Summary
Unlike a static and immobile skeleton, the actin cytoskeleton is a highly dynamic network of filamentous actin (F-actin) polymers that continuously turn over. In addition to generating mechanical forces and sensing mechanical deformation, dynamic F-actin networks serve as cellular tracks for myosin motor traffic. However, much of our mechanistic understanding of processive myosins comes from in vitro studies where motility was studied on pre-assembled and artificially stabilized, static F-actin tracks. In this work, we examine the role of actin dynamics in single-molecule myosin motility using assembling F-actin and the two highly processive motors, myosin-5 and myosin-6. These two myosins have distinct functions in the cell and travel in opposite directions along actin filaments [1–3]. Myosin-5 walks towards the barbed ends of F-actin, traveling to sites of actin polymerization at the cell periphery [4]. Myosin-6 walks towards the pointed end of F-actin [5], traveling towards the cell center along older segments of the actin filament. We find that myosin-5 takes 1.3 to 1.5-fold longer runs on ADP•Pi (young) F-actin, while myosin-6 takes 1.7 to 3.6-fold longer runs along ADP (old) F-actin. These results suggest that conformational differences between ADP•Pi and ADP F-actin tailor these myosins to walk farther toward their preferred actin filament end. Taken together, these experiments define a new mechanism by which myosin traffic may sort to different F-actin networks depending on filament age.
Results
Actin Nucleotide Turnover as an Indicator of Filament Age
Actin filaments in eukaryotic cells are typically born near the plasma membrane, and age as they travel inward by retrograde flow [6]. Concomitantly, they acquire diverse actin binding proteins that tune F-actin organization and dynamics, defining subcellular actin compartments. These actin binding proteins could alter myosin motility, direct myosins to separate compartments, and ultimately serve to organize the cell. One example is tropomyosins, which differentially direct myosin-1s and myosin-2s [7, 8] and are essential for the processivity of budding yeast myosin-5 [9].
Here we consider a simple process that might generate distinct F-actin populations independent of actin binding proteins, namely actin filament aging. As new ATP-actin monomers add to the barbed end of a nascent filament, aging begins with ATP turnover serving as an internal molecular clock. Filament aging is a process that occurs in three sequential events: addition of an ATP actin monomer to the barbed end of the filament, ATP hydrolysis, and phosphate release [10]. ATP hydrolysis occurs ~3 s after monomeric ATP-actin adds to the F-actin barbed end. At typical growth rates of ~10 monomers/s at 1.0 μM globular actin (G-actin), the barbed-end ATP cap has 30 monomers and is ~100 nm long. Release of inorganic phosphate (Pi) from ADP•Pi F-actin is 100-fold slower, taking ~380 s and yielding ADP F-actin that persists for the lifetime of the filament. ATP hydrolysis and Pi release are stochastic processes, so the ATP and ADP•Pi populations decay from the barbed to the pointed end as approximately single exponential functions. While ATP and ADP•Pi F-actin are structurally similar, ADP F-actin is less stable and more flexible [11, 12]. Thus, we focus here on the transition from ADP•Pi to ADP actin.
Reconstituting Myosin Motility on Growing Actin Filaments
Standard motility assays use phalloidin-stabilized actin filaments that are prepared in advance [13, 14]. Phalloidin is typically added after the assembly reaction reaches steady-state, although most of the myosin motility studies do not indicate when phalloidin was added, only that it was present. Here we examined the motile properties of myosin-5 and myosin-6 on growing filaments with an ADP•Pi population gradient (“ADP•Pi decay”), compared to aged, phalloidin-stabilized actin that is in a uniform ADP state (Figure 1A). In both cases, the actin was polymerized with 5% TMR-actin for visualization. Dual-color TIRF imaging shows the continuous growth of Factin at the barbed end and directed myosin movements along the filament (Figure 1B, Movie S1).
Figure 1.
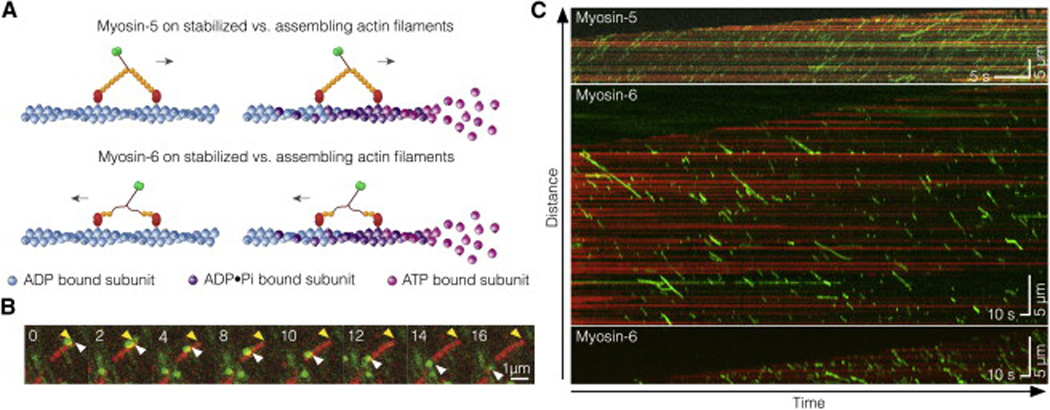
In vitro reconstitution of myosin-5 and myosin-6 motility on assembling F-actin. (A) Schematic of the experiments. Fluorescently labeled myosin-5 (top) and myosin-6 (bottom) walk along two kinds of actin tracks: phalloidin stabilized F-actin (left) and assembling F-actin (right). Nucleotide turnover on actin is illustrated by the transition from pink to blue subunits. (B) Time-lapse fluorescence micrographs of a single myosin-6 motor (green) moving along a single growing actin filament (red). Yellow arrowheads mark the growing F-actin barbed end, white arrowheads mark a single myosin traveling away from the growing end. Time stamp is in s. (C) Representative kymographs showing processive motility of 5 nM myosin-5 and myosin-6 on growing F-actin. Processive runs of myosins appear as green diagonal lines. Actin is shown in red, illustrating elongation of the barbed end toward the kymograph top.
Myosin Runlengths Are Sensitive to Growing or Static F-Actin
We projected actin filament tracings along the time axis to generate kymographs (Figure. 1C). We also separately traced the filament barbed and pointed ends in the kymographs. Myosin runs appear as diagonal lines with a slope that reports the speed and a projected y-axis displacement that reports the runlength. As expected, myosin-5 travels toward the growing barbed end at the top of kymographs, while myosin-6 travels toward the relatively static pointed end at the bottom of kymographs (Figure. 1C).
There is no significant change in either motor's speed (Figure S1AB, Table S1). Likewise, F-actin assembly rates are unaffected by either myosin motor (Figure S1C). However, elongating filaments affected myosin runlengths with approximately equal magnitude, but in opposite directions for the two myosins. Myosin-5 mean runlengths are 1.4-fold longer on growing filaments (ADP•Pi Decay) compared to phalloidin-stabilized filaments (ADP) (Figures 2Ai, iii and 2B, Table S2). Conversely, Myosin-6 mean runlengths are 1.7-fold longer on phalloidin-stabilized filaments (ADP) compared to growing actin (ADP•Pi Decay) (Figures 2Ai, iii and 2C, Table S2).
Figure 2.
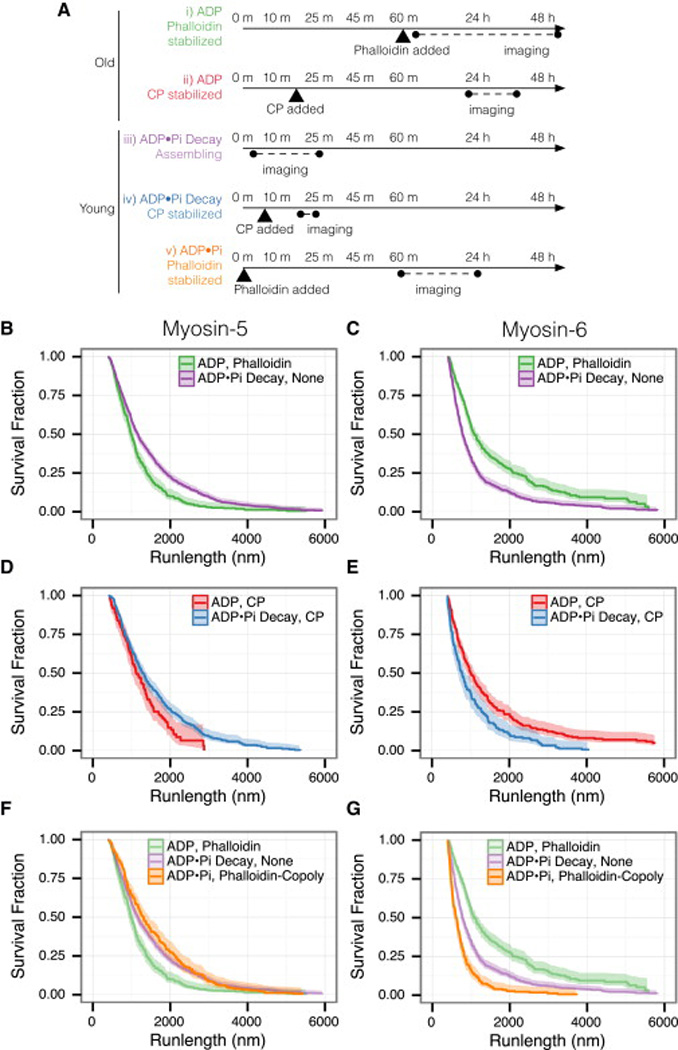
Myosin-5 and myosin-6 runlengths respond to actin nucleotide state in opposite ways. Myosin-5 prefers young filaments, while myosin-6 prefers old. (A) Experimental timecourse. Actin polymerization begins at time zero. Experimental conditions are listed that yield: ADP F-actin (i, ii), mixed ADP and ADP•Pi F-actin (iii, iv), and uniform ADP•Pi F-actin (v). See Supplemental Experimental Details for exact conditions. (B–G) Runlengths of myosins along the F-actins listed in (A). The nucleotide state of the actin and the actin stabilizer are indicated. (B) Runlengths of myosin-5 on filaments assembled without stabilizer (iii) or stabilized with phalloidin after aging (i). Myosin-5 runs 1.4-fold farther on assembling actin (p = 2 × 10−8). (C) Runlengths of myosin-6, as in (B). Myosin-6 runs 1.7-fold farther on aged, phalloidin-stabilized actin (p = 0). (D) Runlengths of myosin-5 on capped (iv) or capped and aged (ii) F-actin. Myosin-5 runs 1.3-fold farther on younger filaments (p = 7 × 10−4). (E) Runlengths of myosin-6, as in (D). Myosin-6 runs 2-fold farther on the older filaments (p = 1 × 10−7). (F) Myosin-5 runlengths on F-actin co-polymerized with phalloidin to trap the ADP•Pi state (v). Myosin-5 runs 1.5-fold farther on the trapped ADP•Pi F-actin than on the ADP phalloidin F-actin (p = 3 × 10−9). Runlength curves from (B) are shown for comparison. (G) Runlengths of myosin-6, as in (F). Myosin-6 runs 3.6-fold farther on the ADP phalloidin F-actin (p = 0). All curves show the Kaplan-Meier estimator of the runlength survivor function; bands report the 0.95 CI. Events are left truncated at 400 nm and are right censored at filament ends. Reported fold-differences apply to mean runlengths, and p-values report the log-rank test. See Table S2 for summary statistics.
Myosin Runlengths Correlate with Filament Age, but not the Stabilizer
Two primary features differ between the growing and stabilized filaments: the nucleotide state of the filament, and the presence or absence of phalloidin. To determine the basis of the runlength effect, we stabilized actin filaments without phalloidin. We added 10 nM capping protein after initial filament assembly to prevent the depolymerization of aged filaments upon dilution. Although elongation is terminated, this arrangement produces F-actin with an ADP•Pi decay along its length (Figure 2Aii, iv). To emulate the phalloidin-stabilized filaments in the ADP state, we simply aged the capped filaments before imaging (Figure 2Aii, ii).
We observed the same trends on capped and aged filaments as on phalloidin-stabilized filaments. Myosin-5 runs 1.3-fold farther on capped ADP•Pi actin decay filaments (Figure 2D), whereas myosin-6 runs 2-fold farther on capped ADP actin filaments (Figure 2E). We expect weaker runlength effects here, because after capping the filament decays to the ADP state without replenishment of ATP-monomers at the barbed end. Myosin runlengths on freshly capped filaments resemble the runlengths on growing actin tracks, while aged and capped filaments resemble aged and phallodin-stabilized filaments (Figure S2). We conclude that phalloidin does not directly alter myosin runlengths.
The Actin Nucleotide State Dictates Myosin Runlengths
Our results suggest that the actin nucleotide state itself influences myosin-5 and myosin-6 processivity. We tested this hypothesis using phalloidin to generate actin filaments containing exclusively ADP•Pi or ADP subunits. Phalloidin dramatically slows Pi release, inhibiting the transition from ADP•Pi to ADP actin (for >20 h), while leaving ATP-hydrolysis unaffected [15]. Thus, copolymerization of actin with phalloidin yields uniform ADP•Pi filaments (Figure 2A, v). In contrast, addition of phalloidin after filaments have assembled and aged yields exclusively ADP filaments (Figure 2A, i). We rejected alternative approaches involving nucleotide analogs or Pi competition because of their direct effects on myosin activity.
Myosin motility on phalloidin-stabilized ADP•Pi or ADP actin filaments show that runlength trends follow the nucleotide state. Myosin-5 mean runlengths are 1.5-fold longer on ADP•Pi than on ADP phalloidin filaments. Conversely, myosin-6 runlengths are 3.6-fold longer on ADP than on ADP•Pi phalloidin filaments (Figure 2F and G, Table S2). Because these F-actin tracks have a uniform nucleotide state, rather than a mixture in our other experiments, this phalloidin experiment should more closely report the intrinsic discrimination ability of each myosin for ADP•Pi vs. ADP actin filaments.
If the nucleotide state of actin dictates myosin runlength, the transition to aged filaments should be detectable on the minute timescale. We therefore performed a time-course experiment in which myosin-5 runlengths were measured on assembling F-actin tracks where polymerization was quenched zero or five minutes prior to imaging. As predicted, myosin-5 runs 1.6-fold farther on zero minute vs. five minute old F-actin (1.6-fold, Figure S3).
Nucleotide State Preferences Are Apparent on Individual Growing Filaments
Given their preference for different actin nucleotide states, we looked at myosin-5 and myosin-6 runlengths on different regions of individual growing filaments. Myosins that travel near the growing barbed end sample predominantly ADP•Pi-actin, while those near the pointed end sample predominantly ADP-actin. To correlate myosin runlengths to actin nucleotide state, we associated each point in a myosin run with the age of the actin monomer at that point. From the actin age and the reported phosphate release decay rate, we calculated the probability of each actin subunit being in the ADP•Pi-actin state (P(ADP•Pi)). The schematic kymograph in Figure 3A of a representative assembling F-actin track shows P(ADP•Pi) along an aging filament.
Figure 3.
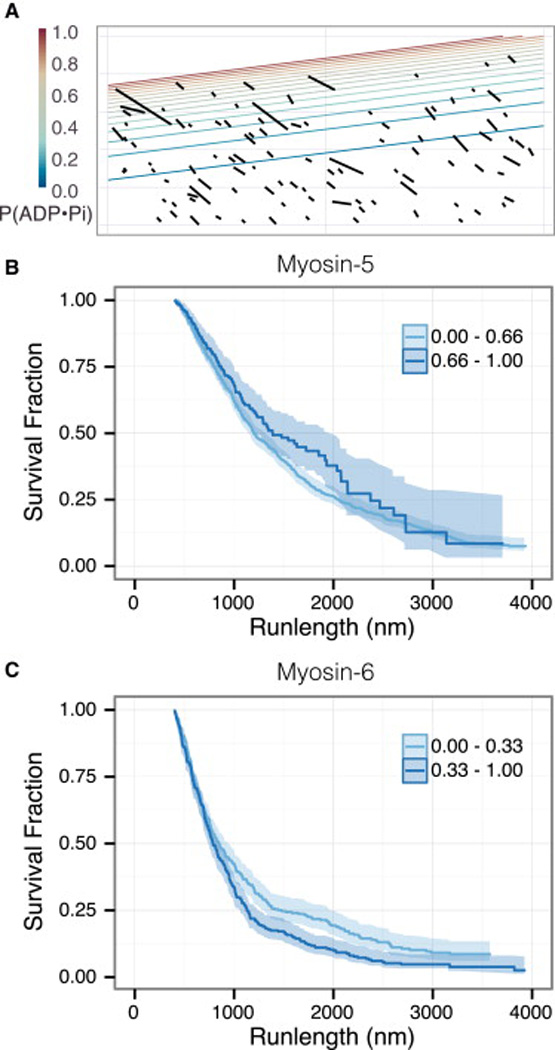
Myosin-5 and myosin-6 runlengths respond to nucleotide state gradients within growing filaments. (A) A schematic kymograph of myosin-6 runs on a growing filament (Figure 2Aiii), based on the kymograph in Figure 1C. Black lines indicate motor runs, contour lines indicate the nucleotide state probabilities along the actin filament. The P(ADP•Pi) values decay from one to zero from the barbed to the pointed end. (B) Myosin-5 runlengths, separated into two classes of P(ADP•Pi) values. Myosin-5 runs farther along stretches of F-actin in the upper third of P(ADP•Pi) values (p = 0.05, log-rank test). (C) Myosin-6 runlengths, separated into two classes. Myosin-6 moves farther along the stretches of actin in the lower third of P(ADP•Pi) values (p = 0.002, log-rank test).
We grouped myosin-5 and myosin-6 runs along growing filaments into two categories, low or high P(ADP•Pi) actin. Consistent with our earlier findings, the myosins exhibit opposite runlength preferences. Myosin-5 takes 1.1-fold longer runs in the actin zone with the upper 33% of P(ADP•Pi) values (Figure 3B, Table S3), where myosin-6 runs are 1.8-fold longer in the bottom 33% of P(ADP•Pi) values (Figure 3C, Table S3). Thus, these two myosins can sense the F-actin nucleotide state in different regions of the same set of filaments. This finding rules out a concerted, all-or-none conformational change in the filament; instead, the structural features of Factin that these two myosins sense must be local in nature.
The Actin Nucleotide State Also Affects Myosin Landing Rates
Overall cellular transport may be regulated by controlling either the runlength or the frequency of myosin runs. To determine if actin nucleotide state also regulates run frequency, we examined the rate at which myosins encounter F-actin and start a processive run, also known as the landing rate (Figure 4). In general, conditions that favor increased runlengths also favor increased landing rates. For example, median myosin-5 landing rates are 4-fold enhanced on ADP•Pi, Phalloidin-Copoly F-actin over ADP, Phalloidin F-actin (Figure 4A). Likewise, median myosin-6 landing rates are 2-fold enhanced on ADP, Phalloidin filaments, compared to ADP•Pi, Phalloidin-Copoly filaments (Figure 4B). Increased landing rates drive myosins on to filament populations that they are tailored for longer runlengths, leading to an overall increase in trafficking capacity.
Figure 4.
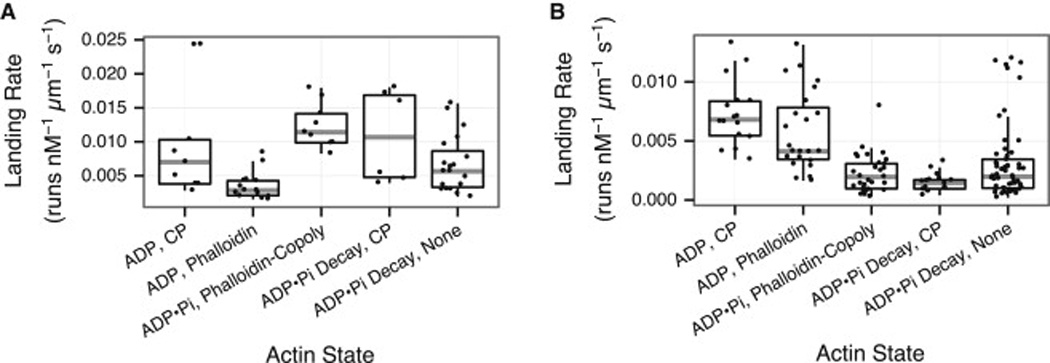
Myosin-5 lands more frequently on ADP•Pi F-actin, while myosin-6 lands more frequently on ADP F-actin. Landing rates (the rate of initiating a processive run) are shown for myosin-5 (A) and myosin-6 (B). Myosin-5 lands significantly more often on ADP•Pi, Phalloidin-Copoly F-actin vs. ADP, Phalloidin F-actin (Figure 2Av vs. 2Ai, p = 5 × 10−6, Wilcoxon rank-sum test). Conversely, myosin-6 lands significantly more often on ADP, Phalloidin F-actin vs. ADP•Pi, Phalloidin-Copoly F-actin (Figure 2Ai vs. 2Av, p = 2 × 10−10, Wilcoxon rank-sum test).
Discussion
We find that myosin-5 runs farther on the younger, ADP•Pi-rich actin, while myosin-6 travels farther on older, ADP-rich actin. Interestingly, one study found that the myosin-5 runlength in untreated cells was 1.5-fold higher than in cells with blocked F-actin dynamics [16]. This difference is strikingly similar to what we find here, under conditions where F-actin growth rates are also similar (20 monomers/s vs. 10 monomers/s). Our results are also consistent with a 20% reduction of kinesin processivity on GMPCPP microtubules vs. taxol-stabilized GDP microtubules [17], illustrating that similar principles apply to microtubule tracks and motors.
Coupling of myosin motor activity to actin’s nucleotide state suggests two guidelines for future work. First, because order of addition matters, it is important to state when phalloidin is added in F-actin polymerization. We suspect that phalloidin is typically added after completion of polymerization. Second, when myosin motility is examined at high Pi concentrations, secondary actin effects should be considered alongside the primary myosin effects on force generation and stiffness [18].
We find that myosin runlengths are affected by the actin nucleotide state, while myosin speeds are not. Myosin runlengths increase with the duty ratio, defined as fraction of the ATPase cycle time when the motor is bound to its filament. An increased duty ratio decreases the period in which the myosin is vulnerable to detach. The two ways to increase the myosin duty ratio are to accelerate rebinding to F-actin or to inhibit F-actin detachment. Because the speed does not change, and because ADP release gates detachment while being rate-limiting for both of these myosins [19, 20], the F-actin nucleotide state likely affects the overall rate of strong binding to actin. This accelerated binding scenario is consistent with the increases in myosin landing rates in Figure 4, on opposite types of actin for myosin-5 and myosin-6.
Myosins may sense the actin nucleotide state in two ways. The first is a classical molecular recognition mechanism involving actin subdomain 2 (SD2), a key determinant of filament flexibility and stability. Release of Pi causes SD2 to rotate 15°, closing the nucleotide-binding cleft [21, 22]. In addition, the DNAse I binding loop folds into an alpha helix upon Pi release [23], changing the longitudinal subunit contacts within the filament and affecting filament bending stiffness [24]. Myosin-5 and myosin-6 may directly sense the orientation of actin SD2. Recent work revealed that strong binding of myosin to F-actin involves interactions between actin SD2 and myosin loop 2, loop 3, and the cardiomyopathy loop [25]. There are many sequence differences between myosin-5 and myosin-6 at the actin binding interface that could influence sensing of the SD2 orientation. In particular, loop 2 is longer in myosin-5, and changes in the loop 2 region are known to affect actin binding [26, 27].
Alternatively, these two myosins may recognize flexibility differences between the stiffer ADP•Pi state and the softer ADP state of F-actin. Stiffened F-actin inhibits gliding filament motility on skeletal myosin-2, without affecting myosin enzymatic activity [28]. Furthermore, both myosin-5 and myosin-6 decoration increase the torsional dynamics of Factin, while myosin-2 has the opposite effect [29, 30]. By thermodynamic linkage, we expect that changes in filament dynamics also affect myosin binding in an isoform specific manner. One difficulty with this argument is that myosin-5 and myosin-6 both increase torsional dynamics. However, one significant difference is that these studies used monomeric motor domain “S1” fragments, while ours used dimeric motors. Dimeric myosin-5 may prefer stiffer filaments, because its long and stiff lever arms would serve to buttress the actin filament. In contrast, dimeric myosin-6 has more flexible lever arms that are oriented nearly parallel to the filament [31] and therefore provide little additional stiffening.
Regulation of myosin motility by the nucleotide state of F-actin is expected to be important in cells. Although the nucleotide state of actin filaments in cells has not been experimentally determined, different F-actin networks are enriched for ADP•Pi or ADP F-actin [32]. Because Pi release takes ~6 min [33], networks that turnover in seconds at the leading edge of migrating cells consist primarily of ADP•Pi filaments. Remodeled networks farther from the leading edge turn over slowly and are likely to have ADP filaments. Given that the severing protein ADF/Cofilin binds tighter to ADP F-actin than ADP•Pi F-actin [34], ADF/Cofilin localization might indicate the cellular nucleotide state of F-actin. As predicted, ADF/Cofilin is absent from F-actin at the leading edge of migrating cells, but strongly localizes to F-actin deeper into the cell [35]. Actin binding proteins may also regulate myosin activity in cells by indirectly affecting the nucleotide state of Factin. For example, ADF/Cofilin allosterically increases the amount of ADP F-actin by accelerating the rate of Pi release from unoccupied filament subunits [36].
Because myosin-5 moves 10-fold faster than the elongation rate of the barbed end (~300 nm/s vs. ~30 nm/s), myosin-5 is able to overtake the F-actin ADP•Pi decay and reach increasingly younger actin subunits as it walks. Myosin-6 travels in the direction of aging, and therefore reaches older actin subunits with each step. We find it interesting that each myosin has evolved to move in a direction that carries it to more favorable transport conditions. Thus, myosin-5 and myosin-6 both head toward greener pastures, allowing enhanced transport of myosin-5 cargoes to the cell periphery and myosin-6 cargoes to the cell interior. Although we fully expect that ABPs in the cell further modulate myosin activity, such effects would be in addition to the inherent ability of these myosins to discriminate filaments based on age.
Experimental Procedures
Proteins and imaging conditions are described in the Supplemental Experimental Procedures. Myosin runlengths were analyzed using Kaplan-Meier survival curves to correct for the finite length of the F-actin track [37]. Motor runlengths were left truncated at 400 nm to handle missed events that were too short to detect. Myosin runs known to underestimate the true runlength were treated as right-censored in the Kaplan-Meier estimator. These artificially short runs occur when a myosin reaches the end of a filament, myosins that start at the beginning of a movie or terminate at the end, or myosins that cross the P(ADP•Pi) threshold indicated in Figure 3.
Supplementary Material
Acknowledgments
This work was supported by NIH R01 GM078450 (to R.S.R.), and NIH R01 GM079265 and Human Frontier Science Program grant RGY0071/2011 (to D.R.K.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
D.R.K. and R.S.R. conceived of the experiment. D.Z. performed experiments on myosin-5. A.S. performed experiments on myosin-6. All authors analyzed the data and wrote the manuscript.
References
- 1.Berg JS, Powell BC, Cheney RE. A millennial myosin census. Mol Biol Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buss F, Spudich G, Kendrick-Jones J. Myosin VI: cellular functions and motor properties. Annu Rev Cell Dev Biol. 2004;20:649–676. doi: 10.1146/annurev.cellbio.20.012103.094243. [DOI] [PubMed] [Google Scholar]
- 3.Sellers JR, Goodson HV. Motor proteins 2: myosin. Protein Profile. 1995;2:1323–1423. [PubMed] [Google Scholar]
- 4.Reck-Peterson SL, Provance DW, Mooseker MS, Mercer JA. Class V myosins. Biochim Biophys Acta. 2000;1496:36–51. doi: 10.1016/s0167-4889(00)00007-0. [DOI] [PubMed] [Google Scholar]
- 5.Wells AL, Lin AW, Chen LQ, Safer D, Cain SM, Hasson T, Carragher BO, Milligan RA, Sweeney HL. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 6.Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- 7.Tang N, Ostap EM. Motor domain-dependent localization of myo1b (myr-1) Curr Biol. 2001;11:1131–1135. doi: 10.1016/s0960-9822(01)00320-7. [DOI] [PubMed] [Google Scholar]
- 8.Fanning AS, Wolenski JS, Mooseker MS, Izant JG. Differential regulation of skeletal muscle myosin-II and brush border myosin-I enzymology and mechanochemistry by bacterially produced tropomyosin isoforms. Cell Motil. Cytoskel. 1994;29:29–45. doi: 10.1002/cm.970290104. [DOI] [PubMed] [Google Scholar]
- 9.Hodges AR, Krementsova EB, Bookwalter CS, Fagnant PM, Sladewski TE, Trybus KM. Tropomyosin is essential for processive movement of a class V myosin from budding yeast. Curr Biol. 2012;22:1410–1416. doi: 10.1016/j.cub.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kudryashov DS, Reisler E. ATP and ADP actin states. Biopolymers. 2013;99:245–256. doi: 10.1002/bip.22155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orlova A, Egelman EH. A conformational change in the actin subunit can change the flexibility of the actin filament. J Mol Biol. 1993;232:334–341. doi: 10.1006/jmbi.1993.1393. [DOI] [PubMed] [Google Scholar]
- 12.De La Cruz EM, Roland J, McCullough BR, Blanchoin L, Martiel J-L. Origin of twist-bend coupling in actin filaments. Biophys J. 2010;99:1852–1860. doi: 10.1016/j.bpj.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kron SJ, Toyoshima YY, Uyeda TQ, Spudich JA. Assays for actin sliding movement over myosin-coated surfaces. Methods Enzymol. 1991;196:399–416. doi: 10.1016/0076-6879(91)96035-p. [DOI] [PubMed] [Google Scholar]
- 14.Sellers JR. In vitro motility assay to study translocation of actin by myosin. Chapter 13. Curr Protoc Cell Biol. 2001;(Unit 13.2) doi: 10.1002/0471143030.cb1302s00. [DOI] [PubMed] [Google Scholar]
- 15.Dancker P, Hess L. Phalloidin reduces the release of inorganic phosphate during actin polymerization. Biochim Biophys Acta. 1990;1035:197–200. doi: 10.1016/0304-4165(90)90116-e. [DOI] [PubMed] [Google Scholar]
- 16.Semenova I, Burakov A, Berardone N, Zaliapin I, Slepchenko B, Svitkina T, Kashina A, Rodionov V. Actin dynamics is essential for myosin-based transport of membrane organelles. Curr Biol. 2008;18:1581–1586. doi: 10.1016/j.cub.2008.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McVicker DP, Chrin LR, Berger CL. The nucleotide-binding state of microtubules modulates kinesin processivity and the ability of Tau to inhibit kinesin-mediated transport. J Biol Chem. 2011;286:42873–42880. doi: 10.1074/jbc.M111.292987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caremani M, Dantzig J, Goldman YE, Lombardi V, Linari M. Effect of inorganic phosphate on the force and number of myosin cross-bridges during the isometric contraction of permeabilized muscle fibers from rabbit psoas. Biophys J. 2008;95:5798–5808. doi: 10.1529/biophysj.108.130435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De La Cruz EM, Wells AL, Rosenfeld SS, Ostap EM, Sweeney HL. The kinetic mechanism of myosin V. Proc Natl Acad Sci U S A. 1999;96:13726–13731. doi: 10.1073/pnas.96.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De La Cruz EM, Ostap EM, Sweeney HL. Kinetic mechanism and regulation of myosin VI. J Biol Chem. 2001;276:32373–32381. doi: 10.1074/jbc.M104136200. [DOI] [PubMed] [Google Scholar]
- 21.Isambert H, Venier P, Maggs AC, Fattoum A, Kassab R, Pantaloni D, Carlier MF. Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J Biol Chem. 1995;270:11437–11444. doi: 10.1074/jbc.270.19.11437. [DOI] [PubMed] [Google Scholar]
- 22.Belmont LD, Orlova A, Drubin DG, Egelman EH. A change in actin conformation associated with filament instability after Pi release. Proc Natl Acad Sci U S A. 1999;96:29–34. doi: 10.1073/pnas.96.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graceffa P, Dominguez R. Crystal structure of monomeric actin in the ATP state. Structural basis of nucleotidedependent actin dynamics. J Biol Chem. 2003;278:34172–34180. doi: 10.1074/jbc.M303689200. [DOI] [PubMed] [Google Scholar]
- 24.Chu J-W, Voth GA. Allostery of actin filaments: molecular dynamics simulations and coarse-grained analysis. Proc Natl Acad Sci U S A. 2005;102:13111–13116. doi: 10.1073/pnas.0503732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorenz M, Holmes KC. The actin-myosin interface. Proc Natl Acad Sci U S A. 2010;107:12529–12534. doi: 10.1073/pnas.1003604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy CT, Spudich JA. The sequence of the myosin 50-20K loop affects Myosin's affinity for actin throughout the actin-myosin ATPase cycle and its maximum ATPase activity. Biochemistry. 1999;38:3785–3792. doi: 10.1021/bi9826815. [DOI] [PubMed] [Google Scholar]
- 27.Yengo CM, Sweeney HL. Functional role of loop 2 in myosin V. Biochemistry. 2004;43:2605–2612. doi: 10.1021/bi035510v. [DOI] [PubMed] [Google Scholar]
- 28.Prochniewicz E, Yanagida T. Inhibition of sliding movement of F-actin by crosslinking emphasizes the role of actin structure in the mechanism of motility. J Mol Biol. 1990;216:761–772. doi: 10.1016/0022-2836(90)90397-5. [DOI] [PubMed] [Google Scholar]
- 29.Prochniewicz E, Chin HF, Henn A, Hannemann DE, Olivares AO, Thomas DD, De La Cruz EM. Myosin isoform determines the conformational dynamics and cooperativity of actin filaments in the strongly bound actomyosin complex. J Mol Biol. 2010;396:501–509. doi: 10.1016/j.jmb.2009.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prochniewicz E, Pierre A, McCullough BR, Chin HF, Cao W, Saunders LP, Thomas DD, De La Cruz EM. Actin filament dynamics in the actomyosin VI complex is regulated allosterically by calcium-calmodulin light chain. J Mol Biol. 2011;413:584–592. doi: 10.1016/j.jmb.2011.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menetrey J, Llinas P, Mukherjea M, Sweeney HL, Houdusse A. The structural basis for the large powerstroke of myosin VI. Cell. 2007 Oct 19;131:300–308. doi: 10.1016/j.cell.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 32.Bindschadler M, Osborn EA, Dewey CF, Jr, McGrath JL. A mechanistic model of the actin cycle. Biophys J. 2004;86:2720–2739. doi: 10.1016/S0006-3495(04)74326-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melki R, Fievez S, Carlier MF. Continuous monitoring of Pi release following nucleotide hydrolysis in actin or tubulin assembly using 2-amino-6-mercapto-7-methylpurine ribonucleoside and purine-nucleoside phosphorylase as an enzymelinked assay. Biochemistry. 1996;35:12038–12045. doi: 10.1021/bi961325o. [DOI] [PubMed] [Google Scholar]
- 34.Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suarez C, Roland J, Boujemaa-Paterski R, Kang H, McCullough BR, Reymann A-C, Guérin C, Martiel J-L, De la Cruz EM, Blanchoin L. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr Biol. 2011;21:862–868. doi: 10.1016/j.cub.2011.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. Journal of the American Statistical Association. 1958;53:457–481. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


