ABSTRACT
Clostridium difficile is the cause of most frequently occurring nosocomial diarrhea worldwide. As an enteropathogen, C. difficile must be exposed to multiple exogenous genetic elements in bacteriophage-rich gut communities. CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) systems allow bacteria to adapt to foreign genetic invaders. Our recent data revealed active expression and processing of CRISPR RNAs from multiple type I-B CRISPR arrays in C. difficile reference strain 630. Here, we demonstrate active expression of CRISPR arrays in strain R20291, an epidemic C. difficile strain. Through genome sequencing and host range analysis of several new C. difficile phages and plasmid conjugation experiments, we provide evidence of defensive function of the CRISPR-Cas system in both C. difficile strains. We further demonstrate that C. difficile Cas proteins are capable of interference in a heterologous host, Escherichia coli. These data set the stage for mechanistic and physiological analyses of CRISPR-Cas-mediated interactions of important global human pathogen with its genetic parasites.
IMPORTANCE
Clostridium difficile is the major cause of nosocomial infections associated with antibiotic therapy worldwide. To survive in bacteriophage-rich gut communities, enteropathogens must develop efficient systems for defense against foreign DNA elements. CRISPR-Cas systems have recently taken center stage among various anti-invader bacterial defense systems. We provide experimental evidence for the function of the C. difficile CRISPR system against plasmid DNA and bacteriophages. These data demonstrate the original features of active C. difficile CRISPR system and bring important insights into the interactions of this major enteropathogen with foreign DNA invaders during its infection cycle.
INTRODUCTION
Clostridium difficile is one of the major pathogenic clostridia. This Gram-positive, strictly anaerobic, spore-forming bacterium is found in soil and aquatic environments and in mammalian intestinal tracts. C. difficile became one of the key public health problems in industrialized countries and one of the major nosocomial enteropathogens. C. difficile-associated diarrhea is currently the most frequently occurring nosocomial diarrhea in Europe and worldwide (1, 2). Over the last decade, the proportion of severe C. difficile infections has risen due to the emergence of epidemic PCR ribotype 027 strains, such as the R20291 strain (3). Two major risk factors for contracting C. difficile infections are the age of the individual and exposure to antibiotics. Antibiotic therapy causes alterations in the colonic microflora, allowing the development of C. difficile from preexisting or acquired spores (4, 5). The pathogen synthesizes two major toxins, TcdA and TcdB, which glucosylate host GTPases, resulting in alterations in the enterocyte cytoskeleton (6). This induces intestinal cell lysis and inflammation, resulting in diarrhea, pseudomembranous colitis, and even death (7). Many aspects of the C. difficile infection cycle, including molecular mechanisms of its adaptation to changing conditions still remain poorly understood (8, 9).
During its infection cycle, C. difficile survives within bacteriophage-rich gut communities and is therefore expected to possess efficient systems to control genetic exchanges favored in such environments. The highly mobile and mosaic genome of C. difficile (10) could reflect the continuous balance between the acquisition of adaptive traits for gastrointestinal lifestyle and the efficient defense against abundant invaders, such as phages and plasmids. The CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) systems are found in about half of sequenced bacterial genomes and in almost all archaeal genomes and can provide defense against mobile genetic elements (11, 12). CRISPR loci are arranged in arrays of almost identical direct repeats of ~30 bp separated by similarly sized variable sequences called spacers. Some spacers match viral or plasmid DNA and must have been acquired during prior encounters with mobile genetic elements in a process referred to as “adaptation.” A CRISPR array is transcribed as a single RNA transcript (pre-crRNA) that is processed to generate small CRISPR RNAs (crRNAs), each containing one spacer and flanking repeat fragments. These crRNAs, in complex with Cas proteins, serve as guides to recognize foreign nucleic acids by complementary base pairing. The recognition leads to degradation of targeted nucleic acids, during a process referred to as “interference,” thus protecting cells from invasion by foreign genetic elements.
The cas gene clusters are often associated with CRISPR arrays, and the Cas proteins are involved in all stages of CRISPR-Cas activity (13). According to the analysis of cas gene sets, the most recent classification defines three major types of CRISPR-Cas systems (types I, II, and III) that can be further divided into 12 subtypes (14, 15). Cas1 and Cas2 proteins required for the adaptation step are universal components of all CRISPR-Cas systems. Endoribonucleases of the Cas6 family in type I and type III systems cleave the pre-crRNA, while RNase III is required for crRNA processing in type II systems. For the interference step, mature crRNA is bound by a Cas protein complex known as Cascade (cas complex for antiviral defense) in type I systems or a distinct multisubunit complex in type III systems, whereas type II systems use a single Cas9 protein (16). Type I systems rely on an additional Cas3 protein for the degradation of foreign DNA. The important determinants for self/nonself discrimination by type I and type II systems are the protospacer-adjacent motifs (PAMs). PAMs are located either at the 3′ or 5′ end of the protospacer (a fragment of invader DNA that was earlier acquired as a spacer into CRISPR arrays) (16).
The type I CRISPR-Cas systems are the most diverse and have been classified into subtypes. Archaea contain mainly subtypes I-A, I-B, and I-D, whereas bacteria contain mainly subtypes I-C, I-E, and I-F (14). In contrast to several detailed studies of representatives of subtypes I-A, I-C, and I-E, comparatively little is known about subtypes I-B and I-D. The subtype I-B CRISPR-Cas system is found in clostridia as well as in methanogenic and halophilic archaea and is defined by a subtype-specific protein Cas8b. The analysis of active haloarchaeal subtype I-B systems revealed several distinct features, including multiple PAMs and 9-nucleotide (nt) noncontiguous seed regions for target DNA recognition (17–19). The experimental evidence for the role of Cas8 protein from archaeal subtype I-B system in Cascade targeting to invader DNA has been recently reported (20). Bacterial subtype I-B systems remain less characterized. On the one hand, the possibility of CRISPR-Cas system acquisition by horizontal gene transfer from Archaea has been suggested for some clostridial strains (21, 22). On the other hand, recent characterization of crRNA processing by Cas6b endonuclease in Clostridium thermocellum (22) and results of analysis of CRISPR systems in industrially relevant clostridia (23) are in good agreement with the data obtained for other type I subtypes, suggesting that at least some haloarchaeal subtype I-B systems features may be specific to this ecological group with its long-term independent evolution.
Our recent deep sequencing and Northern blot analysis revealed the active expression of crRNA from multiple subtype I-B CRISPR arrays in C. difficile reference strain 630, including some located within prophage regions (24). In the present study, we show by transcriptome sequencing (RNA-seq) analysis that nine CRISPR arrays are also actively transcribed in the PCR ribotype 027 epidemic C. difficile strain R20291. To demonstrate the functionality of this CRISPR system in C. difficile, we compared spacers of C. difficile strain 630 and epidemic R20291 strains to phage sequences, including several newly sequenced C. difficile phages, and determined the host range for phages harboring sequences matching spacers. A survey of occurrence and cas operon structures in published C. difficile genomes and extended spacer homology search in nine C. difficile strains suggested that the C. difficile CRISPR-Cas system has extensive potential for targeting of phage and prophage genes. The functionality of the C. difficile CRISPR-Cas system and the predicted 5′ CCW PAM could be directly demonstrated using plasmid conjugation efficiency assays in C. difficile and interference assay in a heterologous Escherichia coli host.
RESULTS
Identification of actively expressed CRISPR arrays in C. difficile.
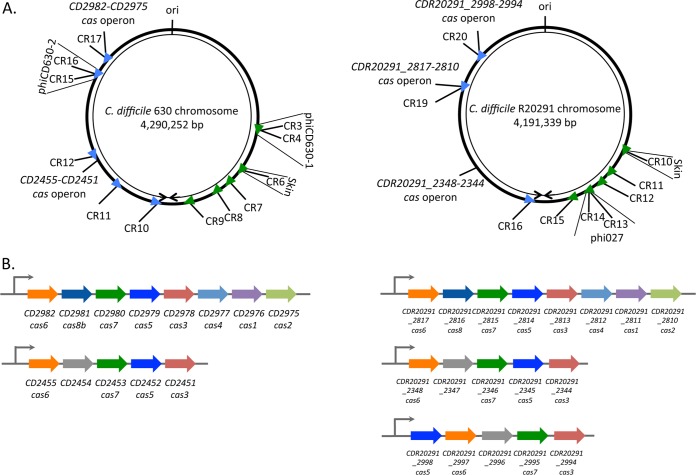
Comparative analysis of subtype I-B CRISPR-Cas systems in the C. difficile reference strain 630 and the PCR ribotype 027 epidemic strain R20291 was performed. According to the CRISPRdb database, C. difficile strain 630 carries genes that encode 12 potential CRISPR arrays, 5 of which are located within prophage regions (Fig. 1; see Table S1 in the supplemental material) (11). For further reference, we used the CRISPRdb numbering for the C. difficile strain 630 CRISPR arrays and numbered spacers within each array according to the identified transcriptional order. All 12 arrays are actively expressed and processed into crRNA under laboratory conditions (24) (see Fig. S1 in the supplemental material). The distribution of CRISPR arrays throughout the chromosome and their orientation appear to be nonrandom (Fig. 1A). CRISPR 3/4, 6, 7, 8, and 9 are transcribed from one DNA strand in the clockwise direction, and CRISPR 10, 11, 12, 15/16, 17 are transcribed from a different strand in the counterclockwise direction. The cas gene cluster CD2982-CD2975 encoding Cas1 to Cas8 proteins is located near the CRISPR 17 array. An additional incomplete cas operon CD2455-CD2451 lacking the universal cas1 component as well as the cas2 and cas4 genes (Fig. 1B) is found close to the CRISPR 12 array (Fig. 1A). Both cas operons are transcribed according to our RNA-seq data.
FIG 1 .
Positions of CRISPR-Cas I-B loci in C. difficile strains 630 and R20291. (A) Schematic view of the genomic locations of expressed CRISPR arrays in strains 630 and R20291. CRISPR arrays (CR) are numbered according to the CRISPRdb database. Arrowheads indicate the array position, and transcriptional orientation is indicated by colors as follows: green for the plus or coding strand and blue for the minus or noncoding strand. The locations of associated cas operons, prophage regions, and replication origin (ori) are indicated. The right and left replichores are shown by black arrowheads. (B) Organization of the operons for the complete (CD2982-CD2975) and partial cas operon (CD2455-CD2451) from C. difficile strain 630 (left) and for the complete (CDR20291_2817-2810) and partial cas operons (CDR20291_2348-2344 and CDR20291_2998-2994) from C. difficile strain R20291 (right). The same color was used for homologous cas genes.
In the epidemic R20291 strain, CRISPRdb predicted 13 CRISPR arrays. Four of the CRISPR arrays are located within the tcdA toxin-encoding gene and contain a 24-bp repeat different from the 29-bp repeat present in reference strain 630 arrays and in the remaining strain R20291 arrays. To investigate the expression of predicted R20291 arrays, we extracted RNA from cultures at late exponential phase and subjected it to RNA-seq analysis. Active expression of 9 CRISPR arrays carrying the 29-bp direct repeats was detected (Fig. 1A; see Fig. S2 in the supplemental material). The highest level of expression was observed for the CRISPR 11, 13, 15, and 16 arrays (Fig. S2). Similar to our previous results obtained with strain 630 (24) (Fig. S1) and in agreement with the data from other bacterial systems (22, 25), the most-abundant sequence reads mapped to leader-proximal regions (Fig. S2). CRISPR “leader” is defined as the region between the transcriptional start site (TSS) and the first repeat of CRISPR arrays. Some genomic sequences coding for proteins containing repetitive motifs may erroneously be considered CRISPR arrays. However, the transcription profiles for such regions and sequence analysis of putative spacers can disprove the prediction. As described in Materials and Methods, we concluded from our RNA-seq data and bioinformatics analysis that predicted arrays with 24-bp repeats within the tcdA gene are not real CRISPR arrays, and they were excluded from further analysis.
Several CRISPR arrays are located in prophages in both C. difficile strains. In strain 630, CRISPR 6 is located within the skin element, and CRISPR 3/4 and CRISPR 15/16 are located within the homologous phiCD630-1 and phiCD630-2 prophages, respectively. The CRISPR 3/4 and CRISPR 15/16 arrays are identical to each other, and each appear to constitute a single array, i.e., are cotranscribed (24). These duplicated arrays are parts of larger duplicated regions corresponding to phiCD630-1 and phiCD630-2 prophages (Fig. 1A). Likewise, in the R20291 strain, CRISPR 10 is located within the skin element, and CRISPR 13 and CRISPR 14 arrays are located within the phi027 prophage (Fig. 1A). Similarly to strain 630, one complete R20291 subtype I-B cas operon CDR20291_2817-CDR20291_2810 (CDR20291_2817-2810) is located near the CRISPR 19 array. The partial operon CDR20291_2348-2344 homologous to the CD2455-CD2451 operon in strain 630 is not associated with a CRISPR array. An additional divergent CDR20291_2998-2994 operon is associated with CRISPR 20 and also lacks cas1, cas2, and cas4 (Fig. 1A and B). Expression of all three operons is detected by RNA-seq analysis.
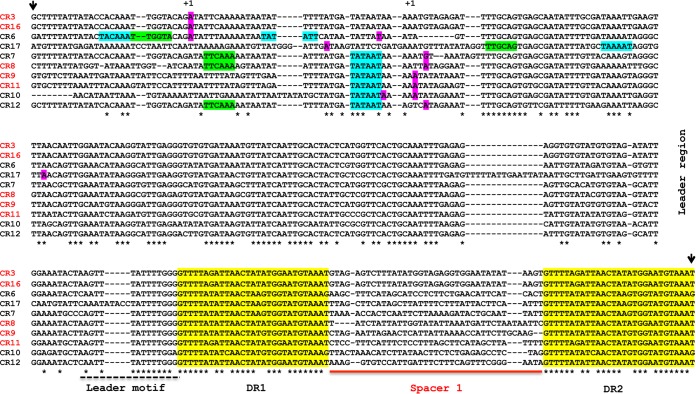
Our recent TSS mapping experiment in strain 630 revealed the presence of 170- to 200-nt transcribed 5′ “leader” regions (24) (see Fig. S1 in the supplemental material). The alignment of expressed CRISPR loci in strain 630 revealed conserved leader elements immediately upstream of the first repeat (Fig. 2). In addition, upstream of the TSS, consensus elements of sigma A-dependent promoters could be identified (Fig. 2). These functional elements are also conserved for the expressed CRISPR arrays from strain R20291 (data not shown).
FIG 2 .
Alignment of CRISPR regions from C. difficile strain 630. The sequences of 10 independent CRISPR arrays (CR) numbered according to the CRISPRdb database were aligned using the CLUSTALW program, and the upstream part of the alignment is shown. The names of the highly expressed arrays are shown in red. The positions of TSS “+1” identified by 5′-end RNA-seq are highlighted in magenta (24). The potential −35 and −10 promoter elements corresponding to sigma A-dependent consensus sequences are indicated by blue and green background, respectively. Direct repeats (DR) are highlighted in yellow and are numbered according to the transcriptional order (DR1, DR2). The sequences of the first spacer (Spacer 1) from each array and a conserved leader motif are indicated by a solid red line and broken black line, respectively. The region used for artificial CRISPR array engineering in the E. coli chromosome is delimited by short black vertical arrows.
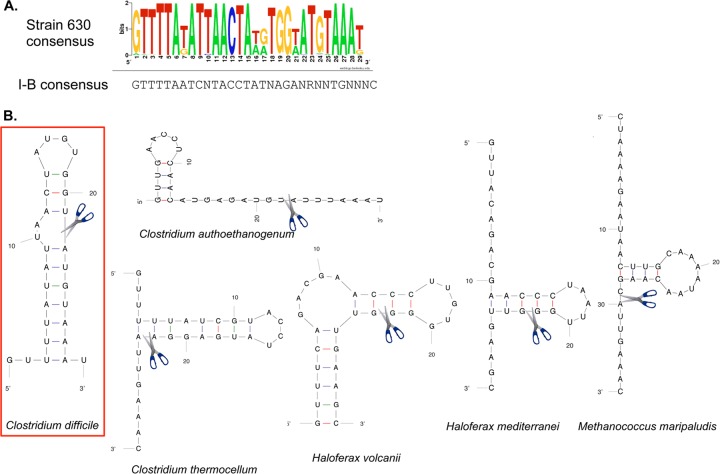
A total of 119 29-bp repeat sequences were collected from the expressed CRISPR loci of strain 630, and aligning these sequences allowed us to establish a consensus sequence shown in Fig. 3A. A total of 105 29-bp repeat sequences were collected from the CRISPR loci expressed in strain R20291 with a consensus sequence similar to that for the strain 630 CRISPR repeats. For some CRISPR arrays in both strains, the leader-distal repeats had a variant sequence that was different from the rest of repeats. The overall high sequence conservation among direct repeats suggests that the same set of Cas proteins processes all expressed pre-crRNAs in strains 630 and R20291. Analysis of repeat consensus sequence using Mfold suggests that in RNA this sequence could form a characteristic stem-loop secondary structure similar to the predicted structures for repeats from other subtype I-B CRISPR-Cas systems (Fig. 3B) (17, 22, 23, 26).
FIG 3 .
C. difficile strain 630 CRISPR repeat consensus sequence in comparison with other CRISPR-Cas I-B systems. (A) The 29-bp direct repeats from all expressed CRISPR arrays of strain 630 were aligned, and a consensus sequence was established on the basis of this alignment using WebLogo (http://weblogo.berkeley.edu). A consensus repeat sequence for subtype I-B CRISPR-Cas systems is shown below the WebLogo sequence and was determined by the method in reference 16 (R means A or G). (B) Predicted RNA secondary structure for repeat sequence of C. difficile strain 630 compared to other repeat sequences for subtype I-B CRISPR-Cas systems (17, 22, 23, 26). The RNA secondary structure was predicted using the Mfold software (66). The proposed position of pre-crRNA cleavage by the Cas6 protein that generates the 8-nt 5′ tag of crRNA during processing is indicated.
Within 12 CRISPR arrays expressed in strain 630, 107 spacers (98 unique spacers) with an average length of 37 bp (individual spacers range from 35 to 39 bp) were identified. The number of spacers ranges from three in CRISPR 4 and CRISPR 15 to nineteen in CRISPR 17 (see Table S1 in the supplemental material). In strain R20291, 96 spacers were identified within nine expressed CRISPR arrays with lengths ranging from 33 to 41 bp (37-bp average length). The number of spacers ranged from four in CRISPR 12 to twenty-six in CRISPR 19 (Table S1). In both the 630 and R20291 strains, the complete cas operons (CD2982-CD2975 and CDR20291_2817-CDR20291_2810) are associated with a CRISPR array containing the largest number of spacers. This may suggest that the longest arrays created by numerous spacer acquisition events and coexpressed with a complete functional set of cas genes might constitute an active CRISPR functional unit within the bacterial genome. We can hypothesize that other CRISPR arrays could use the same set of cas genes for functioning.
In silico analysis of C. difficile CRISPR system targeting. (i) Phage genome sequencing.
To identify sequences targeted by C. difficile CRISPR spacers, we performed a systematic search for sequence similarities in the NCBI database using BLASTN (27) and the CRISPRTarget program (28). The search showed that some C. difficile spacers matched known clostridial phage sequences. However, because of the limited number of available genome sequences from clostridial phages, we set up a C. difficile-specific phage sequencing project. For this parallel study, 10 different temperate phages isolated from human and animal samples and belonging to the Myoviridae or Siphoviridae family were selected (29). Among new Myoviridae phages, the genome sequence of phiCD481-1 was found to be similar to the previously published phiCDHM13 and phiCDHM14 genomes (30), and to a lesser extent to the phiCD506 and phiMMP04 genomes, while the genome sequence of phiCD505 phage was similar to the genome sequence of phiMMP02 (31); the phiMMP01 phage is highly similar to phiMMP03, while the phiMMP03 phage is related to phiC2 (32). Among three new Siphoviridae phages, the genome sequences of phiCD111 and phiCD146 are highly similar to the previously characterized phiCD38-2 (33). The phiCD24-1 genome sequence is unique.
(ii) C. difficile CRISPR spacer homology analysis.
At the time of this writing, the total number of available clostridial phage genome sequences is 22. We performed extensive homology analysis of 819 CRISPR spacers from the available genome sequences of nine C. difficile strains. Four strains of PCR ribotype 027 and multilocus sequence type 3 (MLST 3) (strains 2007855, BI1, CD196, and R20291) have similar spacer contents, reflecting their evolutionary relationships (see Fig. S3 in the supplemental material). The presence of three unique spacers was detected in the R20291 CRISPR arrays compared to the spacers from three other PCR ribotype 027 strains. The CF5 and M68 strains of PCR ribotype 017 belonging to different MLST groups shared several CRISPR spacers but differed greatly in the spacer number within their CRISPR arrays. The M68 strain shows the lowest number of spacers (27 spacers) among analyzed strains. Some nearly identical CRISPR arrays were found in different C. difficile strains, while BI9 and M120 strains possess unique CRISPR arrays (Fig. S3). Overall, this analysis revealed several spacer deletion and acquisition events, suggesting that dynamic changes in the CRISPR array content had occurred, possibly through interactions with foreign DNA elements.
More than one-third of all analyzed spacers targeted the Clostridium phages, and about half of the hits to the Clostridium chromosome corresponded to the prophage regions (Fig. 4; see Table S2A in the supplemental material). For example, among 66 spacers from strain CF5, 29 matched chromosome sequences of which 17 were from prophages. These observations suggest that all the C. difficile strains analyzed had intensive interactions with phages. We also detected hits to Clostridium plasmids especially for CRISPR spacers from strain M120. Interestingly, this strain of PCR ribotype 078 possesses the largest number of unique spacers (153 spacers) that extensively target foreign DNA elements and are distributed in the six CRISPR arrays (Fig. 4, Fig. S3, and Table S2A).
FIG 4 .
Spacer homology analysis of CRISPR arrays for nine C. difficile strains. The spacer content of each CRISPR array is shown. The names of CRISPR arrays transcribed on the plus or minus strand are shown on green and red, respectively. The same number was assigned to identical spacers within CRISPR arrays from different strains. Color was used to show the spacer matching the clostridial phage genome sequence (red), plasmid (dark green), chromosomal prophage region (yellow), other chromosomal region (light green), chromosome and phage or plasmid (taupe), both phage and plasmid (blue), phage and prophage (mauve), and three groups (chromosome, plasmid, and phage0 (gray). Potential spacer deletion events are shown in bold type.
We next evaluated the potential functionality of CRISPR spacers of strains 630 and R20291 targeting phages. Perfect and imperfect matches to phage or plasmid sequences were found for 39 spacers (36% of the total number of spacers) of strain 630 and for 38 spacers (40%) of strain R20291. Seventeen spacers from strain 630 and ten spacers from strain R20291 perfectly matched the foreign DNA sequences, while remaining spacers contained between 1 and 10 mismatches with the targeted sequences (Table 1; see Table S3 in the supplemental material). Remarkably, for each CRISPR array from both analyzed strains, we found at least one spacer targeting a clostridial phage sequence (Fig. 4), arguing in favor of the functionality of each of the numerous C. difficile CRISPR arrays. Previous studies of CRISPR interference efficiency in Escherichia coli, Pseudomonas aeruginosa, and Haloferax volcanii carrying subtype I-E, I-F, and I-B CRISPR-Cas systems, respectively, identified the sequence requirements for the CRISPR targeting, in particular, a perfect match between the 5′ end of the spacer and the target DNA protospacer within up to a 10-nt “seed” sequence. A single mismatch at position 6 and up to five mismatches outside the “seed” are tolerated without an apparent decrease in the interference efficiency (26, 34, 35). We analyzed the locations of mismatches between the C. difficile CRISPR spacers and the targeted protospacers. Spacers without mismatches in the potential “seed” region (first 8 nt of the protospacer except for position 6) and carrying up to five mismatches outside the “seed” region were considered functional. In this way, six additional spacers with permissive mismatches were identified in CRISPR arrays from each strain (Table 1 and Table S3), raising the proportion of potentially functional CRISPR spacers matching the foreign DNA sequences to 59% and 42% in strains 630 and R20291, respectively.
TABLE 1 .
Phage sensitivity of C. difficile strains compared to CRISPR spacer homology to phage protospacers
| Phage | Strain | No. of spacers with exact or allowed matcha | PAMb | Expected phage sensitivity | Experimental phage sensitivityc |
|---|---|---|---|---|---|
| phiCD27 | 630 | 8 (6 leader-proximal) | CCT/A | Expected resistant | Resistant |
| R20291 | 2 | CCA/T | Expected resistant | Resistant | |
| phiC2 | 630 | 2 | CCA | Expected resistant | ND |
| 630 | 1 | ACA | |||
| R20291 | 2 | CCA/T | Expected resistant | ND | |
| phiCD38-2 | 630 | 3 | CCA | Expected resistant | Resistant |
| 630 | 1 | TCC | |||
| R20291 | 3 (leader-distal) | CCA | Possibly sensitive | Sensitive (29) | |
| phiCD119 | 630 | 1 | ACA | Expected sensitive | ND |
| R20291 | 1 | CCA | Expected resistant | ND | |
| phiCD6356 | 630 | 2 | CCA/T | Expected resistant | ND |
| R20291 | 4 | CCA/T | Expected resistant | ND | |
| phiMMP01 | 630 | 3 | CCA/T | Expected resistant | Resistant |
| R20291 | 1 | CCA | Expected resistant | Resistant (29) | |
| phiMMP03 | 630 | 2 | CCA | Expected resistant | Resistant |
| 630 | 1 | ACA | |||
| R20291 | 2 | CCA/T | Expected resistant | Resistant (29) | |
| phiCD24-1 | 630 | 0 | Expected sensitive | Resistantd | |
| R20291 | 0 | Expected sensitive | Resistantd | ||
| phiCD52 | 630 | 0 | Expected sensitive | Resistantd | |
| R20291 | 2 | CCA/T | Expected resistant | Resistant (29) | |
| phiCD111 | 630 | 3 | CCA | Expected resistant | Resistant |
| 630 | 1 | TCC | |||
| R20291 | 1 | CCC | Expected sensitive | Resistantd (29) | |
| phiCD146 | 630 | 3 | CCA | Expected resistant | Resistant |
| 630 | 1 | TCC | |||
| R20291 | 2 (leader-distal) | CCA | Possibly sensitive | Sensitive (29) | |
| R20291 | 1 | CTT | |||
| phiCD211 | 630 | 4 | CCT/A | Expected resistant | Resistant |
| R20291 | 1 | GCT | Expected sensitive | Resistantd | |
| phiCD481-1 | 630 | 1 | TCG | Expected sensitive | Resistantd |
| R20291 | 1 | CCG | Expected sensitive | Sensitive (29) | |
| phiCD505 | 630 | 5 (4 leader-proximal) | CCA/T | Expected resistant | Resistant |
| R20291 | 3 | CCA/T | Expected resistant | Resistant (29) | |
| phiCD506 | 630 | 1 | CCA | Expected resistant | Resistant |
| 630 | 1 | CCG | |||
| R20291 | 1 | CCG | Expected sensitive | Resistantd (29) | |
| phiMMP02 | 630 | 5 (leader-proximal) | CCT/A | Expected resistant | Resistant |
| 630 | 1 | ACA | |||
| R20291 | 2 | CCA | Expected resistant | Resistant (29) | |
| R20291 | 1 | TCA | |||
| phiMMP04 | 630 | 0 | Expected sensitive | Resistantd | |
| R20291 | 1 | CCG | Expected sensitive | Resistantd (29) | |
| phiCDMH1 | 630 | 2 | CCA | Expected resistant | ND |
| 630 | 1 | CTA | |||
| R20291 | 1 | CCA | Expected resistant | ND |
For an allowed match between the CRISPR spacer and phage protospacer, we accepted up to five mismatches outside the potential “seed” region (first 8 nt of protospacer except for position 6).
The consensus PAM motif is CCA or CCT. Mismatches in this motif are shown in bold type.
Phage sensitivity was examined by a phage infection spot assay (29). ND, not determined.
Discrepancies between expected and observed resistance probably due to the existence of other resistance mechanisms. The complete genome sequences of Siphoviridae phages phiCD24-1, phiCD111, and phiCD146 and Myoviridae phages phiCD481-1, phiCD505, phiCD506, phiMMP01, phiMMP03, and phiCD52 were deposited in European Nucleotide Archive under accession no. LN681534, LN681535, LN681536, LN681538, LN681539, LN681540, LN681541, LN681542, and PRJEB7856, respectively. The complete genome sequence of phiCD211 was deposited in European Nucleotide Archive under accession no. LN681537.
(iii) Nature and distribution of CRISPR-targeted genes.
CRISPR spacer homology analysis identified a total of 676 hits to clostridial phage genome sequences with about 40 hits per phage distributed throughout the genome on both DNA strands (Fig. 5). The most “popular” were phiCD38-2 among siphophages with 54 hits and phiMMP04 among myophages with 42 hits. The phiCD24-1 phage contained the lowest number of CRISPR spacer hits (two hits). Interestingly, this analysis revealed several spacers simultaneously targeting the related phage genomes (Fig. 5; see Table S3 in the supplemental material). The majority of potential protospacers were found within genes encoding structural proteins with tail tape measure and capsid protein-encoding genes being most frequently targeted (Table S2B and Table S3). Protospacers were also present within genes of unknown function and in intergenic regions (Table S3). Multiple targeting is observed for the CRISPR spacers from all analyzed C. difficile strains (Table S2C and Table S3) and could provide C. difficile with an efficient and economical defense against several related phages by the same crRNA.
FIG 5 .
CRISPR spacer targeting of C. difficile phages. The CRISPR spacer hits are indicated by red flag symbols on the genomes of representative members of clostridial phage groups: myophages phiMMP02 (for phiMMP02/phiCD505 group), phiCD27, phiCDHM1, phiMMP03 (for phiMMP01/phiMMP03 group), phiC2, phiCD119, phiCD481-1 (for phiMMP04/phiCD481-1/phiCD506 group), phiCD211, and siphophages phiCD38-2 (for phiCD38-2/phiCD111/phiCD146 group) and phiCD6356. Late phage genes (blue), early middle genes (pink), and integrase/resolvase genes (yellow) are indicated. One flag symbol corresponds to either a single hit or multiple hits.
(iv) Identification of PAM.
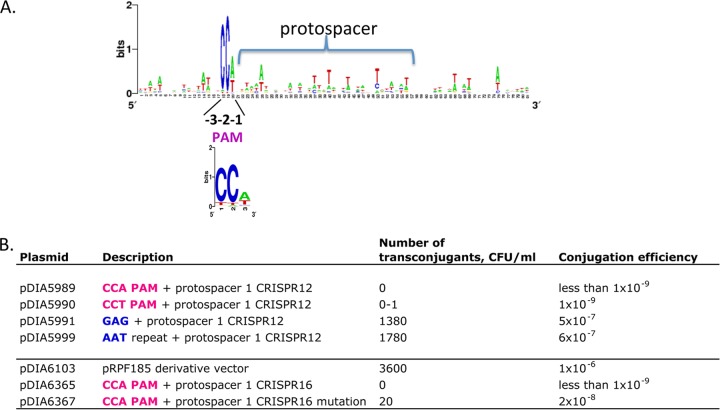
Conserved sequence motifs (PAMs) in the regions flanking the protospacer were shown to be important for the recognition by type I and II CRISPR-Cas systems (12). The alignment of phage sequences flanking protospacers targeted by the C. difficile CRISPR system revealed a conserved 3-nucleotide 5′ PAM motif CCT or CCA (Fig. 6A) and no motif in the region downstream of protospacers. This conserved 5′ motif is in accordance with the putative PAM sequences identified in the previous in silico analyses (30) and is characteristic of subtype I-B bacterial CRISPR-Cas systems (16) but differs from PAMs recognized by the haloarchaeal subtype I-B CRISPR-Cas systems (26). Among the alternative motifs, CCC, CCG, and TCA trinucleotides were detected most frequently. In particular, the TCA motif is associated with the majority of protospacers targeted by strain M120 CRISPR 8 spacers (see Table S2B in the supplemental material).
FIG 6 .
PAM identification for CRISPR system in C. difficile. (A) The alignment of regions flanking protospacers targeted by the CRISPR system was used to create the sequence logo by WebLogo for CRISPR spacers from strains 630 and R20291. The 5′ PAM (protospacer adjacent motif) at positions −3, −2, and −1 relative to the first position of the protospacer is indicated. The PAM 3-nucleotide WebLogo created on the basis of potential protospacer flanking regions from nine C. difficile strains is shown below. (B) Efficiency of conjugation of pMTL84121-derived plasmids (carrying p15a Gram-negative bacterial replicon and pCD6 Gram-positive bacterial replicon) and pRPF185-derived plasmids (carrying ColE1 Gram-negative bacterial replicon and pCD6 Gram-positive bacterial replicon). Protospacer 1 CRISPR 16 mutation indicates a G-to-A substitution at the first position. The representative results of three independent experiments are shown.
Functionality of the C. difficile CRISPR system for interference. (i) Analysis of plasmid conjugation efficiency.
We next set out to investigate the functional importance of the 5′ CCW 3-nucleotide PAM. For this purpose, we compared the conjugation efficiencies of artificial plasmids containing different nucleotides upstream of the protospacer corresponding to the first spacer within the strain 630 CRISPR 12 array. Plasmids pDIA5989 and pDIA5990 carrying the CCA or CCT PAM upstream of the protospacer did not give any transconjugants. In contrast, the replacement of PAM by a GAG trinucleotide in pDIA5991 or an AAT trinucleotide in pDIA5999 corresponding to the 3′ end of CRISPR repeat led to efficient conjugation (Fig. 6B).
It was recently reported that the sequence of protospacer and the nature of the plasmid used for conjugation can influence the CRISPR-mediated interference process (19). We prepared constructs derived from the pRPF185 vector carrying the same pCD6 Gram-positive bacterial replicon but a different Gram-negative bacterial replicon and used them to monitor the plasmid DNA targeting by another CRISPR spacer. In agreement with the results for the pDIA5989 plasmid, no transconjugants were obtained with the pDIA6365 plasmid carrying a CCA PAM upstream of the protospacer corresponding to the first spacer within the CRISPR 16 array from strain 630. Detectable conjugation was observed with the pDIA6367 plasmid carrying the same protospacer mutated at the first position within the potential “seed” region. However, this mutation did not completely abolish the interference compared to the control pDIA6103 plasmid (Fig. 6B). Overall, these results provide the first experimental evidence that the C. difficile CRISPR-Cas system is naturally capable of interfering with horizontal gene transfer and confirm the importance of the PAM region for targeting foreign DNA.
(ii) Comparison of predicted CRISPR-mediated resistance and susceptibility to phage infection.
To assess the ability of the C. difficile CRISPR-Cas system to interfere with phage infection, we analyzed the correlation between the phage susceptibility profiles and the CRISPR spacer homology to the corresponding phage sequences. Generally, the presence of a spacer matching the incoming phage DNA is expected to contribute to the resistance to infection by the corresponding phage provided that a CRISPR-Cas system is functional. In contrast, in the absence of an exact match, the host cell should be phage sensitive.
We deduced the expected phage susceptibility phenotypes of strains 630 and R20291 on the basis of the following criteria. We checked for the following: (i) the presence of CRISPR spacers exactly matching the corresponding phage sequences or carrying only the allowed mismatches in position 6 of the potential “seed” region or up to five mismatches outside the “seed” (the first 8 nucleotides of the protospacer) and (ii) the presence of a CCW 5′ PAM. If several spacers targeted the same phage, the number of leader-proximal spacers meeting the above criteria was considered. The different interference efficiencies of spacers according to their position with respect to the leader region were suggested in previous studies and are likely caused by different abundance, stability, and/or processing efficiency of the corresponding crRNA (26, 36). To compare the predicted phage susceptibility phenotype with the experimental results, we performed phage infection assays with strains 630 and R20291 and phages available to us. In addition, we used our recently reported data on the host range analysis for some newly described temperate phages infecting C. difficile (29).
The results of this comparative analysis are summarized in Table 1, and the complete data set is presented in Table S3 in the supplemental material. Overall, a good correlation between the presence of CRISPR spacer-targeting phage sequences and the corresponding phage susceptibility phenotype was observed. The genome of C. difficile strain 630 carries the spacers targeting nearly all isolated clostridial phages identified thus far. Accordingly, this strain was resistant to all tested phages (Table 1; see Table S3 in the supplemental material). Spacer sequences from strain R20291 contained more mismatches with phage sequences, including multiple mismatches inside the potential “seed” region and noncanonical PAMs in targeted protospacers (Table S3). These could explain the sensitivity of strain R20291 toward at least three of the phages tested: phiCD38-2, phiCD146, and phiCD481-1.
Of note, several CRISPR spacers from strain 630 targeted the same phage. For example, eight different spacers within several CRISPR arrays targeted the myophage phiCD27. Six of them are leader-proximal spacers (first or second position) within their respective CRISPR arrays, including the most highly expressed CRISPR 4 and CRISPR 15 (Fig. 1A). This is consistent with the resistant phenotype observed with phiCD27 (Table 1). In addition, some identical spacers targeted two other related myophages, phiCD505 and phiMMP2 (Table 1; see Table S3 in the supplemental material). The leader-proximal positions of these spacers could reflect relatively recent expansion of the corresponding CRISPR arrays. Another example of simultaneous targeting by the overlapping set of CRISPR spacers from both 630 and R20291 strains is the targeting of three related Siphoviridae phages phiCD38-2, phiCD111, and phiCD146 (Table S3). We also observed the presence of several spacers within the same CRISPR array that target the same phage, suggesting repeated interactions between this C. difficile strain and the corresponding phage (Table S3). This seems to be a general feature of C. difficile CRISPR arrays observed for all nine analyzed C. difficile strains (Table S2C) and may reflect a specific characteristic of interaction with phage, such as primed spacer adaptation (37). In several cases, within long CRISPR arrays (e.g., in M120 strain), spacers that were more distal presented larger numbers of mismatches with phage sequences and/or were more frequently associated with noncanonical PAMs than leader-proximal spacers targeting the same phage (Table S2C). For the phage genome hits, about half of the noncanonical PAMs were associated with at least one mismatch within the potential “seed” region, also suggesting a mutational process to escape CRISPR defense.
Our analysis also revealed some discrepancies between expected and observed phage susceptibility phenotypes. The majority of cases concern the phage sensitivity that could be predicted while the phage resistance phenotype was observed. One opposite example is the R20291 arrays containing spacers targeting phiCD38-2 and phiCD146 phages. The mismatches inside the “seed” or PAM region in some cases and the leader-distal position of other spacers possibly associated with lower abundance and/or stability of corresponding crRNAs could explain the observed sensitivity of R20291 strain to phiCD38-2 and phiCD146 phages (Table 1; see Table S3 in the supplemental material). The R20291 strain was resistant to infection by phiCD111, phiCD506, and phiMMP04 (29). However, in each case, only one potential active spacer was detected within CRISPR arrays of this strain, and a noncanonical CCC or CCG PAM preceded the corresponding protospacer within the phage genomes. Interestingly, these CCC/CCG motifs were also revealed as overrepresented alternative motifs by general analysis of protospacer-flanking regions (Table S2B). This could suggest that a single mismatch within PAM does not disturb the CRISPR-mediated interference, as observed for multiple PAMs in the Haloarchaeal CRISPR I-B system (26), or that other mechanisms could be involved. Interestingly, despite very similar CRISPR array contents in strains CD196 and R20291 of the PCR ribotype 027 (Fig. S3), there are apparent differences in phage susceptibility profiles of these strains that could also be explained by other, non-CRISPR, defense mechanisms. Similarly, a homology search with CRISPR arrays in strain 630 revealed no spacers targeting phiCD24-1, phiCD52, and phiMMP04, while strain R20291 CRISPR arrays contained no spacers targeting phiCD24-1 (Table 1). However, these strains were resistant to the corresponding phages, likely due to CRISPR-independent defense mechanisms.
To conclude, the phage infection assays support the functionality of the C. difficile CRISPR system for protection from phage infection. Possible interference with resident prophages, differences in spacer efficiency, and the existence of other mechanisms of phage resistance like receptor modifications and restriction/modification systems could explain the observed discrepancies with phage sensitivity predictions (12).
(iii) C. difficile cas genes function in CRISPR interference in E. coli heterologous system.
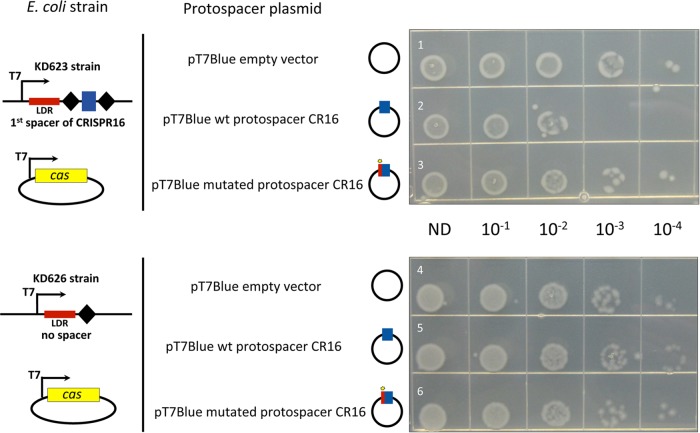
As a first step to mechanistic studies of the C. difficile CRISPR-Cas system, we established a heterologous system in a surrogate E. coli host that had its own CRISPR-Cas system removed. E. coli was chosen as a host that is easier to manipulate genetically than C. difficile. E. coli plasmids expressing the conserved and complete cas operon from C. difficile strain 630 containing eight cas genes (CD2982-CD2975) were created. The first part of the C. difficile cas operon (from CD2982 to CD2977 encoding the interference components) was cloned into the pCDF-1b expression vector (pDIA6351), and the rest of the operon (cas1 [CD2976] and cas2 [CD2975] genes) was cloned into the pRSF-1b vector (pDIA6349) under the control of T7 RNA polymerase (T7 RNAP) promoter. Next, E. coli host strains containing minimized C. difficile CRISPR arrays were created. Sequences of the highly expressed C. difficile 630 CRISPR 12 or CRISPR 16 arrays (Fig. 2; see Fig. S1 in the supplemental material) were selected for this purpose. The third “miniarray” containing only the leader region with direct repeat but without the spacer sequence was used as a negative control. These CRISPR arrays, flanked by a T7 RNAP promoter and transcriptional terminator sequences, were introduced into the genome of the E. coli BL21-AI_ΔCRISPR strain lacking endogenous cas genes and carrying the T7 RNAP-encoding gene under the control of the arabinose-inducible araBAD promoter (strains KD620, KD623, and KD626 [Table S4]). To monitor the CRISPR interference, strains KD620, KD623, and KD626 harboring the C. difficile cas expression plasmids were transformed with the compatible pT7Blue-based plasmids containing the protospacer-matching spacers within CRISPR “miniarrays.” Each strain was transformed with the pT7Blue derivatives containing the CCA PAM followed by either a protospacer perfectly matching the CRISPR spacer (pDIA6361 or pDIA6363), a protospacer with a single mismatch at the first position (pDIA6362 or pDIA6364), or an empty control pT7Blue vector (Table S4). Upon induction of C. difficile subtype I-B CRISPR-Cas in E. coli in the presence of l-arabinose, we observed a decrease in the transformation efficiency of plasmids containing protospacers fully matching the CRISPR array spacers and no difference in the transformation efficiency with a control strain carrying a CRISPR array without a spacer. Mutation in the first position of the protospacer “seed” region abolished the observed interference leading to the transformation efficiencies similar to those obtained with the empty vector (Fig. 7).
FIG 7 .
Functionality of C. difficile cas genes for plasmid interference in E. coli. The transformation efficiency was estimated with pT7Blue derivative plasmids carrying the wild-type (wt) protospacer corresponding to the first spacer of the CRISPR 16 array (CR16) (rows 2 and 5) or a mutated protospacer CR16 (rows 3 and 6) compared to the pT7Blue empty vector used as a negative control (rows 1 and 4). The protospacer plasmid used is indicated to the left of the photographs together with schematic representation of E. coli strains carrying engineered CRISPR arrays with the corresponding spacer under the control of T7 RNAP promoter (T7). E. coli KD623 strain (rows 1 to 3) carries C. difficile CRISPR “miniarray” with the first spacer of CRISPR 16 array flanked by repeats, and E. coli KD626 strain (rows 4 to 6) carries reduced “miniarray” with one repeat lacking spacer sequence. The CRISPR “leader” region (LDR) is indicated. Both strains were transformed with pCDF1-b vector derivative, allowing the expression of C. difficile cas gene set lacking cas1 and cas2 (from CD2982 to CD2977). The Cas protein production and crRNA expression were induced by the addition of 1 mM l-arabinose and 1 mM IPTG. The serial dilutions of transformation mixtures deposited on LB plates with ampicillin are indicated (ND, not diluted).
The pDIA6351 plasmid carrying the first part of the operon (cas6, cas8, cas7, cas5, cas3, and cas4 genes) lacking cas1 and cas2 genes was sufficient to observe the interference, and the presence of both pDIA6349 and pDIA6351 plasmids carrying the entire cas gene set did not lead to improved interference efficiency (data not shown). A representative result obtained with the pDIA6351 plasmid is shown in Fig. 7. Together, these results confirm the functionality of the C. difficile CRISPR system and demonstrate the role of specific cas genes in the plasmid interference process.
DISCUSSION
C. difficile is an emergent human enteropathogen that must cope with abundant bacteriophages and other exogenous genetic elements during its development inside the host. Horizontal gene transfer would be beneficial for the acquisition of new adaptive traits (38); however, foreign invaders could also cause damage. Efficient defense systems could thus be important for C. difficile during the interactions with other members of the gut microbiota. In the present paper, we demonstrate the functionality of C. difficile CRISPR system using plasmid conjugation efficiency assays and interference assays in E. coli as a heterologous host. We show that multiple CRISPR arrays in C. difficile strains carry specific spacers that can be considered “memories” of past C. difficile encounters with foreign genetic elements, including clostridial phages. In addition, our new phage genome sequences provide essential information for detailed functional spacer analysis. Only a fraction of spacers match known sequences, which suggests that numerous other foreign invaders interact with C. difficile and remain to be discovered.
Our work provides evidence for an active bacterial CRISPR system of subtype I-B. In agreement with findings for other CRISPR type I systems, we show that a specific PAM sequence is necessary for self and nonself discrimination for clostridial systems. In addition, the extent of spacer sequence matching the targeted protospacer seems to be critical for CRISPR interference; in particular, an exact match at the first position of the protospacer is needed for CRISPR interference, as observed in other systems (19, 34). We also provide experimental evidence for the role of the C. difficile Cas proteins in the interference process in the heterologous host, E. coli. The universal CRISPR-Cas system components Cas1 and Cas2 important for the adaptation process in other bacterial systems (39, 40) seem to be dispensable for interference mediated by the C. difficile CRISPR-Cas I-B system.
The originality of the CRISPR-Cas system in C. difficile is the presence of multiple active CRISPR arrays, which is in contrast to the presence of silent or barely expressed CRISPR loci in some other bacteria such as Streptococcus pyogenes and E. coli (41, 42). Interestingly, we observed a similar bias on the transcriptional orientation of CRISPR arrays in the direction of replication as a previously reported strong coding bias for the leading strand of the chromosome and GC skew (Fig. 1) (10, 43). This could reflect selection for optimal CRISPR array orientation with respect to chromosome replication as generally observed for the direction of transcription of rRNA genes and other essential and/or highly expressed bacterial genes (44–46). The results of analyzing the CRISPRdb data for seven other C. difficile strains suggest general transcriptional orientation for CRISPR arrays on a leading DNA strand (data not shown). Only one array in both strains 630 and R20291 is associated with a complete cas operon. The presence of one or two additional incomplete cas operons composed of five genes, including cas3, cas5, cas6, and cas7 in both strains, could suggest their possible accessory role in CRISPR function. Less-efficient interference in a surrogate host driven by the major set of C. difficile Cas proteins observed in E. coli can be also taken as evidence for the requirement of other cas gene products (or additional C. difficile functions) for efficient interference.
To obtain a global view of the occurrence of cas operons in C. difficile, we evaluated the presence of homologs of the two cas operons CD2982-CD2975 and CD2455-CD2451 in the published genomes of 2,207 C. difficile strains (see Table S5A in the supplemental material). We found that the majority of sequenced C. difficile strains contain both cas gene sets, the homologs of CD2982-CD2975 cas locus being present in about 90% of strains, while the homologs of CD2455-CD2451 cas locus were detected in almost all strains analyzed. Interestingly, analysis of the MLSTs of strains lacking the homologs of CD2982-CD2975 cas locus revealed a strong correlation between the absence of the complete cas locus and the MLST of C. difficile strains (Table S5B). This could reflect an evolutionary history of cas gene loss or acquisition. The loss of a CD2975-like locus seems to be extremely rare in the group of strains belonging to MLST 3 (PCR ribotype 027 for the majority of strains, including strain R20291) and 23. About half of strains of MLST 19 and 25 lack this locus. In other MLST groups like MLST 9 and 45, the absence of the CD2975-like locus is frequently observed (Table S5B). We have also analyzed the presence of an additional partial cas operon homologous to the CDR20291_2998-2994 operon from strain R20291. The nonrandom distribution according to the MLST groups was observed for this operon as well. It was associated with the majority of the MLST 3 group, including the PCR ribotype 027 strains, and absent in several MLST groups like MLST 19, 33, 39, and 49 (Table S5C). Thus, our analysis showed a strong correlation between the evolutionary relationships of the C. difficile strains and their CRISPR array content and cas operon occurrence. This may be related to the epidemiological context of different C. difficile strains (isolation of the strains), reflecting the intensity of their interactions with foreign DNA elements.
The prophage localization of CRISPR arrays in several strains and a large proportion of prophage targeting by CRISPR spacers are other peculiarities of the C. difficile CRISPR system (30) (Fig. 1A and Fig. 4; see Table S2 in the supplemental material). These two factors raise questions about the role of such spacers in preventing infection by other competing phages and the spread of CRISPR arrays by horizontal gene transfer by lysogenic phages. Our RNA-seq analysis demonstrated that some prophage-related CRISPR RNAs were among the most abundant transcripts detected in both strains 630 and R20291 (Fig. S1 and Fig. S2) (24). Remarkably, none of the CRISPR arrays located within prophages possess associated cas operons, indicating that these prophage-related CRISPR arrays rely on a common Cas protein set encoded by the host. In general, prophages have played an important role in the evolution and virulence of pathogenic bacteria (47). Recent data for C. difficile suggest that prophages can modulate the toxin production, mediate the transmission of antibiotic resistance genes between different strains, and influence the sporulation process and bacterial gene regulation (33, 47, 48). In addition, the phage-related pathogenicity locus (PaLoc) can be transferred by transduction between C. difficile strains converting nontoxigenic strains into toxin producers (49). In Vibrio cholerae, the existence of a phage-encoded CRISPR system targeting a bacterial phage-inhibitory locus was recently uncovered (50). Prophages within C. difficile genomes could also influence the interactions with other phages. The possible transfer of active CRISPR arrays between different C. difficile strains would be important for bacterial fitness within the gut environment, enriching the diversity of spacers in the repertoire by acquisition of entire CRISPR arrays. In relation to the C. difficile infection cycle, it is interesting to note that stress conditions, including antibiotic treatments, could induce prophages and lead to the release of phage particles and infection of neighboring bacteria, thus contributing to the CRISPR spreading within C. difficile populations (31). Together with dysbiosis, this can increase the rigor of this pathogen.
The detailed comparison of CRISPR spacer homology to phage sequences with corresponding phage resistance phenotypes provides important insights on CRISPR system function. The results of this analysis revealed targeting by several spacers of the same phage, which should increase the efficiency of CRISPR defense and suggest a mutational process of the phage to escape the CRISPR system or primed acquisition of spacers (or both). In addition, some overrepresented spacers target conserved genes within related phages leading to economical and efficient defense against several phages. Recent long-term phage infection studies in Streptococcus thermophilus showed preferential selection of specific highly represented spacers within bacterial populations, suggesting enhanced defense capacities of the corresponding crRNA based on location and effectiveness (51). The relatively high frequency of noncanonical PAM sequences observed in our study may suggest that single mismatches within PAM could be tolerated by the CRISPR I-B system, as previously suggested in studies of archaeal systems with multiple PAMs as target interference motifs and a reduced number of PAMs as spacer acquisition motifs (19). Alternatively, other active defense mechanisms against phages working independently against the CRISPR system (12) could also interfere with C. difficile interactions with phages. It should be noted that for the moment only temperate phages infecting C. difficile have been isolated and tested in the present study (29, 52). The possibility that in some cases, the apparent phage resistance could be related to immunity conferred by related endogenous prophages due to the use of temperate phages cannot be excluded.
In conclusion, this study significantly expands our knowledge on C. difficile interactions with specific phages, highlighting specific features of CRISPR-Cas adaptive immune system in this important enteropathogen. Numerous actively expressed CRISPR arrays might provide C. difficile strains with the extended defense capacities against foreign invaders, including phages abundant within the gut communities. Further studies on the role of CRISPR system during infection will bring light on these important aspects of C. difficile adaptation inside the host.
MATERIALS AND METHODS
Plasmid and bacterial strain construction and growth conditions.
C. difficile and E. coli strains and plasmids used in this study together with detailed descriptions of plasmid and bacterial strain construction are presented in Table S4 in the supplemental material. C. difficile strains were grown anaerobically (5% H2, 5% CO2, and 90% N2) in TY (53) or brain heart infusion (BHI) (Difco) medium in an anaerobic chamber (Jacomex). When necessary, cefoxitin (Cfx) (25 µg·ml−1) and thiamphenicol (Tm) (15 µg·ml−1) were added to C. difficile cultures. E. coli strains were grown in LB broth (54), and when needed, ampicillin (100 µg·ml−1), chloramphenicol (15 µg·ml−1), tetracycline (15 µg·ml−1), kanamycin (50 µg·ml−1), or streptomycin (50 µg·ml−1) was added to the culture medium. All primers used in this study are listed in Table S6.
RNA-seq analysis.
Total RNA was isolated from late-exponential-growth-phase cultures (grown for 6 h) of C. difficile R20291 grown in TY medium as previously described (55), and mRNA was enriched from total RNA using MicrobExpress kit (Ambion). Nonoriented RNA-seq library construction was performed with the TruSeq RNA sample prep kit from Illumina as previously described (56) and subjected to Illumina HiSeq 2000 sequencing.
In silico analysis of CRISPR array spacer content and cas locus.
CRISPRdb tools (11), the CRISPRTarget program (28), or BLASTN (27) were used for spacer homology search in the available sequences (April 2015). Several potential CRISPR arrays predicted by CRISPRdb within the tcdA coding region were excluded from further analysis for the following reasons: (i) their location within the toxin-encoding genes in all analyzed C. difficile strains as a part of the cell wall-binding repeat regions within the TcdA amino acid sequence; (ii) the prediction of the corresponding arrays as “questionable sequences” by the CRISPRdb program for several analyzed C. difficile strains; (iii) the absence of a characteristic RNA-seq profile for CRISPR arrays within the tcdA coding region; (iv) the absence of potential targeting of corresponding spacers for the known sequences; (v) the differences in the length and sequence of associated direct repeats with those of active CRISPR arrays. Thus, in such particular cases, the CRISPRdb predictions within repeated coding regions should be considered with caution and would need experimental confirmation.
For general CRISPR spacer homology search, the sequences presented ≤7 single nucleotide polymorphisms (SNPs) (80% match or ≥30/37 nucleotides) were considered positive hits. The raw sequencing read data of published genome sequences from 2,207 C. difficile strains (57–62) were used to search for the presence of cas loci homologous to the CD2982-CD2975 and CD2455-CD2451 cas operons from strain 630 and the CDR20291_2998-2994 operon from strain R20291. For each strain, the sequencing reads were mapped on the sequence of corresponding cas locus using Bowtie (63). Coverage values of ≥80% were considered positive hits for the presence of corresponding cas loci in a given strain. The multilocus sequence typing (MLST) scheme of Lemee et al. (64) has also been inferred from raw sequencing read data.
Phage infection assays.
Phage host range determination was done as previously described (29) using spot tests with diluted phage lysates and exponentially grown C. difficile cultures.
Plasmid conjugation efficiency assays.
Derivatives of pMTL84121 and pRPF185 plasmids used to estimate the conjugation efficiency were transformed into the E. coli HB101 (RP4) and subsequently mated with C. difficile 630Δerm mutant strain on BHI agar plates for 24 h at 37°C. The proportion of C. difficile transconjugants was estimated by subculturing the cell conjugation mixture on BHI agar containing Tm (15 µg·ml−1) and Cfx (25 µg·ml−1) and comparing the number of CFU obtained after plating serial dilutions on BHI agar plates with Cfx lacking Tm.
Plasmid-based interference assays in E. coli.
The transformation efficiency of pT7Blue derivative plasmids (see Table S4 in the supplemental material) was monitored with E. coli strains lacking endogenous cas genes and carrying C. difficile CRISPR “miniarray” within their genome, including a CRISPR spacer targeting pT7Blue constructs and the plasmids pRSF-1b and/or pCDF-1B expressing C. difficile cas genes under the control of T7 RNAP promoter. Cas protein and crRNA production was induced with 0.5 to 1 mM l-arabinose and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and electrocompetent cells were prepared under these inducing conditions. Plasmids were then introduced by electroporation, and undiluted and serially diluted aliquots of the transformation mixture were spotted on LB agar plates containing ampicillin and either streptomycin or kanamycin/streptomycin to measure the transformation efficiency.
Data access.
The complete genome sequences of phiCD24-1, phiCD111, phiCD146, phiCD211, phiCD481-1, phiCD505, phiCD506, phiMMP01, phiMMP03, and phiCD52 were deposited in EMBL-EBI database under accession no. LN681534, LN681535, LN681536, LN681537, LN681538, LN681539, LN681540, LN681541, LN681542, and PRJEB7856, respectively. RNA-seq coverage visualizations of the CRISPR loci are available for strain 630 through https://mmonot.eu/COV2HTML/visualisation.php?str_id=-17 and for strain R20291 through http://mmonot.eu/COV2HTML/visualisation.php?str_id=-18 (65).
SUPPLEMENTAL MATERIAL
Expression analysis of CRISPR arrays from C. difficile 630 strain by deep sequencing. The TAP−/TAP+ profile comparison for 5′-end RNA-seq is aligned with RNA-seq data for the corresponding genomic region. The TSSs identified by 5′-end sequencing are indicated by red broken arrows in accordance with the positions of 5′-transcript ends shown by vertical green lines on the sequence read graphs corresponding either to TSSs (broken arrows) or to processing sites. 5′-end sequencing data show 51-bp reads matching the 5′-transcript ends, while RNA-seq data show reads covering the whole transcript. Black diamonds indicate the positions of the first direct repeat of the CRISPR array. The transcript covering the CRISPR array is indicated by a gray arrow below the deep sequencing data. Deep sequencing data are available at https://mmonot.eu/COV2HTML/visualisation.php?str_id=-17. Download
Expression analysis of CRISPR arrays from C. difficile R20291 strain by RNA-seq. Sequence reads covering the whole transcript from RNA-seq data are shown according to genomic position. Black diamonds indicate the positions of the first direct repeat of the CRISPR array. The transcript covering the CRISPR array is indicated by a gray arrow below deep sequencing data. Deep sequencing data are available at https://mmonot.eu/COV2HTML/visualisation.php?str_id=-18. Download
Spacer content of CRISPR arrays for nine C. difficile strains. CRISPRdb numbering for CRISPR arrays was used, and spacers within each CRISPR array were numbered according to transcriptional order. The same number was assigned to identical spacers within CRISPR arrays from different strains. Colors were used to indicate related CRISPR arrays or spacers. Potential spacer deletion events are shown in bold type. Download
Expressed CRISPR arrays in C. difficile 630 and R20291 strains
General analysis of CRISPR spacer matches for nine C. difficile strains. (A) CRISPR spacer hits to chromosome, phage, and plasmid sequences. (B) CRISPR spacer matching of phage, chromosome, and plasmid sequences. (C) Phage targeting by several CRISPR spacers.
CRISPR spacer homology analysis in comparison with phage sensitivity for strains 630 and R20291
Strains and plasmids used in this study
C. difficile cas locus analysis
Oligonucleotides used in this study
ACKNOWLEDGMENTS
We thank Melinda Mayer for providing us with phiCD27 phage lysate. We are grateful to Odile Sismeiro and Jean-Yves Coppée for help with the RNA-seq experiment and to Laurence Ma and Christiane Bouchier for phage genome sequencing.
This work was supported by grants from the Institut Pasteur, University Paris Diderot, Agence Nationale de la Recherche (“CloSTARn,” ANR-13-JSV3-0005-01), Pasteur-Weizmann Council, National Institutes of Health (NIH R01 GM10407), Natural Sciences and Engineering Research Council of Canada (Discovery grant, no. 341450-2010), and Fonds de la Recherche du Québec - Santé (FRQS) (Junior 2 salary award). O.S. is an assistant professor at the University Paris Diderot. P.B. has a Ph.D. fellowship from the University Paris Diderot.
Footnotes
Citation Boudry P, Semenova E, Monot M, Datsenko KA, Lopatina A, Sekulovic O, Ospina-Bedoya M, Fortier L-C, Severinov K, Dupuy B, Soutourina O. 2015. Function of the CRISPR-Cas system of the human pathogen Clostridium difficile. mBio 6(5):e01112-15. doi:10.1128/mBio.01112-15.
REFERENCES
- 1.Carroll KC, Bartlett JG. 2011. Biology of Clostridium difficile: implications for epidemiology and diagnosis. Annu Rev Microbiol 65:501–521. doi: 10.1146/annurev-micro-090110-102824. [DOI] [PubMed] [Google Scholar]
- 2.Kuijper EJ, Coignard B, Tüll P, ESCMID Study Group for Clostridium difficile, EU Member States, European Centre for Disease Prevention and Control . 2006. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect 12(Suppl 6):2–18. doi: 10.1111/j.1469-0691.2006.01580.x. [DOI] [PubMed] [Google Scholar]
- 3.Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, Frost E, McDonald LC. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084. doi: 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 4.Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol 7:526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 5.Walters BA, Roberts R, Stafford R, Seneviratne E. 1983. Relapse of antibiotic associated colitis: endogenous persistence of Clostridium difficile during vancomycin therapy. Gut 24:206–212. doi: 10.1136/gut.24.3.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Just I, Selzer J, Wilm M, von Eichel-Streiber C, Mann M, Aktories K. 1995. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature 375:500–503. doi: 10.1038/375500a0. [DOI] [PubMed] [Google Scholar]
- 7.Voth DE, Ballard JD. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev 18:247–263. doi: 10.1128/CMR.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denève C, Janoir C, Poilane I, Fantinato C, Collignon A. 2009. New trends in Clostridium difficile virulence and pathogenesis. Int J Antimicrob Agents 33(Suppl 1):S24–S28. doi: 10.1016/S0924-8579(09)70012-3. [DOI] [PubMed] [Google Scholar]
- 9.Dupuy B, Govind R, Antunes A, Matamouros S. 2008. Clostridium difficile toxin synthesis is negatively regulated by TcdC. J Med Microbiol 57:685–689. doi: 10.1099/jmm.0.47775-0. [DOI] [PubMed] [Google Scholar]
- 10.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeño-Tárraga AM, Wang H, Holden MT, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 11.Grissa I, Vergnaud G, Pourcel C. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. 2012. The CRISPRs, they are a-changin. How prokaryotes generate adaptive immunity. Annu Rev Genet 46:311–339. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 13.Bhaya D, Davison M, Barrangou R. 2011. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet 45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 14.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makarova KS, Wolf YI, Koonin EV. 2013. The basic building blocks and evolution of CRISPR-CAS systems. Biochem Soc Trans 41:1392–1400. doi: 10.1042/BST20130038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorek R, Lawrence CM, Wiedenheft B. 2013. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem 82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 17.Li M, Liu H, Han J, Liu J, Wang R, Zhao D, Zhou J, Xiang H. 2013. Characterization of CRISPR RNA biogenesis and Cas6 cleavage-mediated inhibition of a provirus in the haloarchaeon Haloferax mediterranei. J Bacteriol 195:867–875. doi: 10.1128/JB.01688-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maier LK, Dyall-Smith M, Marchfelder A. 2015. The adaptive immune system of Haloferax volcanii. Life 5:521–537. doi: 10.3390/life5010521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoll B, Maier LK, Lange SJ, Brendel J, Fischer S, Backofen R, Marchfelder A. 2013. Requirements for a successful defence reaction by the CRISPR-Cas subtype I-B system. Biochem Soc Trans 41:1444–1448. doi: 10.1042/BST20130098. [DOI] [PubMed] [Google Scholar]
- 20.Cass SD, Haas KA, Stoll B, Alkhnbashi O, Sharma K, Urlaub H, Backofen R, Marchfelder A, Bolt EL. 2015. The role of Cas8 in type I CRISPR interference. Biosci Rep 35:e00197. doi: 10.1042/BSR20150043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng L, Pei J, Pang H, Guo Y, Lin L, Huang R. 2014. Whole genome sequencing reveals a novel CRISPR system in industrial Clostridium acetobutylicum. J Ind Microbiol Biotechnol 41:1677–1685. doi: 10.1007/s10295-014-1507-3. [DOI] [PubMed] [Google Scholar]
- 22.Richter H, Zoephel J, Schermuly J, Maticzka D, Backofen R, Randau L. 2012. Characterization of CRISPR RNA processing in Clostridium thermocellum and Methanococcus maripaludis. Nucleic Acids Res 40:9887–9896. doi: 10.1093/nar/gks737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown SD, Nagaraju S, Utturkar S, De Tissera S, Segovia S, Mitchell W, Land ML, Dassanayake A, Köpke M. 2014. Comparison of single-molecule sequencing and hybrid approaches for finishing the genome of Clostridium autoethanogenum and analysis of CRISPR systems in industrial relevant clostridia. Biotechnol Biofuels 7:40. doi: 10.1186/1754-6834-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soutourina OA, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, Semenova E, Severinov K, Le Bouguenec C, Coppée JY, Dupuy B, Martin-Verstraete I. 2013. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet 9:e1003493. doi: 10.1371/journal.pgen.1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, Resch AM, Glover CV III, Graveley BR, Terns RM, Terns MP. 2012. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell 45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maier LK, Lange SJ, Stoll B, Haas KA, Fischer S, Fischer E, Duchardt-Ferner E, Wöhnert J, Backofen R, Marchfelder A. 2013. Essential requirements for the detection and degradation of invaders by the Haloferax volcanii CRISPR/Cas system I-B. RNA Biol 10:865–874. doi: 10.4161/rna.24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Biswas A, Gagnon JN, Brouns SJ, Fineran PC, Brown CM. 2013. CRISPRTarget: bioinformatic prediction and analysis of crRNA targets. RNA Biol 10:817–827. doi: 10.4161/rna.24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekulovic O, Garneau JR, Néron A, Fortier LC. 2014. Characterization of temperate phages infecting Clostridium difficile isolates of human and animal origins. Appl Environ Microbiol 80:2555–2563. doi: 10.1128/AEM.00237-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hargreaves KR, Flores CO, Lawley TD, Clokie MR. 2014. Abundant and diverse clustered regularly interspaced short palindromic repeat spacers in Clostridium difficile strains and prophages target multiple phage types within this pathogen. mBio 5(5):e01045-13. doi: 10.1128/mBio.01045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meessen-Pinard M, Sekulovic O, Fortier LC. 2012. Evidence of in vivo prophage induction during Clostridium difficile infection. Appl Environ Microbiol 78:7662–7670. doi: 10.1128/AEM.02275-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goh S, Ong PF, Song KP, Riley TV, Chang BJ. 2007. The complete genome sequence of Clostridium difficile phage phiC2 and comparisons to phiCD119 and inducible prophages of CD630. Microbiology 153:676–685. doi: 10.1099/mic.0.2006/002436-0. [DOI] [PubMed] [Google Scholar]
- 33.Sekulovic O, Meessen-Pinard M, Fortier LC. 2011. Prophage-stimulated toxin production in Clostridium difficile NAP1/027 lysogens. J Bacteriol 193:2726–2734. doi: 10.1128/JB.00787-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. 2011. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A 108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJ, Boekema EJ, Dickman MJ, Doudna JA. 2011. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci U S A 108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richter H, Lange SJ, Backofen R, Randau L. 2013. Comparative analysis of Cas6b processing and CRISPR RNA stability. RNA Biol 10:700–707. doi: 10.4161/rna.23715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. 2012. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun 3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 38.Roberts AP, Allan E, Mullany P. 2014. The impact of horizontal gene transfer on the biology of Clostridium difficile. Adv Microb Physiol 65:63–82. doi: 10.1016/bs.ampbs.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Li M, Wang R, Zhao D, Xiang H. 2014. Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res 42:2483–2492. doi: 10.1093/nar/gkt1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nunez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. 2014. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol 21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pougach K, Semenova E, Bogdanova E, Datsenko KA, Djordjevic M, Wanner BL, Severinov K. 2010. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol Microbiol 77:1367–1379. doi: 10.1111/j.1365-2958.2010.07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, Gerding DN, Gibert M, Popoff MR, Parkhill J, Dougan G, Wren BW. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol 10:R102. doi: 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arakawa K, Tomita M. 2007. Selection effects on the positioning of genes and gene structures from the interplay of replication and transcription in bacterial genomes. Evol Bioinform Online 3:279–286. [PMC free article] [PubMed] [Google Scholar]
- 45.Guy L, Roten CA. 2004. Genometric analyses of the organization of circular chromosomes: a universal pressure determines the direction of ribosomal RNA genes transcription relative to chromosome replication. Gene 340:45–52. doi: 10.1016/j.gene.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 46.Rocha EP, Danchin A. 2003. Gene essentiality determines chromosome organisation in bacteria. Nucleic Acids Res 31:6570–6577. doi: 10.1093/nar/gkg859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fortier LC, Sekulovic O. 2013. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 4:354–365. doi: 10.4161/viru.24498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goh S, Hussain H, Chang BJ, Emmett W, Riley TV, Mullany P. 2013. Phage phiC2 mediates transduction of Tn6215, encoding erythromycin resistance, between Clostridium difficile strains. mBio 4(6):e00840-13. doi: 10.1128/mBio.00840-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brouwer MS, Roberts AP, Hussain H, Williams RJ, Allan E, Mullany P. 2013. Horizontal gene transfer converts non-toxigenic Clostridium difficile strains into toxin producers. Nat Commun 4:2601. doi: 10.1038/ncomms3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seed KD, Lazinski DW, Calderwood SB, Camilli A. 2013. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature 494:489–491. doi: 10.1038/nature11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paez-Espino D, Morovic W, Sun CL, Thomas BC, Ueda K, Stahl B, Barrangou R, Banfield JF. 2013. Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nat Commun 4:1430. doi: 10.1038/ncomms2440. [DOI] [PubMed] [Google Scholar]
- 52.Hargreaves KR, Clokie MR. 2014. Clostridium difficile phages: still difficult? Front Microbiol 5:184. doi: 10.3389/fmicb.2014.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dupuy B, Sonenshein AL. 1998. Regulated transcription of Clostridium difficile toxin genes. Mol Microbiol 27:107–120. doi: 10.1046/j.1365-2958.1998.00663.x. [DOI] [PubMed] [Google Scholar]
- 54.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.André G, Even S, Putzer H, Burguière P, Croux C, Danchin A, Martin-Verstraete I, Soutourina O. 2008. S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res 36:5955–5969. doi: 10.1093/nar/gkn601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosinski-Chupin I, Soutourina O, Martin-Verstraete I. 2014. Riboswitch discovery by combining RNA-seq and genome-wide identification of transcriptional start sites. Methods Enzymol 549:3–27. doi: 10.1016/B978-0-12-801122-5.00001-5. [DOI] [PubMed] [Google Scholar]
- 57.Dingle KE, Elliott B, Robinson E, Griffiths D, Eyre DW, Stoesser N, Vaughan A, Golubchik T, Fawley WN, Wilcox MH, Peto TE, Walker AS, Riley TV, Crook DW, Didelot X. 2014. Evolutionary history of the Clostridium difficile pathogenicity locus. Genome Biol Evol 6:36–52. doi: 10.1093/gbe/evt204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eyre DW, Cule ML, Wilson DJ, Griffiths D, Vaughan A, O’Connor L, Ip CL, Golubchik T, Batty EM, Finney JM, Wyllie DH, Didelot X, Piazza P, Bowden R, Dingle KE, Harding RM, Crook DW, Wilcox MH, Peto TE, Walker AS. 2013. Diverse sources of C. difficile infection identified on whole-genome sequencing. N Engl J Med 369:1195–1205. doi: 10.1056/NEJMoa1216064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, D’Arc S, Brazier J, Brown D, Coia JE, Douce G, Gerding D, Kim HJ, Koh TH, Kato H, Senoh M, Louie T, Michell S, Butt E, Peacock SJ, Brown NM, Riley T, Songer G, Wilcox M, Pirmohamed M, Kuijper E, Hawkey P, Wren BW, Dougan G, Parkhill J, Lawley TD. 2013. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet 45:109–113. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, Martin MJ, Holt KE, Seth-Smith HM, Quail MA, Rance R, Brooks K, Churcher C, Harris D, Bentley SD, Burrows C, Clark L, Corton C, Murray V, Rose G, Thurston S, van Tonder A, Walker D, Wren BW, Dougan G, Parkhill J. 2010. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci U S A 107:7527–7532. doi: 10.1073/pnas.0914322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kurka H, Ehrenreich A, Ludwig W, Monot M, Rupnik M, Barbut F, Indra A, Dupuy B, Liebl W. 2014. Sequence similarity of Clostridium difficile strains by analysis of conserved genes and genome content is reflected by their ribotype affiliation. PLoS One 9:e86535. doi: 10.1371/journal.pone.0086535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moura I, Monot M, Tani C, Spigaglia P, Barbanti F, Norais N, Dupuy B, Bouza E, Mastrantonio P. 2014. Multidisciplinary analysis of a nontoxigenic Clostridium difficile strain with stable resistance to metronidazole. Antimicrob Agents Chemother 58:4957–4960. doi: 10.1128/AAC.02350-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lemée L, Bourgeois I, Ruffin E, Collignon A, Lemeland JF, Pons JL. 2005. Multilocus sequence analysis and comparative evolution of virulence-associated genes and housekeeping genes of Clostridium difficile. Microbiology 151:3171–3180. doi: 10.1099/mic.0.28155-0. [DOI] [PubMed] [Google Scholar]
- 65.Monot M, Orgeur M, Camiade E, Brehier C, Dupuy B. 2014. COV2HTML: a visualization and analysis tool of bacterial next generation sequencing (NGS) data for postgenomics life scientists. OMICS 18:184–195. doi: 10.1089/omi.2013.0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression analysis of CRISPR arrays from C. difficile 630 strain by deep sequencing. The TAP−/TAP+ profile comparison for 5′-end RNA-seq is aligned with RNA-seq data for the corresponding genomic region. The TSSs identified by 5′-end sequencing are indicated by red broken arrows in accordance with the positions of 5′-transcript ends shown by vertical green lines on the sequence read graphs corresponding either to TSSs (broken arrows) or to processing sites. 5′-end sequencing data show 51-bp reads matching the 5′-transcript ends, while RNA-seq data show reads covering the whole transcript. Black diamonds indicate the positions of the first direct repeat of the CRISPR array. The transcript covering the CRISPR array is indicated by a gray arrow below the deep sequencing data. Deep sequencing data are available at https://mmonot.eu/COV2HTML/visualisation.php?str_id=-17. Download
Expression analysis of CRISPR arrays from C. difficile R20291 strain by RNA-seq. Sequence reads covering the whole transcript from RNA-seq data are shown according to genomic position. Black diamonds indicate the positions of the first direct repeat of the CRISPR array. The transcript covering the CRISPR array is indicated by a gray arrow below deep sequencing data. Deep sequencing data are available at https://mmonot.eu/COV2HTML/visualisation.php?str_id=-18. Download
Spacer content of CRISPR arrays for nine C. difficile strains. CRISPRdb numbering for CRISPR arrays was used, and spacers within each CRISPR array were numbered according to transcriptional order. The same number was assigned to identical spacers within CRISPR arrays from different strains. Colors were used to indicate related CRISPR arrays or spacers. Potential spacer deletion events are shown in bold type. Download
Expressed CRISPR arrays in C. difficile 630 and R20291 strains
General analysis of CRISPR spacer matches for nine C. difficile strains. (A) CRISPR spacer hits to chromosome, phage, and plasmid sequences. (B) CRISPR spacer matching of phage, chromosome, and plasmid sequences. (C) Phage targeting by several CRISPR spacers.
CRISPR spacer homology analysis in comparison with phage sensitivity for strains 630 and R20291
Strains and plasmids used in this study
C. difficile cas locus analysis
Oligonucleotides used in this study