Summary
We show that DNA methyltransferase inhibitors (DNMTis) upregulate immune signaling in cancer through the viral defense pathway. In ovarian cancer (OC), DNMTis trigger cytosolic sensing of double-stranded RNA (dsRNA) causing a Type I Interferon response and apoptosis. Knocking down dsRNA sensors TLR3 and MAVS reduces this response twofold, and blocking interferon beta or its receptor abrogates it. Upregulation of hypermethylated endogenous retrovirus (ERV) genes accompanies the response and ERV overexpression activates the response. Basal levels of ERV and viral defense gene expression significantly correlate in primary OC and the latter signature separates primary samples for multiple tumor types from The Cancer Genome Atlas into low versus high expression groups. In melanoma patients treated with an immune checkpoint therapy, high viral defense signature expression in tumors significantly associates with durable clinical response and DNMTi treatment sensitizes to anti-CTLA4 therapy in a pre-clinical melanoma model.
Introduction
DNA methyltransferase inhibitors (DNMTis), such as 5-azacytidine (Aza) and 5-aza-2’-deoxycytidine (Dac) are effective cancer therapies in hematologic neoplasms (Tsai et al., 2012) (Matei et al., 2012) and are FDA approved for the pre-leukemic disorder myelodysplasia (MDS) (Kaminskas et al., 2005). These cytidine analogues incorporate into DNA, block catalytic actions of DNA methyltransferases (DNMTs), and trigger their degradation (Stresemann et al., 2006). Preclinically, low doses avoid early cytotoxicity and DNA damage, allowing cells to exhibit apparent reprogramming and blunting of tumorigenicity (Tsai et al., 2012). Mechanisms can include reversal of abnormal promoter DNA methylation, re-expression of silenced genes including tumor suppressors (Baylin and Jones, 2011), and changes to cancer signaling pathways including apoptosis, cell cycle activity, and stem cell functions (Tsai et al., 2012).
A long recognized activity of DNMTis described by others (Karpf et al., 2004; Karpf et al., 1999), and our group (Li et al., 2014; Wrangle et al., 2013), is induction of immune responses in cancer cells. In recent clinical trials for non-small cell lung cancer (NSCLC) (Juergens et al., 2011; Wrangle et al., 2013) a small number of patients had remarkably robust and durable responses to immune checkpoint blockade therapy after first receiving Aza (Wrangle et al., 2013). This immune therapy alone also has activity against NSCLC (Brahmer et al., 2010; Brahmer et al., 2012; Topalian et al., 2012). A larger trial is now ongoing to determine if Aza can indeed prime patients for sensitization to checkpoint inhibition (Brahmer, 2015). For NSCLC and other tumor types, Aza induces interferon signaling and concordant upregulation of surface antigens and their assembly proteins, viral defense pathways, and transcript and surface protein levels of PD-L1, the key checkpoint ligand targeted in the above immunotherapy (Li et al., 2014; Wrangle et al., 2013). Indeed, we have defined a 300 gene expression signature we termed Aza-Induced iMmune genes or AIM (Li et al., 2014) for which activation is greatest for epithelial ovarian cancer (EOC) and NSCLC (Li et al., 2014). Genome-wide expression of AIM separates primary EOC, NSCLC, and other cancers into high and low expression groups (Li et al., 2014). We hypothesize the low group may represent an “immune evasion/ immune editing” pattern (Drake et al., 2006) (Schreiber et al., 2011) that Aza could reverse to sensitize patients to subsequent immune therapy (Li et al., 2014).
We now show that a major mechanism underlying the Aza-triggered immune response is induction of a cytosolic double-stranded RNA (dsRNA) sensing pathway used by epithelial and other cell types as a viral defense mechanism that triggers a Type I interferon response (Kulaeva et al., 2003; Sistigu et al., 2014). A key contributor is induction of increased expression of multiple DNA hypermethylated endogenous retroviruses (ERVs). In The Cancer Genome Atlas (TCGA), the viral defense gene expression separates primary EOC and other cancers into high and low expression and high tumor expression strongly associates with clinical benefit in a trial of immune checkpoint therapy for advanced melanoma. Aza sensitizes to immune checkpoint blockade in a pre-clinical model of melanoma. We thus define a potential approach in which an epigenetic therapy may sensitize cancer cells to various immunotherapies.
Results
DNMTis trigger viral defense and type I interferon signaling
Induction of AIM in a previous study of 23 EOC cell lines (Li et al., 2014) included, in addition to previously reported DNA hypermethylated cancer testis antigens (MAGEA4, MAGEA9, NY-ESO-1) (James et al., 2013; Karpf et al., 2009; Karpf et al., 2004; Odunsi et al., 2014), interferon/viral defense, antigen processing and presentation, and host immune cell attraction genes (Figure 1a). Direct Aza targeting of DNMTs for these changes is suggested by similar findings in DKO colon cancer cells genetically disrupted for two major DNMTs (DNMT1−/−, DNMT3B −/−) versus parental, wild type HCT116 cells (Figure 1a). The induced responses may not be a general stress phenomenon as they do not occur with carboplatin, a cytotoxic agent commonly used in EOC treatment (Figure 1b). Aza and Dac incorporate into DNA, inhibiting 3 DNA methyltransferases (Schaefer et al., 2009), but Aza also incorporates into RNA, inhibiting the RNA methyltransferase DNMT2. Aza thus can demethylate RNA, and unmethylated RNA may activate TLR3 and the interferon response (Kariko et al., 2005). However, Dac and Aza both mimic the DKO cell line results (Figure 1a, c, d; S1a, b) strongly suggesting that the drugs directly target DNA methylation to trigger the interferon response.
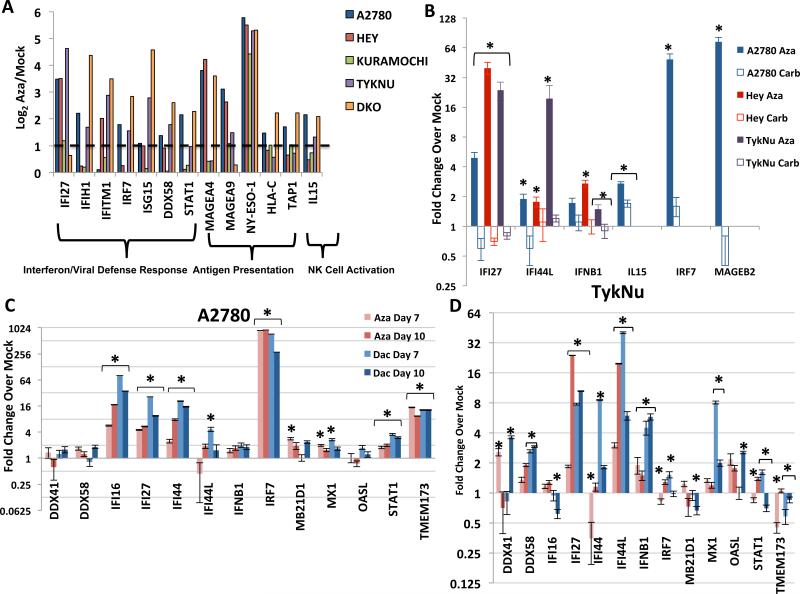
Figure 1. DNMT inhibitors upregulate immune genes in ovarian cancer cell lines.
A) Levels of immune genes in four EOC cell lines and DKO colon cancer cell line (DNMT1−/−, DNMT3B−/−) relative to its parental HCT116 line. Y-axis = log2 Aza/Mock fold change from microarray data. Dotted line denotes twofold change. B) qRT-PCR validation of immune genes in EOC cells treated for 72 hours: Mock, 500 nM Aza, or 500 nM− 3 μM carboplatin and rested for 7 days before assaying (Day 10). IC50s: A2780 (Aza = 848 nM, Carb = 457 nM), Hey (Aza = 4.1 μM, Carb = 12.2 μM), TykNu (Aza = 491 nM, Carb = 986.2 nM). C-D. qRT-PCR validation of interferon response genes in the A2780 (C) and TykNu (D) EOC lines treated with no drug (Mock), 500 nM Aza (Aza), or 100 nM Decitabine (Dac) for 3 days, and rested for 4 (Day 7) or 7 (Day 10) days before assaying. Y-axis = fold change over mock. Data in B-D are represented as mean +/− S.E.M of 3 biological replicates. * = p≤0.05. See also Figure S1.
Aza and Dac similarly trigger an interferon response that includes interferon beta (IFNβ1) and a panel of Interferon Stimulated Genes (ISGs; IFI16, IFI27, IFI44, IFI44L, MX1, OASL) (Figure 1c,d, Figure S1a, b). Each ISG functions predominantly in anti-viral and anti-proliferative signaling (Figure 2a). In four EOC cell lines, key upstream genes in the Type I Interferon pathway (IFNβ, IRF7, and STAT1) were generally upregulated at the 4th day following the end of Aza treatment (“Day 7”) and further increased by Day 10 (Figure 1c, d, S1a,b). Importantly, cytosolic sensors for DNA (MB21D1/CGAS and TMEM173/STING) and RNA (DDX41 and DDX58) were also variably upregulated (Figure 1c, d, Figure S1a, b). In A2780 and TykNu, but not Hey or Kuramochi, cell lines, variable increases occured in Type III interferon signaling genes, also involved in response to viruses (Ding and Robek, 2014). These included IFNL1 (IL28A) and IFNL3 (IL29) ligands (Figure S1c) and especially the IFN III receptor IFNLR1 (Figure S1c), known to be methylated and activated by epigenetic therapy (Ding et al., 2014).
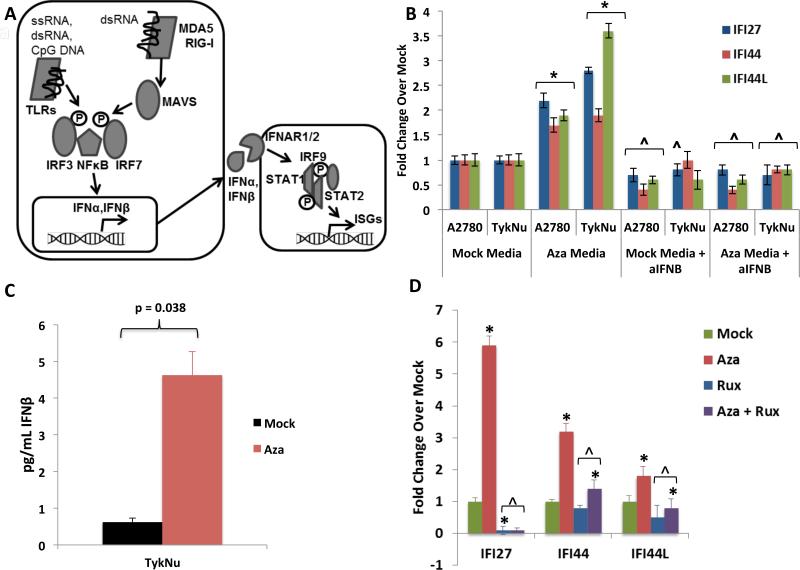
Figure 2. DNMTis upregulate immune signaling through secreted interferon.
A) Schematic of interferon pathway. Protein symbols outlined in text. B) Treatment of recipient A2780 or TykNu cells with media from cells treated with Mock or Aza, +/− addition of anti-IFNβ. Y-axis = qRT-PCR fold Aza/Mock of ISGs. * = p≤0.05 and ^ = p≤0.05, respectively, for Mock or Aza media with versus without anti-IFNβ versus Mock or Aza media plus anti-IFN β; C) ELISA of IFNβ in media from TykNu cells at Day 10 from studies in B. D) Treatment of EOC cells with Aza as in Figure 1c,d, but in the presence of 2 μM Ruxolitinib (Rux). qRT-PCR fold changes for ISGs. * = p≤0.05 for fold Aza over Mock and ^ = p≤0.05, Mock or Aza versus Mock + Rux or Aza + Rux. Data = mean +/− S.E.M. of three (B, D) or four (C) biological replicates. See also Figure S2.
Key viral RNA sensing proteins include TLR3 on the endosomal membrane and MDA5, PKR, and RIG-I in the cytoplasm (Figure 2a). These induce IRF3, IRF7, and NF-κB to translocate to the nucleus and activate transcription of IFNβ1 (Ivashkiv and Donlin, 2014). IRF7 is frequently promoter DNA hypermethylated in cancer and the associated low basal expression can be reversed by Aza in squamous NSCLC (Wrangle et al., 2013). Among 23 EOC lines examined, IRF7 was hypermethylated in only one, A2780 (Li et al., 2014) (Figure S3a), potentially not a classic high-grade EOC (Anglesio et al., 2013; Domcke et al., 2013). Aza induces partial IRF7 demethylation and increased expression in this cell line at Days 7 and 10 while carboplatin did not (Figures S3a, 1b,c) and IRF7 knockdown significantly reduced the Aza interferon response (Figure S3b,c). Such IRF7 induction does not occur in two EOC lines or the HCT116 colon cancer cell line where the gene is not hypermethylated (Figures 1d, S1a,b, S3a).
When IRF7 is not silenced, other mechanisms must then be operative for Aza to trigger viral defense signaling. Secreted IFNβ is critical to this signaling and, through interaction with surface receptors IFNAR1/2, activates JAK/STAT signaling, transcription of ISGs, and resultant translation inhibition and apoptosis (Platanias, 2005) (Ivashkiv and Donlin, 2014) (Figure 2a). Indeed, media transferred to untreated cells from Aza-treated cells 7 days after drug withdrawal causes an interferon response with increased expression of ISGs IFI27, IFI44, and IFI44L (Figure 2b). Moreover, Aza treatment induced secreted IFNβ in media (Figure 2c, Figure S2a) and an IFNβ blocking antibody significantly blocked the Aza induced ISG media response (Figure 2b). Like Type I IFN signaling, Type III IFN signaling can be activated by viral infection (Robek et al., 2005) (Ding and Robek, 2014). However, even though we observed upregulation of Type III ligand transcripts IFNL1 (IL28A) and IFNL3 (IL29) (Figure S1c), secreted Type III interferon proteins are undetectable by ELISA (Figure S2b).
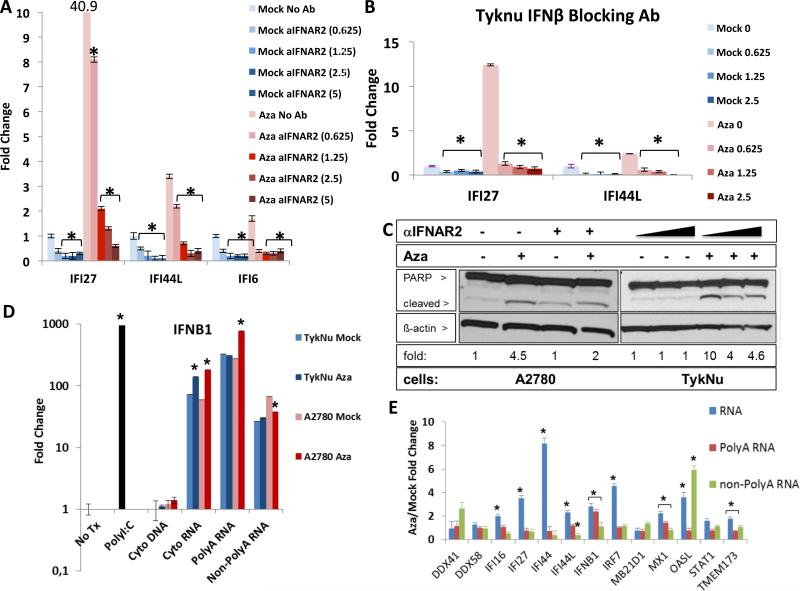
Aza appears to activate Type I, IFNβ mediated signaling through JAK/STAT, as the JAK/STAT inhibitor ruxolitinib strongly reduces ISG responses (Figure 2d, S2c). Further, antibody blocking of IFNAR2, the IFNβ receptor, abrogates Aza induction of IFI27, IFI44L, and IFI6 transcription (Figure 3a, S2d), as does inhibition of IFNβ itself (Figure 3b, S2e). In contrast, blocking the Type III interferon, IL10RB receptor, gives only a modest block of IFI27 increase (Figure S2f). IFNβ binding to IFNAR2 also may contribute to late, Aza induced apoptosis that peaks at 4-7 days after Aza withdrawal, since anti-IFNAR2 leads to a lower ratio of cleaved/total PARP (Figure 3a,c). (Figures 3c, S2f).
Figure 3. Aza induces immune signaling through dsRNA activation of secreted interferon.
A) Blocking IFNAR2 (αIFNAR2) or B) IFNβ in TykNu cells treated vs. non-treated with Aza as in Figures 1c,d; parentheses = U/mL of antibody. Y-axis = qRT-PCR for ISGs. * = p≤0.05 for A), Mock or Aza - anti-IFNAR2 versus + anti-IFNAR2, B) Mock or Aza with no anti-IFNβ versus with anti-IFNβ. C) Immunoblotting for cleaved PARP with β-actin loading control. Fold change shown for cleaved/total PARP ratio, normalized to β-actin for each dose of anti-IFNAR2 (triangles = 0-1.25 U/mL). Aza compared to Mock = 1. D,E) Indicated nucleic acids from the cytoplasm of A2780 or TykNu treated cells with no drug (Mock) or 500 nM Aza (Aza) for 3 days and rested without drug for 4 days before transfection into recipient HT29 cells. Y-axis = fold change, Aza/Mock, for IFNβ1 transcript (D) or ISG transcripts (E) induced in HT29s. No Tx = no transfection, Cyto DNA = Cytoplasmic DNA, Cyto RNA = Cytoplasmic RNA excluding ribosomal RNA. * = p≤0.05 for D) fold change Aza vs Mock, E) Aza / Mock. Data in A,B,D,E is mean +/− S.E.M. of three biological replicates. See also Figure S3.
DNMTis trigger viral defense through induction of dsRNA
Aza-induced viral defense genes and IFNβ1 are not generally DNA methylated at promoter regions (Li et al., 2014), thus Aza may activate the pathway upstream of these genes. We considered increases in dsRNA, viral ssRNA, and unmethylated CpG DNA that might trigger cytosolic sensors (Sun et al., 2013). Indeed, 3 days after ending Aza treatment of A2780 and TykNu ovarian cancer cells and subsequent transfection into HT29 colon cancer cells, known to have a robust interferon response (Chiappinelli et al., 2012), cytoplasmic total RNA (without rRNA) and PolyA+ RNA, but not PolyA− RNA or DNA, increased IFNβ1 transcripts (Figure 3d). Further, RNaseIII treatment of the cytosolic nucleic acids, which specifically digests dsRNA, eliminated the IFNβ1 upregulation (Figure 4a), but this was not seen with RNaseH treatment that digests DNA-RNA hybrids (Figure S4a).
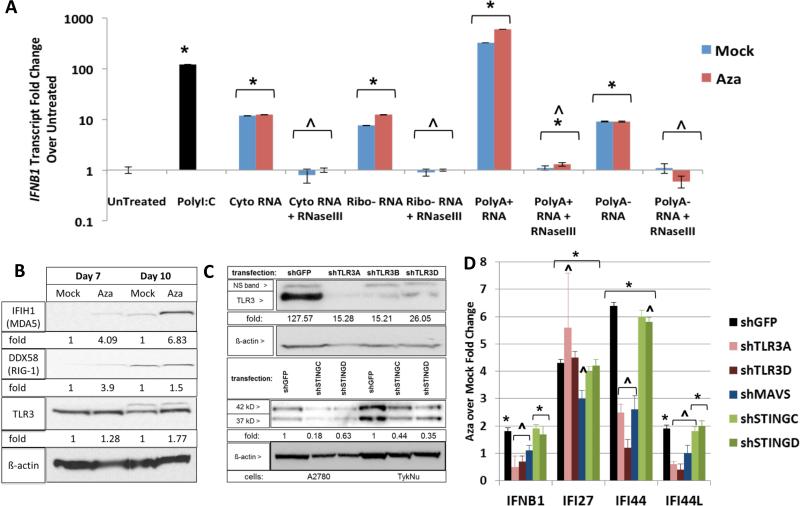
Figure 4. Aza activates dsRNA sensors to induce interferon signaling.
A) Effects on IFNβ1 transcripts, at 24 hours, in HT29 recipient cells transfected with nucleic acid fractions, treated with RNaseIII, from A2780 as in Figure 3d,e. IFNβ1 transcripts were measured at 24 hours. * = p≤0.05 for fold change over untreated; ^ = p≤0.05 for Mock or Aza + versus - RNAseIII. B) Western blots for MDA5, RIG-I, and TLR3 in A2780 cells at four (Day 7) and seven (Day 10) days after Mock vs 500 nM Aza (Aza) for 3 days. C) Knockdown upon lentiviral infection, with puromycin selection, of A2780 and TykNu with shGFP, shTLR3, and shSTING hairpins. Immunoblotting with anti-TLR3, anti-STING and anti-β-Actin. Densitometry fold change, normalized to mock or shGFP, shown at the bottom of the gels. D) q-RT-PCR for ISGs from B and C. * = p≤0.05 Aza over Mock; ^ = p≤0.05 shGFP versus each shRNA sensor with mean fold change +/− S.E.M. of three biological replicates. See also Figure S4.
If dsRNA is required for the above Aza effects, then the key cytosolic sensors, TLR3, MDA5 (IFIH1) and RIG-I (DDX58), the latter two signaling through the mitochondrial protein, MAVS, should be involved in subsequent IFNβ1 induction (Figure 2a). Aza increased transcript (Figure 1a) and protein levels for these (Figure 4b). However, RIG-I, which requires a 5’ triphosphate group on RNA for activation, is likely not a key player since alkaline phosphatase treatment of cytosolic nucleic acid fractions did not abolish IFNβ1 upregulation (Figure S4d). In contrast, knockdown of TLR3 and MAVS in A2780 cells (Figure 2a) decreased by twofold Aza upregulation of interferon genes IFNB1, IFI44, IFI44L and IFI27 as did MAVS knockdown (Figures 4c,d; S4b,c). In TykNu, knockdown of TLR3 and MAVS significantly blunted Aza induction of these gene responses (Figure S4b,c). Importantly, knockdown of STING, the cytosolic DNA sensor (Mankan et al., 2014) did not blunt Aza induced interferon signaling (Figure 4c,d). A previous report had implicated STING in viral cytosolic sensing in B cells, but this was dependent upon viral reverse transcriptase activity, likely to be low in our cells (Mankan et al., 2014). We thus conclude that MAVS and TLR3 are centrally involved in Aza triggering cytosolic sensors to induce an interferon response.
Aza-induced human endogenous retrovirus (ERV) transcripts can activate viral defense responses in EOC
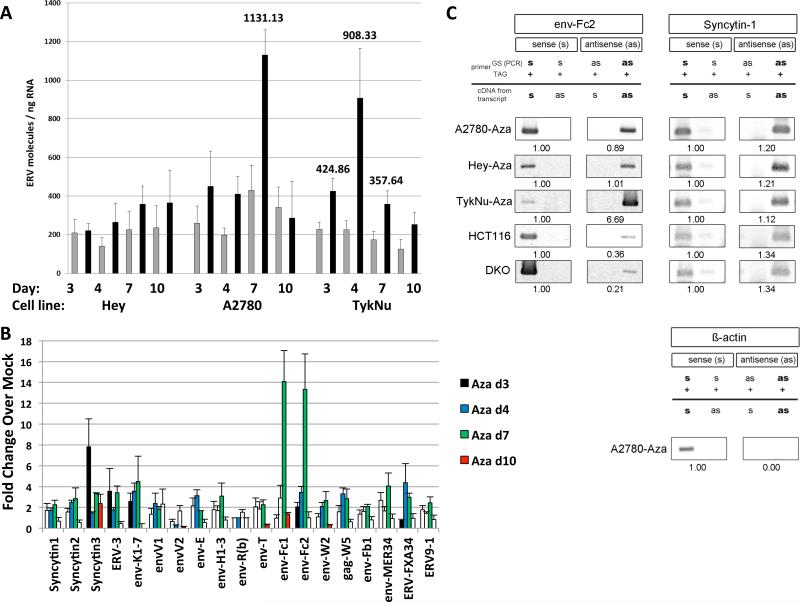
The above data suggests Aza might activate endogenous retroviral sequences (ERVs) that constitute more than 8% of the human genome, can activate cytosolic RNA sensors, and are silenced in normal somatic cells by promoter DNA methylation (Bannert and Kurth, 2004) (Tristem, 2000) (Hurst and Magiorkinis, 2014) (Mankan et al., 2014). Some cancers lose ERV DNA methylation and aberrantly overexpress ERVs (Chen et al., 2004; Cohen et al., 1988; Larsen et al., 2009; Larsson et al., 2007; Rycaj et al., 2014; Strick et al., 2007; Strissel et al., 2012; Wang-Johanning et al., 2001; Wang-Johanning et al., 2007) while others maintain silencing. Aza can induce specific ERV transcripts in melanoma, choriocarcinoma, and endometrial cancer cells (Laska et al., 2013; Ruebner et al., 2013; Stengel et al., 2010) (Strissel et al., 2012). Indeed, in initial testing, the ERVK subfamily (Wang-Johanning et al., 2003) transcripts increased 2.5-fold in the A2780 cell line upon Aza treatment (data not shown). Upregulation of individual ERVs (22 full length env, 6 partial coding env, one full length gag, and two partial coding pols) (Tables S1-S2), in PCR assays for non-repeat sequences, occur especially, at Day 7, coinciding with ISG expression, in three EOC lines following Aza and Dac treatment (Figure 5a,b, Figure S5a,b,c). These include several known ERV env genes like Syncytin-1, ERV-3, env-K and env-H (Blond et al., 1999; Lower et al., 1993; Mi et al., 2000; Rote et al., 2004) and at especially high levels, env-Fc2, a less well characterized gene (Benit et al., 2003). Finally, in DKO as well as Aza treated A2780 and TykNu cells, loss of env-Fc2 promoter methylation correlates with increased Fc2 expression (Figures 5b, 6a-c, S6a) but not in Hey cells (Figure S6a).
Figure 5. Aza upregulates sense and antisense ERV transcripts.
RNA was isolated from cells at last (Day 3), one (Day 4), three (Day 7) and seven (Day 10) days after Mock or 500 nM Aza (Aza) for 3 days. A) Total number of molecules for all ERV genes; error bars = S.E.M for 4 independent experiments. Numbers above bars = significant data for indicated days. Gray = Mock, Black = Aza. B) qRT-PCR of ERV genes in A2780 cells for 4 independent experiments. Y-axis= fold increases for Aza/Mock +/− S.E.M and normalized to Mock = 1. White bars = non-significant and colored bars = significant ERV gene induction (p<0.05). C) TASA-TD PCR amplified sense and antisense transcripts of the env-Fc2 (731 bp) and Syncytin-1 (202 bp) genes from first strand cDNA. Aza treated A2780, Hey, and TykNu, and HCT116 and DKO cells are indicated. Ratios of sense (s) and antisense (as) determined by ImageJ. PCR primers = gene specific (GS); TAG. β-actin sense 399 bp amplification product = negative control for as transcripts (Chen et al., 2004). See also Figure S5.
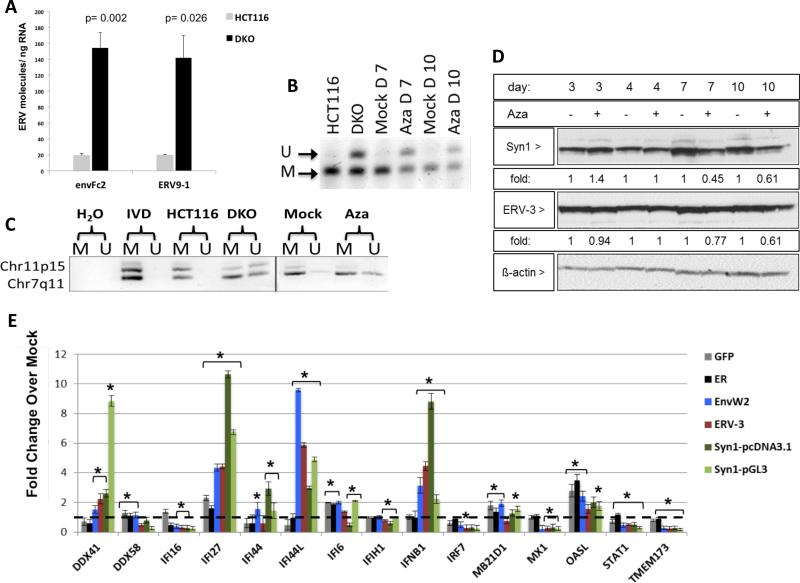
Figure 6. Aza upregulates ERV transcripts, but not proteins, through DNA demethylation.
A) env-Fc2 and erv-9-1 ERV gene total number of molecules, assayed by qRT-PCR, for DKO (DNMT1−/−, DNMT3B−/−) and parental HCT116 cells. Y-axis = mean +/− SEM for n = 6 biological replicates. * = p≤0.05 for DKO versus HCT116. B) DNA methylation changes in ERVs in A2780 cells treated with Mock or 500 nM Aza for 3 days at post-treatment day 4 (Day 7), or 7 (Day 10). Bisulfite treated DNA was amplified and digested with the AciI enzyme producing 155 and 44 bp fragments of methylated DNA while unmethylated DNA does not digest (189 bp fragment). “U” = unmethylated band, “M” = methylated band. C) DNA from B) was subjected to Methylation-specific PCR for Fc2 family members on chromosomes 7 and 11. U” = unmethylated, “M” = methylated. IVD = in vitro methylated DNA. D) Syncytin-1 and ERV-3 protein levels in EOC cells treated as in B). Fold change for densitometry by ImageJ for Aza vs Mock cells normalized to β-actin protein levels E) Transfection of full-length env genes from EnvW2, ERV-3, or Syncytin-1 or EGFP and ER controls in TykNu cells. qRT-PCR was performed for ISGs 7 days after transfection. Dotted black line indicates 1. Y-axis = mean +/− S.E.M fold change of three biological replicates for overexpression/ Mock. * = p≤0.05. See also Figure S6.
Further linking ERVs with a dsRNA-triggered IFN response, bidirectional transcription producing sense and anti-sense transcripts occurred for Syncytin-1 and five env-Fc2 gene loci, but not β -actin, in three EOC lines and HCT 116 and DKO cells (Figure 5c, Table S2), analyzed by the TAG-aided sense/antisense transcript detection (TASA-TD) technique (Henke et al., 2015). Such sense and antisense transcripts can form dsRNA (Faghihi et al., 2008; Su et al., 2012). Interestingly for TykNu there was a 6.69-fold increase of env-Fc2 antisense transcript levels compared to the sense transcript (Figure 5c) but substantially lower antisense transcripts were seen in both HCT116 and DKO cells (Figures 5c, 6a). Disrupting DNMTs seems integral to the above ERV upregulation since increases of env-Fc2 and erv9-1 occurred in DKO versus wild type HCT 116 cells (Figure 6a).
ERV transcripts seem directly involved in the Aza responses in that, first, although drug-induced upregulation of ERV transcripts begins early after Aza, both ERVs and viral defense gene increases generally peak by day 7 (Figures 5, S5). Second, ERV env proteins such as Syn1 and ERV-3 are not increased after Aza treatment, supporting a dominant role for viral defense signaling via RNA transcripts (Figure 6d, Figure S6b, and c). Third, overexpression of ERV-3, EnvW2, and Syncytin-1 in TykNu (Figure 6e-j), A2780 (Figure S6d), and Hey cells (Figure S6e), as compared to control genes, increase, at the transcript but not the protein levels, the same interferon genes induced by Aza (IFNβ1, IFI27, and IFI44L). The increases often exceeded that for the drug likely because total ERV RNA molecules are higher in the overexpression experiments (Figure S6f-i). Finally, although siRNA knockdown of individual ERVs (Syncytin-1, ERV-3) during Aza treatment produced more complex results, targeting two ERVs significantly blunts the Aza induced gene expression of IFI27, IFI44L, and IFI6 in TykNu cells, but not A2780 or Hey cells (Figure S7a).
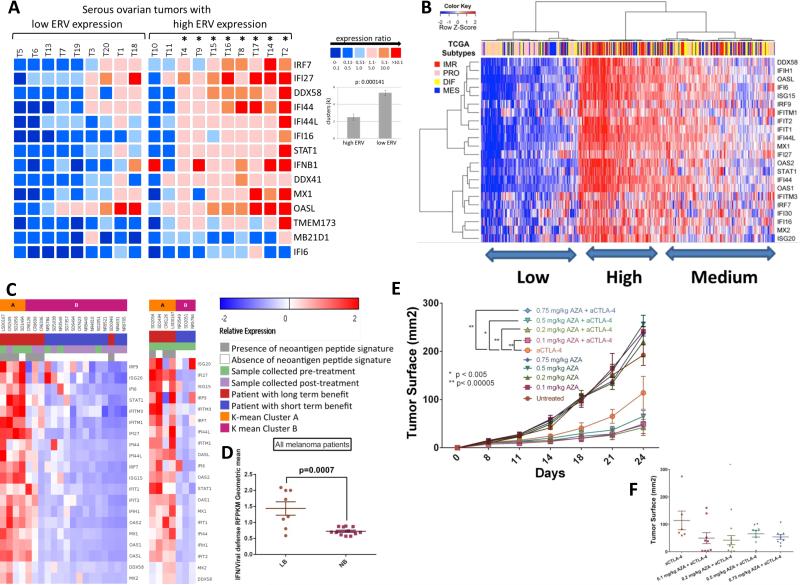
Importantly, a driving role for ERV transcripts in triggering Aza-induced viral defense gene responses is evidenced by a high correlation of basal levels of both in 19 primary EOC. Total molecules of 22 ERV env genes queried are increased (p<0.05) in tumor versus normal (n=9) and divide tumors into lower (n=9) and higher ERV (n=10) expression groups as compared to normal controls. High ERV tumors have significantly higher viral defense response gene expression (p = 0.000141) (Figure 7a).
Figure 7. Aza-upregulated viral defense genes are significantly correlated with ERVs in primary tumors and correlate with sensitivity to immune therapy.
A) Heatmap comparing basal levels of viral defense genes and ERVs in primary EOC. The cut-off for lower or higher ERVs was the mean control tissue value of 237.57 +/− 83.05 molecules/ng RNA. Mean ISGs of the high ERV ovarian tumor (T) cohort (n=10) is 12.65-fold higher than the mean of ISGs of the low ERV cohort (n=9). The (*) denotes that 8 of 10 high ERV tumors had significantly higher ISG expression compared to the low ERV tumors. ISG expression is organized according to low and high ERV expression cohorts in arbitrary units; color code from blue to red shows increasing ISG expression. For clusters (k=6), differences are significant between the high ERV expression (2.5 +/− 0.37) and the low ERV expression cohort (5.33 +/− 0.28). B) Interferon stimulated, viral defense genes upregulated at least twofold by Aza in EOC cell lines (right y-axis) were used to cluster EOC tumors for RNA-Seq data (blue = low; red=high) from The Cancer Genome Atlas (TCGA). EOC TCGA subtypes are shown: DIF (differentiated), IMR (immune reactive), MES (mesenchymal), and PRO (proliferative) C), D) Viral defense gene signature is upregulated in tumors from anti-CTLA-4 treated metastatic melanoma patients who derived durable clinical benefit (complete response, partial response, or progression free-survival > 6 months as previously described (Snyder et al., 2014)) compared to those without benefit. Tumors collected pre-CTLA-4 treatment and shortly post-treatment are shown. E), F) Tumor responses of mice injected with B16-F10 cells and treated with either PBS, anti-CTLA-4, Aza, or both anti-CTLA-4 and Aza. Data represent results from one of two independent experiments with identical results, each with n = 10 per arm.
Viral defense gene levels divide human tumors into high and low expression groups that track with responses to immune checkpoint therapy
Human cancers can evolve immune evasion to become less responsive to immune modulation (Drake et al., 2006) (Schreiber et al., 2011). In this regard, basal transcript levels for the Aza-induced viral defense genes group primary EOC, breast, colon, and lung cancers, and melanoma from The Cancer Genome Atlas (TCGA) studies into high and low groups (Figure 7b, Figure S7c-f). For EOC, this basal expression divides tumors into high, medium, and low expression groups and the former two encompass virtually all of the TCGA (Verhaak et al 2013) immune reactive (IMR), good prognosis tumors. The Low group encompasses the PRO (high proliferative), poor prognosis subtype (p<0.001 to .0001 –Figures 7b, S7b)). Interestingly, virtually all of the right sided colon cancers with a high DNA hypermethylation frequency phenotype (p<.002), termed CIMP, which have a high burden of DNA mutations and respond robustly to immune checkpoint therapy (Le et al., 2015), are in the High and Intermediate groups (Figure S7c). High mutation burden has recently been defined as a key correlate to response to immune checkpoint therapy (Rizvi et al., 2015) (Snyder et al., 2014). Similar sharp high versus low clustering is seen for subgroups of breast and lung cancers and melanoma (Guan et al., 2015) (Figure S7d-f), the last being very responsive to immune checkpoint therapy (Topalian et al., 2015) (Hodi et al., 2010) (Weber et al., 2015).
Could the levels of viral defense pathway signaling correlate with improved responses to immune checkpoint therapy? Indeed, for RNASeq transcriptomes of melanoma patients treated with anti-CTLA-4, high levels of the viral defense signature expression in tumor samples correlate with long term benefit (disease control [stable disease or better] >6mo as measured radiographically) in patients treated with anti-CTLA-4 therapy (Snyder et al., 2014) (Figure 7c,d; Tables S5, S6). Importantly, high viral defense signature again correlates with high mutational burden (Figure 7c).
Aza treatment potentiates immune checkpoint therapy in a mouse model of melanoma
In the B16-F10 mouse melanoma model, multiple combinations of low dose Aza directly enhance tumor responses to anti-CTLA4 immune checkpoint therapy (Figures 7e,f, S7g). Further, B16 cells treated in vitro with Aza, then injected into mice and treated with anti-CTLA-4, are even cleared completely (data not shown). Thus DNMTis can potentiate the anti-tumor effects of immune checkpoint inhibitors.
Discussion
Our present data now provide functional context for our earlier reports that DNMTis induce a complex set of immune pathway responses in tumor cells (Li et al., 2014; Wrangle et al., 2013). DNMTis trigger cytoplasmic dsRNA sensing, central to cellular viral defense responses, and activate interferon in EOC and colon cancer cells by disrupting DNMTs. This activation could induce tumor attraction of lymphocytes (Ivashkiv and Donlin, 2014). There are some important implications for one of the most exciting new developments in cancer treatment, immune checkpoint therapy (Brahmer et al., 2012) (Berger et al., 2008; Brahmer et al., 2010; Leach et al., 1996; Topalian et al., 2015) (Hodi et al., 2010) (Weber et al., 2015) and for underlying mechanisms inherent to both tumor and host cells for reversal of immune tolerance in tumor infiltrating T-lymphocytes (Pardoll, 2012). Indeed, our basal levels of viral defense gene transcripts divide EOC, and other major cancer types in TCGA, into low and high expression subgroups. Perhaps most intriguingly, such high basal expression in tumors tracks with favorable patient responses in a trial of immune checkpoint therapy for advanced melanoma, and strong Aza sensitization to immune checkpoint therapy is seen in a pre-clinical mouse melanoma model.
A major trigger of the Aza-induced viral defense response appears to be bidirectional transcription of ERVs that are known to fold into dsRNA secondary structures. ERVs, representing more than 8% of the human genome (Bannert and Kurth, 2004; Tristem, 2000), integrated into the genome of mammals between 0.1 and 40 million years ago via exogenous retroviral infections of germ cells (Egan et al., 2004; Turner et al., 2001). Most ERV genes are non-functional due to DNA recombination, mutations and deletions, but some produce functional proteins including group-specific antigen (gag), polymerase (pol) with reverse transcriptase (RT) and the envelope (env) surface unit (SU) with a transmembrane immunosuppressive-like peptide (Mi et al., 2000) (Blaise et al., 2005; de Parseval et al., 2003; Villesen et al., 2004). The env gene of ERVW-1 (chromosome 7q21.2) called Syncytin-1 has an essential role in placentogenesis (Blond et al., 1999; Mi et al., 2000).
Importantly, and key to our findings, a major function of DNA methylation in humans is silencing of ERVs and other viral sequences in the human genome; up to 90% of methylated CpGs are located in 45% of the human genome harboring repetitive elements like ERVs (Walsh et al., 1998) (Bestor and Tycko, 1996). However, ERV genes are unmethylated and expressed in embryonic stem cells (Santoni et al., 2012) and especially Syncytin-1 is epigenetically regulated throughout placentogenesis (Matouskova et al., 2006). Some tumors have ERV demethylation and increased expression such as the ERV-K (HML-2) 5’LTR-UTR in melanoma (Stengel et al., 2010) and the 5’-LTR region of several ERVs in testicular cancer (Gimenez et al., 2010). A 20% overall mean demethylation of single CpGs in the ERVW-1 5’ LTR regulating Syncytin-1 correlates with increased expression in endometrial cancer (Strissel et al., 2012). Indeed, ERVs can be targeted as tumor-associated antigens on melanoma cells (Cooper et al., 2015). In contrast, as in our present data, and those of others (Maksakova et al., 2008), in some cancers, individual ERVs can maintain full or partial promoter DNA methylation and low expression and DNMTis can induce ERV demethylation and viral defense signaling in human embryonic stem cells (Grow et al., 2015).
In addition to ERVs, other noncoding RNAs could contribute to the Aza-induced immune response, such as repetitive Alu elements (Tarallo et al., 2012). UV light can damage small nucleolar RNA and activate an interferon response via TLR3 (Bernard et al., 2012) and very high dose (10 μM) Dac can induce an interferon response, apoptosis, increased ERVs and repetitive satellite RNAs in p53-null mouse fibroblasts (Leonova et al., 2013). We suspect, however, that such high Dac doses induce DNA damage rather than simply epigenetic effects. Half of the ovarian cancer lines we studied (Li et al., 2014) have wild-type P53 but we see no differences in Aza interferon response between these and those with mutant P53
The high translational connotations of our findings, including the small number of patients in clinical trials for NSCLC who may have been sensitized by epigenetic priming to immune therapy (Wrangle et al., 2013), remain to be validated in larger clinical trials. These are ongoing for NSCLC (Brahmer, 2015) and planned for advanced ovarian cancer. Moreover, ERV-K env proteins have been shown to increase immunotherapeutic potential of melanoma, breast, and ovarian cancer patients (Rycaj et al., 2014; Wang-Johanning et al., 2012) (Cooper et al., 2015). Also, our hypotheses that drugs like Aza might sensitize patients with multiple cancer types to immune checkpoint blockade and other immunotherapies are further strengthened by the data in our pre-clinical melanoma model. For immune checkpoint therapy, in addition to the functional significance of our data, a potential biomarker strategy is suggested by our findings in a melanoma trial. The high correlation of viral defense signaling with mutational burden suggests that genetic changes, increases in ERVs, and viral defense genes could predict response to immune checkpoint and other immunomodulatory approaches. Finally, our drug approach to upregulate viral defense signaling might be compared to the use of oncolytic viruses to induce inflammatory immune infiltrates at tumor sites to sensitize to immunomodulation (Zamarin et al., 2014).
Experimental Procedures
Detailed materials and methods can be found in Supplemental Experimental Procedures.
Cell Line Treatments
Cell lines were treated with 500 nM Aza, 100 nM Dac, or 500 nM− 3 μM carboplatin (Sigma, St. Louis, Missouri) for 72 hours, and DNA and RNA were isolated using standard methods at 1, 3, or 7 days following removal of drug. 2 μM ruxolitinib (Invivogen #tlrl-rux), 0.625-5 U/mL of anti-IFNAR2 antibody (PBL Interferon Source #21385-1), 0.625-2.5 U/mL of anti-IFNB antibody (PBL Interferon Source #31400-1), or 1.25-5 U/mL of anti-IL10RB antibody (Abcam # ab89884) were added during DNMTi treatment. Preparation of nuclear and cytoplasmic fractions of cultured cells was performed as described (O'Hagan et al., 2011). Ribosomal RNA was depleted using the Ribominus kit (Invitrogen), and PolyA+ and PolyA− RNA were isolated using the Oligotex Direct mRNA Mini Kit (Invitrogen). Nucleic acids were treated with 1 U/μg of RNase III (Ambion), 10 U/μg of RNaseH (Invitrogen), or 3 U/ 1 μg calf intestine alkaline phosphatase (New England Biolabs) according to manufacturer's instructions and 400 ng of each nucleic acid was transfected into HT29 cells.
DNA Methylation Analysis
DNA was bisulfite converted and subjected to Methylation-Specific PCR (Herman et al., 1996) for IRF7 and Fc2, and COBRA (Xiong and Laird, 1997) for the Fc2 locus on chromosome 11.
Transcript Abundance
Real-time RT-PCR was performed with an Applied Biosystems 7500 Fast Real-Time PCR machine by the 2−ΔΔCT method and TASA-TD strand-specific PCR by the method of (Henke et al., 2015).
Protein Analysis
Western blot analyses employed antibodies against ERV-3 (1:1000, Everest), B-Actin (1:5000, Sigma), MDA5 (1:1000, Cell Signaling #5321), PARP (#9542, 1:1000; Cell Signaling Technology, Inc.), RIG-I (1:1000, Cell Signaling #4200), STING (1:1000, Abcam #ab82960), Syncytin-1 (1:350, Dr. Hervé Perron, Geneuro, Geneva Switzerland), and TLR3 (1:1000, Cell Signaling #6961). IFNB ELISA utilized the Verikine-HS™ Human Interferon Beta Serum ELISA kit (PBL Interferon Source) and IFNL ELISA the DuoSet ELISA for Human IL-29/IL28-B (IFNL 1/3) kit (R & D Systems).
Knockdown and Overexpression Experiments
Syncytin-1, ERV-3 and ERV-W2 env, ER, and E-GFP vectors and siRNAs targeting Syncytin-1, ERV-3, or a scrambled control were transfected using the JetPei or Hyperfect transfection reagents, respectively. TLR3, MAVS, and STING shRNA were performed according to established methods (Stewart et al., 2003).
RNAseq Expression Analysis of Tumors from Anti-CTLA-4-Treated Patients
Patients were described previously (Snyder et al., 2014) and samples were obtained with written informed consent per approved institutional review board (IRB) protocols. Expression data were obtained using RNASeq with all data deposited at GEO (accession number pending).
B16-F10 Melanoma Mouse Model
C57BL/6J mice were subcutaneously injected with 1×105 B16-F10 tumor cells. On days 4, 8, 11, 14, 18, mice were treated intraperitoneally with 100 μg anti-ctla-4. Mice received two cycles of intraperitoneal injection of 0.1 to 0.75 mg/kg Aza in PBS for 5 consecutive days followed by 7 days off treatment, starting at day 8 after developing palpable tumors, with control groups receiving corresponding doses of non-specific isotype antibody control and PBS intraperitoneally. Tumor surface was measured with a caliper using the ellipse surface formula (Length*Width*π)/400.
Statistical Analysis
Mean +/− SEM qRT-PCR results were considered statistically significant with p values ≤ 0.05 by Mann-Whitney U test or Student t-tests and 2-tailed p values are reported. Tumor growth was assessed by two way ANOVA between each of the mouse treatment groups with p values adjusted by the Dunnett multiple comparison test (df=512).
Normalized, level 3 Agilent expression data were obtained from The Cancer Genome Atlas data portal (https://tcga-data.nci.nih.gov/tcga/) and analyzed by R statistical software (www.r-project.org) with existing packages and customized routines. Consensus hierarchical clustering was performed with the ConsensusClusterPlus R-package (Wilkerson and Hayes, 2010) and data analyzed by the Fisher exact p value test for association between clusters (p<0.05 *; p<0.01 **; p<0.001 ***).
Supplementary Material
Acknowledgements
Research was supported by grants from The National Cancer Institute (NCI) CA058184 (SBB), Stand Up To Cancer (SU2C) Epigenetic Dream Team (SBB), the Hodson Trust (SBB), the Samuel Waxman Cancer Research Foundation (SBB), The Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (SBB, DJS), Department of Defense (DOD) Teal Award #BC031272 (SBB), the Pershing Square Sohn Cancer Research Alliance (TAC), the STARR Cancer Consortium (TAC), the Ludwig Foundation (JDW), and National Institutes of Health (NIH) Award# F32CA183214 (KBC), and German Cancer Aid (Deutsche Krebshilfe #108215) (RS). We acknowledge Mrs. Elizabeth Stiegler and Florentine Koppitz for their expert technical help and Kathy Bender for manuscript preparation. ABI TLDA qRT-PCR was conducted at the Genetic Resources Core Facility, Johns Hopkins Institute of Genetic Medicine, Baltimore, MD. We thank Ms. Jennifer Meyers and the Next Generation Sequencing Center at the Sidney Kimmel Comprehensive Cancer Center for Agilent Bioanalyzer analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
KBC, PLS, AD, TM, JW, TAC, SBB, and RS designed experiments, performed data analyses, and wrote the manuscript. KBC, PLS, CH, AD, AH, BA, and SB performed experiments. NSR provided ERV-3 env cDNA plasmid and DS provided ovarian cancer cell lines. HL, AS, VM, DMP, LMC, MWB, and CAZ assisted with data analyses. TM, TAC, SBB, and RS contributed equally to this work and are co-senior authors.
References
- Anglesio MS, Wiegand KC, Melnyk N, Chow C, Salamanca C, Prentice LM, Senz J, Yang W, Spillman MA, Cochrane DR, et al. Type-specific cell line models for type-specific ovarian cancer research. PLoS One. 2013;8:e72162. doi: 10.1371/journal.pone.0072162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci U S A 101 Suppl. 2004;2:14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benit L, Calteau A, Heidmann T. Characterization of the low-copy HERV-Fc family: evidence for recent integrations in primates of elements with coding envelope genes. Virology. 2003;312:159–168. doi: 10.1016/s0042-6822(03)00163-6. [DOI] [PubMed] [Google Scholar]
- Berger R, Rotem-Yehudar R, Slama G, Landes S, Kneller A, Leiba M, Koren-Michowitz M, Shimoni A, Nagler A. Phase I safety and pharmacokinetic study of CT-011, a humanized antibody interacting with PD-1, in patients with advanced hematologic malignancies. Clin Cancer Res. 2008;14:3044–3051. doi: 10.1158/1078-0432.CCR-07-4079. [DOI] [PubMed] [Google Scholar]
- Bernard JJ, Cowing-Zitron C, Nakatsuji T, Muehleisen B, Muto J, Borkowski AW, Martinez L, Greidinger EL, Yu BD, Gallo RL. Ultraviolet radiation damages self noncoding RNA and is detected by TLR3. Nat Med. 2012;18:1286–1290. doi: 10.1038/nm.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor TH, Tycko B. Creation of genomic methylation patterns. Nat Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- Blaise S, de Parseval N, Heidmann T. Functional characterization of two newly identified Human Endogenous Retrovirus coding envelope genes. Retrovirology. 2005;2:19. doi: 10.1186/1742-4690-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blond JL, Beseme F, Duret L, Bouton O, Bedin F, Perron H, Mandrand B, Mallet F. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J Virol. 1999;73:1175–1185. doi: 10.1128/jvi.73.2.1175-1185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer J. Phase II Anti-PD1 Epigenetic Priming Study in NSCLC. (NA_00084192) 2015 [Google Scholar]
- Brahmer JR, Drake CG, Wollner I, Powderly JD, Picus J, Sharfman WH, Stankevich E, Pons A, Salay TM, McMiller TL, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. 2010;28:3167–3175. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Shi RZ, Rowley JD. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli KB, Haynes BC, Brent MR, Goodfellow PJ. Reduced DICER1 elicits an interferon response in endometrial cancer cells. Mol Cancer Res. 2012;10:316–325. doi: 10.1158/1541-7786.MCR-11-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper LJ, Krishnamurthy J, Rabinovich B, Mi T, Switzer K, Olivares S, Maiti S, Plummer JB, Singh H, Kumaresan P, et al. Genetic Engineering of T cells to Target HERV-K, an Ancient Retrovirus on Melanoma. Clin Cancer Res. 2015 doi: 10.1158/1078-0432.CCR-14-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Parseval N, Lazar V, Casella JF, Benit L, Heidmann T. Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope proteins. J Virol. 2003;77:10414–10422. doi: 10.1128/JVI.77.19.10414-10422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, Khoury-Hanold W, Iwasaki A, Robek MD. Epigenetic reprogramming of the type III interferon response potentiates antiviral activity and suppresses tumor growth. PLoS Biol. 2014;12:e1001758. doi: 10.1371/journal.pbio.1001758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, Robek MD. Peroxisomal MAVS activates IRF1-mediated IFN-lambda production. Nat Immunol. 2014;15:700–701. doi: 10.1038/ni.2924. [DOI] [PubMed] [Google Scholar]
- Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake CG, Jaffee E, Pardoll DM. Mechanisms of immune evasion by tumors. Advances in immunology. 2006;90:51–81. doi: 10.1016/S0065-2776(06)90002-9. [DOI] [PubMed] [Google Scholar]
- Egan MF, Straub RE, Goldberg TE, Yakub I, Callicott JH, Hariri AR, Mattay VS, Bertolino A, Hyde TM, Shannon-Weickert C, et al. Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia. Proc Natl Acad Sci U S A. 2004;101:12604–12609. doi: 10.1073/pnas.0405077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G,, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez J, Montgiraud C, Pichon JP, Bonnaud B, Arsac M, Ruel K, Bouton O, Mallet F. Custom human endogenous retroviruses dedicated microarray identifies self-induced HERV-W family elements reactivated in testicular cancer upon methylation control. Nucleic Acids Res. 2010;38:2229–2246. doi: 10.1093/nar/gkp1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015 doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J, Gupta R, Filipp FV. Cancer systems biology of TCGA SKCM: efficient detection of genomic drivers in melanoma. Scientific reports. 2015;5:7857. doi: 10.1038/srep07857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke C, Strissel P, Schubert M-T, Mitchell M, Stolt C, Faschingbauer F, Beckmann M, Strick R. Selective expression of sense and antisense transcripts of the sushi ichi-related retrotransposon - derived family during mouse placentogenesis. Retrovirology. 2015;12:9. doi: 10.1186/s12977-015-0138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst T, Magiorkinis G. Activation of the innate immune response by endogenous retroviruses. J Gen Virol. 2014 doi: 10.1099/jgv.0.000017. [DOI] [PubMed] [Google Scholar]
- Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SR, Cedeno CD, Sharma A, Zhang W, Mohler JL, Odunsi K, Wilson EM, Karpf AR. DNA methylation and nucleosome occupancy regulate the cancer germline antigen gene MAGEA11. Epigenetics. 2013;8:849–863. doi: 10.4161/epi.25500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, Sebree R, Rodgers K, Hooker CM, Franco N, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminskas E, Farrell A, Abraham S, Baird A, Hsieh LS, Lee SL, Leighton JK, Patel H, Rahman A, Sridhara R, et al. Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res. 2005;11:3604–3608. doi: 10.1158/1078-0432.CCR-04-2135. [DOI] [PubMed] [Google Scholar]
- Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Karpf AR, Bai S, James SR, Mohler JL, Wilson EM. Increased expression of androgen receptor coregulator MAGE-11 in prostate cancer by DNA hypomethylation and cyclic AMP. Mol Cancer Res. 2009;7:523–535. doi: 10.1158/1541-7786.MCR-08-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpf AR, Lasek AW, Ririe TO, Hanks AN, Grossman D, Jones DA. Limited gene activation in tumor and normal epithelial cells treated with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine. Mol Pharmacol. 2004;65:18–27. doi: 10.1124/mol.65.1.18. [DOI] [PubMed] [Google Scholar]
- Karpf AR, Peterson PW, Rawlins JT, Dalley BK, Yang Q, Albertsen H, Jones DA. Inhibition of DNA methyltransferase stimulates the expression of signal transducer and activator of transcription 1, 2, and 3 genes in colon tumor cells. Proc Natl Acad Sci U S A. 1999;96:14007–14012. doi: 10.1073/pnas.96.24.14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaeva OI, Draghici S, Tang L, Kraniak JM, Land SJ, Tainsky MA. Epigenetic silencing of multiple interferon pathway genes after cellular immortalization. Oncogene. 2003;22:4118–4127. doi: 10.1038/sj.onc.1206594. [DOI] [PubMed] [Google Scholar]
- Larsen JM, Christensen IJ, Nielsen HJ, Hansen U, Bjerregaard B, Talts JF, Larsson LI. Syncytin immunoreactivity in colorectal cancer: potential prognostic impact. Cancer Lett. 2009;280:44–49. doi: 10.1016/j.canlet.2009.02.008. [DOI] [PubMed] [Google Scholar]
- Laska MJ, Nissen KK, Nexo BA. (Some) cellular mechanisms influencing the transcription of human endogenous retrovirus, HERV-Fc1. PLoS One. 2013;8:e53895. doi: 10.1371/journal.pone.0053895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015 doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271:1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- Leonova KI, Brodsky L, Lipchick B, Pal M, Novototskaya L, Chenchik AA, Sen GC, Komarova EA, Gudkov AV. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci U S A. 2013;110:E89–98. doi: 10.1073/pnas.1216922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chiappinelli KB, Guzzetta AA, Easwaran H, Yen RW, Vatapalli R, Topper MJ, Luo J, Connolly RM, Azad NS, et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget. 2014;5:587–598. doi: 10.18632/oncotarget.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower R, Lower J, Tondera-Koch C, Kurth R. A general method for the identification of transcribed retrovirus sequences (R-U5 PCR) reveals the expression of the human endogenous retrovirus loci HERV-H and HERV-K in teratocarcinoma cells. Virology. 1993;192:501–511. doi: 10.1006/viro.1993.1066. [DOI] [PubMed] [Google Scholar]
- Maksakova IA, Mager DL, Reiss D. Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cell Mol Life Sci. 2008;65:3329–3347. doi: 10.1007/s00018-008-8494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankan AK, Schmidt T, Chauhan D, Goldeck M, Honing K, Gaidt M, Kubarenko AV, Andreeva L, Hopfner KP, Hornung V. Cytosolic RNA:DNA hybrids activate the cGAS-STING axis. EMBO J. 2014;33:2937–2946. doi: 10.15252/embj.201488726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matei D, Fang F, Shen C, Schilder J, Arnold A, Zeng Y, Berry WA, Huang T, Nephew KP. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012;72:2197–2205. doi: 10.1158/0008-5472.CAN-11-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouskova M, Blazkova J, Pajer P, Pavlicek A, Hejnar J. CpG methylation suppresses transcriptional activity of human syncytin-1 in non-placental tissues. Exp Cell Res. 2006;312:1011–1020. doi: 10.1016/j.yexcr.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- Nelson PN, Carnegie PR, Martin J, Davari Ejtehadi H, Hooley P, Roden D, Rowland-Jones S, Warren P, Astley J, Murray PG. Demystified. Human endogenous retroviruses. Mol Pathol. 2003;56:11–18. doi: 10.1136/mp.56.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, Clements EG, Cai Y, Van Neste L, Easwaran H, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606–619. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odunsi K, Matsuzaki J, James SR, Mhawech-Fauceglia P, Tsuji T, Miller A, Zhang W, Akers SN, Griffiths EA, Miliotto A, et al. Epigenetic potentiation of NY-ESO-1 vaccine therapy in human ovarian cancer. Cancer immunology research. 2014;2:37–49. doi: 10.1158/2326-6066.CIR-13-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Ogasawara H, Kaneko H, Hishikawa T, Sekigawa I, Hashimoto H, Maruyama N, Kaneko Y, Yamamoto N. Role of DNA methylation in transcription of human endogenous retrovirus in the pathogenesis of systemic lupus erythematosus. J Rheumatol. 2002;29:1678–1682. [PubMed] [Google Scholar]
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79:3851–3854. doi: 10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rote NS, Chakrabarti S, Stetzer BP. The role of human endogenous retroviruses in trophoblast differentiation and placental development. Placenta. 2004;25:673–683. doi: 10.1016/j.placenta.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Ruebner M, Strissel PL, Ekici AB, Stiegler E, Dammer U, Goecke TW, Faschingbauer F, Fahlbusch FB, Beckmann MW, Strick R. Reduced syncytin-1 expression levels in placental syndromes correlates with epigenetic hypermethylation of the ERVW-1 promoter region. PLoS One. 2013;8:e56145. doi: 10.1371/journal.pone.0056145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rycaj K, Plummer JB, Yin B, Li M, Garza J, Radvanyi LG, Ramondetta LM, Lin K, Johanning GL, Tang DG, et al. Cytotoxicity of Human Endogenous Retrovirus K Specific T Cells Toward Autologous Ovarian Cancer Cells. Clin Cancer Res. 2014 doi: 10.1158/1078-0432.CCR-14-0388. [DOI] [PubMed] [Google Scholar]
- Santoni FA, Guerra J, Luban J. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology. 2012;9:111. doi: 10.1186/1742-4690-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Hagemann S, Hanna K, Lyko F. Azacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell lines. Cancer Res. 2009;69:8127–8132. doi: 10.1158/0008-5472.CAN-09-0458. [DOI] [PubMed] [Google Scholar]
- Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- Sistigu A, Yamazaki T, Vacchelli E, Chaba K, Enot DP, Adam J, Vitale I, Goubar A, Baracco EE, Remedios C, et al. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat Med. 2014;20:1301–1309. doi: 10.1038/nm.3708. [DOI] [PubMed] [Google Scholar]
- Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel S, Fiebig U, Kurth R, Denner J. Regulation of human endogenous retrovirus-K expression in melanomas by CpG methylation. Genes Chromosomes Cancer. 2010;49:401–411. doi: 10.1002/gcc.20751. [DOI] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stresemann C, Brueckner B, Musch T, Stopper H, Lyko F. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006;66:2794–2800. doi: 10.1158/0008-5472.CAN-05-2821. [DOI] [PubMed] [Google Scholar]
- Strick R, Ackermann S, Langbein M, Swiatek J, Schubert SW, Hashemolhosseini S, Koscheck T, Fasching PA, Schild RL, Beckmann MW, et al. Proliferation and cell-cell fusion of endometrial carcinoma are induced by the human endogenous retroviral Syncytin-1 and regulated by TGF-beta. Journal of molecular medicine. 2007;85:23–38. doi: 10.1007/s00109-006-0104-y. [DOI] [PubMed] [Google Scholar]
- Strissel PL, Ruebner M, Thiel F, Wachter D, Ekici AB, Wolf F, Thieme F, Ruprecht K, Beckmann MW, Strick R. Reactivation of codogenic endogenous retroviral (ERV) envelope genes in human endometrial carcinoma and prestages: Emergence of new molecular targets. Oncotarget. 2012;3:1204–1219. doi: 10.18632/oncotarget.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su WY, Li JT, Cui Y, Hong J, Du W, Wang YC, Lin YW, Xiong H, Wang JL, Kong X, et al. Bidirectional regulation between WDR83 and its natural antisense transcript DHPS in gastric cancer. Cell Res. 2012;22:1374–1389. doi: 10.1038/cr.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, Kim Y, Cho WG, Kaneko H, Fowler BJ, Bogdanovich S, et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149:847–859. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Drake CG, Pardoll DM. Immune Checkpoint Blockade: A Common Denominator Approach to Cancer Therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristem M. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J Virol. 2000;74:3715–3730. doi: 10.1128/jvi.74.8.3715-3730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HC, Li H, Van Neste L, Cai Y, Robert C, Rassool FV, Shin JJ, Harbom KM, Beaty R, Pappou E, et al. Transient Low Doses of DNA-Demethylating Agents Exert Durable Antitumor Effects on Hematological and Epithelial Tumor Cells. Cancer Cell. 2012;21:430–446. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol. 2001;11:1531–1535. doi: 10.1016/s0960-9822(01)00455-9. [DOI] [PubMed] [Google Scholar]
- Villesen P, Aagaard L, Wiuf C, Pedersen FS. Identification of endogenous retroviral reading frames in the human genome. Retrovirology. 2004;1:32. doi: 10.1186/1742-4690-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Wang-Johanning F, Liu J, Rycaj K, Huang M, Tsai K, Rosen DG, Chen DT, Lu DW, Barnhart KF, Johanning GL. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer. 2007;120:81–90. doi: 10.1002/ijc.22256. [DOI] [PubMed] [Google Scholar]
- Wang-Johanning F, Rycaj K, Plummer JB, Li M, Yin B, Frerich K, Garza JG, Shen J, Lin K, Yan P, et al. Immunotherapeutic potential of anti-human endogenous retrovirus-K envelope protein antibodies in targeting breast tumors. J Natl Cancer Inst. 2012;104:189–210. doi: 10.1093/jnci/djr540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JS, D'Angelo SP, Minor D, Hodi FS, Gutzmer R, Neyns B, Hoeller C, Khushalani NI, Miller WH, Jr., Lao CD, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2015;16:375–384. doi: 10.1016/S1470-2045(15)70076-8. [DOI] [PubMed] [Google Scholar]
- Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrangle J, Wang W, Koch A, Easwaran H, Mohammad HP, Vendetti F, Vancriekinge W, Demeyer T, Du Z, Parsana P, et al. Alterations of immune response of Non-Small Cell Lung Cancer with Azacytidine. Oncotarget. 2013;4:2067–2079. doi: 10.18632/oncotarget.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. NucleicAcidsRes. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamarin D, Holmgaard RB, Subudhi SK, Park JS, Mansour M, Palese P, Merghoub T, Wolchok JD, Allison JP. Localized oncolytic virotherapy overcomes systemic tumor resistance to immune checkpoint blockade immunotherapy. Sci Transl Med. 2014;6:226ra232. doi: 10.1126/scitranslmed.3008095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.