.1126/science.1234848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duquette ML, et al. AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene. 2005;24:5791–5798. doi: 10.1038/sj.onc.1208746. [DOI] [PubMed] [Google Scholar]
- 29.Duquette ML, et al. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes & Development. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mischo HE, et al. Yeast Sen1 Helicase Protects the Genome from Transcription-Associated Instability. Mol Cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powell WT, et al. R-loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation. Proc Natl Acad Sci USA. 2013;110:13938–13943. doi: 10.1073/pnas.1305426110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wahba L, et al. The homologous recombination machinery modulates the formation of RNA–DNA hybrids and associated chromosome instability. eLIFE. 2013 doi: 10.7554/eLife.00505.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–70. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heyer WD, et al. Regulation of Homologous Recombination in Eukaryotes. Annu Rev Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kasahara M, et al. RecA protein-dependent R-loop formation in vitro. Genes & Development. 2000;14:360–365. [PMC free article] [PubMed] [Google Scholar]
- 36.Fairman-Williams ME, et al. SF1 and SF2 helicases: family matters. Current Opinion in Structural Biology. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatchi E, et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Molecular Cell. 2015;57:636–647. doi: 10.1016/j.molcel.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sordet O, et al. Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks. Embo rep. 2009;10:887–893. doi: 10.1038/embor.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El Hage A, et al. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes & Development. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y, et al. Arginine Methylation Facilitates the Recruitment of TOP3B to Chromatin to Prevent R Loop Accumulation. Mol Cell. 2014;53:484–497. doi: 10.1016/j.molcel.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, et al. The RNA binding protein RNPS1 alleviates ASF/SF2 depletion-induced genomic instability. RNA. 2007;13:2108–2115. doi: 10.1261/rna.734407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu K, et al. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 43.Basu U, et al. The RNA Exosome Targets the AID Cytidine Deaminase to Both Strands of Transcribed Duplex DNA Substrates. Cell. 2011;144:353–363. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiarle R, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burns MB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2014;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian M. Transcription-induced Cleavage of Immunoglobulin Switch Regions by Nucleotide Excision Repair Nucleases in Vitro. Journal of Biological Chemistry. 2000;275:24163–24172. doi: 10.1074/jbc.M003343200. [DOI] [PubMed] [Google Scholar]
- 48.Gan W, et al. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes & Development. 2011;25:2041–2056. doi: 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wellinger RE, et al. Replication Fork Progression Is Impaired by Transcription in Hyperrecombinant Yeast Cells Lacking a Functional THO Complex. Molecular and Cellular Biology. 2006;26:3327–3334. doi: 10.1128/MCB.26.8.3327-3334.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alzu A, et al. Senataxin Associates with Replication Forks to Protect Fork Integrity across RNA-Polymerase-II-Transcribed Genes. Cell. 2012;151:835–846. doi: 10.1016/j.cell.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen YZ, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am J Hum Genet. 2004;74:1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moreira MC, et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat Genet. 2004;36:225–227. doi: 10.1038/ng1303. [DOI] [PubMed] [Google Scholar]
- 53.Gostissa M, et al. Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annu Rev Immunol. 2011;29:319–350. doi: 10.1146/annurev-immunol-031210-101329. [DOI] [PubMed] [Google Scholar]
- 54.Helmrich A, et al. Collisions between Replication and Transcription Complexes Cause Common Fragile Site Instability at the Longest Human Genes. Mol Cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 55.Naim V, et al. ERCC1 and MUS81–EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nature Cell Biology. 2013;15:1008–1015. doi: 10.1038/ncb2793. [DOI] [PubMed] [Google Scholar]
- 56.Boerma EG, et al. Translocations involving 8q24 in Burkitt lymphoma and other malignant lymphomas: a historical review of cytogenetics in the light of todays knowledge. Leukemia. 2008;23:225–234. doi: 10.1038/leu.2008.281. [DOI] [PubMed] [Google Scholar]
- 57.Shou Y, et al. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc Natl Acad Sci USA. 2000;97:228–233. doi: 10.1073/pnas.97.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gotlib J, Cools J. Five years since the discovery of FIP1L1-PDGFRA: what we have learned about the fusion and other molecularly defined eosinophilias. Leukemia. 2008;22:1999–2010. doi: 10.1038/leu.2008.287. [DOI] [PubMed] [Google Scholar]
- 59.Rozovski U, et al. The significance of spliceosome mutations in chronic lymphocytic leukemia. Leuk Lymphoma. 2013;54:1364–1366. doi: 10.3109/10428194.2012.742528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2012;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 61.Quesada V, et al. Exome sequencing identifies recurrent mutations of the splicing factor. Nat Genet. 2011;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 62.Ford D, et al. Genetic Heterogeneity and Penetrance Analysis of the BRCA1 and BRCA2 Genes in Breast Cancer Families. The American Journal of Human Genetics. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barlow JH, et al. Identification of Early Replicating Fragile Sites that Contribute to Genome Instability. Cell. 2013;152:620–632. doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Le May N, et al. XPG and XPF Endonucleases Trigger Chromatin Looping and DNA Demethylation for Accurate Expression of Activated Genes. Mol Cell. 2012;47:622–632. doi: 10.1016/j.molcel.2012.05.050. [DOI] [PubMed] [Google Scholar]
- 66.Gifford CA, et al. Transcriptional and Epigenetic Dynamics during Specification of Human Embryonic Stem Cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Groh M, et al. R-loops Associated with Triplet Repeat Expansions Promote Gene Silencing in Friedreich Ataxia and Fragile X Syndrome. PLoS Genet. 2014;10:e1004318. doi: 10.1371/journal.pgen.1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Castellano-Pozo M, et al. R-loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol Cell. 2013;52:583–590. doi: 10.1016/j.molcel.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 69.Zhu X, Schatz GC. Molecular Dynamics Study of the Role of the Spine of Hydration in DNA A-Tracts in Determining Nucleosome Occupancy. J Phys Chem B. 2012;116:13672–13681. doi: 10.1021/jp3084887. [DOI] [PMC free article] [PubMed] [Google Scholar]